Abstract
The Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is responsible for the induction of a class of genes referred to as middle sporulation genes. Among the members of this family are the B-type cyclins and other genes whose products are required for meiotic chromosome division and spore morphogenesis. Inactivation of NDT80 leads to a failure to induce the middle sporulation genes and a subsequent arrest in pachytene. The expression of NDT80 is itself highly regulated. The initial transcription of NDT80 is dependent upon the protein kinase Ime2; once Ndt80 protein accumulates, it activates its own promoter, thus generating an autoactivation loop. In addition to being transcriptionally regulated, Ndt80 protein is posttranslationally regulated. Phosphorylation of Ndt80 occurs coincident with its activation as a transcription factor. If expressed prematurely in meiosis, Ndt80 accumulates initially in an unmodified form that is subsequently modified by phosphorylation. In contrast, Ndt80 expressed in ime2 mutant strains does not become modified and has a reduced ability to activate transcription of its target genes. Ime2 can also phosphorylate Ndt80 in vitro, further supporting a direct role for Ime2 in the phosphorylation of Ndt80. These data indicate that Ime2 plays a novel and previously unexpected role in promoting chromosome dissemination and progress through meiotic development by activating Ndt80.
Gametogenesis is a highly specialized developmental pathway in which diploid cells undergo meiosis to produce haploid germ cells. In yeast, gametogenesis involves the formation of four haploid spores from a diploid parental cell. Each spore is capable of germinating and fusing with a haploid of the opposite mating type, a process analogous to the fusion of egg and sperm in metazoans. Orderly progression through the events of meiotic development depends upon the regulated sequential expression of at least four classes of meiosis-specific genes. These are early, middle, mid-late, and late (29). More detailed analysis of meiotic gene expression has revealed that there may be as many as seven classes of genes that are expressed as temporally distinct families (4).
Temporal regulation of gene families helps to ensure that the proteins they encode are coordinately expressed at the time that their functions are required. Expression of the early class of meiotic genes is dependent upon Ime1 and Ume6 transcription factors. These factors interact with the URS1 DNA sequence found upstream of most early meiotic genes (29, 40). Maximal expression of many early meiosis-specific genes is also dependent upon IME2, which is itself activated by Ime1/Ume6 (30). The products of the early genes induced by these factors perform functions involved in DNA replication, chromosome pairing, synaptonemal complex formation, and recombination (22). Activation of the early sporulation genes, in particular IME2, is required for the induction of the middle sporulation genes (30). Products of the middle sporulation genes are essential for chromosome division and the activation of later events involved in spore formation (4, 10). Mid-late genes are required for outer spore wall formation, and the late gene products are important for spore morphogenesis and maturation (3, 25). This integrated pattern of gene expression ensures orderly progression through meiotic development and spore morphogenesis.
The family of genes referred to as middle sporulation genes are regulated by multiple positively and negatively acting transcription factors. The expression of these genes is inhibited during vegetative growth and the early stages of meiosis by repressors Sum1 and the Set3 complex (33, 46). In the middle stages of sporulation, these genes are largely dependent for their expression on the activity of the transcription factor Ndt80, which is itself classed as a middle sporulation gene (5, 18). In the early stages of meiotic development, relatively low levels of Ndt80 mRNA can be detected; this accumulation is dependent upon the activity of meiosis-specific protein kinase Ime2 (5). When Ndt80 protein accumulates sufficiently, it binds to the promoter region of many middle sporulation genes by virtue of its affinity for the DNA sequence referred to as a middle sporulation element (MSE) (5, 17). The NDT80 upstream region contains two MSE sequences, and once activated, Ndt80 induces the expression of its own gene, thus amplifying the amount of Ndt80 available. Other targets of Ndt80 include the B-type cyclin genes CLB1, CLB3, CLB4, CLB5, and CLB6. These cyclin genes encode activators of the cyclin-dependent kinase (Cdk) Cdc28. The kinase activity associated with Clb/Cdc28 is essential for meiotic chromosome division. Cells that lack CLB1, CLB3, and CLB4 or that have a temperature-sensitive allele of Cdc28 arrest at meiosis I (MI) and fail to progress through meiosis (6, 38). Consistent with a role in inducing expression of the CLB genes, cells that lack Ndt80 efficiently complete DNA replication and meiotic recombination but arrest at pachytene with duplicated but unseparated spindle pole bodies and fully assembled synaptonemal complexes (47). These cells never proceed through any meiotic divisions. Thus, effective expression and activity of Ndt80 are specifically required for progression through meiotic chromosome divisions and spore formation.
The protein kinase encoded by IME2 is expressed uniquely in meiosis, where it performs multiple roles in promoting progression through meiotic development. In addition to activating both early and middle sporulation gene transcription, Ime2 kinase activity is required to down regulate early meiosis-specific gene expression as cells progress into the later stages of sporulation (15). Ime2 activity is also required for timely initiation of meiotic S phase (9, 14). Genetic evidence suggests that the major role of Ime2 in promoting meiotic S phase is to phosphorylate the Cdk inhibitor Sic1, thus instigating its degradation (7). Sic1 binds to and inhibits Clb/Cdc28 complexes, and its effective degradation is essential for DNA replication both during meiosis and during mitotic growth (7, 37, 41). In mitotically growing cells, Sic1 is phosphorylated by the G1 cyclins Cln1 and Cln2 complexed with Cdc28 (37, 44). In contrast, the CLN cyclins are not expressed in cells undergoing meiosis and elimination of Sic1 depends upon Ime2 (7). Ime2 kinase has also been shown to be capable of regulating the activity of one form of anaphase-promoting complex (APCCdh1) (2). The Cdh1-regulated form of the anaphase-promoting complex has been shown to be responsible for regulated degradation of mitotic cyclins in mitosis and early G1 phase (36). In mitotically growing cells, phosphorylation of Cdh1 by Cln1, Cln2, or Clb5 inactivates Cdh1 and allows the accumulation of mitotic cyclins (48). The exclusivity of mitotic growth and meiotic development may be enforced, in part, through the use of meiosis-specific Ime2 kinase to perform some of the functions attributed to Cln/Cdc28 kinase in mitotically growing cells.
Eukaryotic cells have surveillance mechanisms, known as checkpoints, that detect DNA damage or the inability to correctly form a mitotic spindle. Invoking a checkpoint mechanism will halt cell cycle progression until the damage can be repaired or simply cause an arrest to prevent damaged genetic information from being passed to progeny (8, 28). Checkpoints also operate during meiosis, where, in response to DNA damage, failure in the completion of recombination or failure to complete DNA replication results in meiotic arrest and a block to spore formation (5, 18, 28, 32, 41). This arrest is accomplished, in part, by repressing the expression of the middle sporulation genes, and it has been proposed that the checkpoint is enforced through inactivation of Ndt80 (5, 18). By inhibiting Ndt80 activity, the checkpoint can cause pachytene arrest by blocking the accumulation of Clbs and Cdc28 kinase activity, along with other gene products required to initiate spore morphogenesis (5, 18). The ability to use the same transcription factor to regulate progression through meiotic chromosome division and spore formation is an elegant mechanism by which to restrict further development and spore formation if cell cycle progression is hindered by defects in chromosome metabolism.
In cells arrested at pachytene by incomplete recombination, NDT80 is expressed at low levels but a detectable amount of protein does accumulate (42). Despite this accumulation of Ndt80 protein, the middle sporulation genes are not effectively activated (42). The inhibition of Ndt80 activity may be accomplished by regulating posttranslational modification. Ndt80 has been shown to be a phosphoprotein in vivo (42). The phosphorylation events do not appear to be checkpoint induced; rather, the appearance of phospho forms is correlated with the activation of Ndt80 target genes. If the phospho forms of Ndt80 are the “activated” species, then checkpoint arrest could be a consequence of failure to phosphorylate Ndt80. Indeed, examination of Ndt80 in cells arrested at the pachytene checkpoint indicated that Ndt80 was strikingly undermodified, suggesting that the pachytene checkpoint may, in fact, inhibit Ndt80 by preventing it from being phosphorylated and activated (42).
We have found that when Ndt80 is ectopically expressed in meiosis, the protein accumulates initially in an unmodified form and then rapidly becomes modified at the time that Ndt80 becomes active in inducing middle sporulation genes. The modification of Ndt80 is dependent upon the activity of meiosis-specific protein kinase Ime2. When Ndt80 is ectopically expressed in an ime2 mutant, the protein accumulates in an unmodified state and fails to activate expression of the middle sporulation genes unless it is overexpressed. In support of the contention that Ime2 is required for phosphorylation of Ndt80, Ime2 either immunoprecipitated from yeast cells or expressed as a recombinant protein in baculovirus-infected insect cells can phosphorylate Ndt80 in vitro. Our observations support a model wherein Ime2-dependent phosphorylation is required for Ndt80 to effectively interact with MSE sites in the promoters of middle sporulation genes.
MATERIALS AND METHODS
Strains and growth conditions.
