Abstract
We previously identified an RNA binding protein, CUGBP1, which binds to GCN repeats located within the 5′ region of C/EBPβ mRNAs and regulates translation of C/EBPβ isoforms. To further investigate the role of RNA binding proteins in the posttranscriptional control of C/EBP proteins, we purified additional RNA binding proteins that interact with GC-rich RNAs and that may regulate RNA processing. In HeLa cells, the majority of GC-rich RNA binding proteins are associated with endogenous RNA transcripts. The separation of these proteins from endogenous RNA identified several proteins in addition to CUGBP1 that specifically interact with the GC-rich 5′ region of C/EBPβ mRNA. One of these proteins was purified to homogeneity and was identified as calreticulin (CRT). CRT is a multifunctional protein involved in several biological processes, including interaction with and regulation of rubella virus RNA processing. Our data demonstrate that both CUGBP1 and CRT interact with GCU repeats within myotonin protein kinase and with GCN repeats within C/EBPα and C/EBPβ mRNAs. GCN repeats within these mRNAs form stable SL structures. The interaction of CRT with SL structures of C/EBPβ and C/EBPα mRNAs leads to inhibition of translation of C/EBP proteins in vitro and in vivo. Deletions or mutations abolishing the formation of SL structures within C/EBPα and C/EBPβ mRNAs lead to a failure of CRT to inhibit translation of C/EBP proteins. CRT-dependent inhibition of C/EBPα is sufficient to block the growth-inhibitory activity of C/EBPα. This finding further defines the molecular mechanism for posttranscriptional regulation of the C/EBPα and C/EBPβ proteins.
The C/EBP family of transcription factors is involved in the regulation of a number of biological processes (9, 13, 23, 34). Two members of this family, C/EBPα and C/EBPβ, are encoded by intronless genes, but a single mRNA of each produces several protein isoforms by alternative translation from in-frame AUG codons (1, 4, 5, 10, 14, 20). The molecular mechanisms that regulate translation of C/EBPα and -β mRNAs are not well understood. It has been suggested that the alternative usage of AUG codons may be controlled by specific RNA binding proteins which bind to mRNAs and increase the interaction of ribosomes with downstream AUG codons (33, 36). Experimental studies of liver regeneration, the acute-phase response to inflammation, and the pathology of myotonic dystrophy (DM) showed that, in all of these situations, CUGBP1 binds to the GCN repeats within the 5′ region of C/EBPβ mRNA and induces downstream translation initiation of a dominant-negative C/EBPβ isoform, LIP (30, 33, 36).
RNA binding proteins play important roles in the regulation of cell growth and differentiation through the control of mRNAs whose products are involved in the cell cycle (2). It has been shown that a family of elav-like proteins binds to an AU-rich sequence located within the 3′ untranslated region (UTR) of certain mRNAs and increases the stability of these mRNAs (6, 11, 12, 26, 35). A member of the elav-like proteins, HuR, also regulates the rate of translation of p27 mRNA by interacting with the 5′ region of p27 mRNA (17). It has recently been found that RNA binding proteins that specifically interact with GC-rich sequences may also regulate the translation of certain mRNAs (28, 30, 31, 33, 36). Regulation of RNA binding proteins occurs at different levels, including transcription, translation, posttranslational modifications, and intracellular localization. A new pathway for the regulation of RNA binding proteins has been suggested by investigations of the molecular alterations observed in DM type 1 (DM1) patients. Several reports have indicated that the activity of CUG repeat binding protein (CUGBP1) is affected by expansion of GCU (CUG) repeats within the 3′ UTR of myotonin protein kinase (DMPK) mRNA (22, 28-31). The mechanism of this effect is complicated and involves the association of CUGBP1 with GCU repeats, forming RNA-protein complexes (30). Experiments with cultured cells showed that CUGBP1 is more stable in the RNA-protein complexes than free CUGBP1 (30). As a result, DM1 cells generate a reservoir of CUGBP1-RNA complexes, which can potentially release the protein and generate a pool of free and active CUGBP1. This finding suggests that the activities of RNA binding proteins in vivo may be altered by overexpression of RNA transcripts containing the corresponding binding sites. Analysis of nucleotide sequences that are necessary for the interaction of CUGBP1 with RNA revealed that the binding of CUGBP1 does not require a strong consensus: CUGBP1 binds to GC-rich regions, preferentially to GCN repeats (28-31, 36). At least four mRNAs have been shown to contain these repeats, and they are able to interact with CUGBP1 (28-31).
In our search for additional RNA binding proteins interacting with GCN repeats, we purified and characterized a new GC-rich binding protein, calreticulin (CRT). Data in this paper show that the RNA binding activity of CRT is masked in total extracts due to association with endogenous transcripts. Similar to CUGBP1, the separation of CRT from RNA significantly increases its RNA binding activity. Both free CUGBP1 and CRT bind strongly to transcripts containing long GCN repeats. Our data demonstrate that CRT interacts with GCN repeats located within the C/EBPβ and C/EBPα mRNAs and that this interaction represses protein translation from these mRNAs.
MATERIALS AND METHODS
RNA probes.
In this study, we used RNA oligomers that cover regulatory regions of the C/EBPα and C/EBPβ mRNAs. The locations of these oligomers on C/EBP mRNAs are shown in Fig. 1. The nucleotide sequences of RNA probes used in these studies are as follows: sORF, 5′-AUGCCUCCCCGCCGCCGCCCGCCGCCUUAG-3′ (GCC repeats are underlined); sORF-mut, 5′-AUGCCUCCCCAUCUACAUCCUACAUCUUAG-3′ (GC islands are replaced with AU/UA, shown in boldface); LAP, 5′-AUGCACCGCCUGCUGGCCUGGGAC-3′ (the CUGBP1 binding site is underlined); FL-LAP, 5′-AUGCACCGCCUGCUGGCCUGGGACGCAGCAUGCCUCCCCGCCGCCGCCCGCGCCUUAG-3′ (two CUGBP1 binding sites are underlined); and GX, an AU-rich oligomer with a random nucleotide composition (33). RNA oligomers covering secondary structures of C/EBPα mRNAs are as follows: 5′ stem-loop (5′SL), 5′-GGAGGUGCGCGGGUAUACCCGCGCAGGCUU-3′; internal loop (oligomer A containing GCG repeats [underlined]), 5′-CCCCACGGGCGGCGGCGGCGGCGGCGACUU-3′; and internal loop (oligomer B containing GCC repeats [underlined]), 5′-UAACCAGCCGCCGCCGCCGCCGCCGCCGCCGCCC-3′. A double-stranded internal stem structure (SL int) was generated by annealing of oligomers A and B. The double-stranded Int stem RNA was separated from single-stranded oligomers by gel electrophoresis and extracted from the gel. All RNA oligomers were labeled in in vitro kinase reactions with T4 kinase and [γ-32P]ATP. GCU repeats of different lengths (6, 39, and 123 repeats) derived from plasmids described earlier (30) were placed under the T7 promoter, and RNA transcripts were synthesized in an vitro transcription reaction as described previously (30, 33).
FIG. 1.
C/EBPα and C/EBPβ mRNAs and RNA probes. (A) C/EBPβ mRNA. The positions of AUG codons for the full-length (FL), LAP, and LIP isoforms of C/EBPβ are shown. FL-LAP, RNA oligomer covering a sequence from the first to the second AUG codon; LAP, RNA oligomer containing GCU repeats (see Materials and Methods) (33); sORF, short out-of-frame open reading frame containing GCC repeats that interact with CUGBP1 (33) and with CRT (this paper); DBD, coding sequence for the DNA binding domain. (B) C/EBPα mRNA. The positions of the AUG codons for 42-, 30-, and 20-kDa isoforms of C/EBPα are shown. The location of a binding site for hnRNP E2 (21) within the C/EBPα mRNA is also shown. 5′SL, A(GCG)8, and B(GCC)8, positions of RNA oligomers used in these studies. Regions A and B form an internal SL structure which interacts with CRT.
Plasmids.
Expression constructs for wild-type C/EBPβ and C/EBPα were described earlier (33, 34). C/EBPα deletion constructs were characterized previously (34) (see Fig. 8A). GC-mut C/EBPβ mRNA was generated using site-specific mutagenesis. In this construct, GC islands within the sORF sequence are replaced with AU or UA. It has been shown that these substitutions abolish the interaction of CUGBP1 with the 5′ region of C/EBPβ mRNA (35). The CRT plasmids GST-CRT and hemagglutinin HA-CRT were kindly provided by David Williams. GST-CRT protein was isolated from bacteria by using glutathione beads. To further purify the protein, an additional step of ion-exchange chromatography on UnoQ (high-performance liquid chromatography [HPLC]; BioLogic high-resolution chromatography system [BioLogic HR]; Bio-Rad) was performed. The purified GST-CRT represented a highly purified (90 to 95%) protein according to Coomassie staining. The identity of GST-CRT was verified by Western blotting with antibodies to CRT (Santa Cruz Biotechnology and Sigma).
