Abstract
Two major xylanases (XYN I and XYN II) of the filamentous fungus Hypocrea jecorina (Trichoderma reesei) are simultaneously expressed during growth on xylan but respond differently to low-molecular-weight inducers. In vivo footprinting analysis of the xylanase1 (xyn1) promoter revealed three different nucleotide sequences (5′-GGCTAAATGCGACATCTTAGCC-3′ [an inverted repeat of GGCTAA spaced by 10 bp], 5′-CCAAT-3′, and 5′-GGGGTCTAGACCCC-3′ [equivalent to a double Cre1 site]) used to bind proteins. Binding to the Cre1 site is only observed under repressed conditions, whereas binding to the two other motifs is constitutive. Applying heterologously expressed components of the H. jecorina cellulase regulators Ace1 and Ace2 and the xylanase regulator Xyr1 suggests that Ace1 and Xyr1 but not Ace2 contact both GGCTAA motifs. H. jecorina transformants containing mutated versions of the xyn1 promoter, leading to elimination of protein binding to the left or the right GGCTAA box revealed either strongly reduced or completely eliminated induction of transcription. Elimination of Cre1 binding to its target released the basal transcriptional level from glucose repression but did not influence the inducibility of xyn1 expression. Mutation of the CCAAT box prevents binding of the Hap2/3/5 complex in vitro and is partially compensating for the loss of transcription caused by the mutation of the right GGCTAA box. Finally, evidence for a competition of Ace1 and Xyr1 for the right GGCTAA box is given. These data prompted us to hypothesize that xyn1 regulation is based on the interplay of Cre1 and Ace1 as a general and specific repressor with Xyr1 as transactivator.
Annually, 830 Gt of renewable plant biomass are formed consisting mainly of cellulose and hemicelluloses such as xylans, mannans, and glucomannans. β-1,4-Xylans are heteropolysaccharides that have a backbone of β-1,4-linked xylopyranosyl residues, to which side groups such as d-glucuronic acid, l-arabinose, p-coumaric acid, and ferulic acid are attached. Various microorganisms produce a wide spectrum of hydrolytic enzymes capable of degrading xylan to the corresponding monomeric sugars.
The filamentous ascomycete Hypocrea jecorina offers one of the best characterized xylanolytic enzyme systems so far due to its broad application in pulp and paper, food and feed, as well as textile industry (for examples, see reference 7). H. jecorina forms two major specific endo-β-1,4-xylanases, XYNI and XYNII (EC 3.2.1.8) (44), which contribute to 90% of the xylanase activity, one β-xylosidase (EC 3.2.1.91) (16), and one endo-β-1,4-glucanase which nonspecifically degrades xylan (EG1, EC 3.2.1.4) (37). Although the biochemistry of xylan degradation by H. jecorina has been studied in detail (for examples, see references 5, 14, 31, and 34) and the mechanisms governing xyn2 gene expression have been elucidated on its molecular level (49, 52), investigations on the regulation of xyn1 gene expression remained fragmentary.
Using promoter deletion analysis, we could previously prove that a 217-bp fragment of the xyn1 5′ noncoding region bears all information necessary for transcriptional regulation of xyn1 gene expression (23). Furthermore, we could demonstrate that a tight carbon catabolite repression of xyn1 expression is mediated by Cre1 via binding to two “SYGGRG” consensus sequences arranged as an inverted repeat in the xyn1 promoter (23). Additionally, some evidence for the involvement of a CCAAT box in transcriptional regulation of xyn1 was given by Zeilinger and coworkers (52). So far, no further cis-acting elements and trans-acting factors of this system have been characterized. In Aspergillus niger, the xylanolytic system is mainly under control of the GAL4-type transcriptional regulator XlnR (46) which, presumably, is a wide domain regulator governing the expression of more than 10 genes involved not only in the degradation of xylan but also in xylose metabolism and cellulose degradation (11, 13, 45).
Lately, the isolation of two transcription factors, Ace1 and Ace2, both being involved in the cellulase expression of Trichoderma reesei, was reported (3, 38). While Ace2 affects regulation of transcription of the xyn2 gene (49), Ace1 was reported as a partial repressor of cellulase and xylanase expression (2).
In this study, we identified the nucleotide sequences within the 217-bp fragment of the xyn1 promoter that are essential for its transcriptional regulation by using in vivo and in vitro techniques. Furthermore, we functionally characterized several DNA-binding factors contacting the aforedescribed motifs within this fragment. Xyr1, a homolog to XlnR of A. niger, was cloned and demonstrated to act as a transactivator mediating both xylose- and xylan-dependent induction. Ace1 is shown to function as a specific repressor of xyn1 transcription by competing with Xyr1 for one of the two corresponding GGCTAA elements arranged as an inverted repeat.
MATERIALS AND METHODS
Microbial strains and plasmids.
H. jecorina (T. reesei) QM9414 (ATCC 26921); RUT C-30 (ATCC 56756), due to a cre1 truncation, a glucose-derepressed strain (18) which could recently be complemented with cre1 to regain carbon catabolite repression (9); and N26, a Δace1 mutant (VTT, Espoo, Finland) were used throughout this study. Strain TU-6 (12), a pyr4-negative mutant of QM9414, was used as a recipient for pyr4-mediated cotransformation experiments. The strains were maintained on malt agar, containing 5 mM uridine in the case of TU-6. Escherichia coli JM109 (39) was used for the propagation of vector molecules, strain BL21-Gold (Stratagene, La Jolla, CA) was used as a host for production of glutathione S-transferase (GST) fusion proteins. Plasmids pFG1 (12), pSJ3 (22), pRAMB1, and pRAMB11 (23) were obtained from our department stock.
Fungal growth, induction of xylanases, and preparation of cell extracts.
For replacement experiments, H. jecorina was precultivated in 1-liter Erlenmeyer flasks on a rotary shaker (250 rpm) at 30°C in 250 ml of the medium described by Mandels and Andreotti (MA) (24), applying 1% (wt/vol) glycerol as a carbon source. 108 conidia per liter (final concentration) were used as inoculum. Pregrown mycelia were washed and resuspended in MA medium containing 1% (wt/vol) xylan from oat spelt (Sigma, Steinheim, Germany), xylose, glucose, or glycerol, and incubation was continued for 12 and 24 h (glucose-free [GOX] activity determination) or for 8 h (preparation of cell extracts). Cell extracts were prepared as described previously (40).
In vivo genomic footprinting via ligation-mediated PCR.
Methylation of genomic DNA was performed at 30°C in a shaking water bath by transferring 18-ml aliquots of H. jecorina cultures pregrown on glucose to media containing glucose, glucose plus xylose, or xylose and incubating them for the period indicated. In vivo methylation, DNA extraction, and HCl cleavage were accomplished as described previously (51). In vitro methylation and cleavage of genomic DNA was performed as described by Mueller and Wold (30). Methylated and cleaved DNA was analyzed by ligation-mediated PCR as described in reference 10 and modified according to the method described in reference 48 using Vent-polymerase (NCB, Beverly, Mass.). For visualizing the noncoding strand, primers Xyn1R-vivo1, 2, and 3 were used, and for visualizing the coding strand, primers Xyn1F-vivo1, 2, and 3 were used (Table 1). Evaluation of the gels was carried out using a Typhoon 8600 variable mode imager (Amersham Bioscience, Buckinghamshire, United Kingdom). Differences in band intensity stronger than 50% were determined to be significant and described as either protected or hypersensitive signals.