All of the strains used in this study are SK1 strains derived from the parents DSY1030 (MATa lys2 ho::LYS2 ura3 leu2::hisG trp1::hisG arg4Bgl his4X) and DSY1031 (MATα lys2 ho::LYS2 ura3 leu2::hisG trp1::hisg arg4Nsp his4B) (28). The strains used in this study and their relevant genotypes are listed in Table 1. All strains were generated by standard genetic procedures (34). Yeast strains were routinely propagated in YEP supplemented with 2% glucose or 2% potassium acetate (34). The sporulation medium (SPM) used in this study was 1% potassium acetate. In experiments in which NDT80 was expressed under the regulation of the CUP1 promoter, CuSO4 was added to the SPM to a final concentration of 100 μM. The NDT80 deletion ndt80::Kanr in DSY1257 replaces the 1.2-kb XhoI-to-BamHI fragment of Ndt80 with a kanamycin resistance-encoding gene isolated from pFA6-KanMX2 (45). In addition to removing much of the promoter, this deletes the first 410 amino acids of the 627-residue protein. The ndt80::Kanr mutation in DSY1342, DSY1344, DSY1347, DSY1349, and DSY1352 is a precise deletion of the NDT80 open reading frame generated by a PCR-based method (45). ime2::TRP1 in DSY1087 is a deletion that replaces the entire open reading frame of IME2 with TRP1. SIC1 was deleted by replacing a 708-bp KpnI-NdeI fragment from within the SIC1 open reading frame with the kanamycin resistance cassette from pFA6-KanMX2 (45). To generate an epitope-tagged version of Ndt80, site-directed mutagenesis was used to create a NotI site at the C terminus immediately prior to the stop codon. A NotI cassette encoding three copies of the hemagglutinin (HA) epitope was inserted in frame with the Ndt80 open reading frame (43). This tagged version of NDT80 was able to rescue the sporulation defect of DSY1257 (ndt80::Kanr/ndt80::Kanr) A similar strategy was used to fuse the C terminus of NDT80 to a green fluorescent protein (GFP) cassette. IME2 was similarly tagged at its C terminus with a cassette that encodes three copies of the MYC epitope (35). A kinase-dead version of Ime2 (Ime2 K97R) was generated by using site-directed mutagenesis to change lysine 97 to arginine in Myc-tagged IME2. This mutation was previously shown to cause a loss of Ime2 activity in vivo (39), and we have found that it has no kinase activity in vitro against any substrate that we have tested. A similar mutation (lysine 97 to alanine) has also been reported to generate a kinase-dead ime2 allele (15). A functional truncated version of Ime2 was generated by introducing three tandem copies of the MYC epitope, followed by a stop codon at amino acid 470 of the IME2 open reading frame. This truncated mutant was ligated with the integrating vector YIplac128 and cleaved with BamHI to direct integration into the IME2 gene. In order to ectopically express Ndt80, the HA-tagged NDT80 open reading frame was placed under the regulation of an IME2 or a CUP1 promoter in integrating vector YIplac211. This plasmid was cleaved with EcoRV (IME2-NDT80HA) or NcoI (CUP1-NDT80HA) to direct integration into the URA3 locus. Similar constructs were generated in centromere plasmid YCplac111 for use in the β-galactosidase assays reported in Fig. 3. The SPS4-LacZ reporter plasmid used in β-galactosidase assays has been described previously (17) and was a generous gift from J. Segall. Quantitative β-galactosidase assays were preformed as previously described (17). The β-galactosidase values reported in Fig. 3 and 6 are derived from the mean activity produced by three independent transformants, and standard deviations are indicated by error bars.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotypea | Source |
|---|---|---|
| DSY1249 | a/α | This study |
| DSY1087 | a/α ime2::TRP1/me2::TRP1 | This study |
| DSY1312 | a/α URA3::IME2-NDT80HA/URA3::IME2-NDT80HA | This study |
| DSY1270 | a/α clb5::Kanr/clb5::Kanrclb6::TRP1/clb6::TRP1 URA3::IME2-NDT80HA/URA3::IME2-NDT80HA | This study |
| DSY1279 | a/α ime2::TRP1/ime2::TRP1 URA3::IME2-NDT80HA/URA3::IME2-NDT80HA | This study |
| DSY1289 | a/α URA3::CUP1-NDT80HA/URA3::CUP1-NDT80HA | This study |
| DSY1299 | a/α ime2::TRP1/ime2::TRP1 URA3::CUP1-NDT80HA/URA3::CUP1-NDT80HA | This study |
| DSY1303 | a/α ime2::TRP1/ime2::TRP1/ sic1::Kanr/sic1::KanrURA3::CUP1-NDT80HA/URA3::CUP1-NDT80HA | This study |
| DSY1257 | a/α ndt80::Kanr/ntd80::Kanr | This study |
| DSY1342 | a/α ndt80::Kanr/ndt80::Kanr | This study |
| DSY1344 | a/α ndt80::Kanr/ndt80::KanrURA3::IME2-NDT80HA/URA3::IME2-NDT80HA | This study |
| DSY1347 | a/α ime2::TRP1/ime2::TRP1 ndt80::Kanr/ndt80::KanrURA3::IME2-NDT80HA/URA3::IME-NDT80/IT"HA | This study |
| DSY1349 | a/α ndt80::Kanr/ndt80::KanrURA3::CUP1-NDT80HA/URA3::CUP1-NDT80HA | This study |
| DSY1352 | a/α ime2::TRP1/ime2::TRP1 ndt80::Kanr/ndt80::KanrURA3::CUP1-NDT80HA/URA3::CUP1-NDT80HA | This study |
| DSY1262 | a/α ime2::IME2-3×myc-LEU2/ime2::IME2-3×mye-LEU2 | This study |
| DSY1264 | a/α ime2::IME2Δ175-3×myc-LEU2/ime2::IME2Δ175-3×myc-LEU2 | This study |
| DSY1355 | a/α URA3::8×Lex°-LacZ/URA3::8×Lex°-LacZ | This study |
| DSY1357 | a/α ime2::TRP1/ime2::TRP1 URA3::8×Lex°-LacZ/URA3::8×Lex°-LacZ | This study |
| DSY1365 | a/α URA3::CUP1-NDT80-GFP/URA3::CUP1-NDT80GFP | This study |
| DSY1367 | a/α URA3::IME2-NDT80-GFP/URA3::IME2-NDT80-GFP | This study |
| DSY1369 | a/α ime2::TRP1/ime2::TRP1 URA3::CUP1-NDT80-GFP/URA3::CUP1-NDT80-GFP | This study |
| DSY1372 | a/α ime2::TRP1/ime2::TRP1 URA3::IME2-NDT80-GFP/URA3::IME2-NDT80-GFP | This study |
| PJ69-4A | atrp1 leu2 ura3 his3 gal4 gal80 LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL1-lacZ | P. James (21) |
All of the yeast strains listed, with the exception of PJ69-4A, are derived from the SK1 genetic background.
FIG. 3.
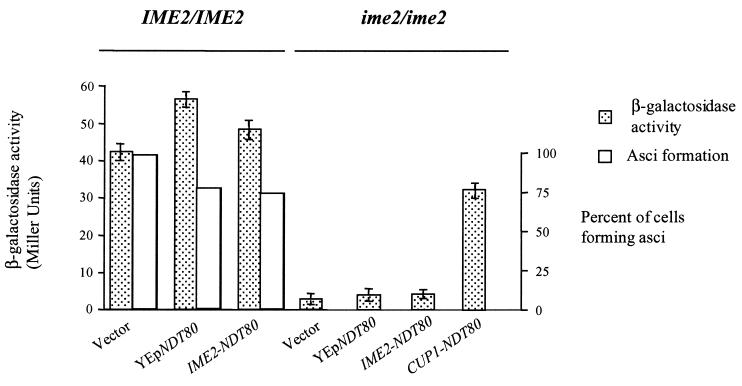
Overproduction of Ndt80 allows activation of middle sporulation gene promoters in an ime2 mutant but does not promote spore formation. Wild-type cells (IME2/IME2) or ime2 mutants (ime2/ime2) harboring an SPS4-LacZ reporter gene were transformed with vector plasmid YCplac111 (vector), a high-copy plasmid carrying NDT80 (YEpNDT80), YCplac111 expressing NDT80 under the regulation of an IME2 promoter (IME2-NDT80), or YCplac111 expressing NDT80 under the regulation of a CUP1 promoter (CUP1-NDT80). The transformed cells were induced to sporulate, and 6 h following the induction of sporulation, β-galactosidase activity was assayed and expressed as Miller units (filled boxes). In cells that carry CUP1-NDT80, the expression of Ndt80 was induced at time zero by the addition of CuSO4 to the medium to a final concentration of 100 μM. After 24 h in SPM, the cultures were microscopically examined for the percentage of cells that had formed asci (open boxes).
FIG.6.
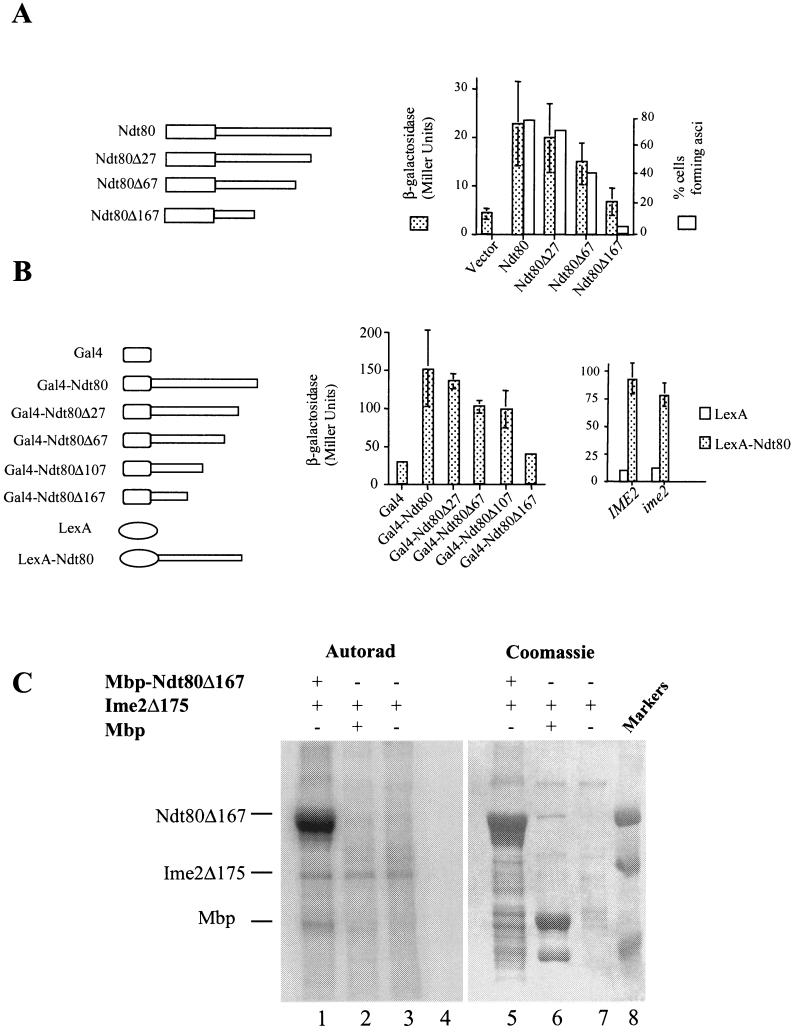
The C terminus of Ndt80 encodes a transactivation domain. (A) The endogenous NDT80 in a diploid yeast strain was replaced by a version of NDT80 encoding either full-length Ndt80 or the indicated C-terminal deletion (Δ27, Δ67, or Δ167). These strains also harbored an SPS4-LacZ reporter gene. The ability of these mutant versions of NDT80 to promote middle sporulation gene expression was assayed by the β-galactosidase activity induced after 6 h in SPM and is expressed in Miller units. The ability of the mutant versions of NDT80 to promote and scored for the formation of asci. (B) IME2 is not required for the function of the NDT80 trans activation domain. The C terminus of NDT80 or the indicated C terminally deleted versions of NDT80 were fused to the Gal4 DNA binding domain and expressed in vegetatively growing cells under the regulation of a CUP1 promoter. β-Galactosidase activity expressed from an integrated GAL1-LacZ reporter gene was measured and is displayed in Miller units. Similarly, the entire Ndt80 C terminus (amino acids 426 and 627) was fused to LexA and expressed under the regulation of a CUP1 promoter in IME2/IME2 or ime2/ime2 diploid cells. Both strains harbor an integrated 8×Lexo-LacZ reporter gene. β-Galactosidase activity induced by the LexA fusions was assayed after the cells had been in SPM for 5 h. (C) Ime2 phosphorylates Ndt80 lacking its transactivation domain. An Mbp-Ndt80 fusion lacking 167 amino acids from its C terminus was produced in E. coli and used as a substrate for immunoprecipitated Ime2Δ175. Lanes 1 to 4 show an autoradiogram (Autorad) of the kinase assay, while lanes 5 to 8 show Coomassie staining of the same gel. Ime2Δ175 kinase activity was assayed against Mbp-Ndt80Δ167 (lanes 1 and 5), Mbp1 (lanes 2 and 6), or no added substrate (lanes 3 and 7). Molecular weight markers are included in lane 8.
