Abstract
Lipid rafts have been identified in the membranes of mammalian cells, the yeast Saccharomyces cerevisiae, and the pathogenic fungus Candida albicans. Formed by a lateral association of sphingolipids and sterols, rafts concentrate proteins carrying a glycosylphosphatidylinositol (GPI) anchor. We report the isolation of membranes with the characteristics of rafts from the fungal pathogen Cryptococcus neoformans. These characteristics include insolubility in Triton X-100 (TX100) at 4°C, more-buoyant density within a sucrose gradient than the remaining membranes, and threefold enrichment with sterols. The virulence determinant phospholipase B1 (PLB1), a GPI-anchored protein, was highly concentrated in raft membranes and could be displaced from them by treatment with the sterol-sequestering agent methyl-β-cyclodextrin (MβCD). Phospholipase B enzyme activity was inhibited in the raft environment and increased 15-fold following disruption of rafts with TX100 at 37°C. Treatment of viable cryptococcal cells in suspension with MβCD also released PLB1 protein and enzyme activity, consistent with localization of PLB1 in plasma membrane rafts prior to secretion. The antioxidant virulence factor Cu/Zn superoxide dismutase (SOD1) was concentrated six- to ninefold in raft membrane fractions compared with nonraft membranes, whereas the cell wall-associated virulence factor laccase was not detected in membranes. We hypothesize that raft membranes function to cluster certain virulence factors at the cell surface to allow efficient access to enzyme substrate and/or to provide rapid release to the external environment.
The rigid lateral aggregation of sphingolipids and sterols forms detergent-resistant plasma membrane microdomains known as rafts. In mammalian cells, rafts are involved in adhesion, signaling, and transport of glycosylphosphatidylinositol (GPI)-anchored proteins to, and localization within, the cell membrane. In the case of GPI-anchored proteins, the functional protein is displayed extracellularly, with the GPI anchor embedded in rafts of the outer membrane layer. Additionally, molecules involved in cell signaling, such as the monomeric G protein H-Ras, localize to rafts of the inner membrane layer via fatty acyl chains (37), where they are positioned to interact with membrane-spanning receptors and appropriate effectors. The model yeast Saccharomyces cerevisiae contains membrane rafts that concentrate the transmembrane channel-forming proteins Can1p (27) and Pma1p and the GPI-anchored protein Gas1p, which is involved in cell wall synthesis. In S. cerevisiae, rafts are also essential for the transport of Gas1P to the cell surface (2, 45).
Little is known about the existence and/or the role of membrane rafts in pathogenic fungi. A recent study of Candida albicans in which membrane sterols were stained with filipin suggested that raft-like domains enriched in sterols polarize to the growing hyphal tips during cell replication, and since hyphae are involved in disease pathogenesis, it was postulated that rafts contribute to fungal virulence (28).
Cryptococcus neoformans is a yeast of medical importance that causes life-threatening neurological infection, especially in immunocompromised hosts (21). Its virulence can be attributed in part to the secretion of the multifunctional enzyme phospholipase B1 (PLB1) (7, 9). PLB1 contains phospholipase B (PLB), lysophospholipase (LPL), and lysophospholipase transacylase (LPTA) activities, and the enzymatic properties of the purified protein have been extensively characterized; however, little is known about the mechanisms regulating its secretion.
Two observations suggest that, prior to secretion, PLB1 is transported to the cell surface as a GPI-anchored protein. First, the primary amino acid sequence contains an N-terminal, hydrophobic motif indicative of a secretory leader peptide that directs the nascent PLB1 to enter the endoplasmic reticulum, where it acquires N-linked glycosylation. Second, there is an additional hydrophobic motif at the C terminus that, in all eukaryotes, is a signal for the attachment of an endoplasmic reticulum membrane-derived GPI anchor. We recently demonstrated, with a heterologous S. cerevisiae expression system, the critical role of these motifs in the secretion of active PLB1. Removal of the N-terminal motif abolished secretion, while removal of the C-terminal motif eliminated PLB1 membrane and cell wall association, resulting in greater PLB1 secretion (16).
Two enzymes that contribute to virulence by protecting cryptococcal cells against oxidative stress are superoxide dismutase (SOD) (19) and laccase (30). SOD is a metalloenzyme of the oxidoreductase family that catalyzes the reduction of toxic host-derived superoxide anions to hydrogen peroxide (12). Gene disruption studies have identified two cryptococcal SOD enzymes involved in virulence (19, 31). SOD1, which is a Cu/Zn-requiring enzyme susceptible to inhibition by cyanide (17, 32, 44, 46, 47), has been purified from cell lysates and has been identified in secretions during the stationary phase of growth (20). SOD2, which is an Mn-requiring and cyanide-insensitive enzyme (5), is predicted to be targeted to the mitochondria by its N-terminal mitochondrial leader peptide. Like PLB1, laccase is transported to the cell surface by the classical secretory pathway, as it possesses a secretory leader peptide and acquires N-linked glycosylation. A large proportion localizes in the cell wall, where it covalently associates with β-glucans (53, 54). It is not known whether either SOD or laccase associate with membranes. However, the possibility of membrane association is supported by the presence in their amino acid sequences of potential sites for the acquisition of N myristoylation and S palmitoylation, fatty acid modifications demonstrated previously to promote membrane raft localization (24, 29, 43).
Given that posttranslational modifications such as GPI anchoring and fatty acylation promote the integration of proteins within membrane rafts in mammalian and yeast systems, we hypothesized and have shown that the membranes of C. neoformans contain raft-like domains that act to concentrate certain virulence factors at the cell surface.
MATERIALS AND METHODS
Reagents and antibodies.
PLB1 antibodies were prepared by immunizing goats (Institute of Medical and Veterinary Science, Gillies Plains, South Australia) with a PLB1 peptide (synthesized by Mimotopes Pty. Ltd., Clayton, Victoria, Australia). Donkey anti-goat horseradish peroxidase (HRP) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Protein G Sepharose was obtained from Amersham Biosciences (Castle Hill, NSW, Australia). PLB1 peptide affinity columns (prepared by Mimotopes Pty. Ltd.) were used to purify PLB1 peptide-specific immunoglobulin G (IgG). Bicinchoninic acid protein assay reagents and Super Signal (enhanced chemiluminescence reagent) were obtained from Pierce Chemical Co. l-α-Phosphatidylcholine, dipalmitoyl (C16:0) (DPPC); l-α-lysophosphatidylcholine, palmitoyl (C16:0) (LysoPC); xanthine oxidase; xanthine; nitroblue tetrazolium; methyl-β-cyclodextrin; and 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) diammonium salt (ABTS) were obtained from Sigma (Castle Hill, NSW, Australia). 1,2-di[1-14C]palmitoyl phosphatidylcholine and 1-[14C]palmitoyl LysoPC were obtained from Amersham Biosciences (Castle Hill, NSW, Australia). The Amplex Red cholesterol assay kit and phosphatidylinositol-phospholipase C were obtained from Molecular Probes (Invitrogen, VIC, Australia). Zirconia/silica beads (0.5 mm) were purchased from Biospec Products, Inc. (Daintree Scientific, TAS, Australia). The SOD assay kit-WST was obtained from Dojindo Laboratories (Kumamoto, Japan). Immobilon-PSQ 0.2-μm membrane was obtained from Millipore Corporation. NuPAGE 4 to 12% bis-Tris gels and loading reagents were all obtained from Invitrogen (VIC, Australia). Coomassie brilliant blue R-250 was obtained from Bio-Rad.
Cryptococcal strains.
A highly virulent, high PLB1-producing strain (H99) of C. neoformans var. grubii serotype A (wild type) and its plb1 and sod1 deletion mutant strains HCM5 (Δplb1) and Δsod1, respectively, were supplied by Gary Cox, Duke University (Durham, NC) (13). The sod2 (19) and lac1 (39) deletion mutant strains Δsod2 and Δlac1, respectively, were supplied by Steven Giles, Duke University.
Detergent-resistant membrane (raft) isolation.