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′)a | Position (promoter or gene)b |
|---|---|---|
| Xyn1F-vivo1 | CTCAAGCAACTACGTAAAACTCCATG | −524 to −499 (xyn1 promoter) |
| Xyn1F-vivo2 | CACTGGAATACAACATCCTCCGCAAG | −481 to −456 (xyn1 promoter) |
| Xyn1F-vivo3 | GGAATACAACATCCTCCGCAAGTCCGAC | −477 to −450 (xyn1 promoter) |
| Xyn1R-vivo1 | ATAGATTGAACGCCACCGCAATATC | −276 to −300 (xyn1 promoter) |
| Xyr1R-vivo2 | TATGGCAGTAATGCATCGAACCCGG | −332 to −356 (xyn1 promoter) |
| Xyr1R-vivo3 | GCAGTAATGCAATCGAACCCGGCGCTT | −336 to −361 (xyn1 promoter) |
| Xyr1-4f | ACIGAC/TGAC/TGTIGTIACITAC/TATA/C/TCA | (xyr1 gene) |
| Xyr1-7r | TAA/G/TATICCA/GAAA/GAAA/GAAIGGCATA/GAA | (xyr1 gene) |
| xyn1outf | GTCGAGGTCGACGCAAATGG | −538 to −526 (xyn1 promoter) |
| xyn1outr | GTGAGTTCAGGCTTTTTCATCTAGAGATG | 20 to 1 (hph gene) |
| xyr1M1f | GATTGGCAGTCTAAATGCGACATCTT | −445 to −420 (xyn1 promoter) |
| xyr1M1r | AAGATGTCGCATTTAGACTGCCAATC | −420 to −445 (xyn1 promoter) |
| xyr1M2f | GCGACATCTTAGACGGATGCAC | −429 to −408 (xyn1 promoter) |
| xyr1M2r | GTGCATCCGTCTAAGATGTCGC | −408 to −429 (xyn1 promoter) |
| xyr1M3f | GCCCCTTGACTTGAAAGGCAGGCT | −457 to −434 (xyn1 promoter) |
| xyr1M3r | AGCCTGCCTTTCAAGTCAAGGGGC | −434 to −457 (xyn1 promoter) |
| xyr1Exf | CGAGGATCCGAGCTTTCGAGTTCACGCATG | 163 to 183 (xyr1 gene) |
| xyr1Exr | TTTCTCGAGCTCAATGTGGCCATGAG | 651 to 635 (xyr1 gene) |
| Prxyn1.1f | TTGGCAGGCTAAATGCGACATCTTAGCCGGA | −430 to −400 (xyn1 promoter) |
| Prxyn1.1r | TGCATCCGGCTAAGATGTCGCATTTAGCCTG | −396 to −426 (xyn1 promoter) |
| Prxyn1.1M1f | TTGGCAGTCTAAATGCGACATCTTAGCCGGA | −430 to −400 (xyn1 promoter) |
| Prxyn1.1M1r | TGCATCCGGCTAAGATGTCGCATTTAGACTG | −396 to −426 (xyn1 promoter) |
| Prxyn1.1M2f | TTGGCAGGCTAAATGCGACATCTTAGACGGA | −430 to −400 (xyn1 promoter) |
| Prxyn1.1M2r | TGCATCCGTCTAAGATGTCGCATTTAGCCTG | −396 to −426 (xyn1 promoter) |
| Prxyn1.2f | CCCCTTGACTTGATTGGCAGGC | −443 to −422 (xyn1 promoter) |
| Prxyn1.2r | GGGAGCCTGCCAATCAAGTCAA | −418 to −439 (xyn1 promoter) |
| Prxyn1.2M3f | CCCCTTGACTTGAAAGGCAGGC | −443 to −422 (xyn1 promoter) |
| Prxyn1.2M3r | GGGAGCCTGCCTTTCAAGTCAA | −418 to −439 (xyn1 promoter) |
| Prcbh1f | ATATACCAGCGGCTAATAATTG | −789 to −768 (cbh1 promoter) |
| Prcbh1r | TGTACAATTATTAGCCGCTGGT | −764 to −785 (cbh1 promoter) |
| Prcbh1mut2D2f | ATATACCAGCTTATAATAATTG | −789 to −768 (cbh1 promoter) |
| Prcbh1mut2D2r | TGTACAATTATTATAAGCTGGT | −764 to −785 (cbh1 promoter) |
Letters underlined by single lines indicate identified regulatory elements, double lines mark introduced restriction sites. Bold characters indicate mutations within identified motifs.
Positions of the oligonucleotides in the respective promoters or genes are given.
Cloning of xyr1 of H. jecorina.
For PCR-based amplification of a xyr1 fragment, degenerate primers xyr-4f and xyr1-7r (Table 1) and H. jecorina QM 9414 chromosomal DNA, as a template, were used. Primers were deduced from sequence alignments of a hypothetical protein of Neurospora crassa (accession no. XP 327257) with the Aspergillus niger xlnR (46) gene. An iCycler (Bio-Rad, Hercules, CA) was used to run 30 cycles of 1 min at 95°C, 1 min at 46°C to 56°C (gradient block), and 1 min at 74°C; the optimal annealing temperature was found to be 52°C. The derived DNA fragment was sequenced and thereafter used as a probe to screen a genomic λ BlueStar (Novagen, Darmstadt, Germany) phage library of H. jecorina QM9414 by plaque lift hybridization. Positive clones were excised from the phages via automatic subcloning by means of E. coli BM25.8.
Construction of pGXI reporter series.
The pGXI reporter vector series was developed from plasmid pSJ3 by fusing the H. jecorina xyn1 5′ noncoding region (−1 to −532) to the goxA (glucose oxidase) gene of A. niger as a reporter. The xyn1 promoter fragment was amplified by PCR either from plasmid pRAMB1 for the wild-type (WT) fragment or from pRAMB11 for fragments bearing a deletion in the Cre1 double site using the primer pair xyn1outf and xyn1outr (Table 1). To yield fragments of the xyn1 5′ noncoding region bearing respective mutations of regulatory elements, a four-primer PCR mutagenesis strategy was followed (17). Briefly, respective mutations were inserted by using the overlapping primer pairs XyrlM1f and Xyr1M1r (mutation of GGCTAA 1 to GTCTAA), XyrlM2f and Xyr1Mr (GGCTAA 2 to GTCTAA), and XyrM3f and XyrlM3r (CCAAT to CCTTT); full-length xyn1 promoter fragments were reamplified using the confining primers xyn1outf and xyn1outr (all primers sequences are listed in Table 1). Combinations of mutations were introduced by consecutive use of the above-described primers and vectors already bearing one or two of the desired mutations as a template for further four-primer PCR mutagenesis approaches. Derived fragments were inserted into plasmid pSJ3 via the restriction enzyme sites SalI/XbaI. All vector constructs were verified by DNA sequencing of the PCR-amplified parts.
Isolation, analysis, and manipulation of nucleic acids.
RNA was isolated as described previously (8). After electrophoretic separation, RNA was blotted onto Biodyne B membranes (Pall, Pensacola, FL) and hybridized at 42°C for 20 h according to standard protocols (39). Washing was performed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% (wt/vol) sodium dodecyl sulfate at 42°C (two times for 5 min). Standard methods were used for plasmid isolation, restriction enzyme digestion, and random priming (39).