FIG. 8.
CRT inhibits translation of C/EBPα mRNA in cultured cells. (A) C/EBPα constructs used in these studies. Gal4-C/EBPα constructs lack the sequence that forms the 5′SL structure. N175 lacks both 5′SL and the C-terminal portion of C/EBPα containing region B. Gal4-C/EBPα-ΔB has an internal deletion and lacks the 5′SL and region B. DBD, coding sequence for the DNA binding domain. (B) HA-CRT inhibits translation of C/EBPα mRNAs containing the internal (SL int) structure. HA-CRT was cotransfected with C/EBPα constructs, and the proteins were isolated 18 h after translation and analyzed by Western blotting. Each filter was probed with Gal-4 or C/EBPα antibodies and then reprobed with antibodies to HA (top) and then with antibodies to β-actin to verify protein loading. The β-actin control is shown for the Gal4-C/EBPα membrane. A representative result of five independent experiments is shown. CRT represses translation of CMV-C/EBPα and Gal4-C/EBPα but does not inhibit translation of C/EBPα mRNAs lacking region B (Gal4-N175 and C/EBPα-ΔB).
Isolation of total protein extracts, fractionation on a DEAE column, and UV cross-link assay.
Whole-cell extracts were isolated from HeLa cells as previously described (29, 33). To separate RNA and proteins, total extracts were loaded on DEAE-Sepharose in DEAE binding buffer (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 5 mM dithiothreitol, 30 mM NaCl, and 10% glycerol). After being intensively washed, the proteins were eluted using 0.1, 0.2, 0.3, and 0.4 M NaCl. The elution fractions were spun through G-50 spin columns to remove high levels of salt. The presence of RNA within DEAE elution fractions was examined by slot hybridization with an 18S rRNA probe. Under the conditions of the experiments, RNA was not detectable within 0.1 to 0.4 M NaCl elution fractions. Therefore, these samples are referred to as RNA free. HeLa whole-cell protein extracts, DEAE protein fractions, or bacterially expressed GST-CRT proteins were incubated with RNA transcripts containing various numbers of GCU repeats. Equal amounts of radioactive RNA probes (50,000 to 200,000 cpm) were incubated with proteins for 30 min at room temperature and subjected to UV treatment for 5 min at 125 mJ. The samples containing long RNA transcripts (DMPK mRNA, GCU39, and GCU123) were treated with RNase A and then loaded on 10% polyacrylamide gel electrophoresis gels containing 0.1% sodium dodecyl sulfate (SDS). After electrophoresis, the proteins were transferred to the membrane and autoradiographed. The same membranes were stained with Coomassie blue to verify protein loading.
Analysis of RNA-protein interactions using HPLC-based size exclusion chromatography.
Whole-cell protein extracts from HeLa cells were fractionated by HPLC-based size exclusion chromatography on an SEC-400 column (BioLogic HR; Bio-Rad). Prior to each run, the column was calibrated with standard protein molecular mass markers. Gel filtration fractions (300 μl) were collected and analyzed by gel shift assay with the sORF or (GCU)8 probe. Where appropriate, protein extracts were treated with RNase A (0.2 to 1.0 U/μl) and then fractionated by HPLC chromatography. To locate 18S rRNA transcripts within gel filtration fractions, 150 μl of each fraction was denatured with 50% formamide, spotted on a nitrocellulose filter, and hybridized with 18S probe as described previously (32). Under conditions optimal for protein isolation, RNA transcripts are partially degraded and are observed in multiple fractions ranging from 450 to 1,000 kDa. The majority of GC-rich binding proteins colocalize with RNA transcripts.
UV cross-link-immunoprecipitation assay (UV-IP).
To identify CUGBP1 within gel filtration fractions, the fractions were incubated with sORF or (GCU)8 probes, the proteins were linked to the probe by UV treatment as described previously (31, 33), and CUGBP1 was precipitated from reaction mixtures by using specific polyclonal antibodies (31). After being intensively washed, the immunoprecipitates were loaded on a denaturing gel, transferred to a membrane, and autoradiographed.
Purification of the GCN binding protein CRT.
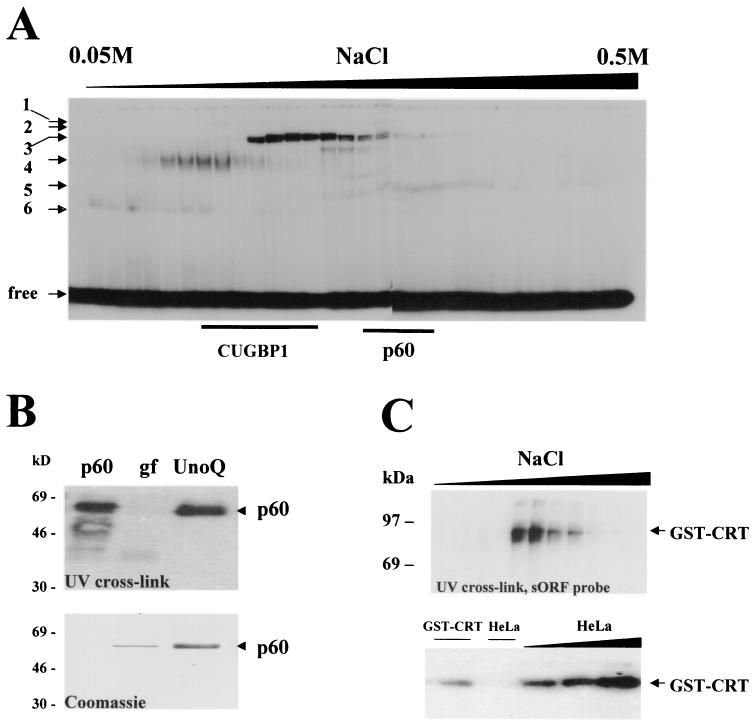
We have previously shown that rat liver contains six or seven proteins that specifically interact with GCN repeats within the 5′ region of C/EBPβ mRNA (33, 36). Therefore, cytoplasmic extracts from rat liver were used for the isolation of GCN binding proteins. Twenty milligrams of protein were first fractionated by ion-exchange chromatography using an UnoQ column (HPLC; Bio-Rad). Proteins were eluted from the column using a linear gradient of NaCl from 0.05 to 0.5 M. Fractions containing GCN binding proteins were identified by gel shift and UV cross-link assays with sORF or (GCU)8 probes. Fractions containing GCN binding protein with a mass of 60 kDa were combined, dialyzed against buffer for UnoQ, and applied for a second round of ion-exchange chromatography by elution with a narrow range of NaCl concentrations (from 0.15 to 0.25 M). Analysis of the elution fractions by silver staining revealed that purified p60 is electrophoretically homogenous and binds to sORF with high affinity (see Fig. 4). The amino acid sequence of p60 was determined by the sequencing core at Baylor College of Medicine. Comparison of the amino acid sequence of p60 with those of known proteins indicated that p60 is identical to CRT.
FIG. 4.
p60 is identical to CRT and binds to the 5′ region of C/EBPβ mRNA. (A) Ion-exchange chromatograph of cytoplasmic proteins on UnoQ column (HPLC; Bio-Rad). Twenty milligrams of cytoplasmic proteins from rat liver were loaded, washed, and eluted with a linear gradient of NaCl (from 0.05 to 0.5 M). Every other fraction was analyzed by gel shift assay with the sORF probe. The positions of six RNA-protein complexes are shown on the left. The locations of CUGBP1 and the p60 protein within gel filtration fractions were determined by UV cross-link assay and are indicated at the bottom. free, position of free sORF probe. (B) (Top) UV cross-link analysis of the purified p60 protein with the sORF probe. p60, fraction from UnoQ chromatography containing p60 RNA binding activity; gf, gel filtration fraction containing p60 activity after the second step of purification; UnoQ, ion-exchange chromatography fraction containing p60 activity after the last purification step. (Bottom) Coomassie staining of the same membrane after UV cross-link analysis. The N-terminal sequence of p60 was determined and was shown to be identical to that of CRT. (C) Bacterially expressed, purified GST-CRT interacts with the 5′ region of C/EBPβ mRNA. (Top) GST-CRT was expressed in bacteria, isolated by pull-down with glutathione beads, and further purified by HPLC-based ion-exchange chromatography. UV cross-link analysis of elution fractions with the sORF probe is shown. (Bottom) Binding activity of bacterially expressed, purified GST-CRT is increased after incubation with cytoplasmic proteins from HeLa cells. In the right-hand three lanes, GST-CRT was incubated with increasing amounts of cytoplasmic proteins from HeLa cells (0.5, 1, and 2 μg) and examined by UV cross-link assay with the FL-LAP probe. Lane GST-CRT, UV cross-link with GST-CRT alone. Lane HeLa, incubation of 2 μg of HeLa proteins alone with the FL-LAP probe.