The method used to prepare rafts from the nonencapsulated yeast S. cerevisiae (2) was adapted to suit the encapsulated C. neoformans. Cryptococci were cultured on Sabouraud's dextrose agar (SAB) for 72 h at 30°C. The cells were scraped into 40 ml of 0.9% (wt/vol) saline, resuspended by vortexing, and pelleted by centrifugation for 15 min at 2,000 × g. The cell pellet was washed once with saline and once with morpholineethanesulfonic acid (MES)-buffered saline (25 mM MES, 150 mM NaCl, 2 mM EDTA [pH 6.5]). All of the following steps were performed at 4°C to minimize disruption of rafts, as this has been reported to occur at higher temperatures (25, 41). The resulting pellet, approximately 5 ml in volume, was snap-frozen in liquid nitrogen and then resuspended in 3 ml of resuspension buffer (MES-buffered saline, 0.1% [vol/vol] Triton X-100 [TX100], and a protease inhibitor cocktail [Sigma]). Aliquots of 1 ml were transferred to tubes containing 1 ml of glass beads. Cells were homogenized in a MiniBeadbeater-8 cell disrupter (Daintree Scientific, Tasmania, Australia) for three cycles of 1 min, alternating with a 1-min cooling period on ice. The cell lysate was aspirated and centrifuged at 3,500 × g for 10 min at 4°C. The supernatant was collected and retained. The pellet was kept cold by placing the tube in an ice-salt slurry, further disrupted by probe sonication (five cycles of 10 s on/10 s off) using a Branson Sonifier 450 set on an output of 2, and then centrifuged at 3,500 × g for 15 min at 4°C. The pellet contained mostly cell walls, organelles, and unbroken cells. This supernatant was combined with the first supernatant and centrifuged at 135,000 × g (45,000 rpm) for 1 h at 4°C in a Beckman bench-top ultracentrifuge fitted with a TLA45 rotor. Lipid droplets were recovered as a milky layer from the top of the supernatant and used for further analyses. The membrane pellet was resuspended in 200 μl of ice-cold 1% (vol/vol) TX100 in MES-buffered saline, probe sonicated for 3 to 4 s, and held on ice for 1 h. It was then diluted with 300 μl of 68% sucrose in resuspension buffer to bring the sucrose concentration to 40% and added to the bottom of a 2.2-ml Beckman ultracentrifuge tube. Resuspension buffer containing 30% sucrose (1.8 ml) was layered over the top, and the tube was centrifuged at 201,000 × g (55,000 rpm) for 18 h at 4°C in a TLS55 rotor. Eleven fractions, each of 200 μl, were collected from the top to the bottom of the gradient for analysis by radiometric enzyme assays and Western blotting, as described below.
Protein visualization and Western blotting.
Proteins were concentrated by trichloroacetic acid precipitation. A volume of 90% (wt/vol) trichloroacetic acid was added to achieve a final concentration of 10%. Following incubation on ice for at least 30 min, the samples were centrifuged at 14,000 × g for 15 min at 4°C. The supernatants were removed. and the pellets were resuspended in 20 μl of sample loading buffer as described by the manufacturer, heated for 10 min at 75°C, and then loaded onto a 4 to 12% NuPAGE bis-Tris gradient gel. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (100 mA, 1 h), proteins were visualized by Coomassie brilliant blue R-250 staining.
For Western analysis, equal gradient volumes were subjected to SDS-PAGE as described above and then transferred to a polyvinylidene difluoride membrane (Millipore) in ice-cold Tris-glycine-methanol buffer (25 mM Tris base, 192 mM glycine, 20% [vol/vol] methanol [pH 8]) containing antioxidant at 40 to 50 V constant (∼170 mA) for 1 h. Membranes were removed from transfer buffer, dipped in methanol for 15 s, and allowed to air dry on a double layer of filter paper for 15 to 30 min. All antibodies were diluted in 2% (wt/vol) bovine serum albumin in Tris-buffered saline (77 mM Tris base, 0.14 M NaCl, 27 mM KCl [pH 8]) containing 0.1% Tween 20. For PLB1 and laccase detection, membranes were incubated with anti-PLB1 peptide antibody (1 μg/ml) or laccase antibody (2 μg/ml [a kind gift from Peter Williamson, University of Illinois, Chicago]), respectively, for 1 h followed by a 1-h incubation period with secondary antibody diluted 1:2,000 (donkey anti-goat IgG-HRP [Santa Cruz Biotechnology] or rabbit anti-mouse IgG-HRP [Amersham], respectively). Bands were visualized with enhanced chemiluminescence on Hyper film (Amersham).
Lipid extraction, separation, and identification.
For lipid analysis, sucrose gradient fractions 3 to 6 (rafts) and fractions 9 to 11 (nonraft membranes) were each pooled. The resulting raft and nonraft fractions were dialyzed against water for 18 h, and the lipids from 0.5 ml of each pooled fraction were extracted by adding methanol (3.5 ml)-chloroform (7 ml). The addition of 2.21 ml of water and standing overnight at 4°C achieved phase separation. The organic phase was dried under nitrogen. Lipids were resuspended in 100 μl of chloroform-methanol (6:1 [vol/vol]) prior to thin-layer chromatography (TLC) on silica plates. Neutral lipids were separated in petroleum ether (bp 60 to 80°C)-diethyl ether-acetic acid (80:20:1 [vol/vol/vol]). Polar lipids were separated by two-dimensional TLC. The first solvent system contained chloroform-methanol-7 M ammonium hydroxide (65:30:4 [vol/vol/vol]) followed by chloroform-methanol-acetic acid-water (170:25:25:4 [vol/vol/vol/vol]) in the second dimension. Total lipids were visualized with iodine or Coomassie blue, and identification was performed with 0.25% ninhydrin in acetone for aminophospholipids, with 0.2% orcinol in H2SO4-H2O (3:1 [vol/vol]) for glycolipids, and with 0.005% FeCl3 in H2O-H2SO4-glacial acetic acid (90:5:5 [vol/vol/vol]) for sterols. Identification was also achieved by comparison of the Rf values of unknown spots with authentic standards. In the case of two-dimensional TLC, a plate containing standards only was run in the same tanks at the same time as the test plates. Total lipids were also visualized by charring after the sterol spray.
Sterol analysis.
Extracted lipids were assayed for sterol content with the Amplex Red assay kit (Molecular Probes) as previously described (1, 13). The active component of the kit, cholesterol oxidase, catalyzes the oxidation of sterols with a free C-3β hydroxyl group (10). The two major sterols in C. neoformans, ergosterol (27%) and obtusifoliol (44%) (6), both contain this hydroxyl group. Briefly, lipids were resuspended in methanol, and 5 to 10 μl was combined with 40 to 45 μl of reaction buffer (supplied by the manufacturer). The reaction was initiated by adding 50 μl of Amplex Red reaction mixture without the esterase. Samples were measured in a FLUOstar Optima fluorescence microplate reader (BMG Labtech) with a 530- to 560-nm excitation range and emission detection at 590 nm. The sterol content of each sample was determined with an ergosterol standard curve that was linear over a concentration range of 0.5 to 5 μg/ml.
Fatty acid analysis.
Lipids were extracted by the MIDI Inc. method, version 5. Fatty acid analyses were conducted with saponified lipid extracts that had been converted to fatty acid methyl esters by acid methanolysis. These were separated and identified by an HP 5890 Series II gas chromatography system with an Agilent Ultra 2 capillary column 19091B-102 (7673 auto sampler and flame ionization detector) and Sherlock MIDI System software (YST28 library). Results are expressed as percentages of the sum of the areas of all peaks identified by comparison with standards with the same retention times.
Radiometric assays for phospholipase activities.
PLB1 (LPL, LPTA, and PLB) activities in secretions and subcellular fractions were assayed radiometrically. Unless specified otherwise, a unit of activity was defined as micromoles of the product, DPPC, formed in the LPTA assay or as micromoles of substrate, DPPC or LysoPC, degraded in the PLB and LPL assays, respectively, per minute per milligram of protein at 37°C and pH 4.0 (8).
SOD assay.