DNA transformations.
E. coli transformations were carried out according to standard techniques (39). H. jecorina TU-6 was transformed according to an optimized protocol for particle bombardment (15), applying cotransformation of pFG1 and the respective reporter constructs. For each construct, at least 20 uridine prototrophs were purified to mitotic stability and their genomic DNA was isolated and examined for integration and copy number by Southern analysis. Cotransformation frequency varied from 90 to 100%, yielding copy numbers between 1 and 3 integrated at ectopic loci.
Determination of gene copy number and integration locus in transformants.
Genomic DNA was isolated as described previously (12). Southern hybridization was carried out as described in reference 39. Chromosomal DNA was digested with PstI, and the obtained blot was hybridized with an [α-32P]dCTP-labeled 660-bp fragment bearing 532 bp of the xyn1 promoter and 128 bp of the goxA gene. The endogenous xyn1 gene and the integrated vector copies were quantified using a Typhoon 8600 variable mode imager (Amersham Bioscience) to measure the corresponding amounts of radioactivity; values obtained were thereafter normalized to the length of the labeled probe.
Glucose oxidase assay.
Glucose oxidase activity was assayed as described previously (22). One unit (1 U) of activity is defined as the amount of enzyme which oxidizes 1 μmol of glucose per min at pH 5.8 and 25°C.
Production and purification of the DNA-binding domain of Ace1, Ace2, Hap2/3/5, and Xyr1 as GST fusion proteins.
All heterologously expressed proteins used throughout this study were produced as GST fusions by applying the pGEX system (Amersham Bioscience) according to the manufacturer's guidelines. Construction of the expression vectors for Ace1, Ace2, and Hap2/3/5 essentially followed the protocol described by Saloheimo et al. (38), Aro et al. (3), and Zeilinger et al. (50). Due to the intron (bp 355 to bp 422) positioned in the zinc cluster domain of Xyr1, a first-strand cDNA was used as a template and primers xyrExf and xyrExr (Table 1) were used to amplify a 422-bp fragment, thereby generating additional BamHI and XhoI terminal sites at the 5′ and 3′ ends for ligation of the fragment into plasmid pGEX-2T. First-strand cDNA synthesis was performed using the DNA-free kit system (Ambion, Austin, Texas) for RNA purification and the reverse transcription-PCR kit (Promega, Madison, Wis.) combined with xyrExr as a specific primer. All expression products were purified using glutathione-Sepharose columns (Amersham Bioscience), verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE)-Coomassie blue staining and thereafter cleaved for 16 h at 4°C, applying 1 U of thrombin (Amersham Bioscience) to remove the GST moiety.
Electrophoretic mobility shift assay (EMSA).
Double-stranded oligonucleotides used throughout this study were gained by annealing the respective synthetic oligonucleotides (Table 1) by heating them to 95°C and letting them cool to room temperature. Labeling of double-stranded oligonucleotides was carried out with [α-32P]dCTP and Klenow polymerase (Promega), followed by purification via nondenaturing PAGE. The binding assay and PAGE were performed as essentially described in reference 40. Binding was achieved by incubating 100 μg of cell extract protein with 5 ng of labeled fragment (15 min, 0°C). For competition experiments, unlabeled synthetic oligonucleotides were used in a 50- to 100-fold molar excess. Unlabeled oligonucleotides were annealed with the complementary synthetic oligonucleotide as described in reference 41. After annealing, double strands were filled in by Sequenase, version 2.0 (Amersham Bioscience). For binding analysis of Xyr1, Ace1, Ace2, and Hap2/3/5 to the respective DNA fragments, thrombin-cleaved GST fusion proteins were used at concentrations of 0.2 μg per binding assay, except where indicated differently.
RESULTS
Identification of cis-acting elements involved in transcriptional regulation of the xyn1 gene.
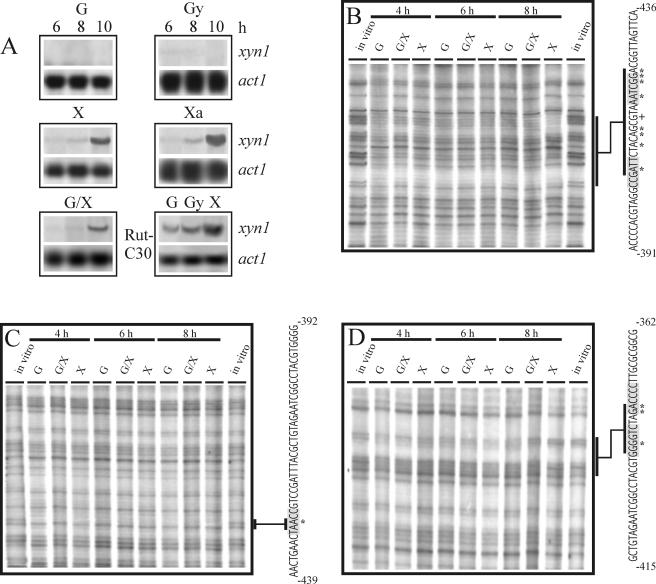
xyn1 transcript is abundantly accumulated during growth on xylose and xylan. The basal transcription of xyn1 is subject to carbon catabolite repression mediated by carbon sources such as glucose and glycerol. As described previously (9, 23), transcriptional activation can overcome carbon catabolite repression as xylose triggers induction even in the presence of glucose. A Cre1-dependent repression/derepression system is not involved in this activation, as the basal transcriptional level of xyn1 constitutively appearing in a Cre1-negative background (H. jecorina RUT-C30) still remains inducible by xylose (Fig. 1A). Promoter deletion analysis revealed that a 217-bp fragment (−538 to −321) of the 5′ noncoding region of xyn1 contains all cis-acting motifs necessary for this regulation (23, 52). To further identify these motifs, we performed in vivo genomic footprinting of the respective nucleotide area by using mycelia pregrown on glycerol and thereafter replaced on glucose (repressing conditions), glucose plus xylose, and xylose (both inducing conditions) in a time course experiment of 4, 6, and 8 h, respectively. Under all conditions tested, a distinct protection pattern could be observed on the coding as well as on the noncoding strand (Fig. 1B, C, D). On the coding strand, several nucleotides (methylation protected, A425, G424, G423, A420, G414, A413, A411, A406; methylation hypersensitive, G416) within a 23-bp fragment circumscribed by an inverted repeat of 5′-GGCTAA-3′ exhibited an identical protection pattern under all carbon sources tested. On the noncoding strand, A429 within a CCAAT motif is protected under all conditions, thus corroborating previous in vitro data indicating an involvement of the CCAAT box in xyn1 regulation (52) (Fig. 1C). Further downstream, the nucleotides G392, G386, and A385 are protected under repressing (glucose) and early inducing conditions (4, 6 h; glucose plus xylose, xylose), whereas a clear release from tight methylation protection can be observed under late inducing conditions (8 h; glucose plus xylose, xylose) (Fig. 1D). All of these nucleotides are positioned within two Cre1 binding sites arranged as an inverted repeat which have previously been shown to cause carbon catabolite repression of xyn1 expression (23).
FIG. 1.