A truncation of Ndt80 lacking the C-terminal 167 amino acid residues was created by PCR amplification of the NDT80 open reading frame. The PCR products were sequenced, and a fragment with no sequence errors was placed under the regulation of an IME2 promoter in integrating plasmid YIplac211. Three copies of the HA epitope were inserted into this truncated open reading frame immediately following the start codon. A series of C-terminal deletions was generated in NDT80 by PCR amplification with a common upstream oligonucleotide primer (5′-GAG ATC TAT GAA TGA AAT GGA AAA CAC A-3′) and a series of downstream primers (5′-CGG TCT AGA TTA GAC CCC TTT CGA ATA TAC CC-3′, 5′-CGG TCT AGA TTG AAA AGT GCA GTT TCC AAG TG-3′, 5′-CGG TCT AGA TTT GGT CTT AGA TAA TTC TGT TC-3′, and 5′-CGG TCT AGA TTA ACT AAA TCC TCC TTT TCT AA-3′). These downstream oligonucleotides create C-terminal deletions of 83, 203, 323, and 503 nucleotides, respectively, in the NDT80 open reading frame and correspond to deletions of 27, 67, 107, and 167 amino acids, respectively. Each of the mutated NDT80 open reading frames was placed under the regulation of 700 bp of the NDT80 upstream DNA sequence inserted into centromere plasmid YCplac22; in addition, a 300-bp fragment containing the NDT80 3′ untranslated sequence was inserted 3′ to the mutated open reading frames. These constructs allowed expression of truncated versions of NDT80 under the regulation of the native NDT80 promoter. The ability of the NDT80 C terminus to function as an independent transactivating domain was tested by fusing the complete Ndt80 C-terminal region (codons 426 to 628) to the GAL4 DNA binding domain in pGBDC-3 (21). In addition, the same series of C-terminal deletions described above were fused to GAL4 starting from NDT80 codon 426. These plasmids were introduced into a yeast strain that harbors an integrated GAL1-regulated LacZ reporter gene, PJ69-4a (21), and β-galactosidase activity expressed in vegetatively growing cells was assayed as described above. To assay the ability of the NDT80 C terminus to function as a transactivator in sporulating cells, the entire C terminus (codons 426 to 628) was fused in frame to LexA and expressed under the regulation of a CUP1 promoter from centromere plasmid YCplac111. The transactivating potential of the NDT80 C terminus in this fusion was assayed in sporulating DSY1355 and DSY1357 cells that also harbor a LacZ reporter gene that is regulated by eight tandem repeats of a LexA binding site.
Expression and purification of recombinant proteins.
Ndt80 or Ndt80Δ167 was produced in Escherichia coli by ligating the open reading frame into pMALC2 (New England Biolabs) and expressing the protein as a fusion with maltose binding protein (Mbp). One liter of E. coli ER2508 (New England Biolabs) that had been induced to express the Mbp-Ndt80 fusion was lysed by sonication in 20 mM Tris-HCl (pH 7.4)-200 mM NaCl-1 mM EDTA in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysate was clarified by centrifugation at 12, 000 × g for 15 min. The fusion proteins were then purified by chromatography on amylose resin.
A recombinant baculovirus expressing Myc-tagged Ime2 was produced by ligating the epitope-tagged open reading frame with the vector pFAST-bac1. This vector was used to generate a recombinant virus by transposition as directed by the manufacturer (Gibco-BRL). Recombinant Ime2 was produced by infecting monolayers of Sf9 cells with virus at a multiplicity of infection of 10. Infections were allowed to proceed for 48 h, after which the cells were harvested and lysed by Dounce homogenization in 20 mM Tris-HCl (pH 8.0)-0.5% NP-40-2 mM dithiothreitol (DTT)-1 mM PMSF-20 μg (each) of leupeptin, pepstatin, and aprotinin per ml-1 mM sodium orthovanadate. Ime2 was immunoprecipitated with anti-MYC antibodies. RNA preparation from sporulating cells and Northern blot analysis were performed as described previously (41).
Protein extraction, immunoprecipitation, and Western blotting.
Protein extracts were prepared from yeast as previously described (13). For analysis by immunoblot assay, 50 μg of total protein for each sample was separated by electrophoresis in 10% polyacrylamide gels. Protein was transferred to Immobilon membrane, and blots were probed with either monoclonal antibody 12CA5 (1:10,000) to detect the HA epitope, 9E10 (1:5,000) to detect the MYC epitope, or affinity purified polyclonal anti-Ndt80 (N terminal; 1:1,000). TATA box binding protein Tbp1 was detected with anti-Tbp1 serum as previously described (11). The anti-Tbp1 serum used was a generous gift from Michael Schultz. A monoclonal antibody that recognizes the PSTAIRE epitope in Cdc28 and Pho85 was provided by Steve Reed and was used at a 1:10,000 dilution. Antitubulin monoclonal antibody MAS078 (1:2,000) was used to detect tubulin as a control for protein loading on gels. Primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse, anti-rabbit, or anti-rat secondary antibodies used at a 1:5,000 dilution (Jackson Immunoresearch). Immunoprecipitation was carried out by lysing cells in 50 mM Tris-HCl (pH 7.4)-250 mM NaCl-0.1% NP-40 plus phosphatase inhibitors (5 mM EDTA, 5 mM EGTA, 10 mM sodium pyrophosphate, and 1 mM sodium orthovanadate) and protease inhibitors (20 μg [each] of leupeptin, pepstatin, and aprotinin per ml and 1 mM PMSF). Following lysis, 10 mg of protein was diluted with 2 volumes of IP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.5% Triton X-100 plus protease and phosphatase inhibitors) and samples were precleared by the addition of 40 μl of protein A Sepharose beads. Ndt80HA was immunoprecipitated by incubation with 12CA5 (1:1,000) for 2 h at 4°C, and then immune complexes were isolated by addition of protein A Sepharose and incubation for 1 h at 4°C. Samples that were analyzed by Western blot assay were washed three times with IP buffer and then resuspended in 1× sample buffer (2% sodium dodecyl sulfate [SDS], 100 mM DTT, 50 mM Tris-HCl [pH 7.4], 10% glycerol, 0.1% bromophenol blue) prior to electrophoresis. Samples to be treated with calf intestinal phosphatase (CIP) were washed three times with IP buffer and then two times with CIP buffer (20 mM MgCl2, 40 mM KCl, 50 mM Tris-HCl [pH 8.0]). Immunoprecipitates were then resuspended in 300 μl of CIP buffer and divided into three 100-μl aliquots. One sample was untreated, while the other two were treated with 100 U of CIP for 15 min at 37°C. One of these two samples included 5 mM β-glycerophosphate, which acted as a phosphatase inhibitor. The samples were then processed and analyzed by Western blot assay as described above.
Ime2 kinase was immunoprecipitated from sporulating yeast. Ten-milliliter culture samples were harvested at the indicated time points and frozen in liquid nitrogen. The cell pellets were broken by vortexing with glass beads in the presence of protein extraction buffer as indicated above. Ime2-Myc was immunoprecipitated as described above, except that immune complexes were washed three times with IP buffer and then two times with 2× kinase buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, 10 μM ATP) before being resuspended in 1× kinase buffer with either 150 μg of histone H1 per ml or 100 μg of Mbp, Mbp-Ndt80, or Mbp-Ndt80Δ167 fusion protein per ml. Kinase reaction mixtures were incubated at 30°C for 15 min, and the reactions were stopped by addition of an equal volume of 2× sample buffer. Kinase reaction mixtures were electrophoresed in 10% gels, and the dried gels were visualized by autoradiography.
Chromatin immunoprecipitation assays.
The ability of Ndt80 to interact with the MSE sites in the endogenous NDT80 promoter was assayed by chromatin immunoprecipitation as previously described (16). Briefly, 200-ml cultures of IME2/IME2 (DSY1344 and DSY2349) or ime2/ime2 (DSY1347 and DSY1352) diploid cells with integrated IME2-NDT80HA or CUP1-NDT80HA were induced to sporulate by inoculation into SPM. The CUP1-NDT80HA gene was induced at time zero by the addition of CuSO4 to a final concentration of 100 μM. At the indicated time points, 25-ml samples of cells were fixed with 1% formaldehyde. The cell samples were lysed by bead beating, and the extracts were sonicated to shear the DNA prior to immunoprecipitation with anti-HA monoclonal antibodies. The immunoprecipitates were washed for 5 × 10 min with IP buffer prior to elution and reversal of cross-linking. Purified DNA from the immunoprecipitates was subjected to PCR amplification with the NDT80 promoter oligonucleotides 5′-TCT AAT AAA ATC GTC TTT GCC C-3′ and 5′-AAT AGG TGA CAC AAA ATG GAG G-3′ or internal control oligonucleotides that amplify DNA at ARS305 (5′-CTC CGT TTT TAG CCC CCC GTG-3′ and 5′-GAT TGA GGC CAC AGC AAG ACC G-3′). The amplifications were carried out for 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The products of these amplification reactions were separated on 1.2% agarose gels. The interaction of Tbp1 with the ACT1 promoter was assayed by immunoprecipitation of Tbp1 from cross-linked cell extracts and, following reversal of the cross-linking, performance of PCR amplification with ACT1 promoter oligonucleotides 5′-CCA TTT TCT TCT TTA CCC GCC-3′ and 5′-TCA GTA AAT TTT CGA TCT TGG G-3′. The same internal control oligonucleotides that hybridize with ARS305 were used to control for nonspecific immunoprecipitation. The amplification reactions were carried out for 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
Other methods.