Western analysis.
Proteins from DEAE or HPLC fractions (50 to 100 μg) were separated by 10 to 12% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked with 10% dry milk prepared in TTBS buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature. Primary monoclonal antibodies against CUGBP1 (3B1) or polyclonal antibodies against EXP42 were added and incubated for 1 h. The filter was washed and incubated with secondary antibodies for 1 h, and then immunoreactive proteins were detected using the enhanced-chemiluminescence method (Pierce). After detection of proteins, the membranes were stripped and reprobed with antibodies against β-actin or stained with Coomassie blue to verify protein loading.
Gel shift analysis.
The conditions for electrophoretic mobility shift assays were described previously (33, 36). Proteins (5 to 25 μg) were incubated with sORF or (GCU)8 labeled with [γ-32P]ATP and T4 kinase. Where appropriate, RNA competitors (100 ng) were added prior to the addition of probe.
Effect of CRT on translation of C/EBPα and C/EBPβ mRNAs in reticulocyte lysate.
To examine the result of the interaction of CRT with C/EBP mRNAs, a cell-free coupled transcription-translation system in rabbit reticulocyte lysate was used. The conditions for this assay were described previously (33, 36). GST-CRT was expressed in bacteria and isolated to homogeneity as described above. C/EBPβ or C/EBPα was translated in reticulocyte lysate containing [35S]methionine from wild-type constructs or from mutant C/EBPβ constructs containing an ATG→TTG mutation of the third initiation AUG codon for the LIP (ATG-3) C/EBPβ isoform (36). Each translation mixture also contained a plasmid coding for USF, which served as a negative control. GST-CRT was added to the mixture before the addition of constructs. After translation, C/EBPβ, C/EBPα, and upstream stimulatory factor (USF) were immunoprecipitated from the same reaction mixtures with specific antibodies and analyzed by electrophoresis and autoradiography. The effect of CRT on the translation of C/EBP proteins was calculated as a ratio of C/EBP to USF. Densitometric analysis was carried out using a Molecular Dynamics Personal Densitometer and ImageQuant software.
Effect of CRT on translation of C/EBPα and C/EBPβ mRNAs in cultured cells.
COS7 or HT1080 cells were cotransfected with C/EBPα plus HA-CRT or C/EBPβ plus HA-CRT constructs using the Fugene 6 transfection reagent (Boehringer Mannheim) according to the manufacturer's protocol. Since CRT is highly expressed in cultured cells, preliminary studies were performed to examine the induction of CRT relative to endogenous levels of the protein. We established conditions for transfection where CRT levels are three- to fivefold induced in each transfected cell. The data in this paper were obtained under the established conditions. The best effect was reproducibly observed when the ratio of transfected C/EBP to CRT plasmids was 1:5. Eighteen hours after transfection, total protein extracts were isolated and the levels of C/EBPα, C/EBPβ, and CRT were examined by Western blotting with specific antibodies as described above. Summaries of three or four independent experiments are presented in this paper.
Immunostaining of HT1 cells and colony formation assay.
To visualize transfected cells, the HA-CRT vector was cotransfected into HT1 cells with a plasmid coding for green fluorescent protein (GFP) (10:1). Control cells were transfected with GFP alone. C/EBPα was induced by the addition of 10 mM IPTG. Sixteen hours after C/EBPα induction, the cells were fixed and stained with antibodies to C/EBPα (14AA; Santa Cruz). The presence of GFP indicated cells transfected with HA-CRT. DAPI (4′,6′-diamidino-2-phenylindole) staining was also performed to visualize all cell nuclei.
To determine whether CRT can abolish C/EBPα growth-inhibitory activity, formation of single- and multiple-cell colonies was performed in HT1 cells as described earlier (34). Briefly, HT1 cells were cotransfected with either an empty vector or HA-CRT and a plasmid expressing β-galactosidase (β-Gal). Cotransfection with β-Gal allows the transfected cells to be easily visualized and counted. C/EBPα expression was induced by IPTG (isopropyl-β-d-thiogalactopyranoside), the cells were fixed on days 1 and 3 after the addition of IPTG, and the β-Gal activity was examined. Dividing cells are present as clusters containing three or more cells by day 3 after transfection, while arrested cells remain as single-cell colonies. The numbers of arrested and dividing cells were calculated, and a summary of five experiments is presented (see Fig. 9C).
FIG. 9.
Overexpression of CRT blocks C/EBPα-mediated growth arrest. (A) Overexpression of CRT in HT1 cells inhibits the expression of C/EBPα. HT1 cells were transfected with HA-CRT plus GFP (10:1) (top) or with GFP alone (bottom). C/EBPα was induced by the addition of 10 mM IPTG, and C/EBPα levels were determined by immunostaining with specific antibodies (14AA). Two fields for CRT-plus-GFP transfections are shown. DAPI (nuclear) staining (blue) shows all of the cells in the field. CRT-plus-GFP cells are green, and C/EBPα expression is shown in red. The table below shows a summary for 84 cells transfected with CRT plus GFP and for 30 cells transfected with GFP alone. (B and C) Overexpression of CRT abolishes C/EBPα-mediated growth arrest. HT1 cells were cotransfected with β-Gal plasmid and an empty vector (pcDNA) or with plasmid expressing HA-CRT. C/EBPα was induced by the addition of 10 mM IPTG, and the cells were stained for β-Gal activity on days 1 and 3 after C/EBPα induction. (B) Typical picture of colonies on day 3 after transfection. CRT-overexpressing cells proliferate and form colonies containing three or more cells, while cells transfected with the empty vector do not proliferate and stay as single cells. (C) Summary of three independent experiments. The error bars indicate standard deviations.
RESULTS
The majority of GCU binding proteins are associated with endogenous transcripts in extracts from HeLa cells.
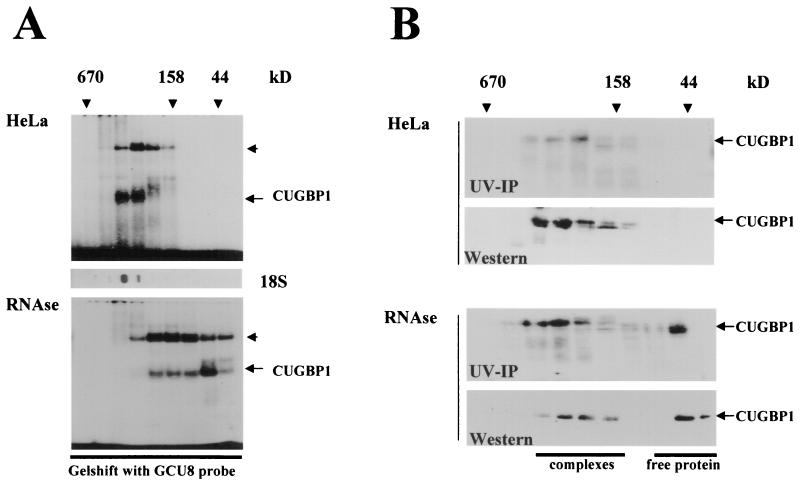
Previous investigations showed that certain RNA binding proteins interact with the C/EBPα and C/EBPβ mRNAs and that these interactions affect the translation of C/EBP proteins (21, 28-31). Figure 1 shows the structures of the C/EBPα and C/EBPβ mRNAs and the locations of the binding sites for RNA binding proteins. Previous studies demonstrated that the 5′ region of C/EBPβ mRNA (sORF) interacts with at least six or seven RNA binding proteins in cytoplasmic extracts from liver (33, 36). We also found that one of these RNA binding proteins, CUGBP1, increases the translation of LIP, a dominant-negative isoform of C/EBPβ, during the acute-phase response (36) and in DM1 patients (30). In DM1 patients, CUGBP1 is associated with GCU repeats, which are expanded within the mutant DMPK mRNA (30). In these studies, the association of other unknown GC-rich RNA binding proteins with endogenous transcripts was also observed (30). To further address the identities of these proteins and to investigate their roles in the translation of C/EBP proteins, we utilized a method which allowed the separation of RNA-protein complexes and free RNA binding proteins. This method utilizes HPLC-based size exclusion chromatography followed by gel shift, UV cross-link, and Western analyses of gel filtration fractions (30). As an initial step, we analyzed whether GC-rich RNA binding proteins are associated with endogenous transcripts in HeLa cells. Free proteins and RNA-protein complexes were separated and analyzed by gel shift assay with a short (GCU)8 probe (Fig. 2A). As can be seen, the majority of GCN-interacting proteins (including CUGBP1, as shown below) are located in fractions containing high-molecular-mass (up to 600-kDa) complexes. To determine whether the gel filtration fractions contained endogenous RNA transcripts, half of each fraction was analyzed by slot hybridization with an 18S rRNA probe (Fig. 2A, 18S). Under conditions optimal for protein isolation, RNA transcripts are partially degraded and are located in fractions with molecular masses smaller than expected. Slot hybridization with the 18S rRNA probe showed that the majority of GC-rich RNA binding proteins colocalize with RNA transcripts in high-molecular-mass fractions. To examine whether the localization of GC-rich RNA binding proteins in high-molecular-mass fractions is due to interaction with RNA, total protein extracts were treated with RNase A prior to gel filtration and then separated by size exclusion chromatography. The digestion of RNA by RNase A prior to fractionation leads to the shift of a significant portion of GCU binding proteins (including CUGBP1 [see below]) to fractions corresponding to the sizes of free, unbound proteins (Fig. 2A). Thus, these data demonstrate that, in HeLa cells, GCU binding proteins are associated with endogenous transcripts.