Total SOD activity was assayed with the SOD assay kit-WST (Dojindo, Japan) (17). Briefly, 20 μl of cryptococcal membrane or cytosolic fractions was incubated with assay reagent containing xanthine, xanthine oxidase, and a water-soluble tetrazolium salt, WST-1. The superoxide free radicals generated from the xanthine substrate by xanthine oxidase reduce WST-1 to WST-1 diformazan, which absorbs maximally at 450 nm. SOD in the cryptococcal fractions inhibits WST-1 reduction as it catalyzes the dismutation of superoxide ions to molecular oxygen and hydrogen peroxide. The reduction of WST-1 was measured spectrophotometrically. SOD activity was calculated as a rate of inhibition at which 1 U was defined as a 50% decrease from the control value over a period of 30 min at 37°C (17, 20). The results were presented as specific activity, which was determined by the total activity per fraction divided by the total amount of protein per fraction (see Table 1 for total protein values). Differentiation between Cu/Zn SOD and Mn/Fe SOD was determined by two experimental approaches. The first approach involved comparing SOD activity assayed in the presence and absence of 0.1 mM KCN (Cu/Zn SOD is sensitive and Mn/Fe SOD is insensitive to KCN). The second approach was to compare wild-type SOD activity with that found in the Δsod1 and Δsod2 deletion mutant strains.
TABLE 1.
Sterol content of cryptococcal cellular fractionsa
| Fraction | Total sterol/ fraction (μg) (mean ± SEM) | Total protein/ fraction (μg) | Sterol/protein ratiob |
|---|---|---|---|
| Raft | 64.2 ± 7 | 233 | 0.280* |
| Nonraft | 20.3 ± 3 | 788 | 0.025 |
| Lipid droplets | 22.5 ± 2.5 | 547 | 0.041 |
| Cytosol | 145.5 ± 10 | 3,640 | 0.040 |
Determined with the Amplex Red sterol detection kit, as described in Materials and Methods.
*, the increase in the sterol/protein ratio, relative to the other fractions, is statistically significant (P < 0.05), as determined by an unpaired two-tailed parametric t test.
Laccase assay.
Antioxidant (including laccase) activity was determined by oxidation of the substrate ABTS (Sigma). The reaction, carried out at room temperature with gentle shaking, contained 0.5 mM ABTS, 0.1 M sodium acetate buffer (pH 5.0), and 200 μl of protein solution and was made up to 1 ml with water. Oxidation of ABTS was observed over a 4-h time course by measuring an increase in absorbance at 420 nm due to the formation of oxidized ABTS (ɛ420=3.6 × 104 M−1 cm−1) (UV-Visible Spectrophotometer Cary 50 Bio; Varian). Laccase activity was expressed as units per milligram of protein, where units are equal to micromoles of substrate oxidized per minute (3).
MβCD disruption of lipid rafts.
Intact cells with a packed volume of approximately 5 ml were washed in 0.9% (wt/vol) saline, resuspended in MES-buffered saline containing 10 mM methyl-β-cyclodextrin (MβCD), and incubated for 1 h at 37°C with agitation. Cells were pelleted by centrifugation, and the supernatant was subjected to lipid extraction and TLC analysis to confirm the presence of sterols. The cells were washed and either used to prepare and fractionate membranes or resuspended intact in 1 ml of PLB1 secretion buffer (10 mM imidazole, 2 mM CaCl2, 2 mM MgCl2, 1% glucose [pH 5.5]). Secretions and membrane fractions were tested for PLB1 protein and enzyme activity by Western analysis and radiometric assays, respectively.
Quantifications of protein in gradient fractions.
The protein content of subcellular fractions was determined using the bicinchoninic acid protein assay kit and the Compat-Able protein assay preparation reagent set with an albumin protein standard kit, all of which were supplied by Pierce Chemical Company.
Statistics.
Statistics were calculated with GraphPad Instat, version 3. The t test used and the P values obtained are indicated in the table and figure legends. P values of <0.05 were considered significant.
RESULTS
PLB1 protein is mainly localized to raft membranes.
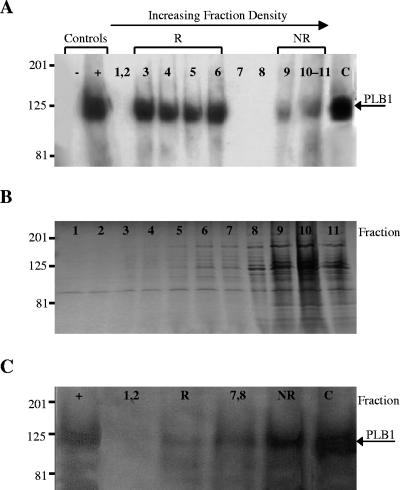
Cryptococcal membranes from H99 (wild type) and a knockout strain, Δplb1, were prepared, incubated with TX100 at 4°C, and fractionated by sucrose density gradient centrifugation as described in Materials and Methods. To determine the location of the PLB1 protein within the sucrose gradient, each fraction was subjected to SDS-PAGE and Western blotting with an anti-PLB1 antibody. In wild-type cells (Fig. 1A), the antibody recognized a major population of PLB1 protein in fractions 3 to 6 (∼90% of the total membrane-associated PLB1) and a minor population in fractions 9 to 11. The PLB1 band had a molecular mass of approximately 120 kDa, corresponding to that of secreted cryptococcal PLB1 (positive control) (51), and was absent in secretions from Δplb1 (negative control). PLB1 protein was also present in the cytosol. No PLB1 band was detected in any of the membrane fractions from Δplb1 (results not shown). Coomassie blue staining of SDS-PAGE-separated gradient fractions that had not been electroblotted (Fig. 1B) showed that most of the membrane protein was localized to the least-buoyant fractions (fractions 9 to 11), indicating that almost all of the PLB1 protein had separated from the bulk of the membrane proteins.
FIG. 1.
Distribution of PLB1 and total protein in fractionated cryptococcal membranes. Total membranes from C. neoformans were prepared and fractionated by sucrose density gradient centrifugation as described in Materials and Methods. Proteins from the 11 membrane fractions and cytosol were subjected to SDS-PAGE and either transferred to a polyvinylidene difluoride membrane and probed with anti-PLB1 antibody (A) or stained with Coomassie brilliant blue for total protein (B). PLB1 protein distributions in Western blots of membranes prepared from cryptococcal cells treated with MβCD are shown in panel C. Secretions from the wild type (H99) and from Δplb1 were used as positive (+) and negative (−) controls, respectively. Fractionated membranes were pooled as follows, starting from the top of the gradient: fractions 1 and 2, raft fractions 3 to 6 (R), intermediate fractions 7 and 8, nonraft fractions 9 to 11 (NR), and cytosol (C). Gradient fractions are shown from least (left) to most (right) dense, as indicated by the arrow at the top (this applies to panels A, B, and C). Molecular mass standards are indicated (in kilodaltons), and the arrows on the right indicate the position of PLB1.
MβCD disrupts PLB1 raft membrane association.
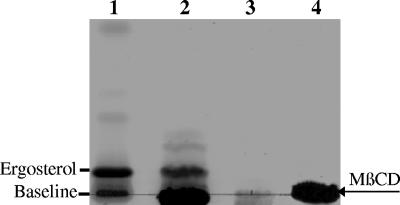
Membranes were prepared from cryptococci that had been incubated with the sterol-sequestering reagent MβCD. To confirm that sterols had been extracted from the cryptococcal cells, the MβCD-containing supernatants were subjected to lipid extraction and TLC analysis. The results in Fig. 2 show that MβCD treatment successfully extracted a sterol (lane 2) with an Rf value similar to the ergosterol standard (lane 1). The MβCD-treated membranes were then fractionated by sucrose gradient centrifugation. Prior to SDS-PAGE and Western analysis, the sucrose gradient fractions were pooled on the basis of PLB1 distribution (see Fig. 1A) as follows: fractions 1 and 2, rafts (fractions 3 to 6), intermediate fractions (fractions 7 and 8), and nonraft fractions (fractions 9 to 11). In contrast to the profile obtained for untreated cells (Fig. 1A), most of the membrane-associated PLB1 protein was now in the nonraft membrane fractions and in the cytosol (Fig. 1C). This redistribution indicates that cryptococcal rafts were disrupted by MβCD treatment and is consistent with findings in S. cerevisiae and mammalian cells (2, 52).
FIG. 2.