Sporulation was assayed by microscopic examination of cultures that had been incubated in SPM for 24 h. Two hundred cells per culture were counted, and the percentage of cells that had formed asci was scored. Progression through MI and meiosis II (MII) was assayed by staining nuclear DNA with 4′,6-diamidino-2-phenylindole (DAPI) as previously described (41). The cellular localization of an Ndt80-GFP fusion was determined by fluorescence microscopy as previously described (5). Unfixed cells from sporulating cultures were examined with a Zeiss Axioskop II, and 200 cells per culture were scored for the presence of a nuclear GFP fluorescence signal.
RESULTS
Phosphorylation of Ndt80 is dependent upon Ime2.
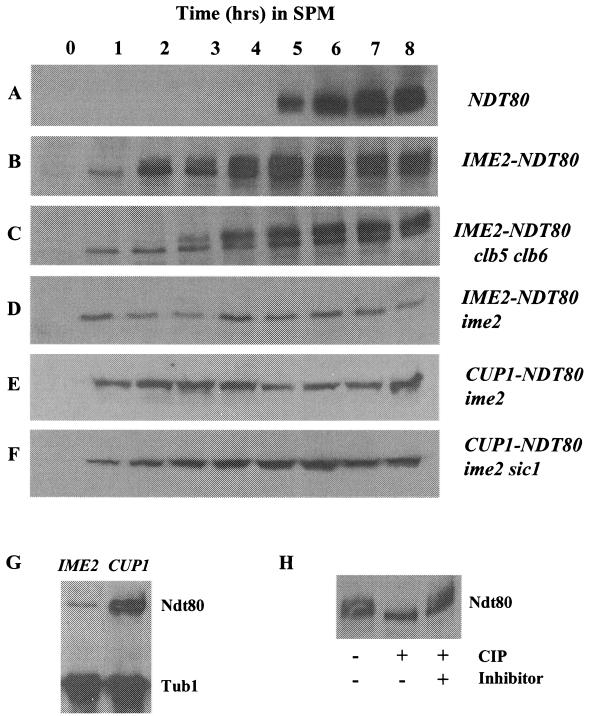
During the course of normal meiotic development, NDT80 is induced and accumulates just prior to the other middle sporulation genes. Ndt80 protein accumulates as a series of phospho species that display retarded mobility in SDS-polyacrylamide gel electrophoresis (Fig. 1) (42). Ndt80 typically becomes detectable and begins to accumulate about 5 h after SK1 strains are induced to initiate synchronous sporulation (Fig. 1A). It is notable that the protein is immediately detectable in a smear of modified forms that have previously been shown to be due to phosphorylation. When NDT80 is prematurely expressed under the regulation of an IME2 promoter, the protein can be detected within 1 h of induction of sporulation. This is consistent with the early time at which IME2 is induced in meiosis. In this case, Ndt80 accumulates first as a single unmodified species (Fig. 1B). As sporulation progresses and the cells enter S phase and MI, Ndt80 begins to appear in multiple more slowly migrating forms. As is the case for Ndt80 expressed under the regulation of its own promoter, these more slowly migrating species can be collapsed into a single band that comigrates with the unmodified form of Ndt80 if the protein is treated with CIP (Fig. 1H). Our analysis cannot distinguish whether the phosphorylated species of Ndt80 that appear after 3 h represent protein that was synthesized earlier and is being phosphorylated or if only the nascent protein synthesized after 3 h can be subjected to this modification. These data show that Ndt80 can be phosphorylated even at early times in meiosis, indicating that the kinase responsible is active during both the early and middle stages of sporulation.
FIG. 1.
Phosphorylation of Ndt80 is dependent upon IME2. The expression of Ndt80 during sporulation was monitored by Western blot assay in wild-type SK1 diploids (A and B), a clb5 clb6 mutant diploid (C), an ime2 mutant diploid (D and E), and an ime2 sic1 double mutant (F). Ndt80 was expressed under the regulation of its endogenous promoter (A) or ectopically expressed under the regulation of an IME2 promoter (B, C, and D) or a copper-inducible CUP1 promoter (E and F). The same amount of protein extract was loaded in each lane, and the time points at which samples were taken from the sporulating cultures are indicated in hours postinduction of sporulation. To induce Ndt80 expression in cells that carry NDT80 under the regulation of the CUP1 promoter, CuSO4 was added to the medium to a final concentration of 100 μM at time zero. The abundance of Ndt80 expressed under the regulation of an IME2 promoter or a CUP1 promoter was compared (G). Equal amounts of protein isolated from ime2 IME2-NDT80 and ime2 CUP1-NDT80 homozygous diploid strains 6 h after the induction of sporulation were probed for the abundance of Ndt80. Tubulin served as a loading control. (H) The species of Ndt80HA that display retarded mobility when expressed under the regulation of an IME2 promoter in IME2/IME2 cells are due to phosphorylation. DSY1312 IME2-NDT80HA was induced to sporulate, and after 6 h in SPM, cells were harvested and Ndt80HA was isolated by immunoprecipitation. The sample was divided into three aliquots. One sample was untreated, the second was incubated with 100 U of CIP, and the third was incubated with CIP in the presence of 5 mM β-glycerophosphate to inhibit the CIP activity.
Ndt80 and cdc28 mutants have similar arrest phenotypes; this is likely because Ndt80 is necessary to induce the expression of CLB1, CLB3, and CLB4, which activate Cdc28 to drive cells through pachytene and into MI and MII (5, 18). However, it has been shown that Cdc28 kinase activity is not responsible for the phosphorylation and activation of Ndt80 (42). Consistent with those observations, we find that Ndt80 is expressed and phosphorylated in clb5 clb6 double mutants that both lack detectable S-phase-dependent Cdc28 kinase activity and fail to effectively activate DNA replication (Fig. 1C). It is notable that these cells arrest in G1 because of a failure to replicate DNA, yet the timing of the appearance of Ndt80 phospho species is similar to the timing of their appearance in an otherwise wild-type cell (compare Fig. 1B and C). The 1-h delay in the appearance of phospho-Ndt80 in the clb5 clb6 mutants may simply reflect a difference in synchrony between the wild-type and clb5 clb6 strains. In addition, Ndt80 continues to accumulate in the clb5 clb6 cells, consistent with the inability to effectively down regulate the IME2 promoter in arrested cells (41). These results indicate that the phosphorylation of Ndt80 is independent of both S-phase cyclin activity and normal progression through meiotic S phase.
Ime2 protein kinase is expressed early in meiosis and is required for maximal induction of the early meiotic genes, as well as for progression into meiotic S phase and for activation of the middle sporulation genes. Although the early meiotic functions of Ime2 are most clearly limiting in ime2 mutants, there is evidence that Ime2 functions at later times in meiosis (39). The critical function of Ime2 early in sporulation has been proposed to be the phosphorylation of Sic1 (7). In ime2 mutants, Sic1 is stable and cells have a very long delay before entering meiotic S phase (7). In contrast, deletion of both ime2 and sic1 allows DNA replication but the cells arrest in a pachytene-like state resembling the arrest displayed by ndt80 mutants (7). We thought it possible that these cells might arrest because of an inability to phosphorylate and activate Ndt80. To test this idea, we ectopically expressed NDT80 in an ime2 mutant diploid. Since the NDT80 promoter is normally not activated in these cells, we placed NDT80 under the regulation of an IME2 promoter to ensure meiosis-specific expression. When NDT80 is expressed under the regulation of an IME2 promoter in an ime2 mutant diploid, the Ndt80 protein accumulates to relatively low levels (Fig. 1D). The reduced abundance of Ndt80 in ime2/ime2 cells compared to that in IME2/IME2 cells was expected, since Ime2 activity is required for maximal activation of the IME2 promoter. However, unlike the Ndt80 protein expressed in IME2 cells, the protein that accumulates in ime2 mutants appears to be strikingly undermodified and displays no retarded mobility on SDS-polyacrylamide gel electrophoresis (Fig. 1D). Wild-type cells that have been arrested at the pachytene checkpoint also display undermodification of Ndt80. However, when Ndt80 was overexpressed in these checkpoint-arrested IME2 cells, modified forms of Ndt80 accumulated (42). We were able to drive the production of much higher levels of Ndt80 in ime2 mutant cells by placing NDT80 under the regulation of a CUP1 promoter (Fig. 1E and F). In the presence of 100 μM CuSO4, Ndt80 was highly overexpressed, but unlike the situation with checkpoint-arrested IME2 cells, the overexpressed Ndt80 was not subjected to modification in an ime2 mutant strain (Fig. 1E). Ime2 is required for timely progression through meiotic S phase and the MI and MII divisions in part because it is required to degrade Sic1 and allow accumulation of S-phase and mitotic cyclins (7). We thought it possible that in our ime2 mutant strain, ectopically expressed Ndt80 might be unmodified because of a failure of these cells to accumulate any Clb/Cdc28 activity. To test this possibility, we expressed Ndt80 under the regulation of a CUP1 promoter in an ime2 sic1 mutant diploid. In this strain, the accumulated Ndt80 displayed the same pattern of undermodified forms produced in the ime2 single mutant (Fig. 1F). All of these data support the proposal that Ime2 is required for modification of Ndt80.
Ime2 is required for Ndt80 to effectively activate middle sporulation genes.