Transcript analysis and in vivo footprinting studies of xyn1. (A) Northern analysis of xyn1 transcript accumulation in H. jecorina strains after replacement of various carbon sources (G, glucose; Gy, glycerol; X, xylose; Xa, xylan; G/X, glucose and xylose) for 6, 8, and 10 h for QM 9414 (panel 1 to 5) and for 6 h for the carbon catabolite derepressed strain RUTC30 (panel 6). Twenty micrograms of RNA was loaded, and hybridizations were performed with a 1.8-kb PstI fragment of the xyn1 gene and a 1.9-kb KpnI fragment of the act1 (actin-encoding) gene of H. jecorina. (B, C, and D) identification of nucleotides contacted by DNA-binding proteins using in vivo footprinting techniques via ligation-mediated PCR of the coding strand (B) and of the noncoding strand (C and D) previously identified to contain all necessary elements for regulation of xyn1 transcription (23). Abbreviations as defined for panel A indicate H. jecorina cultures with different carbon sources replaced and thereafter subjected to in vivo methylation. Isolated DNA was treated with HCl and cleaved with NaOH. Bases involved in protein-DNA contact are indicated by an asterisk, whereas bases showing increased methylation are marked by plus sign. The corresponding parts of the lanes are marked and related to the sequence. Most left and right lanes of each gel show control DNAs methylated in vitro and are indicated “in vitro” (for an example of a detailed evaluation procedure of the in vivo footprint pattern, see Fig. S1 in the supplemental material).
In vitro characterization of proteins binding to the xyn1 transcriptosome.
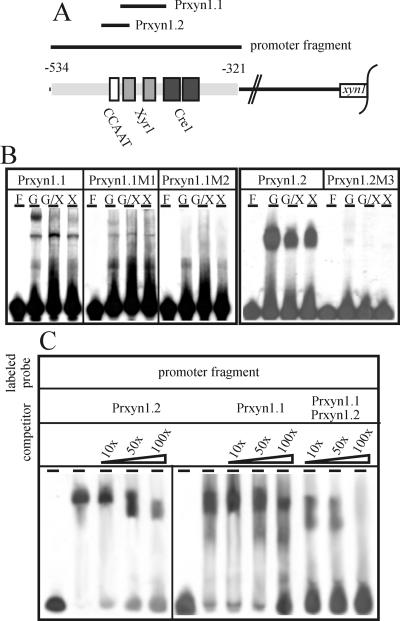
To verify the binding of regulatory proteins to the two motifs identified in addition to the Cre1 sites, EMSAs were carried out using cell-free extracts prepared form H. jecorina mycelia replaced on glucose, glucose plus xylose, or xylose as a carbon source. Synthetic oligonucleotides (Table 1; Fig. 2A) covering either the CCAAT box (Prxyn1.2) or the inverted repeat GGCTAA (Prxyn1.1) and respective oligonucleotides containing point mutations in these motifs were used as probes (Prxyn1.1M1, Prxyn1.2M1, and Prxyn1.2M2) (Table 1; Fig. 2B). Under repressing conditions, Prxyn1.1 yielded two protein-DNA complexes whereof the complex of lower mobility (top most band) is lost during induction. Mutation of GGCTAA positioned on the coding strand (GGCTAA 1) into GTCTAA resulted in a general attenuation of protein-DNA complex formation and in a slight increase in mobility of the repression-specific complex. Insertion of the same mutation into GGCTAA positioned on the noncoding strand (GGCTAA 2) completely impaired protein-DNA interaction (Fig. 2B). Following a similar approach but using oligonucleotides Prxyn1.2 and Prxyn1.2M3, the contribution of the CCAAT box to protein-DNA complex formation under all conditions tested could be demonstrated (Fig. 2B).
FIG. 2.
Protein-DNA interaction in the xyn1 promoter using cell-free extracts. (A) Schematic drawing of the oligonucleotides used in EMSAs. The given sequence spans the whole area of the previously identified xyn1 5′ noncoding region responsible for regulation of transcription (−534 to −321). Potential regulatory elements (CCAAT box, Xyr1 element, and Cre1 element) are boxed. (B) EMSA analysis using 100 μg cell-free extract derived from H. jecorina mycelia replaced on either 1% (wt/vol) glucose (G, repressing conditions) or 1% (wt/vol) xylose or xylose + glucose (X, X/G, inducing conditions) and 5 ng of labeled oligonucleotides as probes (Prxyn1.1, Prxyn1.1M1, Prxyn1.1M2, Prxyn1.2, Prxyn1.2M3). (C) EMSA competition experiments using 100 μg cell-free extract derived from H. jecorina mycelia replaced on 1% (wt/vol) xylose, 50 ng labeled promoter fragment (−534 to −321), and the oligonucleotides Prxyn1.2 and/or Prxyn1.1 as competitors in a 10-, 50-, or 100-fold molar excess. F, free probe.
To investigate whether the predetermined motifs (CCAAT, GGCTAA) are sufficient for the formation of the induction-specific complex, EMSA and competition experiments were carried out using a promoter fragment from −534 to −321 as a probe and a 10-, 50-, or 100-fold molar excess of Prxyn1.1, Prxyn1.2, or a combination of both as competitors and cell extracts from a culture replaced on xylose. The protein-DNA complex generated without competitor DNA gained mobility and lost its intensity when adding either Prxyn1.1 or Prxyn1.2 in increasing amounts to the reaction mixture. However, a complete loss of complex formation only occurred when a combination of both fragments was used in a 100-fold molar excess (Fig. 2C). We conclude from these data that the CCAAT box and the adjacent GGCTAA motifs are the main cis-acting elements involved in induction-specific protein-DNA complex formation in the investigated xyn1 promoter fragment.
Cloning of xyr1 encoding the H. jecorina homologue of the A. niger transactivator XlnR.
Most of the investigated zinc binuclear cluster proteins bind as homodimers to a direct or inverted repeat of sequences comprising three of five C/G combinations (for examples, see references 1, 25, 26, and 43). The fact that the inverted repeat of GGCTAA within the xyn1 5′ noncoding region perfectly fits this pattern and that it is bound by the heterologously expressed zinc finger domain of A. niger XlnR (data not shown) prompted us to isolate the corresponding factor of H. jecorina. A protein sequence alignment of the A. niger transactivator XlnR with its putative N. crassa homologue served as the basis for the design of two degenerated primers (Table 1) thereafter used in a PCR-based approach to clone the respective orthologue of H. jecorina. An 800-bp amplification product was obtained which, in the following, was applied as a probe to screen an H. jecorina genomic library. A 4,323-bp SacII fragment was found to contain an open reading frame of 2,931 bp initialized by a postulated translation start codon positioned 5′ of a zinc binuclear cluster domain. The putative gene was consequently named xyr1 (xylanase regulator 1; GenBank accession no. AF479644). The coding sequence is interrupted by two introns of 67 and 62 bp, conserved in positioning compared to the Aspergillus xlnR and the putative N. crassa homologue. It encodes a deduced protein (Xyr1) of 934 amino acids with a predicted molecular mass of 102 kDa. Sequence alignments (see Fig. S2 in the supplemental material) showed that the entire Xyr1 protein comprises significant sequence similarity to the hypothetical N. crassa factor NcXlnR (55%) and to the transcription activators AoXlnR from Aspergillus oryzae (48%) (27), AkXlnR from Aspergillus kawachii (50%) (accession no. AB064658), and XlnR from A. niger (49%) (46). The highest similarities were found in the N-terminal part of the deduced protein, where it forms a GAL4-like Zn(II)2Cys6 binuclear cluster domain complying exactly with the Cys-X2-Cys-X6-Cys-X5-Cys-X2-Cys-X6-Cys type where six cysteines ligate to zinc ions (25). C-terminally adjacent to the putative DNA-binding domain, a clustering of the basic amino acids lysine and arginine, which have been considered to take part in nuclear localization in other cases (for examples, see reference 35), can be observed. Previously described coiled coil domains in the C-terminal region of XlnR, which have been postulated to act in homodimerization, and, especially, a leucine within this domain (Leu-650) essential for its functionality (46) are highly conserved in Xyr1 of H. jecorina. Two amino acids found to be absolutely vital for gene activation by XlnR (Leu-823, Tyr-864) (46) are also conserved in Xyr1 (Leu-882, Tyr-923) and the putative N. crassa protein (Leu-892, Tyr-933). They are located in the last 160 amino acids of the C-terminal region, which is considered the transactivating domain of XlnR (46) and is almost identical in all three proteins.