Extraction of sterols from cryptococcal cells with MβCD. Cells were incubated with or without 10 mM MβCD for 1 h and pelleted by centrifugation. Supernatants were collected and subjected to lipid extraction as described in Materials and Methods. Extracted lipids were fractionated by TLC and stained for neutral lipids. A lipid band comigrated with the ergosterol standard (lane 1) when MβCD was present in the cell incubation buffer (lane 2) but not when it was absent (lane 3). MβCD that had been taken through the extraction procedure in the absence of cells appears as an intense band on the baseline in lane 4 and is also present in lane 2.
Effect of raft membrane localization on PLB activity.
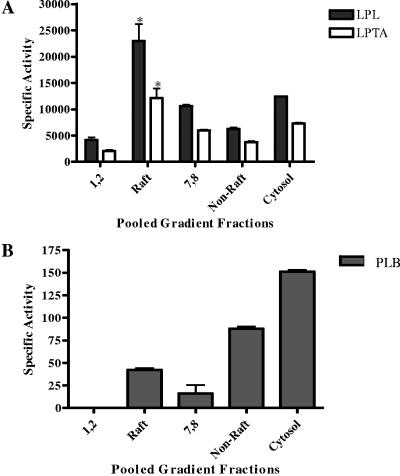
The membrane fractions from the wild type and Δplb1 were assayed radiometrically for phospholipase activities (LPL, LPTA, and PLB). None of the three activities were detected in pooled raft and nonraft membrane fractions prepared from Δplb1 (data not shown). Raft membranes prepared from wild-type cells contained approximately threefold more LPL and LPTA specific activities than nonraft membrane fractions (Fig. 3A), which supports the PLB1 protein distribution results presented in Fig. 1A. In contrast, twofold more PLB specific activity was present in nonraft fractions (Fig. 3B). Notably, LPL/LPTA activity was detected in nonraft membrane fractions 7 and 8 when no PLB1 protein was identified by Western analysis (compare Fig. 3A to Fig. 1A). This discrepancy is most likely due to the greater sensitivity of the activity assay. To explain the inconsistency in the levels of raft-associated PLB activity and protein, we hypothesized that the raft environment inhibits the PLB activity of raft-associated enzyme.
FIG. 3.
Distributions of phospholipase activity in fractionated cryptococcal membranes. Total membranes from C. neoformans were prepared, fractionated by sucrose density gradient centrifugation, pooled as described in the legend to Fig. 1, and assayed for LPL/LPTA (A) and PLB (B) activities. Activities are expressed as micromoles of LysoPC substrate degraded (LPL) or DPPC formed (LPTA) per minute per milligram of protein (A) or nanomoles of DPPC degraded (PLB) per minute per milligram of protein (B). *, the increase in LPL/LPTA activity of rafts is statistically significant relative to that of nonraft membranes. P < 0.05 (using an unpaired, nonparametric t test).
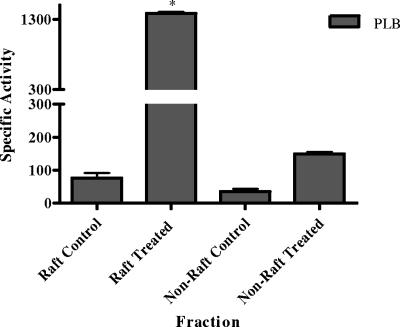
We first tested this hypothesis by disrupting raft membranes with MβCD but found that the MβCD was inhibitory to phospholipase activity. We then disrupted rafts with 1% TX100 at 37°C for 1 h to determine whether raft disruption led to increased release of PLB activity. Nonraft membranes were taken through the same procedure as controls. The results in Fig. 4 show that raft-associated PLB activity increased approximately 15-fold following raft disruption. This was not due to stimulation of enzyme activity, since the detergent concentration in the assay was <0.1%, a concentration at which we have shown previously causes no stimulation of PLB activity (8, 40). In contrast, only a fivefold increase in PLB activity was obtained following disruption of nonraft membranes. The disruption experiment indicated that approximately 90% of the membrane-associated PLB activity was derived from the raft fraction, correlating with the PLB1 protein distribution presented in Fig. 1A. No difference in LPL/LPTA activity was observed in intact and disrupted rafts (not shown).
FIG. 4.
Comparison of PLB activities in intact and disrupted raft membranes. Pooled gradient fractions containing raft and nonraft membranes were incubated with TX100 at 37°C for 1 h to disrupt membrane lipids and then assayed for PLB activity. Activity is expressed as nanomoles of DPPC degraded per minute per milligram of protein. The increase in the activity derived from the treated raft membranes was statistically significant relative to both the untreated raft membrane control and the treated nonraft membranes. P < 0.05 (using an unpaired, nonparametric t test).
PLB1 localizes in the plasma membrane prior to secretion.
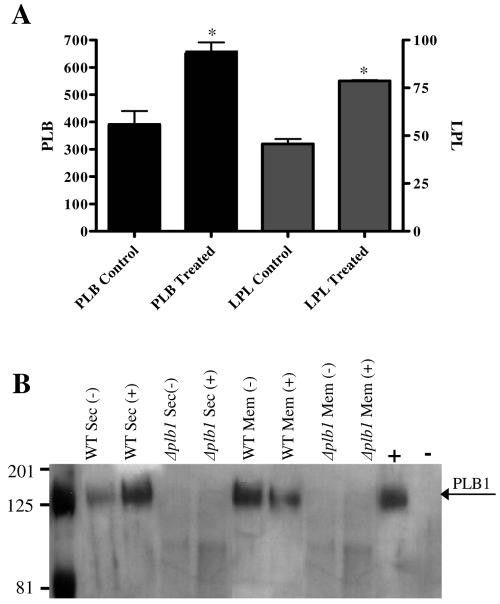
Cryptococcal cells were preincubated for 1 h with MβCD to disrupt membrane rafts, washed, and permitted to secrete PLB1 for 4 h (Fig. 5), at which time PLB1 secretion has been shown to plateau (8). The treated cells secreted almost twofold more LPL, LPTA (not shown), and PLB (Fig. 5A) activities than the untreated cells, presumably due to MβCD-induced disruption of membrane rafts and release of raft-associated enzyme from the membrane surface. The ratio of LPL to LPTA to PLB activity secreted in the presence and absence of MβCD remained constant, consistent with secretion of a single enzyme protein, PLB1, and not additional lysophospholipase enzymes which have been identified previously (51). Western blotting was also performed on the membranes and supernatants (secretions) obtained from the wild-type and Δplb1 strains, treated or not treated with MβCD (Fig. 5B). As expected, no PLB1 protein band was present in either fraction prepared from Δplb1. For the wild-type strain there was a greater-than-twofold increase in PLB1 protein in the secretions from treated cells, correlating with the enzyme activity results. There was a similar decrease in membrane-associated PLB1 protein in treated cells, confirming that the additional secreted protein was derived from the membrane.
FIG. 5.
Effects of sterol depletion on PLB1 secretion and membrane association. Cells, either untreated (control) or treated with MβCD for 1 h at 37°C, were washed and resuspended in secretion buffer. At 4 h, cells were pelleted by low-speed centrifugation and the secretions were assayed for PLB and LPL/LPTA specific activities (A). Specific activity is expressed as nanomoles of DPPC formed (PLB) or micromoles of LysoPC (LPL) degraded per minute per milligram of protein. Data are means and standard errors of triplicate assays (three experiments). Asterisks indicate that the increase in specific activity, relative to the respective control, was statistically significant (P < 0.05) using a two-tailed unpaired parametric t test. LPTA activity is not shown, but the percent increase in MβCD-released activity was similar to that obtained for LPL and PLB. Western analysis of secreted (Sec) and membrane (Mem)-associated PLB1 before (−) and after (+) MβCD treatment is shown in panel B. No bands were observed in either fraction from Δplb1. The arrow indicates the position of the PLB1 band. Secretions from the wild-type (H99) and Δplb1 strains were used as positive (+) and negative (−) controls, respectively.
Location of SOD in membrane rafts.