FIG. 2.
(A) GC-rich RNA binding proteins within total HeLa extracts are associated with endogenous transcripts. (Top) Whole-cell extracts from HeLa cells were fractionated by size exclusion chromatography (SEC-400; HPLC; Bio-Rad). The positions of standard protein markers are shown at the top. The presence of GC-rich RNA binding proteins in each fraction was investigated by gel shift assay with the (GCU)8 probe. The positions of the previously described complex containing CUGBP1, as well as a new complex with slower mobility (short arrow), are shown to the right. (Middle) Slot hybridization of gel filtration fractions with 18S rRNA probe (18S). (Bottom) Protein extracts were treated with 1 U of RNase A/100 μl and fractionated by gel filtration as described above. (B) Analysis of CUGBP1 in size exclusion fractions with or without RNase A treatment prior to fractionation. UV-IP, gel filtration fractions of proteins from HeLa cells analyzed by UV cross-link assay followed by immunoprecipitation with antibodies to CUGBP1. Western blots show the positions of CUGBP1 within gel filtration fractions.
To further examine the association of GC-rich binding proteins with RNA, we investigated the status of CUGBP1 using specific reagents. Gel filtration fractions were examined using two specific assays: (i) UV cross-link analysis followed by immunoprecipitation with antibodies to CUGBP1 (UV-IP) and (ii) Western blotting with antibodies to CUGBP1. Figure 2B shows a reproducible result of these experiments. The UV-IP method is a specific assay which shows three characteristics of the protein: specific binding to the RNA probe, immunoreactivity, and molecular mass. As can be seen in Fig. 2B, both UV-IP and Western blotting analyses revealed that, in total extracts from HeLa cells, CUGBP1 is observed in high-molecular-mass gel filtration fractions. The analysis of the samples treated with RNase A shows that a portion of CUGBP1 is shifted to low-molecular-mass fractions (Fig. 2B) after the digestion of RNA. We reproducibly observed a shift of approximately 30 to 50% of CUGBP1 to low-molecular-mass fractions according to Western analysis. Thus, these studies indicate that, like other GC-rich RNA-interacting proteins, the majority of CUGBP1 is bound to endogenous transcripts in total extracts from HeLa cells.
The separation of proteins from RNA identifies new proteins interacting with the 5′ region of C/EBPβ mRNA.
Given the observation that the majority of GC-rich RNA binding proteins in HeLa cells are bound to endogenous transcripts, we separated these binding proteins from endogenous RNA for further analysis. For this purpose, we carried out ion-exchange chromatography on a DEAE column using 0.1 to 0.4 M NaCl step elution. Under these conditions, the majority of proteins are eluted within 0.1 to 0.4 M NaCl, while nucleic acids (RNA and DNA) are tightly bound to the DEAE column and require higher salt concentrations to be eluted. Examination of 18S rRNA within elution fractions by slot hybridization showed no RNA in 0.1 to 0.4 M NaCl fractions (data not shown); therefore, we refer to these fractions as RNA free. After DEAE fractionation, the binding activities of GC-rich RNA-interacting proteins within RNA-free fractions were examined using a variety of GC-rich RNA probes, including sORF riboprobe (33, 36) and RNAs containing GCU repeats of different lengths. The left-hand gel in Fig. 3A shows a representative gel shift assay with the sORF probe. We found that the separation of RNA binding proteins from RNA leads to an increase in detection of the binding activities of the proteins and to identification of new GC-rich RNA binding proteins, the activities of which were not detected in total extracts. In addition to CUGBP1, three other RNA-protein complexes are observed in RNA-free fractions by gel shift assay with the sORF probe. To examine whether these complexes are formed by specific interactions, a number of cold competitors were incorporated in the binding reaction with RNA-free fractions. The right-hand gel in Fig. 3A shows the result of competition with the 0.2 M elution fraction. Proteins from this fraction form three major complexes. Two sites were previously identified within C/EBPβ mRNA to which CUGBP1 binds: sORF and LAP sequences (33) (see Materials and Methods). The two slower-migrating complexes are competed by both cold (GCU)8 and LAP probes, while a faster-migrating complex is competed only by cold sORF. This pattern of competition indicates that separation of proteins from RNA does not affect their specificities but increases their detectable binding activities, perhaps due to the removal of endogenous RNA competitors.
FIG. 3.
Identification of additional GC-rich RNA binding proteins after separation from RNA. (A) The left-hand gel shows gelshift analysis of DEAE fractions (shown at the top) with the sORF probe. The positions of GCU- and sORF-specific complexes are indicated. C, unfractionated cytoplasm. −, no protein added. The right-hand gel shows gel shift and competition analyses of the 0.2 M NaCl elution fraction. Cold competitors (indicated at the top) were added to the binding reaction before the addition of probe. GCU, L (LAP), and sORF, competitors that contain GC-rich sequences (see Materials and Methods) and interact with CUGBP1 (35); GX, AU-rich oligomer with random sequence (35). free, position of free sORF probe. (B) (Top) UV cross-link analysis of DEAE fractions with a short (GCU)8 probe. Cytoplasm (C) and elution fractions (shown at the top) were incubated with the (GCU)8 probe, linked by UV treatment, and separated by SDS-gel electrophoresis. The positions of the CUGBP1, p60, and p150 binding proteins are shown on the right. (Bottom) Western blot of DEAE fractions with monoclonal antibodies to CUGBP1 and with antibodies to EXP42. (C) RNA-free CUGBP1 and p60 bind strongly to RNA containing long GCU repeats. Both gels show the interaction of GC-rich RNA binding proteins with RNA transcripts containing 39 GCU repeats (left) or 6, 39, and 123 GCU repeats (right). C, unfractionated cytoplasm. The position of EXP42 was determined by reprobing the membrane with antibodies to EXP42.
Next, we analyzed the RNA-free fractions by UV cross-link assay with the (GCU)8 probe. In agreement with the results of the gel shift assay, UV cross-link and Western blotting analyses (Fig. 3B) showed that the majority of CUGBP1 is located in 0.2 and 0.3 M elution fractions. As can be seen in both the gel shift and UV cross-link assays, the RNA-binding activity of CUGBP1 after separation from RNA is dramatically increased. Although the amounts of CUGBP1 used for the UV cross-link from 0.2 and 0.3 M DEAE elution fractions are smaller than the amounts of CUGBP1 in total extract (Fig. 3B, Western), the binding activity of the RNA-free CUGBP1 is significantly higher (Fig. 3B, UV). These results are consistent with the suggestion that, in HeLa cells, CUGBP1 is associated with endogenous transcripts and that there are limited amounts of free CUGBP1 protein capable of binding to the probe. Incorporation of specific competitors into the binding reaction revealed that CUGBP1 separated from RNA interacts specifically with GCN repeats (data not shown).