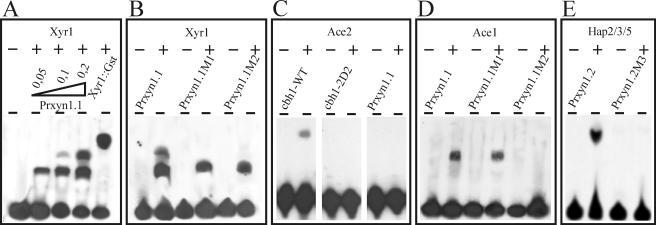
The xyn1 promoter contains regulatory elements that are bound by Xyr1, Ace1, and Hap2/3/5.
Interestingly, the aforementioned inverted GGCTAA motif closely resembles the consensus sequence of not only A. niger XlnR and, therefore, most probably also that of H. jecorina Xyr1 but also that of the transactivator Ace2 (38), recently shown to be involved in regulation of xyn2 expression (49), and that of the previously published cellulase and xylanase repressor Ace1 (3). To investigate which of these factors bind to GGCTAA, EMSAs with thrombin-cleaved, heterologously expressed DNA-binding domains of each of these proteins were performed. Using increasing amounts of the binuclear zinc cluster domain of Xyr1 and oligonucleotide Prxyn1.1 (Table 1), the formation of two complexes could be observed, whereas only one complex with significantly reduced mobility is formed when applying the uncut fusion protein still containing the GST tail (Fig. 3A). The observation that a similar approach employing oligonucleotide Prxyn1.1M1 or Prxyn1.1M2 (Table 1), bearing a point mutation in either one of the GGCTAA motifs, only led to the formation of the complex with highest mobility strongly indicates that both motifs are independently contacted by the Xyr1 DNA-binding domain (Fig. 3B). However, in contrast to a positive-control experiment with oligonucleotide Prcbh1 (38), complex formation was not observed with the Ace2 zinc cluster domain (Fig. 3C). In contrast, by using the overexpressed Cys2His2 zinc finger domain of Ace1, binding to oligonucleotides Prxyn1.1 and Prxyn1.1M1 was noticed by the appearance of complexes with identical mobility, whereas the mutation in Prxyn1.1M2 fully abolished complex formation (Fig. 3D). We conclude that Ace1 binds only to the downstream GGCTAA motif and most probably competes with Xyr1 for this binding site.
FIG. 3.
EMSA using recombinant DNA-binding domains or proteins. Analysis of the involvement of Xyr1 (A, B), Ace2 (C), Ace1 (D), and Hap2/3/5 (E) in binding to the xyn1 5′ noncoding region. The respectively labeled oligonucleotides used are described in detail in Table 1. Thrombin-cleaved fusion protein (0.2 μg, corresponding molar ratio is approximately 1:20 between DNA and recombinant protein) was used, if not indicated differently: panel A, lanes 2, 3, and 4, 0.05, 0.1, and 0.2 μg recombinant protein; lane 5, 0.2 μg uncleaved Xyr1::Gst fusion protein. −, free probe; +, addition of the respective thrombin-cleaved fusion proteins to the binding reaction.
Recently, Zeilinger et al. (50) and Würleitner et al. (49) demonstrated that the CCAAT boxes of the cellulase-activating element (CAE) in the cbh2 promoter and the xylanase-activating element (XAE) in the xyn2 promoter are bound by the Hap2/3/5 complex. To establish if this protein complex also contacts the xyn1 promoter, oligonucleotide Prxyn1.2 (Table 1) and the heterologously expressed Hap subunits 2, 3, and 5 from H. jecorina were subjected to EMSA. DNA-protein complex formation was clearly observed; it was abolished by introduction of a TT to AA mutation within the CCAAT box (Prxyn1.2.M3) (Fig. 3E).
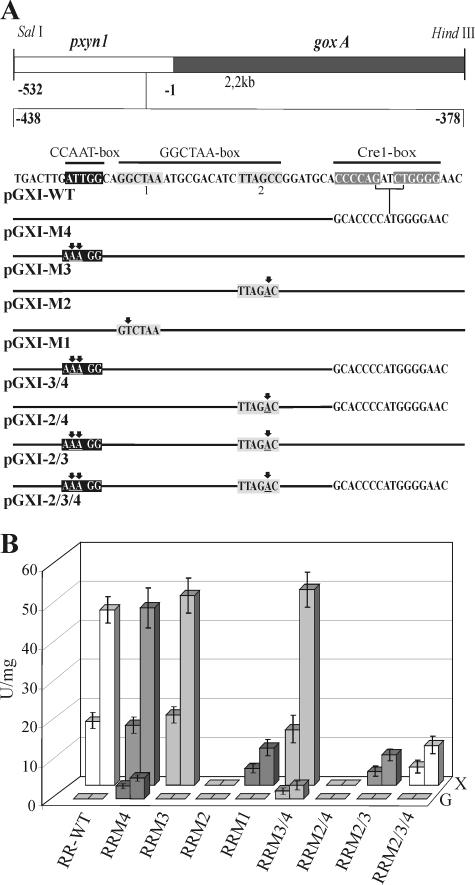
Mutations impairing protein binding in vitro lead to loss of distinct regulatory functions in xyn1 gene expression in vivo.