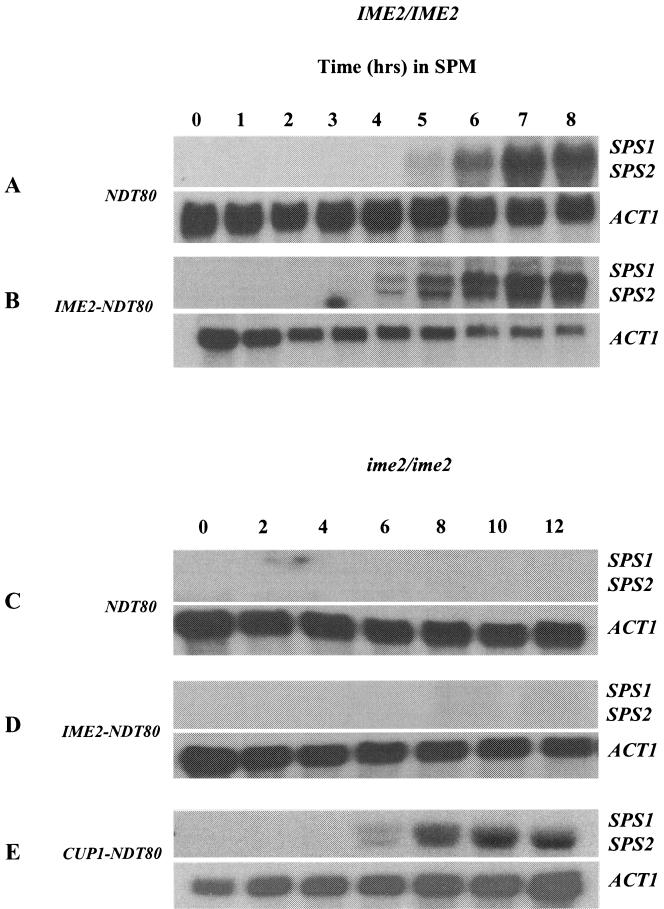
In wild-type cells progressing through meiosis, the accumulation of phospho species of Ndt80 coincides with the activation of middle sporulation genes that are the targets of Ndt80 activity. In these strains, activation of the middle sporulation genes SPS1 and SPS2 occurs at about 5 h, consistent with the accumulation of Ndt80 (Fig. 2A). However, when Ndt80 is expressed under the regulation of the IME2 promoter, the protein appears in an unmodified form that apparently is unable to induce premature expression of its middle sporulation gene targets. Despite the appearance of Ndt80 after only 1 h in an IME2-NDT80 strain, SPS1 and SPS2 transcripts do not accumulate until 4 h, coincident with the accumulation of phospho-Ndt80 (Fig. 2B). Premature expression of Ndt80 does modestly accelerate the expression of middle gene transcripts; however, none of these transcripts was induced until modified forms of Ndt80 accumulated to account for approximately 50% of the total detectable protein, suggesting that unmodified Ndt80 may be less effective in activating transcription. To further investigate the possibility that phosphorylation of Ndt80 might be essential for its transcriptional activation function, we monitored the effects of ectopically expressing Ndt80 in an ime2 mutant strain. The middle sporulation genes SPS1 and SPS2 are not activated in an ime2 mutant (Fig. 2C). The simplest explanation for this is that NDT80 is not expressed in an ime2 mutant. However, when NDT80 was ectopically expressed under the regulation of an IME2 promoter, a condition under which we can detect accumulation of Ndt80, SPS1 and SPS2 are still not induced, implying that unmodified Ndt80 may not be capable of activating transcription of the middle sporulation genes (Fig. 2D). The requirement for phosphorylation may not be absolute however, since when Ndt80 was highly overexpressed from a CUP1 promoter in an ime2 mutant, SPS1 and SPS2 transcripts could be detected (Fig. 2E). It is interesting that, despite the elevated and premature expression of Ndt80 in this strain, SPS1 and SPS2 were not prematurely induced. This observation implies that some form of negative regulation that cannot be directly overridden by Ndt80 is being imposed on these genes in the early stages of meiotic development.
FIG. 2.
Ectopic expression of Ndt80 does not profoundly accelerate the induction of middle sporulation gene transcription. RNA samples (10 μg) obtained at the indicated time points from wild-type NDT80-expressing cells (A) or wild-type cells that also express NDT80 under the regulation of an IME2 promoter, IME2-NDT80 (B), were probed for the middle sporulation gene transcripts SPS1 and SPS2. RNA samples (10 μg) from a homozygous ime2 mutant expressing NDT80 from its endogenous promoter, NDT80 (C), an IME2 promoter, IME2-NDT80 (D), or a CUP1 promoter, CUP1-NDT80 (E), were probed for the middle sporulation gene transcripts SPS1 and SPS2. The ACT1 transcript served as a loading control.
The regulation of SPS1 and SPS2 was largely mirrored in quantitative β-galactosidase assays that measured the response of the SPS4 promoter to ectopically expressed NDT80. Overexpression of NDT80 (YEp-NDT80) or ectopic expression of NDT80 (IME2-NDT80) in otherwise wild-type cells resulted in a modest increase, 33 or 14%, respectively, in the activity of the SPS4-LacZ reporter gene (Fig. 3). However, neither YEp-NDT80 nor IME2-NDT80, both of which produced very little Ndt80 protein in an ime2 mutant, could increase expression of the reporter plasmid in an ime2 mutant (Fig. 3). In contrast, CUP1-NDT80, which is capable of producing much greater amounts of Ndt80 protein, was able to induce the SPS4-LacZ reporter gene, even in an ime2 mutant. Despite the ability of CUP1-NDT80 to induce middle sporulation gene promoters in the absence of Ime2, it was not able to promote spore formation (Fig. 3). Indeed, microscopic examination of DAPI-stained cells indicated that ime2 CUP1-NDT80 diploid cells were unable to proceed through MI and remained arrested with a single nucleus (data not shown). Thus, although high levels of Ndt80 can bypass the requirement for IME2 in middle sporulation gene expression, it cannot suppress the requirement for IME2 at earlier stages of meiotic development.
Ime2 kinase activity accumulates early in meiosis and persists through the early and middle stages of sporulation.
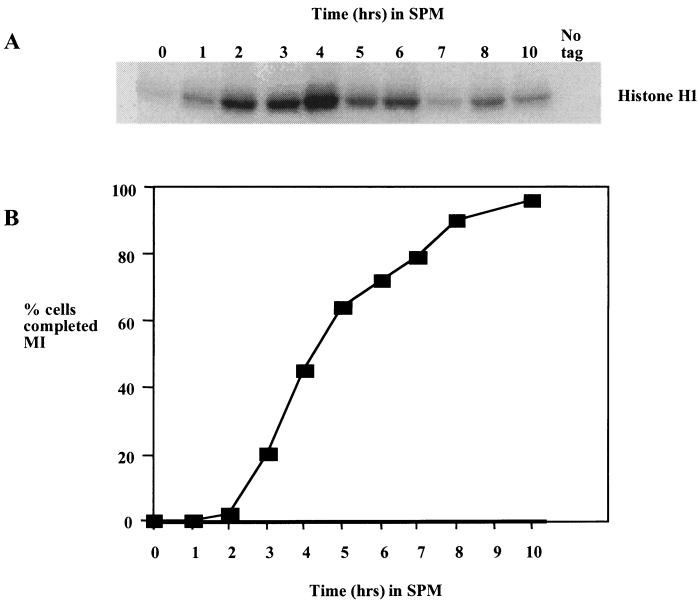
The most well-characterized function of IME2 is its role in activating the early meiotic genes prior to meiotic S phase and recombination (29, 30). However, there is some evidence that Ime2 has a direct role in activation of the middle sporulation gene family (39). Indeed, we find that Ime2 kinase activity peaks at the time that cells are proceeding through MI and MII (Fig. 4). The timing with which the kinase activity accumulates is thus consistent with Ime2 playing a role in MI when Ndt80 activity is required. It may also be significant that Ime2 kinase activity is reduced at later times in the sporulation time course when middle sporulation gene expression is reduced.
FIG. 4.
Ime2 kinase accumulates early in meiosis and persists throughout MI and MII. (A) Ime2-Myc was immunoprecipitated at the indicated times from a culture of sporulating yeast cells, and the kinase activity was assayed by using histone H1 as a substrate. (B) Samples of the culture were stained with the DNA binding dye DAPI, and the number of cells that had completed MI was assayed by fluorescence microscopy. Two hundred cells were counted at each time point, and cells displaying more than one DAPI-staining body were counted as past MI.
Ndt80 is an in vitro substrate for Ime2 kinase.
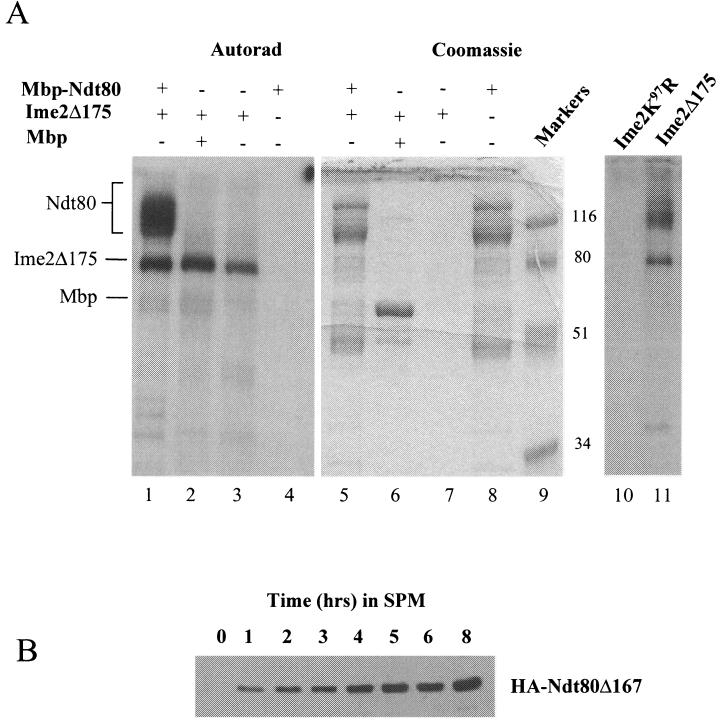
Ime2 is a key regulator of meiosis and appears to have many potential substrates required for the activation of early and middle sporulation genes, as well as substrates required for progression through meiotic chromosome divisions. So it is possible that Ime2 kinase is not directly responsible for the phosphorylation of Ndt80 but that another kinase encoded by an early meiotic gene is activated by Ime2 and then phosphorylates nascent Ndt80. To determine if it is possible that Ime2 directly phosphorylates Ndt80, we treated the recombinant Mbp-Ndt80 fusion with Ime2 kinase that was immunoprecipitated from sporulating yeast cells. In the experiment presented, we made use of a truncated but fully functional form of Ime2 (Ime2Δ175). The use of this truncated mutant was necessitated by the fact that full-length Ime2 migrates in the same position as the recombinant Mbp-Ndt80 fusion protein; Ime2 autophosphorylates and thus partially obscures the signal from phosphorylated Ndt80. Figure 5A shows that Ime2Δ175 will phosphorylate Mbp-Ndt80 fusion protein (lane 1) but not Mbp alone (lane 2). Ime2Δ175 autophosphorylates, but none of the phospho products comigrate with the Mbp-Ndt80 substrate (lane 3). In addition, the recombinant Ndt80 preparation has no intrinsic autophosphorylating capacity (lane 4). Identical results were observed when full-length Ime2 precipitated from either yeast or baculovirus-infected insect cells was used to phosphorylate recombinant Ndt80 (data not shown). The phosphorylation observed with immunoprecipitated Ime2Δ175 is due to Ime2 kinase activity and not to a spuriously associated kinase, since no phosphorylation of Ndt80 is observed when kinase reactions are performed with a kinase-dead Ime2 mutant (lane 10). The Mbp-Ndt80 fusion proteins detected by Coomassie staining in lane 5 consist of not only full-length Mbp-Ndt80 but also series of truncated Mbp-Ndt80 species. These proteins are C terminally degraded species of the Mbp-Ndt80 fusion, as all can be rebound to amylose resin by virtue of the amino-terminal Mbp affinity tag, and they react with an anti-Ndt80 polyclonal antibody that recognizes the Ndt80 amino terminus (data not shown). The failure of Ime2 to phosphorylate fusion products that are smaller than approximately 80 kDa in size suggested that Ime2 might be phosphorylating Ndt80 on its C terminus in vitro.