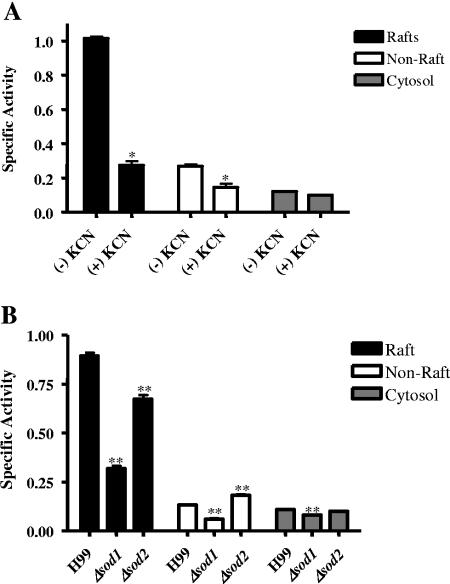
The specific activity of SOD was determined in the cryptococcal membrane fractions and cytosol in the presence and absence of KCN. Total SOD activity (assayed in the absence of KCN) was four-and eightfold greater in raft membrane fractions than in nonraft membrane fractions and cytosol, respectively (Fig. 6A). Approximately 70% of raft-associated SOD activity and 40% of non-raft-associated SOD activity was sensitive to 0.1 mM KCN and therefore attributable to Cu/Zn SOD. Consequently, Cu/Zn SOD activity was enriched at least sixfold in raft membranes compared with nonraft membranes. KCN-insensitive SOD activity, which may be due to Mn- or Fe-requiring SOD, comprised most of the activity in the cytosolic fraction.
FIG. 6.
Membrane distribution of cryptococcal SOD activity. Pooled raft and nonraft membrane fractions and cytosol were prepared from the wild type (H99) and assayed in the presence (+) and absence (−) of KCN, an inhibitor of Cu/Zn SOD (A). Subcellular fractions from H99, Δsod1, and Δsod2 strains were assayed for SOD (B). Data are means and standard errors of the means of triplicate assays (three experiments). *, significant decrease in activity of the wild type in the presence of KCN compared with untreated cells; **, significant change in activity of deletion mutants compared with wild-type cells. One unit of SOD was defined as a 50% decrease in activity relative to controls.
To confirm that the Cu/Zn SOD activity was the product of sod1 and to determine whether KCN-insensitive activity was due to SOD2 or another SOD, we compared the cellular distributions of SOD activity in the Δsod1 and Δsod2 mutants with that in wild-type cells. In Δsod1, SOD activities were reduced in raft, nonraft membrane, and cytosolic fractions by 70%, 47%, and 25%, respectively (Fig. 6B), analogous to the effect of KCN on wild-type SOD activity (Fig. 6A). In contrast, in the Δsod2 mutant, raft-associated SOD activity was reduced by only 25%, while nonraft membrane activity increased by 38% and cytosolic activity was unchanged. SOD1 was calculated to be enriched ninefold in the raft fractions (cf. the sixfold enrichment based on KCN sensitivity data, above).
The specific activities of SOD in the nonraft membrane and cytosolic fractions of Δsod2 were either higher than or the same as in the wild type (Fig. 6B), indicating that neither of these fractions contain Mn-requiring SOD activity. However, residual SOD activity was present in these fractions in Δsod1. This suggests that an additional SOD gene(s) and enzyme(s) are present in cryptococci.
Laccase and membrane rafts.
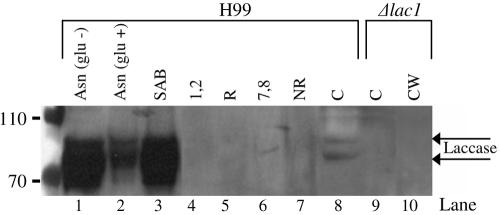
Although laccase protein (product of the lac1 gene) is mainly localized to the cryptococcal cell wall, we considered the possibility that, analogous to some GPI-anchored proteins, it may be transported there via the cell membrane. Thus, we looked for its presence in fractionated membranes by Western blotting and by performing an antioxidant activity assay. Notably, the laccase protein was not present in raft or nonraft membranes (Fig. 7A, lanes 4 to 7) but was present in the supernatants of β-glucanase-disrupted cell walls (lane 3) prepared from cells grown under the same conditions (SAB plates for 3 days) as those used to prepare raft membranes. Despite the presence of 4% glucose in the SAB plates, which reportedly suppresses laccase production (53), we still found a high concentration of laccase in the cell wall. This was presumably due to glucose depletion from the medium during the long period of cryptococcal growth. The level of cell wall-associated laccase was comparable to that obtained for cells that were cultured on asparagine minimal plates lacking glucose (Fig. 7, lane 1). As expected from previous work, the addition of glucose to the medium reduced the intensity of laccase (lane 2) (54), which now appeared to be two bands. A similar laccase banding pattern was also observed in the cytosol prepared from the wild-type strain, H99, but was absent from Δlac1 (Fig. 7; compare lane 8 with lane 9), confirming that the bands were attributable to lac1 gene products. As expected, laccase was not detected in the cell walls prepared from the Δlac1 mutant (lane 10). The antioxidant assay results indicate that there is minimal activity (which includes laccase activity) in the membrane fractions (not shown). This correlates with the absence of laccase by Western analysis.
FIG. 7.
Subcellular distribution of laccase.β-Glucanase-treated cell wall preparations (lanes 1 to 3 and 10), pooled gradient fractions (lanes 4 to 7), and cytosol (C) (lanes 8 and 9), prepared from either H99 or Δlac1 as indicated in the figure, were subjected to SDS-PAGE and Western blotting with a monoclonal antibody against laccase. Control conditions on the Western blot showed that laccase expression is repressed when glucose is present (lane 2). Arrows indicate the positions of bands attributable to products of the lac1 gene.
Characterization of cryptococcal lipid rafts. (i) Sterol content.
Since raft membranes are characterized by a higher proportion of sterols than nonraft membranes, the sterol content of each fraction was determined as described in Materials and Methods and the results are presented in Table 1 and Fig. 8. As expected, the raft fractions contained significantly more sterol, with a sterol to protein ratio 11 times greater than in the nonraft membrane fractions. This agrees with findings in S. cerevisiae and mammalian lipid rafts (33). Sterol was also present in lipid droplets (which were obtained from the surface layer following high-speed centrifugation to separate total membranes; see Materials and Methods). The presence of sterol in the cytosol may be attributed to residual membrane fragments or vesicles and TX100 solubilization of nonraft membrane-associated ergosterol during cellular disruption.
FIG. 8.
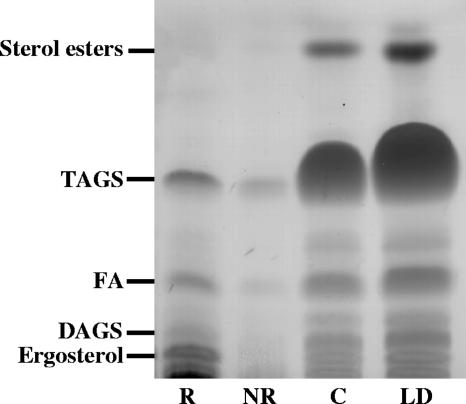
Neutral lipid composition of subcellular fractions from C. neoformans. Lanes were loaded with equal volumes of total lipids from raft fractions (R), nonraft fractions (NR), cytosol (C), and lipid droplets (LD). Lipid separation was achieved with a petroleum ether-diethyl ether-acetic acid (80:20:1 [vol/vol/vol]) solvent system. Separated lipids were visualized with iodine vapor and then sterol spray, followed by charring to reveal all lipid species. Lipids were identified by comparing Rf values with authentic standards. These plates are representative of three separations. The positions of TAGs, diacylglycerols (DAGS), fatty acids (FA), sterol esters, and ergosterol are indicated.
(ii) Neutral lipid composition.
Triacylglycerols (TAGs) were identified in raft membranes (Fig. 8). This is in accordance with previous studies of THP-1 and other mammalian cell lines (50). As expected, the TAGs were most prominent in lipid droplets. They were also observed in the cytosolic fraction, most likely due to contamination from the lipid droplet layer and some lipid extraction with TX100 used in raft preparation. More total neutral lipids, including diacylglycerols, TAGs, and free fatty acids (FFA), were observed in the raft fractions than in the nonraft fractions.
Extraction and separation of lipids from control gradient fractions containing no cryptococcal membranes revealed that there were no artifactual compounds present in the raft fractions (results not shown). Similar blank gradients were used as controls for polar lipid separations and for fatty acid analyses, below.