To investigate whether the identified protein-binding motifs indeed confer regulation of xyn1 transcription in vivo, the mutations studied above and a deletion in the Cre1 consensus sequences were introduced either alone or in different combinations into a 540-bp fragment of the xyn1 5′ noncoding region fused to the A. niger goxA (glucose oxidase-encoding) gene as a reporter (Fig. 4A). The respective constructs were inserted into H. jecorina TU-6 by cotransformation with plasmid pFG1 (12) employing biolistic bombardment (15). Three transgenic strains bearing ectopic single-copy integrations of the respective reporter vectors (including the WT construct) were investigated under inducing (xylan) and repressing (glycerol) conditions (Fig. 4B). The tested modifications of the GGCTAA boxes (Fig. 4A) perfectly reflected the results obtained by EMSA, as a mutation in the 5′ motif (M1) yielded an approximately fivefold decrease of GOX activity on xylan. Transcription was totally impaired when the downstream motif (M2) was changed to GTCTAA, indicating that both sites are involved in xyn1 gene expression but only motif 2 is essential (Fig. 4B). A deletion within the double Cre1-binding site of the xyn1 promoter (M4) produced no significant effect under inducing conditions, while a slight increase in reporter gene activity (10%) was obtained when the CCAAT box was mutated (M3) or when both the CCAAT box and the Cre1-binding sites were mutated simultaneously (M3/4) (Fig. 4B). As expected, no expression on xylan was obtained for combined mutations in GGCTAA 2 and both Cre1-binding sites (M2/4); however, mutations within CCAAT and GGCTAA 2 (M2/3) restored GOX activity at a low level. As a consequence of the above-described effect of the mutation in the CCAAT box, a comparable low level of GOX activity on xylan was also measured when all three motifs (M2/3/4) were mutated (Fig. 4B). Under repressing conditions, low levels of reporter gene expression could only be observed for strains bearing the deletion in the Cre1-binding sites (M4, M3/4), with only the exception of combined mutations of the Cre1 sites and GGCTAA 2 (M2/4, M2/3/4), where formation of GOX activity is completely abolished (Fig. 4B). Summarizing these data, we conclude that Xyr1 is essential for both the derepressed as well as the induced expression of xyn1. Whereas the Hap proteins only affect the mechanism of induction, Cre1 mediates its repressor function merely on the basal repressed/derepressed level of expression.
FIG. 4.
(A) Functional analysis of the identified DNA elements using a reporter gene system. Schematic representation of the reporter vectors used for transformation and the respective mutations in the xyn1 5′ noncoding region. Arrows indicate point mutations; putative motifs are boxed. (B) Comparison of GOX activity given in mU/mg NaOH-soluble protein measured after cultivation of the strains RR-WT, RRM1, RRM2, RRM3, RRM4, RRM3/4, RRM2/4, RRM2/3, and RRM2/3/4 (M# indicates the respective mutation as given in panel A) on glycerol (G, repressing conditions) and xylan (X, inducing conditions) for 8 h (left column) and 24 h (right column). Data are means of results from two independent experiments performed with three different single-copy integration strains per mutation; error bars indicate standard deviations. As the observed standard deviations are very low, significant positioning effects of the integrated vector copies can be excluded.
Ace1 participates in the repression-specific complex formed with the GGCTAA motif.
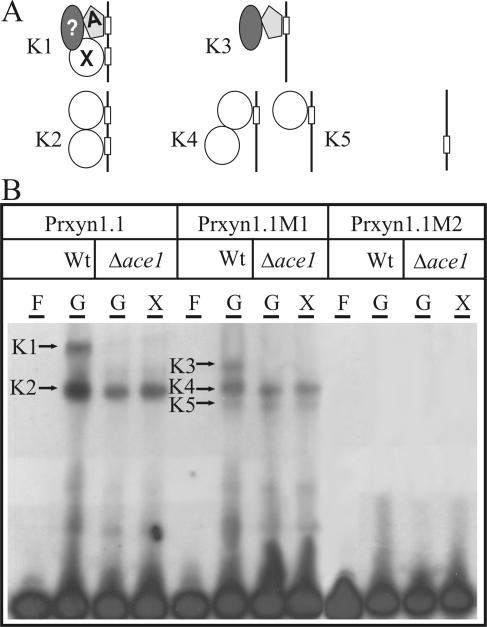
To further prove the supra vide postulated hypothesis that Ace1 competes with Xyr1 for the GGCTAA 2 site, cell-free extracts from H. jecorina N26 (Δ ace1) and oligonucleotide Pxyr1.1 (Table 1) were used for EMSA (Fig. 5). Contrary to the case of wild-type H. jecorina QM9414, where two protein-DNA complexes (K1, K2) were observed under repressing conditions, only the induction-specific complex (K2) was formed using extracts from glucose-grown mycelia of strain N26. Applying oligonucleotide Prxyn1.1M1 (Table 1) in addition to a general attenuation of complex formation, the repression-specific complex gained mobility (K3), most probably due to loss of Xyr1 binding, whereas the induction-specific complex remained unaltered (K4). An additional complex showing the highest mobility could be observed for all conditions tested (K5). In agreement with complete loss of the protein binding ability of oligonucleotide Prxyn1.1M2 (Table 1), no protein-DNA complex formation was observed with extracts from the Δace1strain. These data strongly indicate that Ace1 participates in repression-specific complex formation with the GGCTAA motif and that it competes with Xyr1 for the essential GGCTAA 2 part of it. A possible model presenting the composition of the complexes observed, which is addressed in detail in Discussion, is given in Fig. 5.
FIG. 5.
(A) EMSA demonstrating the involvement of Ace1 in the repression-specific protein-DNA complex formation: model of the factors contacting the two Xyr1-boxes positioned as an inverted repeat in the xyn1 promoter. The line with boxes indicates oligonucleotides Prxyn1.1, Prxyn1.1M1, and Prxyn1.1M2, respectively. A, Ace1; X, Xyr1; ?, a yet unknown factor; K1 to K5, complexes formed in EMSA. (B) EMSA analysis using 100 μg cell-free extract derived from H. jecorina wild-type (Wt) and the Δace1 strain N26 (Δace1) mycelia replaced on either 1% (wt/vol) glucose (G, repressing conditions) or 1% (wt/vol) xylose (X, inducing conditions) and 5 ng of labeled oligonucleotides as probes (Prxyn1.1, Prxyn1.1M1, Prxyn1.1M2). F, free probe.
DISCUSSION
In the present study, we have identified all cis-acting elements and their respective trans-acting factors within the 217-bp upstream regulatory fragment (−538 to −321) of the xyn1 promoter governing the transcriptional regulation of this gene in H. jecorina. In addition to the previously described repressor function of Cre1 (23), we could identify the general activator Xyr1 as essential for xyn1 transcription. Furthermore, we could demonstrate that the previously described repressor Ace1 (2) directly antagonizes Xyr1 function by competing for one of its binding sites.
A combination of in vivo and in vitro techniques led to the identification of three adjacent cis-acting elements in a 217-bp fragment of the xyn1 promoter which was previously shown to be sufficient for regulation of xyn1 gene expression (23). The GGCTAA motif present as an inverted repeat in the xyn1 promoter (Fig. 4, compare GGCTAA box 1 and 2) closely resembles the consensus sequence for binding of the XlnR regulatory protein from A. niger (46). XlnR is a central regulator controlling the expression of more than 10 genes involved not only in the degradation of xylan but also in xylose metabolism and cellulose degradation (11, 13, 45). Similarly, the homologous factor AoXlnR of A. oryzae mediates the expression of at least four xylanolytic and four cellulolytic genes (27). Both XlnR and AoXlnR belong to the GAL4 family of zinc binuclear cluster transcriptional regulators (27, 46).