FIG. 5.
Ime2 phosphorylates Ndt80 in vitro. (A) A C terminally truncated but functional version of Ime2 (Ime2Δ175) was immunoprecipitated from sporulating yeast cells, and its kinase activity was assayed in vitro. Lanes 1 to 4 show an autoradiogram (Autorad) of the kinase reactions, while lanes 5 to 8 show Coomassie staining of the same gel. Ime2Δ175 kinase activity was assayed against E. coli-expressed Mbp-Ndt80 (lanes 1 and 5), Mbp (lanes 2 and 6), or no added substrate (lanes 3 and 7). The substrate, Mbp-Ndt80, was also assayed for intrinsic kinase activity (lanes 4 and 8). Molecular weights of markers (lane 9) are indicated in thousands. Mbp-Ndt80 was not phosphorylated by a kinase-dead mutant form of Ime2 (Ime2K97R) that was immunoprecipitated from yeast. Mbp-Ndt80 was treated with Ime2K97R (lane 10) or Ime2Δ175 (lane 11), and the kinase assays were separated on a 10% polyacrylamide gel and visualized by autoradiography. (B) Ndt80 lacking 167 amino acids from its C terminus was expressed under the regulation of an IME2 promoter in otherwise wild-type diploid cells. Expression of the truncated protein was monitored by Western blot analysis.
The C terminus of NDT80 encodes a potential transactivating domain.
To determine if the C-terminal region of Ndt80 might be an in vivo target of Ime2, a truncated version of Ndt80, lacking 167 C-terminal amino acids was expressed in yeast under the regulation of an IME2 promoter. This truncated protein accumulated but failed to display the full array of slower-mobility forms that we observed with full-length NDT80 (Fig. 5B). This observation suggested that the Ndt80 C terminus might be the target of Ime2 phosphorylation. To further investigate the possibility that the C terminus of Ndt80 might encode an important functional domain that is phosphorylated by Ime2, we generated a series of C-terminal deletions in NDT80 ranging from the full-length version to a deletion predicted to remove 167 amino acids. The full-length and truncated versions of NDT80 were expressed in an ndt80Δ diploid strain (DSY1257) under the regulation of an NDT80 promoter to ensure that they were expressed appropriately in sporulating cells. The ability of the mutants to function in vivo was assayed by testing their ability to induce the expression of an MSE-regulated LacZ reporter gene (SPS4-LacZ). The increasing length of deletions in NDT80 produced a graded loss of the ability to induce the expression of the reporter gene (Fig. 6A). A deletion of 167 amino acids resulted in near-background levels of β-galactosidase activity induced in the sporulating cells, suggesting that this mutation severely reduced the ability of NDT80 to induce transcription in vivo (Fig. 6A). This graded loss of activity was mirrored by the ability of the wild-type or mutant version of NDT80 to promote spore formation. While versions of NDT80 with small C-terminal deletions (Δ27) were able to effectively promote spore formation, larger C-terminal deletions like Δ167 prevented NDT80 from effectively promoting the completion of meiosis and spore formation (Fig. 6A). These data indicate that The C-terminal domain of NDT80 is required for NDT80 to effectively activate middle sporulation gene promoters. The critical function of the NDT80 C terminus was revealed when this region of the protein was fused to the heterologous DNA binding protein Gal4. When fused to the GAL4 DNA binding domain, NDT80 amino acids 426 to 627 can effectively promote expression of a GAL1-regulated LacZ reporter gene, indicating that the C-terminal domain of NDT80 has potential to function as an effective transactivation domain (Fig. 6B). The same C-terminal deletions that caused a graded reduction in NDT80 activity in sporulating cells (Fig. 6A) also resulted in a graded loss of trans-activating ability in the context of the hybrid GAL4-NDT80 construct (Fig. 6B). The ability of the NDT80 C terminus to act as a transactivation domain when expressed in vegetative cells suggested that this domain might be able to function as a transactivator independently of phosphorylation by Ime2. To confirm that the C terminus of NDT80 has the potential to act as a transactivation domain in sporulating cells, we fused the NDT80 C terminus (residues 426 to 627) to the LexA DNA binding domain. This hybrid LexA-NDT80 fusion was expressed in either IME2/IME2 or ime2/ime2 diploids that had been induced to initiate sporulation. The ability of the fusion to activate gene expression was monitored by the accumulation of β-galactosidase activity produced from an integrated LacZ-encoding gene that was regulated by eight tandem LexA binding sites. LexA alone does not activate the expression of this reporter in either IME2 or ime2 diploids that have been induced to sporulate (Fig. 6B). In contrast, LexA-Ndt80 is an effective activator of this reporter independent of IME2 (Fig. 6B). These observations suggest that the transactivating potential of the Ndt80 C-terminal domain may not require Ime2 activity. Consistent with this, we observed that in vitro Ime2 can phosphorylate Ndt80 that lacks the C-terminal transactivation domain (Fig. 6C). In this case, Mbp-Ndt80Δ167 (lanes 1 and 5) was produced in E. coli and used as a substrate for Ime2Δ175 as described in the legend to Fig. 5. Ime2Δ175 effectively phosphorylated Mbp-Ndt80Δ167 (lane 1) but did not phosphorylate Mbp1 alone (Fig. 6C). From these data, we conclude that the C-terminal domain of Ndt80 is required for the function of NDT80 in vivo and that this domain of the protein has the potential to act as a transactivation domain. In addition, this domain can function as a transactivator independent of IME2, suggesting that modification of Ndt80 by Ime2 is not required for the transactivation activity of Ndt80.
Ime2 is not required for Ndt80 to enter the nucleus.
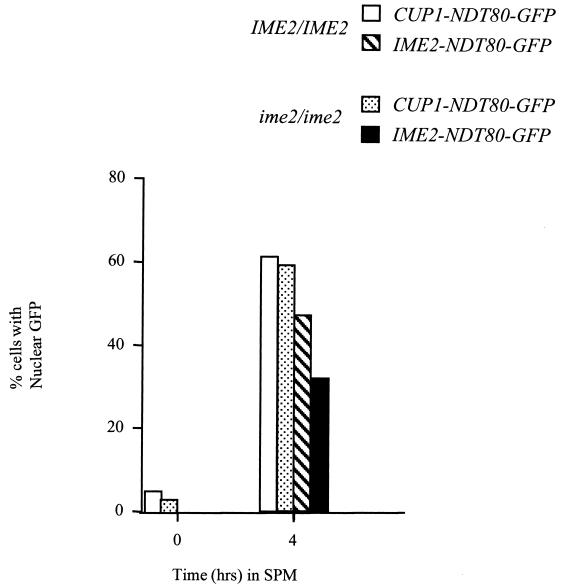
Another potential regulatory function of phosphorylation is to regulate nuclear localization. To determine if IME2 activity is required to promote nuclear entry of Ndt80, we generated fusions of the NDT80 open reading frame to GFP and expressed these fusions under the regulation of a CUP1 promoter or an IME2 promoter in IME2/IME2 or ime2/ime2 diploids. The cells were induced to sporulate (cells carrying a CUP1-NDT80-GFP fusion were induced to sporulate in the presence of 100 μM CuSO4 to induce the CUP1 promoter), and samples of the culture were taken at the indicated times. Samples of the unfixed cells were examined by fluorescence microscopy to determine the localization of the fusion proteins. After 4 h in SPM, a large proportion of the cells expressing NDT80-GFP under the regulation of a CUP1 promoter displayed nuclear GFP fluorescence (greater than 60% of the cells examined) (Fig. 7). A somewhat smaller proportion of cells expressing the fusions from an IME2 promoter displayed nuclear fluorescence, but this is likely due to the higher levels of expression driven by the CUP1 promoter. NDT80-GFP expressed from the IME2 promoter in ime2 mutants accumulated to much lower levels, but in most cases, this protein accumulated in the nucleus (Fig. 7). These observations indicate that Ime2 activity is not required to regulate the nuclear entry of Ndt80.
FIG. 7.
Phosphorylation of Ndt80 by Ime2 is not required for Ndt80 to enter the nucleus. An NDT80-GFP fusion was expressed under the regulation of an IME2 promoter in an IME2/IME2 strain or an ime2/ime2 mutant strain or from a CUP1 promoter in an IME2/IME2 strain or an ime2/ime2 mutant strain. Samples of the cultures were taken at 0 and 4 h after induction of sporulation, and unfixed cells were examined by fluorescence microscopy to determine the localization of Ndt80-GFP fusions. Two hundred cells per sample were scored. CUP1-NDT80 was induced at time zero by addition of CuSO4 to a final concentration of 100 μM.