(iii) Polar lipid composition.
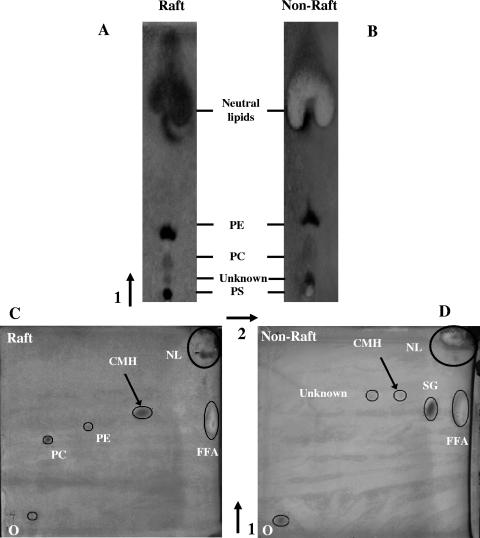
Separation of raft and nonraft polar lipids by one- and two-dimensional TLC is shown in Fig. 9. One-dimensional TLC (Fig. 9A and B) detected phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in both fractions. However, much more lipid had to be loaded on the plate (saturation is indicated by the white staining of the neutral lipids) to reveal these phospholipids in the nonraft fraction. This was due to the nonraft membranes having a low lipid to protein ratio compared with the raft membranes, which are relatively protein depleted (see Fig. 1B). Both fractions contained neutral lipid. The polar lipids (PC and PE), however, were not detectable in the nonraft fraction by two-dimensional TLC due to the reduced amount of total lipid, which must be loaded to achieve adequate separation when this method is used (Fig. 9C and D). Because of the better resolution, a new glycolipid spot with an Rf value consistent with ceramide monohexosides (CMH) was revealed. This was detected in both raft and nonraft fractions but only faintly in nonraft fractions. A faint unknown spot and a spot with an Rf value consistent with steryl glycosides (SG) were also detected in nonraft fractions. Due to the unavailability of SG and CMH standards, these putative identifications were made by comparing our Rf values with those in reference 36.
FIG. 9.
One- and two-dimensional TLC separations of polar lipids from raft and nonraft fractions. The solvent system used for one-dimensional lipid separation in panels A and B was chloroform-methanol-7 M ammonium hydroxide (65:30:4 [vol/vol/vol]). A two-dimensional separation using the above solvent system in the first dimension and chloroform-methanol-acetic acid-water (170:25:25:4 [vol/vol/vol/vol]) in the second dimension is shown for raft (C) and nonraft (D) membranes. Spots were revealed with iodine vapor or Coomassie blue, which stains FFA white. Aminophospholipids (PE and PS) were identified by using ninhydrin, sterols were identified with sterol spray, and glycolipids were identified with orcinol spray (see Materials and Methods). Spots were also identified by comparison with the Rf values of standard lipids run at the same time as the test plates and using the same solvent system. Where no standards were available (SG and CMH), comparison was made with Rf values of total cryptococcal lipids as used in reference 36. O, origin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; CMH, ceramide monohexosides; SG, steryl glycosides; FFA, free fatty acids; NL, neutral lipids. These plates are representative of two separations. The arrows indicate the direction of solvent flow.
(iv) Fatty acid composition.
The fatty acid compositions of the total lipids extracted from four cryptococcal subcellular membrane fractions (raft and nonraft membranes, cytosol, and lipid droplets) are shown in Table 2. An important observation is the presence of a C12 primary alcohol in the lipids from the raft fraction. The origin of this product is unknown, but it may be the breakdown product of a sphingolipid species, such as a ceramide. Higher quantities (though not statistically significant) of the saturated fatty acids palmitic and stearic acid and the monounsaturated fatty acid oleic acid were present in the lipids from the nonraft membrane fractions than in the rafts. There appeared to be more of the di-unsaturated fatty acid species linoleic acid in raft lipids than in nonraft lipids, but again the increase was not statistically significant. The very long chain fatty acids (>20 carbons), identified by others in lipid rafts from mammalian cells and S. cerevisiae were not detectable by the MIDI program we used for fatty acid analysis (see Materials and Methods).
TABLE 2.
Comparison of fatty acid species from fractionated C. neoformans cells
| Fraction | Fatty acid species (%)a
|
|||||
|---|---|---|---|---|---|---|
| C12 1° alcohol | Palmitic | Linoleic | Oleic | Stearic | Myristic | |
| Raft | 9 ± 2.5 | 17 ± 1.2 | 36 ± 4.8 | 36 ± 1.8* | 4 ± 0.2 | ND |
| Nonraft | ND | 20 ± 0.4 | 29 ± 3.3 | 44 ± 1.3 | 5 ± 1.1 | ND |
| Lipid droplets | ND | 19 ± 0.3 | 35 ± 1.5 | 39 ± 2.1 | 3 ± 0.6 | 1 ± 0.06 |
| Cytosol | ND | 19 ± 0.9 | 38 ± 2.3 | 38 ± 0.9 | 3 ± 0.25 | 1 ± 0.01 |
Results are expressed as means ± standard errors of the means (n = 3) of the area under the peak as a percentage of the sum of the areas of all the peaks measured. ND, not detected; *, the decrease in raft oleic acid content, relative to nonraft, is statistically significant as determined by an unpaired two-tailed t test (P = 0.013).
DISCUSSION
Using a recombinant yeast expression system, we have previously demonstrated that the 22 C-terminal amino acids of the cryptococcal PLB1 protein are responsible for its attachment to fungal membranes via a GPI anchor (16). We now demonstrate that native PLB1 produced by C. neoformans is membrane associated and that it is concentrated in detergent-resistant membrane fractions with the lipid and protein characteristics of rafts. This study is the first to establish the presence of lipid rafts in the membranes of C. neoformans and to demonstrate that the virulence factors PLB1 and Cu/Zn SOD are localized within them.
Characterization of cryptococcal rafts.
Lipid rafts in the membranes of eukaryotic cells are detergent resistant and highly enriched in sterols and sphingolipids (2, 26). Consistent with this, detergent-resistant membranes from cryptococci were enriched with a sterol that comigrated on TLC with the fungal sterol ergosterol. A contribution from another major sterol previously identified in C. neoformans, obtusifoliol (18), cannot be ruled out, due to the unavailability of a commercial standard. We obtained indirect evidence for the presence of sphingolipids within our raft membranes by using two-dimensional TLC; the predominant polar lipid had an Rf value consistent with CMH (36). Fungal CMH were nominated as putative raft sphingolipids in C. neoformans when clustering of CMH was observed by fluorescent antibody labeling during cryptococcal budding, and they have been studied in detail (38). In C. neoformans, some CMH contain C16 or C18 hydroxy fatty acids as well as a 4,8-sphingadienine-containing ceramide moiety. A compound of the latter type may give rise to the C12 primary alcohol that we observed by fatty acid analysis of raft lipids.
Neutral lipids, especially TAGs, were also abundant in cryptococcal rafts. Past evidence would suggest that TAGs are not classic membrane lipids; however, they have been identified in detergent-resistant membranes of various mammalian cell lines by lipid analysis and 1H nuclear magnetic resonance spectroscopy (50). This observation is of interest because it suggests that rafts are at least in part derived from lipid droplets, which are rich in TAGs. Moreover, raft-associated proteins such as caveolins and GPI-anchored proteins have been identified in the surface monolayer of lipid droplets (34, 35).
Raft localization and PLB1 activity.
The presence of lipid rafts within cell membranes is thought to allow partitioning and hence concentration of certain proteins in the raft environment, for example, proteins involved in cell adhesion, signal transduction pathways, and GPI-anchored proteins (33). Polarization of lipid rafts to growing hyphal tips was noted in C. albicans and was proposed to contribute to disease pathogenesis, since hyphal growth is required for full virulence (28). A specific association of these rafts with a known virulence determinant was not investigated. Our data demonstrate clearly that the GPI-anchored cryptococcal virulence determinant PLB1 is concentrated in membrane rafts.