The separation of RNA binding proteins from RNA also shows that the binding activities of several other proteins are increased in RNA-free fractions, while in total extracts, the activity is weak or not detected. In particular, we identified a new protein with a molecular mass of 60 kDa (p60), the binding activity of which was not observed in total extracts. However, this protein strongly interacts with the (GCU)8 probe after separation from RNA (Fig. 3B). It is interesting that the 0.1 M elution fraction also contains a protein migrating in the position of p60. To further characterize the activity of p60, we analyzed the interaction of RNA-free p60 with transcripts containing different numbers of GCU repeats and compared the RNA binding activity of p60 with the activities of two other proteins, CUGBP1 and EXP42, which have both been shown to bind to GCU repeats (18). Western blotting with specific antibodies showed that CUGBP1 is enriched in 0.2 and 0.3 M elution fractions, while EXP42 is observed in the 0.4 M elution fraction (Fig. 3B). Three RNA probes containing 6 (DMPK mRNA), 39, and 139 GCU repeats were synthesized in vitro and applied to UV cross-link assays with whole-cell extracts and with DEAE fractions containing RNA-free proteins. The results of these experiments are shown in Fig. 3C. UV cross-link analysis with the (GCU)39 probe showed that both RNA-free p60 and CUGBP1 interact with long GCU repeats. The UV cross-link assay also indicated that, in RNA-free DEAE fractions, p60 and CUGBP1 bind more strongly to RNA containing longer GCU repeats, while in total protein extracts the binding of p60 is not detectable and the binding activity of CUGBP1 is much weaker. Because EXP42 has been identified as a protein that interacts with long GCU repeats (18), we also examined EXP42 in HeLa extracts and compared its activities in total extracts and RNA-free samples. As can be seen in Fig. 3B (Western), EXP42 is observed only in the 0.4 M NaCl fraction. Surprisingly, no binding of EXP42 to GCU repeats was detected either with total protein extracts or with 0.4 M DEAE fractions enriched for the EXP42 protein (Fig. 3C; the position of EXP42 is shown). Consistent with these observations, we also found that bacterially expressed, purified CUGBP1 and p60 bind more strongly to transcripts with longer GCU repeats, while bacterially expressed purified EXP42 does not interact with GCU repeats under the conditions of the experiment (data not shown).
GCN binding protein p60 is identical to CRT.
Previous studies showed that p60 binding activity is also detectable in rat liver after separation from RNA (33, 36). To further characterize and purify p60, cytoplasmic proteins from rat liver were used as a source of the protein. The BioLogic high-resolution chromatography system (BioLogic HR) was used for the isolation of biologically active p60. Using a combination of ion-exchange chromatography and gel filtration (see Materials and Methods), we purified the GC-rich binding protein p60 to homogeneity. In agreement with previous fractionations on DEAE columns, HPLC-based ion-exchange chromatography on an UnoQ column revealed the existence of six proteins (Fig. 4A) interacting with sORF. The positions of CUGBP1 and p60 within gel filtration fractions were determined by Western blotting and UV cross-link assays and are also shown. Fractions containing p60 were used for further purification. Figure 4B shows UV cross-link analysis of proteins after ion-exchange chromatography (the first step of purification; fractions containing p60), the second (gel filtration) step, and the last step (p60 purification; UnoQ). Coomassie staining of the membrane reveals that p60 is purified to homogeneity. The N-terminal sequence of p60 was determined by the Core Facilities of Baylor College of Medicine. Comparison of the determined N-terminal amino acids with known sequences shows that p60 is identical to CRT (DPAIYFKEGFLDGWAWTNRWVES; the determined amino acid sequence of p60 is underlined). CRT has been previously characterized as a multifunctional protein that is involved in a variety of cellular processes (16). One of its functions involves interaction with rubella virus RNA and regulation of the processing of the mRNA (3, 7, 8, 24, 27). Although the regulation of viral RNAs by CRT has been described, endogenous mRNA targets for CRT have not yet been identified. To further investigate the interaction of CRT with GC-rich mRNAs, GST-CRT was expressed in bacteria, isolated by a GST pull-down procedure, and further purified to homogeneity using a combination of ion-exchange and size exclusion chromatography (HPLC). As can be seen, purified GST-CRT interacts with the sORF probe containing GCC repeats (Fig. 4C). In these studies, we found that the interaction of bacterially expressed GST-CRT with RNA can be significantly increased by the incubation of GST-CRT with very small amounts of cytoplasmic proteins from HeLa cells (Fig. 4C, bottom). It is possible that the full RNA binding activity of CRT requires posttranslational modifications or interaction with some other protein(s). This observation is consistent with published data showing that posttranslation modifications of CRT are required for its activation (27).
CRT binds to a stable SL structure within the 5′ region of C/EBPβ mRNA.
Since p60/CRT was first identified by UV cross-link analysis with a GCU probe (Fig. 4), we examined the interaction of bacterially expressed, purified CRT with GCU repeats. These studies showed that, similar to RNA-free p60 (Fig. 4C, CRT), bacterially expressed GST-CRT interacts with GCU repeats, and this interaction is stronger with transcripts containing longer GCU repeats (Fig. 5A). Experiments with rubella virus RNA indicated that CRT preferentially interacts with SL structures that contain a GC-rich sequence within the stem (7, 24, 27). Therefore, we examined the specificity of binding of CRT to the 5′ regions of C/EBPβ mRNA capable of forming the secondary structure. Secondary-structure predictions have shown that the 5′ region of C/EBPβ mRNA is able to form an SL structure in which a potential CRT binding site is located within the stem (Fig. 5B) (25). To examine whether CRT binds to the SL structure, we tested the interaction of CRT with a short sORF oligomer and with a longer oligomer, FL-LAP, which covers the region of C/EBPβ mRNA from the first (FL) to the second (LAP) AUG codons (Fig. 1) (see Materials and Methods). Figure 5C shows the results of these experiments. We found that CRT binds much more strongly to the long FL-LAP oligomer than to the short sORF probe. The binding is specific, since both FL-LAP and sORF compete for binding. Finally, we examined another source of CRT (Sigma) for interaction with the long 5′ region of C/EBPβ mRNA (FL-LAP). CRT from Sigma interacts with the 5′ region of C/EBPβ mRNA, and the interaction is specific, since cold wild-type sORF competes for the interaction but sORF-GC-mut does not (Fig. 5D). In these experiments, we compared our HPLC-purified samples of CRT (Fig. 4B, UnoQ) with CRT from Sigma and found identical electrophoretical mobilities of CRT in both samples. Taken together, these studies show that CRT specifically binds to the GC-rich sequence within the 5′ region of C/EBPβ mRNA.
FIG. 5.
CRT binds to transcripts containing GC-rich sequences. (A) GST-CRT interacts with long GCU repeats. GST-CRT was incubated with RNA transcripts containing 6 or 139 GCU repeats, treated with UV and then with RNase A, and loaded on denaturing (SDS) gel electrophoresis gels. Lane CRT shows the interaction of GST-CRT with the sORF probe. (B) Predicted secondary structure of the 5′ region of C/EBPβ (25). The positions of the AUG codons for the FL and LAP proteins are shown, and the binding site for CRT (CRT BS) is indicated. (C) Interaction of bacterially expressed, purified GST-CRT with the 5′ region of C/EBPβ mRNA determined by UV cross-link assay. GST-CRT (50 ng) was incubated with the sORF or FL-LAP (oligomer from the first to the second AUG codon of C/EBPβ mRNA) RNA probe, treated with UV, and separated by SDS-gel electrophoresis. Cold RNA competitors (shown at the top) were added to the binding reaction mixture before the addition of probe. The competitor oligomers were described previously (33, 36), and their nucleotide sequences are given in Materials and Methods. M, mutant sORF oligomer containing replacements of GC islands with AU or UA (see Materials and Methods). (D) CRT binds to the long 5′ region (FL-LAP) of C/EBPβ mRNA. UV cross-link analysis was performed as described above using the FL-LAP probe and CRT obtained from two different sources: CRT commercially available from Sigma and purified p60/CRT from liver. sORF, mutant sORF (M), and nonspecific (GX) cold competitors (shown at the top) were added to the reaction mixtures prior to the addition of probe. −, no competitors added.
CRT inhibits translation of C/EBPβ mRNA in a cell-free translation system and in cultured cells.