Ime2 activity aids Ndt80 in binding to middle sporulation gene promoters.
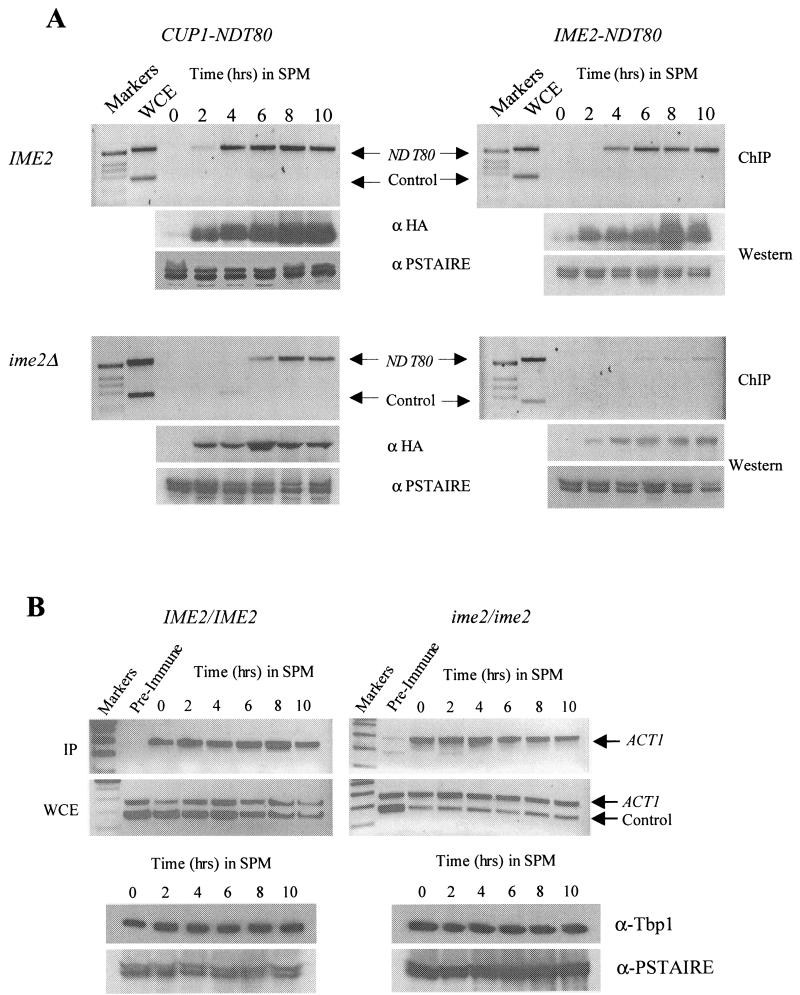
Yet another potential mechanism by which Ime2 activity might regulate Ndt80 is modulation of its ability to interact with MSE target sites in the promoters of middle sporulation genes. Phosphorylation of Ndt80 is not required for the protein to bind to naked DNA in vitro (5) (R.S., unpublished observations). However, binding to chromosomal MSE sites may be a more complex issue because of the presence of repressor proteins that may compete with Ndt80 for binding (33, 46). We used chromatin immunoprecipitation to determine if Ndt80 requires Ime2 to effectively interact with a middle sporulation gene promoter. NDT80HA was expressed under the regulation of either a CUP1 promoter (Fig. 8A, left side) or an IME2 promoter (Fig. 8A; right side) in either IME2/IME2 diploids (Fig. 8A, top) or ime2/ime2 diploids (Fig. 8A, bottom). The cells were induced to sporulate, and at the indicated times, samples were cross-linked and Ndt80HA was immunoprecipitated. The immunoprecipitates were assayed for the presence of DNA corresponding to the NDT80 promoter, which encodes two MSE sites to which Ndt80 has been proposed to bind. In IME2 cells, Ndt80HA effectively binds to the NDT80 promoter (Fig. 8A, top). Although considerable Ndt80HA protein accumulates within 2 h of induction of the CUP1 promoter (Fig. 8A, anti-HA Western blot), very little of this protein is found interacting with the target MSEs until 4 h into the sporulation time course (Fig. 8A, upper left). CUP1-regulated Ndt80HA also binds to the target MSE DNA in ime2 mutant cells, although the interaction is delayed relative to that in IME2 cells (Fig. 8A, lower left). This apparent defect is amplified when Ndt80HA is expressed under the regulation of an IME2 promoter, which produces lower levels of the tagged protein (Fig. 8A, right). To ensure that the apparent ime2-dependent defect in Ndt80 binding to chromosomal MSE sequences was not an artifact of our protocol, we assayed the interaction of TATA box binding protein Tbp1 with the ACT1 TATA box in IME2/IME2 and ime2/ime2 cells that had been induced to sporulate in the presence of CuSO4. We chose this specific interaction as a control because it is well characterized in mitotically growing cells (26) and because we have consistently observed stable steady-state levels of ACT1 mRNA throughout sporulation. Figure 8B shows that the Tbp1 protein is stable throughout the sporulation time course and that it can be found interacting with the ACT1 TATA box at all of the time points we assayed during sporulation in both IME2/IME2 and ime2/ime2 cells. These observations suggest that IME2 activity is required for Ndt80 to effectively interact with MSE DNA in the context of chromatin.
FIG.8.
Chromatin immunoprecipitation (ChIP) indicates that IME2 is required for Ndt80 to effectively interact with a middle sporulation gene promoter. (A) NDT80-HA was expressed under the regulation of a CUP1 promoter that was induced at the zero time point by addition of 100 μM CuSO4 (left side) or an IME2 promoter (right side) in either an IME2/IME2 diploid strain (top) or an ime2/ime2 diploid strain (bottom). The strains were inoculated in SPM, and samples of the culture were fixed in formaldehyde at the indicated time points. Following immunoprecipitation with anti-HA antibodies, whole-cell extracts (WCE) and immunoprecipitates (IP) were assayed by PCR for the presence of NDT80 promoter DNA (upper band in all panels) or the ARS305 sequence as a control (lower band). Ndt80-HA accumulation was assayed by probing samples of each extract with anti-HA antibodies. To control for loading, the blots were also probed with an anti-PSTAIRE antibody that recognizes both Cdc28 and Pho85. (B) DSY1289 (IME2/IME2) and DSY1299 (ime2/ime2) were assayed for Tbp1 binding to the TATA box sequence of the ACT1 promoter. Both strains were induced to sporulate in the presence of 100 μM CuSO4. At the indicated time points, cells were fixed with formaldehyde and the extracts were subjected to immunoprecipitation with anti-Tbp1 serum or preimmune serum. Both the immunoprecipitates (IP) and extracts (WCE) were assayed for the presence of ACT1 promoter DNA or the control DNA ARS305. Samples of each extract were assayed for the presence of Tbp1 by Western blotting with anti-Tbp1 antiserum. As in panel A, PSTAIRE antibody was used as a control for sample loading.
DISCUSSION
Progression through meiosis and exit from pachytene are dependent upon the expression of a family of middle sporulation genes that are activated by Ndt80. Given its central role in controlling meiotic gene expression, it is not surprising that Ndt80 is subject to multiple forms of regulation. The initial transcription of NDT80 is dependent on Ime1 and Ime2. Since the accumulation of these proteins is strictly dependent upon conditions that are conducive to sporulation, Ndt80 is induced only when cells have initiated meiotic differentiation. In addition, Ndt80 is posttranslationally modified by phosphorylation. We show that phosphorylation of Ndt80 is correlated with its ability to activate expression of the middle sporulation genes. The significance of this activating phosphorylation is highlighted by the demonstration that undermodified Ndt80 fails to activate the middle sporulation genes unless it is overexpressed. Further evidence of the importance of Ndt80 phosphorylation derives from observations that the DNA recombination checkpoint appears to impose a pachytene arrest by reducing the phosphorylation of Ndt80 and thus inhibiting its ability to induce middle sporulation gene transcription (42).
The role of phosphorylation in promoting Ndt80 activity.
Activation of transcription factors by phosphorylation is a common theme in many biological systems (19). Phosphorylation could, in principle, regulate Ndt80 activity by modifying its transactivation domain, regulating its cellular localization, altering its DNA binding capacity, or affecting its ability to interact with coactivators or repressors. Phosphorylation could have a direct impact on the transactivating ability of Ndt80 by leading to a conformational change that exposes the activation domain or by adding a negative charge to make it a more effective activator. Either effect could potentially facilitate interaction with either the basal transcription machinery or coactivators. Both p53 and CREB are activated by phosphorylation within their transactivation domains (12, 23). However, our analysis of NDT80 indicates that the C terminus encodes a transactivation domain and that this domain can function in either vegetative or sporulating cells in the absence of Ime2. It is possible that, by taking this domain out of its normal context as part of Ndt80 and fusing it to a heterologous DNA binding protein, we have spuriously made it independent of Ime2-dependent phosphorylation. However, when Ndt80 is overexpressed, it can activate target genes independently of modification by Ime2. In addition, we have demonstrated that Ndt80 lacking this C-terminal domain is still a substrate for Ime2 in vitro. These observations make it unlikely that the trans-activating capacity of Ndt80 is dependent upon Ime2.
Modification by phosphorylation also has the potential to regulate nuclear import. Several transcription factors have been shown to be regulated by phosphorylation at or near their nuclear localization signals, which determines how effectively they can enter the nucleus (20, 31). This does not appear to be the case for Ndt80, since an Ndt80-GFP fusion can effectively enter the nuclei of sporulating cells that lack Ime2. In addition, it has previously been demonstrated that overexpressed Ndt80 can enter the nuclei of vegetatively growing cells that do not express Ime2 (5).