Initially we were concerned to note an inconsistency in the distribution of raft-associated PLB1 protein and PLB activity. We first considered the possibility of a contribution from phospholipases other than PLB1, as activity can also arise from the gene product of CnLYSO1 (11) or the CnLPL1 protein (51), as well as any other unidentified phospholipases. However, as none of the three activities were present in membranes isolated from Δplb1, and because the LPL/LPTA activities mirrored the distribution of the PLB1 protein, the activity present in raft and nonraft membranes could be attributed only to PLB1.
We then investigated the possibility that the raft environment was inhibiting PLB activity, since maintaining membrane-associated PLB in an inactive state may be important for membrane integrity. Consistent with this hypothesis, PLB activity was increased to levels that were comparable with those of the PLB1 protein following raft disruption, indicating that the raft environment was indeed inhibiting PLB activity. Furthermore, disruption of membrane rafts by incubation with the sterol-sequestering agent MβCD released equivalent amounts of activity and PLB1 protein, with a similar loss of PLB1 protein from the membrane preparation. Further investigation of the observed inhibition of PLB1 activity in the raft environment is beyond the scope of this study, but several mechanisms are possible. Firstly, the GPI anchor could place conformational constraints on the PLB1 substrate binding and/or active site. Secondly, lipids within the raft environment could impose the inhibition. It has been shown previously that the protein component of GPI-anchored ectoenzymes such as 5′ nucleotidase and human placental alkaline phosphatase lies close to or is in direct contact with the lipid bilayer (4, 23), potentially allowing the membrane lipids to influence the activity of the enzymes. Furthermore, the catalytic properties of 5′ nucleotidase are influenced by the phase state and fluidity of the bilayer (42). In C. neoformans, the sterols concentrated in rafts may inhibit PLB, but not LPL and LPTA, activities directly, or indirectly, via an interaction between PLB1 and the sterol-stabilized lipid structure of the raft. This explanation is consistent with release of 10-fold more PLB activity from rafts than from nonraft membrane fractions after membrane disruption. Alternatively, PLB1 activity may be reduced because of the absence of its preferred enzyme substrate(s) from membrane rafts. However, the phospholipid PC, which we demonstrated previously to be the preferred substrate of cryptococcal PLB1 (8), was readily demonstrated in cryptococcal rafts. PLB1 enzyme activities are also influenced by the fatty acids present in the Sn-1 and Sn-2 positions of the glycerol backbone of phospholipids (8). In the present study we did not determine the fatty acid disposition on the glycerol backbone of raft phospholipids. However, the distributions of fatty acids in lipids extracted from the different cell fractions were determined and were found to be similar (except for a small but significant reduction of 20% in oleic acid in the rafts). Finally, PLB1 activity (PLB to a greater extent than LPL/LPTA) may be inhibited by an association of PLB1 with an inhibitory protein. The observed increase in PLB activity following raft disruption may be due to the dissociation of PLB1 from the putative inhibitory protein or to the release of the enzyme from its GPI anchor.
Membrane localization of SOD and laccase.
SOD and laccase are both virulence factors involved in protection of cryptococci against oxidative stress encountered in vivo. Our data indicate that there is a distinct enrichment of Cu/Zn SOD activity (the product of sod1) in raft membranes, whereas laccase (the product of lac1) is neither raft nor membrane associated.
N- and C-terminal amino acid sequences predicted from the sod1 gene do not conform to all of the currently established criteria for a GPI-anchored protein; thus, it is unlikely that GPI anchoring is the mode of raft membrane attachment. Alternatively, palmitoylation, a reversible posttranslational modification, would allow SOD to switch between a cytosolic and a membrane-bound form, as has been previously proposed for heterotrimeric G proteins (48, 49). Possible advantages of SOD membrane localization are concentration prior to secretion and accessibility to host-derived superoxide anions, allowing rapid neutralization of their cytotoxic effects.
By comparing wild-type SOD activity in the presence and absence of KCN and between the wild type and Δsod deletion mutants, we showed that Cu/Zn-requiring SOD1 activity was concentrated in rafts six- to ninefold compared to nonraft membranes. In contrast, SOD2 activity represented less than 30% of the total SOD activity in the raft fractions. This may reflect the presence of some mitochondrial membranes in our preparation. As expected, there was no SOD2 activity in the cytosol, nor was it present in the nonraft membrane fractions. We were therefore surprised to find residual SOD activity in these fractions in the Δsod1 mutant. We considered the possibility of a “compensatory” increase in sod2 gene expression during cryptococcal cell growth. However, this possibility is unlikely, as the presence of KCN in the assays of all fractions prepared from the wild-type strain resulted in the SOD activities being reduced by levels similar to those obtained for the Δsod1 knockout strain. The most likely explanation is that an additional non-Cu/Zn-requiring SOD enzyme(s) contributes to the membrane and cytosolic activity. Mn-requiring SODs lacking mitochondrial leader peptides are present in other fungi, such as Penicillium chrysogenum (15), Aspergillus fumigatus (14), G. microsporum (GenBank accession number U56403), and C. albicans (22) (sod3). Candidal sod3 is upregulated during the stationary phase of growth (22). If a similar gene were present in C. neoformans, it should be expressed in the stationary-phase cells used in our cryptococcal studies. Although a search of the cryptococcal genome databases did not reveal a protein with homology to Candida SOD3, the existence of additional SODs with low homology to candidal SODs remains a possibility.
In conclusion, we have demonstrated both the presence and functionality of rafts in C. neoformans. In particular, rafts tether the virulence determinants PLB1, most likely via a GPI anchor, and Cu/Zn SOD, by an undetermined mechanism, to the membrane surface. The surface localization of enzymes involved in virulence may provide pathogens with a rapid-release mechanism that is activated when adverse conditions are encountered, as in the mammalian host, and/or may allow them better access to substrate.
Acknowledgments
We thank Peter Williamson (University of Illinois, Chicago) for providing laccase monoclonal antibody and helpful advice on laccase detection, Gary Cox (Duke University, Durham, NC) for supplying Δplb1 and Δsod1 deletion mutants, Steven Giles (Duke University) for providing Δsod2 and Δlac1 cryptococcal deletion mutant strains, Leanne Hicks for work on gas chromatography, and the research members of the Centre for Infectious Diseases and Microbiology (CIDM) for support.
This work was supported by National Health and Medical Research Council of Australia (NHMRC) project grants 352354 and 211040, a University of Sydney Postgraduate Award to A.R.S., a Westmead Millennium Foundation Top-Up scholarship to A.R.S., and a New South Wales Department of Health Stream 1 Infrastructure grant to the CIDM, Westmead Millennium Institute, University of Sydney.
REFERENCES
- 1.Bagnat, M., A. Chang, and K. Simons. 2001. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., S. Keranen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourbonnais, R., and M. G. Paice. 1990. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 267:99-102. [DOI] [PubMed] [Google Scholar]
- 4.Brasitus, T. A., and D. Schachter. 1980. Lipid dynamics and lipid-protein interactions in rat enterocyte basolateral and microvillus membranes. Biochemistry 19:2763-2769. [DOI] [PubMed] [Google Scholar]
- 5.Brouwer, M., T. H. Brouwer, W. Grater, J. J. Enghild, and I. B. Thogersen. 1997. The paradigm that all oxygen-respiring eukaryotes have cytosolic CuZn-superoxide dismutase and that Mn-superoxide dismutase is localized to the mitochondria does not apply to a large group of marine arthropods. Biochemistry 36:13381-13388. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, D.C.
- 7.Chen, S. C., M. Muller, J. Z. Zhou, L. C. Wright, and T. C. Sorrell. 1997. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J. Infect. Dis. 175:414-420. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. C., L. C. Wright, J. C. Golding, and T. C. Sorrell. 2000. Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 347:431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. C., L. C. Wright, R. T. Santangelo, M. Muller, V. R. Moran, P. W. Kuchel, and T. C. Sorrell. 1997. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect. Immun. 65:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, L. C., Jr., and T. A. Grooms. 1980. Substrate specificity of cholesterol oxidase. Clin. Chem. 26:678. [PubMed] [Google Scholar]
- 11.Coe, J. G., C. F. Wilson, T. C. Sorrell, N. G. Latouche, and L. C. Wright. 2003. Cloning of CnLYSO1, a novel extracellular lysophospholipase of the pathogenic fungus Cryptococcus neoformans. Gene 316:67-78. [DOI] [PubMed] [Google Scholar]
- 12.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 14.Crameri, R., A. Faith, S. Hemmann, R. Jaussi, C. Ismail, G. Menz, and K. Blaser. 1996. Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J. Exp. Med. 184:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diez, B., C. Schleissner, M. A. Moreno, M. Rodriguez, A. Collados, and J. L. Barredo. 1998. The manganese superoxide dismutase from the penicillin producer Penicillium chrysogenum. Curr. Genet. 33:387-394. [DOI] [PubMed] [Google Scholar]
- 16.Djordjevic, J. T., M. Del Poeta, T. C. Sorrell, K. M. Turner, and L. C. Wright. 2005. Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem. J. 389:803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dojindo Laboratories. February 2004. SOD assay kit-WST technical manual, p. 4. Dojindo Laboratories, Kumamoto, Japan.