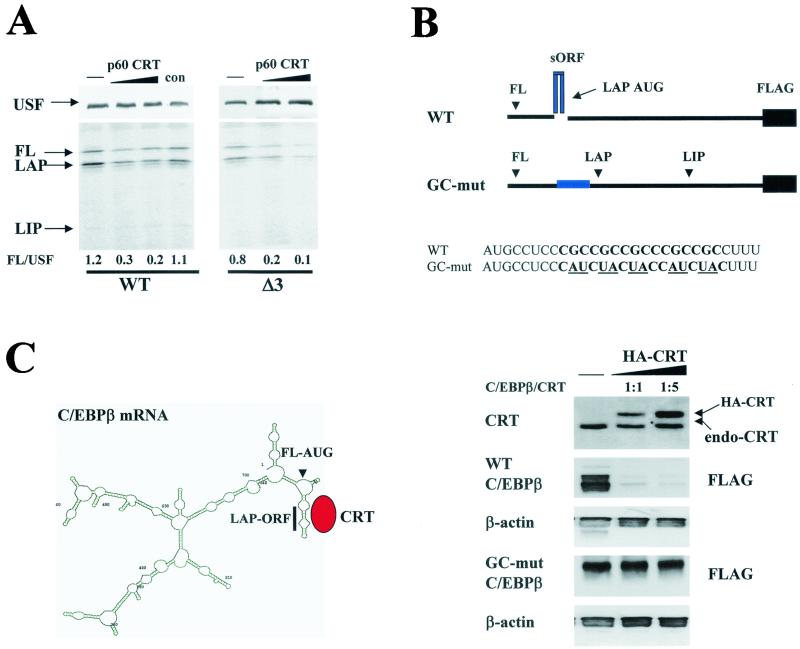
C/EBPα and C/EBPβ play important roles in the regulation of cell growth and differentiation. The mRNAs of both proteins are GC rich and contain several regions which may form SL structures and serve as binding sites for CRT (Fig. 5; also see Fig. 7). We suggested that the interaction of CRT with GCN repeats within C/EBPβ mRNA may stabilize the secondary structure and potentially affect the translation of C/EBPβ. To test this suggestion, we utilized a cell-free translation system programmed with C/EBPβ mRNA. The purified p60/CRT was added to the cell-free translation system programmed with wild-type C/EBPβ mRNA or with an ATG3 mutant construct which cannot translate a dominant-negative isoform, LIP (33, 36). An example of the reproducible results is shown in Fig. 6A. p60/CRT specifically inhibits the translation of C/EBPβ mRNA from the first and second AUG codons. The CRT-dependent inhibition of translation from the first AUG codon is specific to C/EBPβ mRNA, because p60/CRT does not inhibit the translation of the unrelated USF mRNA in the same translation reactions. Similar results were obtained with bacterially expressed, purified GST-CRT (data not shown). To examine whether CRT represses translation of C/EBPβ in cultured cells, a cotransfection approach was applied. For these studies, we incorporated mutations into the sORF sequence within the full-length C/EBPβ mRNA shown in Fig. 6B. The mutations were incorporated based on data presented in Fig. 5 (see Materials and Methods) and on predictions that these mutations abolish the secondary structure. Cotransfection studies are shown in Fig. 6B. Western analysis of the transfected cells with HA antibodies and with anti-CRT showed that CRT is expressed at levels that are two- to threefold higher than the levels of endogenous CRT. Analysis of C/EBPβ expression in cells overexpressing HA-CRT clearly indicates that CRT inhibits translation of wild-type C/EBPβ mRNA but not that of the GC mutant mRNA. Under the conditions of our experiments, the complete inhibition of C/EBPβ translation takes place when the ratio of transfected plasmids is 1:5 (C/EBPβ to CRT). Since CRT binds preferentially to the SL structure within C/EBPβ mRNA and because GC-mut mRNA does not form the secondary structure, we suggest that the CRT-dependent inhibition of C/EBPβ translation may occur through the interaction with and stabilization of the secondary structure (Fig. 6C). Thus, these studies show that CRT specifically interacts with the 5′ sORF region of C/EBPβ mRNA and that this interaction leads to the inhibition of translation of C/EBPβ mRNA in an in vitro translation system and in cultured cells.
FIG. 7.
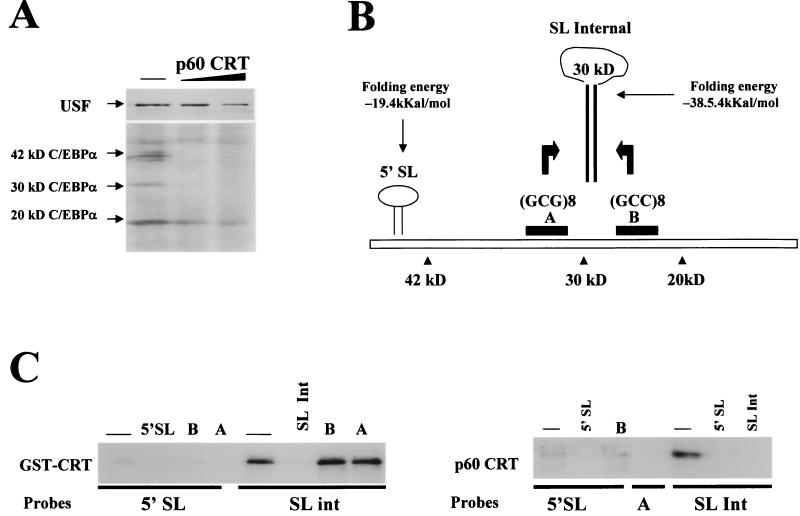
CRT binds to C/EBPα mRNA and represses translation of the 42- and 30-kDa isoforms of C/EBPα. (A) Purified p60/CRT represses translation of C/EBPα in a cell-free translation system. Experiments were performed as described in the legend to Fig. 6A. C/EBPα and USF were translated simultaneously in the presence of increasing amounts of CRT and then immunoprecipitated with specific antibodies. The positions of the 42-, 30-, and 20-kDa isoforms are shown. (B) Putative SL structures within C/EBPα mRNA. RNA secondary-structure predictions revealed two putative SL structures with GC-rich stems. The predicted SL structure in the 5′ region is located upstream of the first AUG codon. Within the coding region, the GCG repeat (region A) and the GCC repeat (region B) can form a stable secondary structure. (C) CRT binds preferentially to the stem structure within the internal SL of C/EBPα mRNA. (Left) UV cross-link reaction using activated GST-CRT (see Materials and Methods) and the 5′ SL or with SL int probes. Cold competitors (shown at the top) were added to the reaction mixture prior to the addition of probe. A and B are single-stranded RNA oligomers covering regions A (GCG repeat) and B (GCC repeat), respectively. CRT does not interact with the single-stranded A and B RNA oligomers but binds strongly to a double-stranded structure formed by annealing these probes (SL int). (Right) p60/CRT was incubated with the 5′ SL or SL int probe in the absence or the presence of RNA competitors (shown at the top) and analyzed by UV cross-link assay.
FIG. 6.
CRT inhibits translation of C/EBPβ proteins. (A) CRT inhibits translation of C/EBPβ mRNA in a cell-free translation system. Purified p60/CRT (Fig. 4) was added to rabbit reticulocyte lysate programmed with USF (control) mRNA and with wild-type (WT) C/EBPβ or Δ3 (mutation of the third AUG codon) mutant C/EBPβ mRNA. The translation mixtures were divided, and USF and C/EBPβ were precipitated with specific antibodies from the same reactions. The levels of C/EBPβ isoforms were calculated as ratios to that of USF. The positions of full-length (FL), LAP, and LIP isoforms are shown. con, an UnoQ fraction which does not contain CRT. (B) CRT inhibits translation of C/EBPβ in cultured cells. (Top) Structure of the 5′ regions of wild-type and GC-mut C/EBPβ mRNAs and mutations incorporated in the 5′region. Replacements of GC islands (shown in bold) with AU or UA (underlined) abolish the formation of an SL structure within GC-mut C/EBPβ mRNA. Both wild-type and mutant C/EBPβ mRNAs were fused to FLAG to distinguish expression of endogenous and exogenous C/EBPβ proteins. The positions of the AUG codons for isoforms of C/EBPβ are shown. The sequences of wild-type and mutant 5′ regions of C/EBPβ mRNA are shown below. (Bottom) Wild-type C/EBPβ-FLAG and GC-mut-C/EBPβ-FLAG were cotransfected with increasing amounts of plasmid expressing HA-CRT into HT1080 cells; 18 h after transfection, total proteins were isolated and examined by Western blotting with antibodies to CRT or to FLAG. The positions of HA-CRT and an endogenous CRT (endo-CRT) are shown. The membranes were reprobed with antibodies to β-actin. (C) Hypothetical model for CRT-dependent inhibition of C/EBPβ translation. The predicted secondary structure of full-length C/EBPβ mRNA revealed a stable SL organization of the CRT binding site (sORF). We suggest that CRT binds to the GC-rich stem of the 5′ region of C/EBPβ mRNA and blocks initiation from the first and second AUG codons.
CRT interacts with C/EBPα mRNA and inhibits translation of C/EBPα in an in vitro cell-free translation system.