Since complex formation could be observed between a heterologously expressed zinc cluster domain of the A. niger XlnR and the GGCTAA motif of the H. jecorina xyn1 promoter (C. Wacenovsky and R. L. Mach, unpublished data), we consequently assumed that an orthologous factor also exists in this fungus. Cloning of the H. jecorina xyr1 gene and application of the DNA-binding domain of the corresponding protein in EMSAs resulted in a specific retardation complex with radiolabeled oligonucleotide fragments of the xyn1 promoter containing only the inverted repeat of the GGCTAA motif (Fig. 4, compare GGCTAA box 1 and 2). No complex formation could be observed when the 3′ part of the motif (GGCTAA box 2) was mutated to GTCTAA, which abolished XlnR binding in A. niger (46), while mutation of the 5′ part (GGCTAA box 1) only weakened Xyr1 binding. These in vitro data were further supported by in vivo experiments using the xyn1p::goxA reporter gene construct, where mutation of the 3′ part of the motif (GGCTAA box 2) led to a complete loss of reporter gene expression and mutation of the 5′ part (GGCTAA box 1) significantly decreased transcription to a very low level. We therefore concluded that Xyr1 contributes to the transcriptional regulation of xyn1.
The GGCTAA motif also closely resembles the consensus sequences of the recently cloned H. jecorina transcriptional regulators Ace1 and Ace2 (2, 38). While deletion of ace2 had no effect on xyn1 expression (3), a Δace1 knockout strain ALKO2221 exhibited an elevated level of xyn1 transcription on cellulose-based media (2). However, a Δace1 mutant of strain QM9414 showed no differences in xyn1 transcription levels when grown on glucose, xylose, or glucose plus xylose (E. Würleitner and R. L. Mach, unpublished data). This could be explained by the different growth conditions and strains used. According to those data, the involvement of Ace2 in xyn1 gene regulation could be ruled out due to a lack of complex formation in gel retardation analysis between the heterologously expressed zinc cluster DNA-binding domain of Ace2 and the xyn1 promoter. Nevertheless, distinct binding of Ace1 to an oligonucleotide containing the GGCTAA inverted repeat was observed in EMSA. Furthermore, EMSAs carried out with the same oligonucleotide and cell-free extracts prepared from H. jecorina QM9414 mycelia grown on glucose as the sole carbon source resulted in a complex of lower mobility than those with cell extracts of H. jecorina QM9414 Δace1. We therefore hypothesize that a competition between the Ace1 repressor and Xyr1 activator for the GGCTAA motif occurs. A similar mechanism was shown for the regulation of the alcA gene of A. niger, where the transcriptional activator AlcR competes with CreA for the corresponding, partially overlapping binding sites (28, 29, 33, 36). As the transcriptional regulation of xyn1 and xyn2 strongly differs according to their respective inducer molecules (52), it might be speculated that, in addition to a general inducer (e.g., Xyr1), specific repressors confer these regulatory differences. Whereas a specific xyn2-repressing element (AGAA box) has recently been identified (49), Ace1 acts as a repressor of xyn1 gene transcription. This assumption is supported by the facts that, vice versa, neither a significant influence of Ace1 on xyn2 expression (2) nor the occurrence of a functional AGAA-like element in the xyn1 promoter can be observed. Furthermore, we recently found that xyn2 expression is also strictly dependent on Xyr1 (A. R. Stricker, K. Großteßner-Hain, and R. L. Mach, unpublished data).
In addition to the GGCTAA boxes, two previously identified consensus sequences were shown by in vivo footprinting to be contacted by DNA-binding proteins. Whereas the CCAAT box was constitutively bound, the functional Cre1-binding site (23) was only occupied under carbon catabolite repressing conditions and released from DNA-binding proteins during induction. These findings are in accordance with our previous observations that Cre1 mediates repression of the low constitutive level of xyn1 transcription but cannot interfere with inducing conditions. Ace1 functions as an additional double lock mechanism by preventing Xyr1 binding to the essential 3′ GGCTAA box under noninducing conditions. This mechanism probably causes the strict shut off of xyn1 transcription under repressing conditions.
EMSAs showed that the Hap2/3/5 complex, whose corresponding genes have already been cloned (50), could be reconstituted on the CCAAT box within this fragment and that a mutation of the CCAAT consensus sequence into CCTTT eliminated complex formation. Although the CCAAT box present in many fungal promoters (6) has been shown to be involved in maintaining a transcriptionally active chromatin structure (32), inhibition of protein binding to this motif in the xyn1 promoter is partially compensating for the loss of transcription caused by the mutation of the 3′ GGCTAA box. Similar examples of filamentous fungi with CCAAT boxes exhibiting repressing functions have been observed for, e.g., the A. nidulans lysF promoter (47) and the H. jecorina xyn2 promoter (49).
EMSAs with cell-free extracts and radiolabeled oligonucleotides bearing the inverted GGCTAA boxes or respective mutations suggested that Xyr1 binds to these motifs as a homodimer under xyn1-inducing conditions (Fig. 5). Whereas many members of GAL4-type zinc cluster proteins known so far bind to a repeat, e.g., GAL4 (25), PPR1 (26), QA1F (4), the UaY repeat (42), HAP1 (53), and AlcR (19-21), the mode of binding of A. niger XlnR is still unclear, but in at least some promoters subject to XlnR control, the binding sites are present as repeats (e.g., spaced by 7 bp in the case of xlnD and 12 bp in the case of xlnA) (45). As mentioned before, we hypothesize that H. jecorina Ace1 occupies the right GGCTAA box under repressing conditions, while the left GGCTAA box is still contacted by Xyr1. This finding seems to contradict the in vivo footprinting data where the DNA methylation pattern is not changed for this region under all conditions tested. This may either be due to an insensitivity of the in vivo footprinting method not allowing identifying differences in binding affinities and/or factors contacting the same motive or to the possibility that Ace1 and Xyr1 contact more or less the same bases, thereby not leading to the occurrence of a different methylation protection pattern. It is important that the reduced mobility of the repression-specific protein-DNA complex can be explained by neither the calculated molecular mass nor the isoelectric point of Ace1. Therefore, we currently follow the working hypothesis that an additional factor is involved. Due to the absence of changes in the in vivo footprinting patterns of this region when comparing repressing and inducing conditions, we assume that this factor contacts the repressor complex via protein-protein interaction.