- 18.Ghannoum, M. A., B. J. Spellberg, A. S. Ibrahim, J. A. Ritchie, B. Currie, E. D. Spitzer, J. E. Edwards, Jr., and A. Casadevall. 1994. Sterol composition of Cryptococcus neoformans in the presence and absence of fluconazole. Antimicrob. Agents Chemother. 38:2029-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giles, S. S., I. Batinic-Haberle, J. R. Perfect, and G. M. Cox. 2005. Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot. Cell 4:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton, A. J., and M. D. Holdom. 1997. Biochemical comparison of the Cu/Zn superoxide dismutases of Cryptococcus neoformans var. neoformans and Cryptococcus neoformans var. gattii. Infect. Immun. 65:488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husain, S., M. M. Wagener, and N. Singh. 2001. Cryptococcus neoformans infection in organ transplant recipients: variables influencing clinical characteristics and outcome. Emerg. Infect. Dis. 7:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamarre, C., J. D. LeMay, N. Deslauriers, and Y. Bourbonnais. 2001. Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon entry and during the stationary phase. J. Biol. Chem. 276:43784-43791. [DOI] [PubMed] [Google Scholar]
- 23.Lehto, M. T., and F. J. Sharom. 2002. Proximity of the protein moiety of a GPI-anchored protein to the membrane surface: a FRET study. Biochemistry 41:8368-8376. [DOI] [PubMed] [Google Scholar]
- 24.Lindwasser, O. W., and M. D. Resh. 2002. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. USA 99:13037-13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.London, E., and D. A. Brown. 2000. Insolubility of lipids in Triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508:182-195. [DOI] [PubMed] [Google Scholar]
- 26.Luberto, C., D. L. Toffaletti, E. A. Wills, S. C. Tucker, A. Casadevall, J. R. Perfect, Y. A. Hannun, and M. M. Del Poeta. 2001. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 15:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malinska, K., J. Malinsky, M. Opekarova, and W. Tanner. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, S. W., and J. B. Konopka. 2004. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melkonian, K. A., A. G. Ostermeyer, J. Z. Chen, M. G. Roth, and D. A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274:3910-3917. [DOI] [PubMed] [Google Scholar]
- 30.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narasipura, S. D., J. G. Ault, M. J. Behr, V. Chaturvedi, and S. Chaturvedi. 2003. Characterization of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol. Microbiol. 47:1681-1694. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, K. M., R. Dirmeier, M. Engle, and R. O. Poyton. 2004. Mitochondrial protein oxidation in yeast mutants lacking manganese- (MnSOD) or copper- and zinc-containing superoxide dismutase (CuZnSOD): evidence that MnSOD and CuZnSOD have both unique and overlapping functions in protecting mitochondrial proteins from oxidative damage. J. Biol. Chem. 279:51817-51827. [DOI] [PubMed] [Google Scholar]
- 33.Pike, L. J. 2004. Lipid rafts: heterogeneity on the high seas. Biochem. J. 378:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pol, A., R. Luetterforst, M. Lindsay, S. Heino, E. Ikonen, and R. G. Parton. 2001. A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J. Cell Biol. 152:1057-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pol, A., S. Martin, M. A. Fernandez, C. Ferguson, A. Carozzi, R. Luetterforst, C. Enrich, and R. G. Parton. 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell 15:99-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad, R. G. M. A. 1996. Lipids of pathogenic fungi. CRC Press, Inc., Boca Raton, Fla.
- 37.Prior, I. A., and J. F. Hancock. 2001. Compartmentalization of Ras proteins. J. Cell Sci. 114:1603-1608. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues, M. L., L. R. Travassos, K. R. Miranda, A. J. Franzen, S. Rozental, W. de Souza, C. S. Alviano, and E. Barreto-Bergter. 2000. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect. Immun. 68:7049-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene CNLAC1 on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santangelo, R. T., M. H. Nouri-Sorkhabi, T. C. Sorrell, M. Cagney, S. C. Chen, P. W. Kuchel, and L. C. Wright. 1999. Biochemical and functional characterisation of secreted phospholipase activities from Cryptococcus neoformans in their naturally occurring state. J. Med. Microbiol. 48:731-740. [DOI] [PubMed] [Google Scholar]
- 41.Schnitzer, E., D. Lichtenberg, and M. M. Kozlov. 2003. Temperature-dependence of the solubilization of dipalmitoylphosphatidylcholine (DPPC) by the non-ionic surfactant Triton X-100, kinetic and structural aspects. Chem. Phys. Lipids 126:55-76. [DOI] [PubMed] [Google Scholar]
- 42.Sharom, F. J., I. Lorimer, and M. P. Lamb. 1985. Reconstitution of lymphocyte 5′-nucleotidase in lipid bilayers: behaviour and interaction with concanavalin A. Can. J. Biochem. Cell Biol. 63:1049-1057. [DOI] [PubMed] [Google Scholar]
- 43.Smotrys, J. E., and M. E. Linder. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73:559-587. [DOI] [PubMed] [Google Scholar]
- 44.Sriranganathan, N., S. M. Boyle, G. Schurig, and H. Misra. 1991. Superoxide dismutases of virulent and avirulent strains of Brucella abortus. Vet. Microbiol. 26:359-366. [DOI] [PubMed] [Google Scholar]
- 45.Sutterlin, C., T. L. Doering, F. Schimmoller, S. Schroder, and H. Riezman. 1997. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 110:2703-2714. [DOI] [PubMed] [Google Scholar]
- 46.Troy, C. M., S. A. Rabacchi, W. J. Friedman, T. F. Frappier, K. Brown, and M. L. Shelanski. 2000. Caspase-2 mediates neuronal cell death induced by beta-amyloid. J. Neurosci. 20:1386-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warnke, E., E. Barker, A. Brilman, C. Young III, and L. Cook. 2002. Inheritance of superoxide dismutase (Sod-1) in a perennial × annual ryegrass cross and its allelic distribution among cultivars. Theor. Appl. Genet. 105:1146-1150. [DOI] [PubMed] [Google Scholar]
- 48.Wedegaertner, P. B., and H. R. Bourne. 1994. Activation and depalmitoylation of Gs alpha. Cell 77:1063-1070. [DOI] [PubMed] [Google Scholar]
- 49.Wedegaertner, P. B., P. T. Wilson, and H. R. Bourne. 1995. Lipid modifications of trimeric G proteins. J. Biol. Chem. 270:503-506. [DOI] [PubMed] [Google Scholar]
- 50.Wright, L. C., J. T. Djordjevic, S. D. Schibeci, U. Himmelreich, N. Muljadi, P. Williamson, and G. W. Lynch. 2003. Detergent-resistant membrane fractions contribute to the total 1H NMR-visible lipid signal in cells. Eur. J. Biochem. 270:2091-2100. [DOI] [PubMed] [Google Scholar]
- 51.Wright, L. C., J. Payne, R. T. Santangelo, M. F. Simpanya, S. C. Chen, F. Widmer, and T. C. Sorrell. 2004. Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii. Biochem. J. 384:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xavier, R., T. Brennan, Q. Li, C. McCormack, and B. Seed. 1998. Membrane compartmentation is required for efficient T cell activation. Immunity 8:723-732. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, X., J. Gibbons, J. Garcia-Rivera, A. Casadevall, and P. R. Williamson. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69:5589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, X., and P. R. Williamson. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5:1-10. [DOI] [PubMed] [Google Scholar]