In the course of our studies with C/EBPβ, we found that, similar to C/EBPβ mRNA, CRT also dramatically represses translation of 42- and 30-kDa isoforms of C/EBPα in the cell-free translation system (Fig. 7A). The inhibition of C/EBPα translation by CRT seems to be specific to full-length (42-kDa) and 30-kDa isoforms, while translation of the 20-kDa isoform is less affected by CRT. This inhibition is specific to C/EBPα mRNA, since the translation of USF mRNA in the same reactions is not inhibited. This specific CRT-dependent inhibition of C/EBPα translation suggested that CRT affects translation through direct interaction with the C/EBPα mRNA rather than with the translation machinery. Given the observation that CRT binds to SL structures with GC-rich stems (7, 16, 24, 27), we searched for the secondary structures with GC-rich stems within the C/EBPα mRNA and found two possible SL organizations containing GC-rich regions within the stem (Fig. 7B). One SL structure is observed in the 5′ region (5′SL) of C/EBPα mRNA in front of the first AUG codon. The second potential SL structure (SL int) can be formed by GCG (region A) and GCC (region B) repeats located upstream and downstream of the second AUG codon (Fig. 7B). To examine whether CRT can interact with the stems of either of these structures, we synthesized a short RNA oligomer (5′SL) which contains a GC-rich stem (see Materials and Methods) and two RNA oligomers (A and B) which, after annealing, mimic the GC-rich stem within the putative internal loop. UV cross-link analysis with bacterially expressed, purified GST-CRT shows that CRT interacts with the 5′SL structure and with the SL int probe. The interactions of CRT with both probes are specific, since cold 5′SL and SL int oligomers compete for binding while the single-stranded oligomers A and B do not (Fig. 7C). Although CRT binds to both probes, we reproducibly observed significantly stronger interaction of GST-CRT with the SL int RNA probe. To verify these data, we examined the interaction of purified p60/CRT (Fig. 4) protein with both probes. As can be seen in Fig. 7C (right), purified p60/CRT also interacts with the 5′ SL and with the SL int probes; however, no binding is detected with the single-stranded probes A and B. Consistent with data for bacterially expressed GST-CRT, the binding of CRT purified from tissues to the SL int probe is stronger than to the 5′SL oligomer. Taken together, these data demonstrate that CRT preferentially interacts with the internal stem structure of C/EBPα mRNA and inhibits the translation of 42- and 30-kDa isoforms when it is added to a cell-free translation system.
CRT inhibits translation of C/EBPα mRNA in cultured cells.
To further examine whether CRT inhibits translation of C/EBPα mRNA in cells, HA-CRT was cotransfected with different C/EBPα deletion constructs into COS7 cells and the expression of C/EBPα was examined in cells expressing high levels of CRT. The CMV-C/EBPα construct codes for the full-length C/EBPα mRNA and contains both 5′SL and internal SL structures, while the Gal4-C/EBPα construct lacks the 5′ region of C/EBPα mRNA (Fig. 8A). As can be seen in Fig. 8B, CRT inhibits translation of both mRNAs. Since Gal4-C/EBPα does not contain the 5′ UTR of C/EBPα mRNA, this result suggests that CRT represses translation via binding to the internal SL structure. This result is also consistent with UV cross-link data showing that CRT displays a stronger interaction with the internal SL structure (Fig. 7). To further address whether the internal GC-rich SL structure of C/EBPα mRNA is required for inhibition by CRT, two additional C/EBPα constructs lacking the second side (region B) of the internal SL structure (N175 and Gal4-C/EBPα-ΔB) were examined. As can be seen in Fig. 8B, CRT does not inhibit translation of these mRNAs. The failure of CRT to inhibit translation of two mRNAs that lack region B suggests that CRT inhibits translation of C/EBPα mRNA via interaction with the internal SL structure, possibly through stabilization of the stem. Thus, these studies demonstrate that CRT inhibits translation of C/EBPα mRNA in cultured cells and that CRT fails to inhibit translation of C/EBPα mRNA lacking the GC-rich region B.
Overexpression of CRT abolishes C/EBPα-mediated growth arrest in cultured cells.
To further investigate the biological consequences of CRT-dependent inhibition of C/EBPα translation, we examined whether overexpression of CRT in cultured cells is sufficient to block the ability of C/EBPα to inhibit cell proliferation. To examine this possibility, a clonal cell line in which induction of C/EBPα causes growth arrest (HT1 [34]) was used. We first tested whether overexpression of HA-CRT inhibits C/EBPα translation in these cells. To visualize cells expressing HA-CRT, HT1 cells were cotransfected with HA-CRT and with a vector expressing GFP at a 10:1 ratio. Under these conditions, each green cell containing GFP also expresses CRT. As a negative control, transfection with GFP alone was performed. C/EBPα expression was induced by IPTG treatment 6 h after transfection, and the cells were fixed 18 h after the addition of IPTG. The expression of C/EBPα in HT1 cells was examined by immunostaining with specific antibodies to C/EBPα. Cells were also stained with DAPI to visualize the nuclei. Figure 9A shows a reproducible result of these studies. Consistent with data obtained in cotransfection studies (Fig. 8), the expression of HA-CRT in HT1 cells inhibits translation of C/EBPα. This CRT-dependent inhibition of C/EBPα translation is specific, since GFP alone does not affect C/EBPα expression (Fig. 9A, bottom). We examined the levels of C/EBPα in 84 cells transfected with CRT and found that C/EBPα expression was inhibited in 82 of the cells. Similar analysis of C/EBPα protein in 30 cells transfected with GFP alone showed only 1 cell with reduced levels of C/EBPα (Fig. 9A, table). Thus, these studies indicated that overexpression of CRT blocks translation of C/EBPα in HT1 cells.
To determine if the CRT-dependent inhibition of C/EBPα translation is sufficient to block the growth-inhibitory activity of C/EBPα, a colony formation assay was performed. HA-CRT or pCDNA6 (control) vector was cotransfected with β-Gal vector into HT1 cells as described above, C/EBPα was induced by IPTG after the transfection, and the formation of single- and multiple-blue-cell colonies was calculated as described earlier (34). A typical picture of colony formation on day 3 after C/EBPα induction is shown in Fig. 9B. As can be seen, HT1 cells transfected with empty vector are arrested by C/EBPα and remain as single-cell colonies (Fig. 9B, vector). On the other hand, HT1 cells overexpressing CRT proliferate and form colonies containing 3 and >3 blue cells per colony (Fig. 9B, HA-CRT). A summary of three independent experiments shows that 90 to 92% of HT1 cells overexpressing CRT proliferate in the presence of C/EBPα mRNA while >90% of control cells are arrested by C/EBPα (Fig. 9C). Thus, experiments with HT1 cells indicated that overexpression of CRT blocks C/EBPα translation and also prevents C/EBPα-mediated growth arrest.
DISCUSSION
C/EBPα and C/EBPβ are involved in the regulation of a variety of biological processes, such as cell growth and differentiation and the acute-phase response (9, 13, 23, 34). Although the transcriptional control of C/EBPα and C/EBPβ expression is an important level of regulation, a number of recent observations revealed several additional pathways by which expression of these proteins is controlled. Given the GC-rich content of both the C/EBPα and C/EBPβ mRNAs, we performed purification and analysis of RNA binding proteins that preferentially bind to GC-rich sequences within these mRNAs and potentially regulate translation of C/EBP proteins. It was previously found that CUGBP1 binds to the 5′ region of C/EBPβ mRNA and that there are several additional RNA binding proteins interacting with C/EBPβ mRNA (33, 36). HPLC-based size exclusion chromatography of HeLa proteins showed that the majority of these GC-rich RNA binding proteins are observed in high-molecular-mass fractions containing endogenous RNA transcripts. Although putative protein-protein interactions of RNA binding proteins may also contribute to their presence in high-molecular-mass fractions, the digestion of endogenous transcripts with RNase A shifts a significant portion of these proteins to low-molecular-mass fractions and demonstrates that GCN binding proteins, including CUGBP1, are associated with endogenous RNA.
Further experiments showed that the activities of the majority of RNA binding proteins that interact with GC-rich sequences are very weak or not detectable within total protein extracts, while they are readily detectable after separation from endogenous RNA by ion-exchange chromatography. Experiments with RNA-free CUGBP1 showed that binding of CUGBP1 to RNA transcripts increases proportionally with the number of GCU repeats. Another family of GCU RNA binding proteins, EXP, was recently identified (18). Based on its preferential binding to long GCU repeats, EXP42 was suggested to be affected in DM1 patients by virtue of its association with GCU repeats, which are expanded within DMPK mRNA (18). Surprisingly, in our studies under several conditions, we were not able to detect interaction of EXP42 with RNA transcripts containing long or short GCU repeats in either total HeLa extracts or RNA-free DEAE fractions. The lack of interaction of EXP42 with GCU repeats is not due to loss of total activity in the DEAE fractions, because CUGBP1 from the same fractions interacts with RNA proportionally to the number of GCU repeats.