Supplementary Material
Acknowledgments
This study was supported by a grant from Fonds zur Förderung Wissenschaftlicher Forschung (FWF) to R.L.M. (P-16401-B07), which is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Andrianopoulos, A., J. Brons, M. A. Davis, and M. J. Hynes. 1997. The amdA regulatory gene of Aspergillus nidulans: characterization of gain-of-function mutations and identification of binding sites for the gene product. Fungal Genet. Biol. 21:50-63. [DOI] [PubMed] [Google Scholar]
- 2.Aro, N., M. Ilmen, A. Saloheimo, and M. Penttila. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 4.Baum, J. A., R. Geever, and N. H. Giles. 1987. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol. Cell. Biol. 7:1256-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biely, P., M. Vrsanska, M. Tenkanen, and D. Kluepfel. 1997. Endo-beta-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 57:151-166. [DOI] [PubMed] [Google Scholar]
- 6.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and M. J. Hynes. 1999. HAP-like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 7.Buchert, J., T. Oksanen, J. Pere, M. Siika-aho, A. Suurnäkki, and L. Viikari. 1998. Application of Trichoderma reesei enzymes in pulp and paper industry, p. 343-357. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Taylor & Francis Ltd., London, United Kingdom. [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Cziferszky, A., R. L. Mach, and C. P. Kubicek. 2002. Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277:14688-14694. [DOI] [PubMed] [Google Scholar]
- 10.Garrity, P. A., and B. J. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 89:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gielkens, M. M., E. Dekkers, J. Visser, and L. H. de Graaff. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require D-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 13.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 14.Havukainen, R., A. Torronen, T. Laitinen, and J. Rouvinen. 1996. Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei. Biochemistry 35:9617-9624. [DOI] [PubMed] [Google Scholar]
- 15.Hazell, B. W., V. S. Te'o, J. R. Bradner, P. L. Bergquist, and K. M. Nevalainen. 2000. Rapid transformation of high cellulase-producing mutant strains of Trichoderma reesei by microprojectile bombardment. Lett. Appl. Microbiol. 30:282-286. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, M. C., M. Vrsanska, M. Jurickova, J. Hirsch, P. Biely, and C. P. Kubicek. 1997. The beta-D-xylosidase of Trichoderma reesei is a multifunctional beta-D-xylan xylohydrolase. Biochem. J. 321(Pt 2):375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols, a guide to methods and applications, vol. 1. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 18.Ilmen, M., C. Thrane, and M. Penttila. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 19.Kulmburg, P., N. Judewicz, M. Mathieu, F. Lenouvel, D. Sequeval, and B. Felenbok. 1992. Specific binding sites for the activator protein, ALCR, in the alcA promoter of the ethanol regulon of Aspergillus nidulans. J. Biol. Chem. 267:21146-21153. [PubMed] [Google Scholar]
- 20.Kulmburg, P., D. Sequeval, F. Lenouvel, M. Mathieu, and B. Felenbok. 1992. Identification of the promoter region involved in the autoregulation of the transcriptional activator ALCR in Aspergillus nidulans. Mol. Cell. Biol. 12:1932-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenouvel, F., I. Nikolaev, and B. Felenbok. 1997. In vitro recognition of specific DNA targets by AlcR, a zinc binuclear cluster activator different from the other proteins of this class. J. Biol. Chem. 272:15521-15526. [DOI] [PubMed] [Google Scholar]
- 22.Mach, R. L., C. K. Peterbauer, K. Payer, S. Jaksits, S. L. Woo, S. Zeilinger, C. M. Kullnig, M. Lorito, and C. P. Kubicek. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 24.Mandels, M. 1985. Applications of cellulases. Biochem. Soc. Trans. 13:414-416. [DOI] [PubMed] [Google Scholar]
- 25.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 26.Marmorstein, R., and S. C. Harrison. 1994. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 8:2504-2512. [DOI] [PubMed] [Google Scholar]
- 27.Marui, J., A. Tanaka, S. Mimura, L. H. de Graaff, J. Visser, N. Kitamoto, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2002. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 35:157-169. [DOI] [PubMed] [Google Scholar]
- 28.Mathieu, M., and B. Felenbok. 1994. The Aspergillus nidulans CREA protein mediates glucose repression of the ethanol regulon at various levels through competition with the ALCR-specific transactivator. EMBO J. 13:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathieu, M., S. Fillinger, and B. Felenbok. 2000. In vivo studies of upstream regulatory cis-acting elements of the alcR gene encoding the transactivator of the ethanol regulon in Aspergillus nidulans. Mol. Microbiol. 36:123-131. [DOI] [PubMed] [Google Scholar]
- 30.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 31.Muilu, J., A. Torronen, M. Perakyla, and J. Rouvinen. 1998. Functional conformational changes of endo-1,4-xylanase II from Trichoderma reesei: a molecular dynamics study. Proteins 31:434-444. [PubMed] [Google Scholar]
- 32.Narendja, F. M., M. A. Davis, and M. J. Hynes. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, is essential for the formation of a DNase I-hypersensitive site in the 5′ region of the amdS gene. Mol. Cell. Biol. 19:6523-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaev, I., F. Lenouvel, and B. Felenbok. 1999. Unique DNA binding specificity of the binuclear zinc AlcR activator of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 274:9795-9802. [DOI] [PubMed] [Google Scholar]
- 34.Ntarima, P., W. Nerinckx, K. Klarskov, B. Devreese, M. K. Bhat, J. Van Beeumen, and M. Claeyssens. 2000. Epoxyalkyl glycosides of D-xylose and xylo-oligosaccharides are active-site markers of xylanases from glycoside hydrolase family 11, not from family 10. Biochem. J. 347(Pt. 3):865-873. [PMC free article] [PubMed] [Google Scholar]
- 35.Osborne, M. A., and P. A. Silver. 1993. Nucleocytoplasmic transport in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 62:219-254. [DOI] [PubMed] [Google Scholar]
- 36.Panozzo, C., E. Cornillot, and B. Felenbok. 1998. The CreA repressor is the sole DNA-binding protein responsible for carbon catabolite repression of the alcA gene in Aspergillus nidulans via its binding to a couple of specific sites. J. Biol. Chem. 273:6367-6372. [DOI] [PubMed] [Google Scholar]
- 37.Penttila, M., P. Lehtovaara, H. Nevalainen, R. Bhikhabhai, and J. Knowles. 1986. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene 45:253-263. [DOI] [PubMed] [Google Scholar]
- 38.Saloheimo, A., N. Aro, M. Ilmen, and M. Penttila. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817-5825. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 40.Stangl, H., F. Gruber, and C. P. Kubicek. 1993. Characterization of the Trichoderma reesei cbh2 promoter. Curr. Genet. 23:115-122. [DOI] [PubMed] [Google Scholar]
- 41.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 42.Suarez, T., M. V. deq Ueiroz, N. Oestreicher, and C. Scazzocchio. 1995. The sequence and binding specificity of UaY, the specific regulator of the purine utilization pathway in Aspergillus nidulans, suggest an evolutionary relationship with the PPR1 protein of Saccharomyces cerevisiae. EMBO J. 14:1453-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd, R. B., J. M. Kelly, M. A. Davis, and M. J. Hynes. 1997. Molecular characterization of mutants of the acetate regulatory gene facB of Aspergillus nidulans. Fungal Genet. Biol. 22:92-102. [DOI] [PubMed] [Google Scholar]
- 44.Törrönen, A., A. Harkki, and J. Rouvinen. 1994. Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 13:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 47.Weidner, G., S. Steidl, and A. A. Brakhage. 2001. The Aspergillus nidulans homoaconitase gene lysF is negatively regulated by the multimeric CCAAT-binding complex AnCF and positively regulated by GATA sites. Arch. Microbiol. 175:122-132. [DOI] [PubMed] [Google Scholar]
- 48.Wolschek, M. F., F. Narendja, J. Karlseder, C. P. Kubicek, C. Scazzocchio, and J. Strauss. 1998. In situ detection of protein-DNA interactions in filamentous fungi by in vivo footprinting. Nucleic Acids Res. 26:3862-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Würleitner, E., L. Pera, C. Wacenovsky, A. Cziferszky, S. Zeilinger, C. P. Kubicek, and R. L. Mach. 2003. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeilinger, S., A. Ebner, T. Marosits, R. Mach, and C. P. Kubicek. 2001. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genomics 266:56-63. [DOI] [PubMed] [Google Scholar]
- 51.Zeilinger, S., R. L. Mach, and C. P. Kubicek. 1998. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273:34463-34471. [DOI] [PubMed] [Google Scholar]
- 52.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., and L. Guarente. 1994. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J. Biol. Chem. 269:14643-14647. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.