Unmodified Ndt80 that has been produced in E. coli can bind to naked MSE DNA in vitro, suggesting that modification is not required for DNA binding (5). However, this should not be completely eliminated as a possible regulatory mechanism because the demands for binding to chromatin, and in particular meiotic chromosomes, may be greater than those for binding to naked DNA. Yet another possibility is that activating phosphorylation may release a transcription factor from an inhibitor or allow it to effectively compete with a repressor. A well-studied example of this is NFκB (1). No inhibitors of Ndt80 have been reported; however, genetic evidence suggests that middle sporulation genes are repressed by Sum1 or components of the Set3 complex that bind to MSE DNA (27, 46). Phosphorylation of Ndt80 may be required for it to effectively compete with the repressors and bind to MSE sites to activate the target genes. Indeed, the presence of Sum1 and its purported ability to act as an Ndt80 antagonist may explain our observation that premature accumulation of Ndt80 does not profoundly accelerate the timing of middle sporulation gene transcription. It may be that at least some members of the middle sporulation gene family are repressed until the Sum1 repressor complex is inactivated, degraded, or displaced, despite the accumulation of active Ndt80. The regulation of Sum1 and the Set3 repressor complex has not been well characterized. Given this uncertainty, we cannot categorically exclude the possibility that Ime2 is required to reciprocally regulate both (i) factors that act as repressors of the middle sporulation gene promoters and (ii) Ndt80.
Ndt80 phosphorylation is dependent on Ime2.
Posttranslational modification by phosphorylation is an important component of the regulation of Ndt80. Therefore, a key component of understanding this regulation is knowing the identity of the kinase responsible for providing the activating phosphorylation to Ndt80 in meiosis. The Cdk Cdc28 was an obvious choice because cdc28 temperature-sensitive mutants arrest at pachytene, similar to ndt80 mutants. In addition, Ndt80 has seven amino acid motifs that loosely match the consensus Cdk phosphorylation site. However, inactivation of Cdc28 or deletion of CLB5 and CLB6 does not prevent phosphorylation of Ndt80 or reduce middle sporulation gene expression. These observations imply that Cdc28 is not required for the modification of Ndt80. Phosphorylation of a transcription factor by a kinase that is expressed exclusively in meiosis enforces a direct linkage between activation of the transcription factor and meiotic progression. Ime2 plays a crucial role in promoting progression through meiosis. The most clearly defined role for Ime2 is in the induction of the early meiosis-specific genes and in promoting the meiotic G1-S transition through the targeting of Sic1 for degradation and possibly promoting the inactivation of APCCdh1 (2, 7, 29). In addition, there is some evidence that Ime2 provides an important function at later times in meiosis (39). One such function may be to down regulate the expression of early meiosis-specific genes (15). However, this has been difficult to clarify, since ime2 mutants arrest early in meiosis. IME2 is transcribed throughout meiosis, and indeed, we have shown that Ime2 kinase activity can be detected early in meiosis and persists until after MII has been completed. The abundance of Ime2 kinase activity during the meiotic divisions is consistent with a role for Ime2 in promoting events involved in chromosome division, including the activation of Ndt80. Cells that have a defect in Ime2 kinase fail to activate the expression of either early or middle sporulation genes. The failure to activate middle sporulation gene transcription is not simply the result of arrest in G1 phase, since ime2 sic1 double mutants progress to pachytene yet still fail to induce the middle sporulation genes (R.S., unpublished). In addition, we have shown that modest levels of Ndt80 are unable to induce middle sporulation gene expression in an ime2 mutant. These observations are consistent with the proposal that Ime2 kinase activity is required not only to induce transcription of NDT80 but also to activate the protein as a transcription factor.
Since Ime2 plays a key role in the regulation of many meiosis-specific genes and events, it remains possible that Ime2 is not directly responsible for Ndt80 phosphorylation but induces the expression or activity of another kinase that can phosphorylate Ndt80. In support of a direct role for Ime2 in phosphorylating and activating Ndt80, we have shown that Ime2 can directly phosphorylate purified Ndt80 or Ndt80 lacking the C-terminal transactivation domain in vitro.
Ndt80 is negatively regulated by the DNA recombination checkpoint. Since Ime2 appears to have a crucial role in the induction of Ndt80 expression and activity, this prompts the following question: is Ime2 regulated by the meiotic checkpoint controls? It has been shown that IME2 is down regulated by the DNA replication checkpoint, which prevents entry into MI in the presence of hydroxyurea (24). A role for Ime2 in the DNA recombination checkpoint is less clear, since IME2 transcription is not reduced in cells arrested at pachytene but NDT80 is poorly expressed and undermodified in this arrest (18, 42). It is possible that Ime2 kinase activity is affected by the checkpoint; however, this seems unlikely, since early meiotic genes are effectively induced in the arrested cells (18). It seems more likely that Ndt80 is directly targeted by some component of the recombination checkpoint, since increased dosage of Ndt80 will bypass the arrest and result in the accumulation of modified forms of Ndt80. In contrast, multicopy Ndt80 fails to suppress an ime2 mutant. Thus, while Ime2 is required for Ndt80 activation, it may not be the conduit by which the pachytene checkpoint inactivates Ndt80.
The precise role of phosphorylation in the regulation of Ndt80 activity is not clear. However, our observations are consistent with a simple model for the relationship between Ime2 and Ndt80. Ime2 kinase is required for the activation of early meiotic gene expression and the initial transcription of NDT80. Ime2 kinase is then responsible for the phosphorylation of Ndt80. Phosphorylated Ndt80 appears to be more effective in activating middle sporulation genes, perhaps because it is more effective in competing with the repressors that interact with MSE DNA during vegetative growth and early sporulation. Phospho-Ndt80 can thus effectively activate the NDT80 promoter and induce elevated levels of NDT80 and other middle sporulation genes. As sporulation progresses, Ime2 kinase activity decreases, which may allow the Sum1 or Set3 repressor complex to again impose repression on the middle sporulation genes, including NDT80. In the absence of Ime2, the initial expression of NDT80 is reduced and any Ndt80 that is synthesized is not modified and thus fails to effectively compete with Sum1 or Set3 and activate its own gene and the other middle sporulation genes. The identification of Ime2 as the kinase responsible for Ndt80 phosphorylation brings Ime2 to prominence as a key regulator of both the G1-S and MI cell cycle transitions in meiosis. The function of Ime2 in promoting Ndt80 activity was not previously anticipated and reveals an unexpected layer of complexity in the transcriptional cascade that regulates progression through meiosis. In particular, the use of Ime2 to induce both transcription and activity of Ndt80 highlights an additional point at which meiotic development can be regulated.
Acknowledgments
Thanks to Jason Lamoureux and Mark Glover for helpful discussions. We thank Jacqueline Segall for the gift of the SPS4-LacZ reporter plasmid, Michael Schultz for the gift of antibodies that recognize Tbp1, and Steve Reed for the anti-PSTAIRE antibody. We offer special thanks to Karen Robinson and Curt Wittenberg for critical reading of the manuscript and advice on purification of the recombinant proteins.
This research was supported by an Alberta Heritage Foundation for Medical Research Establishment grant and a Canadian Institutes of Health Research operating grant to D.S.
REFERENCES
- 1.Beg, A. A., and A. J. Baldwin. 1993. The IkB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 7:2064-2070. [DOI] [PubMed] [Google Scholar]
- 2.Bolte, M., P. Steigemann, G. H. Braus, and S. Irniger. 2002. Inhibition of APC-mediated proteolysis by the meiosis-specific protein kinase Ime2. Proc. Natl. Acad. Sci. USA 99:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 4.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 5.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 6.Dahmann, C., and B. Futcher. 1995. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics 140:957-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S-phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 8.Elledge, S. J. 1996. Cell cycle checkpoints: preventing and identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 9.Foiani, M., E. Nadgar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 10.Friesen, H., R. Lunz, S. Doyle, and J. Segall. 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8:2162-2175. [DOI] [PubMed] [Google Scholar]
- 11.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, G. A., and M. R. Montimony. 1989. cAMP stimulates the somatostatin gene transcription by phosphorylation of CREB at Ser 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 13.Grandin, N., and S. I. Reed. 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman-Raviv, N., E. Boger-Nadjar, I. Edri, and Y. Kassir. 2001. Cdc28 and Ime2 possess redundant functions in promoting entry into premeiotic DNA replication in Saccharomyces cerevisiae. Genetics 159:1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttmann-Raviv, N., S. Martin, and Y. Kassir. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecht, A., B. S. Strahl, and M. Grunstein. 1996. Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 17.Hepworth, S., L. Ebisuzaki, and J. Segall. 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hepworth, S., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of the middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, S. S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80:199-211. [DOI] [PubMed] [Google Scholar]
- 20.Hunter, T., and M. Karin. 1992. The regulation of transcription by phosphorylation. Cell 70:375-387. [DOI] [PubMed] [Google Scholar]
- 21.James, P., J. Halladay, and E. A. Graig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), Molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Lakin, N. D., and S. P. Jackson. 1999. Regulation of p53 in response to DNA damage. Oncogene 18:7644-7655. [DOI] [PubMed] [Google Scholar]
- 24.Lamb, T. M., and A. P. Mitchell. 2001. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics 157:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law, D. T. S., and J. Segall. 1988. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 8:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X.-Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 27.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moll, T., G. Tebb, U. Surana, H. Robitsch, and K. Nasmyth. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor, SWI5. Cell 66:743-758. [DOI] [PubMed] [Google Scholar]
- 32.Murakami, H., and P. Nurse. 2001. Regulation of premeiotic S phase and recombination related double strand DNA breaks during meiosis in fission yeast. Nat. Genet. 28:290-293. [DOI] [PubMed] [Google Scholar]
- 33.Pijnappel, W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose, M. D., F. Winston, and P. Heiter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 36.Schwab, M., A. S. Lutum, and W. Seufert. 1997. Yeast HCT1 is a regulator of CLB2 cyclin proteolysis. Cell 22:683-693. [DOI] [PubMed] [Google Scholar]
- 37.Schwob, E., T. Boehm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1/S transition in Saccharomyces cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 38.Shuster, E. O., and B. Byers. 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123:29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sia, R. A. L., and A. P. Mitchell. 1995. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol. Cell. Biol. 15:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strich, R., C. Surosky, C. Steber, F. Dubois, F. Messenguy, and R. E. Esposito. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796-810. [DOI] [PubMed] [Google Scholar]
- 41.Stuart, D., and C. Wittenberg. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12:2698-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tung, K. S., E.-J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma, R., S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 45.Wach, A., R. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 46.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zacharie, W., K. Schwab, K. Nasmyth, and W. Seufert. 1998. Control of cyclin ubiquitination by Cdk-regulated binding of Hct1 to the anaphase promoting complex. Science 282:1721-1724. [DOI] [PubMed] [Google Scholar]