The RNA binding activities of two additional proteins, p60 and p150, were not detectable within total protein extracts but were identified after separation from endogenous RNA by ion-exchange chromatography (Fig. 3B). Further purification and sequence analysis identified p60 as CRT. Although the majority of CRT is located in the endoplasmic reticulum, where it performs chaperone functions and regulates Ca+ homeostasis (16), CRT is also detected in the cytoplasm, where it interacts with rubella virus RNA and controls its processing (3, 7, 8, 27). Detailed analysis of the interaction of CRT with wild-type and mutant rubella virus RNA revealed that CRT recognizes an SL structure containing a GC-rich sequence within the stem (24, 27). The finding that CRT interacts with GC-rich putative SL sequences of C/EBPα and C/EBPβ mRNAs prompted us to examine the result of this interaction. In vitro experiments revealed that binding of CRT to C/EBPα and C/EBPβ mRNAs inhibits translation of these proteins. We also observed CRT-dependent inhibition of C/EBPα and C/EBPβ translation when CRT was overexpressed in cultured cells. This inhibition is abolished if the CRT binding sites (GC-rich sequences) are mutated or deleted. The failure of CRT to affect the mutant RNAs suggests that the CRT-dependent inhibition of translation occurs via direct interaction with RNAs rather than through an interaction with protein components of translation machinery. Although CRT interacts in vitro with the 5′ region and with the internal stem of C/EBPα mRNA, we reproducibly observed that both GST-CRT purified from bacteria and p60/CRT isolated from liver cells preferentially bind to the internal SL structure of C/EBPα mRNA. In agreement with the preferential binding to the internal SL structure, the deletion of the internal SL abolishes the CRT-dependent repression of C/EBPα, while the C/EBPα mRNA lacking the 5′SL structure is still inhibited by CRT. Our data with HT1 cells demonstrate that when CRT is overexpressed, it is able to block the ability of C/EBPα to inhibit cell proliferation (Fig. 9). Perrotti et al. have recently shown the role of RNA binding proteins in the regulation of translation of C/EBPα mRNA (21). Similar to our results with CRT, the authors show that another RNA binding protein, poly(rC)-binding protein hnRNP E2, interacts with C/EBPα mRNA through a different binding site in the 5′ end and inhibits translation of the C/EBPα protein. This inhibition blocks C/EBPα-dependent regulation of downstream targets and leads to the development of leukemia (21).
In conclusion, we have discovered that CRT interacts with the GC-rich regions of the C/EBPα and C/EBPβ mRNAs and inhibits translation of full-length C/EBP proteins. Although CRT is known to bind and regulate translation of rubella virus RNA, our data identify the first endogenous mRNAs to which CRT binds to regulate translation. Interestingly, deletion of the CRT gene in mice leads to embryonic lethality (15), while overexpression of CRT leads to complete heart block and death (19). Since both C/EBPα and C/EBPβ are involved in cell growth and differentiation, our data suggest that the novel role of CRT in the posttranscriptional processing of certain GC-rich mRNAs may be involved in these processes via regulation of C/EBP protein expression.
Acknowledgments
We thank Mark Entman for critical reading of the manuscript. We thank Triona Goode for helpful suggestions and critical analysis of the data. We thank David Williams for the GST-CRT and HA-CRT plasmids. We thank Gretchen Darlington and members of her laboratory for useful discussion of the results.
This work is supported by NIH grants AR01D44387 and AG16392 (L.T.T.) and GM55188 and AG20752-01 (N.A.T.) and by grants from the Muscular Dystrophy Association (L.T.T.).
REFERENCES
- 1.An, M. R., C. C. Hsieh, P. D. Reisner, J. P. Rabek, S. G. Scott, D. T. Kuninger, and J. Papaconstantinou. 1996. Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol. Cell. Biol. 16:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic, D., and J. D. Keene. 1997. Embryonic lethal abnormal visual RNA binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 61:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atreaya, C. D., N. K. Singh, and H. J. Nakashi. 1995. The rubella virus RNA binding activity of human calreticulin is localized to the N-terminal domain. J. Virol. 69:3848-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkhoven, C. F., P. R. J. Bouwman, L. Snippe, and A. B. Geert. 1994. Translation start site multiplicity of the CCAAT/enhancer binding protein α mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Res. 22:5540-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 6.Campos, A. R., D. Grossman, and K. White. 1985. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects: a developmental genetic analysis. J. Neurogenet. 2:197-218. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M.-H., and T. K. Frey. 1999. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73:3386-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, S. T., T. Q. Nguyen, Y. S. Yang, J. D. Capra, and R. D. Sonheimer. 1996. Calreticulin binds hYRNA and 52-kD polypeptide component of the Ro/SS-A ribonucleoprotein autoantigen. J. Immunol. 156:4484-4491. [PubMed] [Google Scholar]
- 9.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 10.Descombes, P., and U. Schibler. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569-579. [DOI] [PubMed] [Google Scholar]
- 11.Jain, R. G., L. G. Andrews, K. M. McGowan, P. H. Pakela, and J. D. Keene. 1997. Ectopic expression of Hel-N1, an RNA binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol. 17:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph, B., M. Orlian, and H. Furneaux. 1998. p21 mRNA contains a conserved element in its 3′-untranslated region that is bound by Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 273:20511-20516. [DOI] [PubMed] [Google Scholar]
- 13.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 14.Lin, F.-T., O. A. MacDougald, A. M. Diehl, and M. D. Lane. 1993. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha messages: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90:9606-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesaeli, N., K. Nakamura, E. Zvaritch, P. Dickie, E. Dziak, K.-H. Krause, M. Opas, D. H. MacLennan, and M. Michalak. 1999. Calreticulin is essential for cardiac development. J. Cell Biol. 144:857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalak, M., E. F. Corbett, N. K. Mesaeli, K. Nakamura, and M. Opas. 1999. Calreticulin: one protein, one gene, many functions. Biochem. J. 344:281-292. [PMC free article] [PubMed] [Google Scholar]
- 17.Millard, S. S., A. Vidal, M. Markus, and A. Koff. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 20:6947-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. W., C. R. Urbinati, P. Teng-Umnuay, M. G. Strenberg, B. J. Byrne, C. Thornton, and M. S. Swanson. 2000. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19:4439-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura, K., M. Robertson, G. Liu, P. Dickie, J. Q. Guo, H. J. Duff, M. Opas, K. Kavanagh, and M. Michalack. 2001. Complete heart block and sudden death in mice overexpressing calreticulin. J. Clin. Investig. 107:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossipow, V., P. Descombes, and U. Schibler. 1993. CCAAT/enhancer binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA 90:8219-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrotti, D., V. Cesi, R. Trotta, C. Guerzoni, G. Santilli, K. Campbell, A. Iervolino, E. Condorelli, C. Gambacorti-Passerini, M. A. Caligiuri, and B. Calabretta. 2002. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2". Nat. Genet. 30:48-58. [DOI] [PubMed] [Google Scholar]
- 22.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737-741. [DOI] [PubMed] [Google Scholar]
- 23.Poli, V. 1998. The Role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279-29282. [DOI] [PubMed] [Google Scholar]
- 24.Pugachev, K., and T. Frey. 1998. Effects of defined mutations in the 5′ nontranslated region of rubella virus genomic RNA on virus viability and macromolecule synthesis. J. Virol. 72:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raught, B., A.-C. Gingras, A. James, D. Medina, N. Sonenberg, and J. M. Rosen. 1996. Expression of a translationally regulated, dominant-negative CCAAT/enhancer binding protein β isoform and up-regulation of the eukaryotic translation initiation factor 2α are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res. 56:4382-4386. [PubMed] [Google Scholar]
- 26.Robinov, S., A. R. Campos, K.-M. Yao, and K. White. 1988. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science 242:1570-1572. [DOI] [PubMed] [Google Scholar]
- 27.Singh, N. K., C. D. Atreya, and H. L. Nakashi. 1994. Identification of calreticulin as a rubella virus RNA binding protein. Proc. Natl. Acad. Sci. USA 91:12770-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timchenko, L. T., J. W. Miller, N. A. Timchenko, D. R. DeVore, K. V. Datar, L. Lin, R. Roberts, C. T. Caskey, and M. S. Swanson. 1996. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timchenko, L. T., N. A. Timchenko, C. T. Caskey, and R. Roberts. 1996. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet. 5:115-121. [DOI] [PubMed] [Google Scholar]
- 30.Timchenko, N. A., Z.-J. Cai, A. L. Welm, S. Reddy, T. Ashizawa, and L. T. Timchenko. 2001. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 276:7820-7826. [DOI] [PubMed] [Google Scholar]
- 31.Timchenko, N. A., P. Iakova, Z.-J. Cai, J. R. Smith, and L. T. Timchenko. 2001. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 21:6927-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timchenko, N. A., A. L. Welm, X. Lu, and L. T. Timchenko. 1999. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 27:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, H., P. Iakova, M. Wilde, T. Goode, A. L. Welm, W. Roesler, and N. A. Timchenko. 2001. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 35.Wang, W., H. Furnaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holdbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welm, A. L., S. L. Mackey, L. T. Timchenko, G. J. Darlington, and N. A. Timchenko. 2000. Translational induction of liver-enriched transcriptional inhibitory protein during acute phase response leads to repression of CCAAT/enhancer binding protein α mRNA. J. Biol. Chem. 275:27406-27413. [DOI] [PubMed] [Google Scholar]