Abstract
The Rho GTPase family and their effectors are key regulators involved in many eukaryotic cell functions related to actin organization and polarity establishment. Schizosaccharomyces pombe Rho1p is essential, directly activates the (1,3)-β-d-glucan synthase, and participates in regulation of cell wall growth and morphogenesis. Here we describe the characterization of the fission yeast Rho5p GTPase, highly homologous to Rho1p, sharing 86% identity and 95% similarity. Overexpression of the hyperactive allele rho5-G15V causes a morphological effect similar to that of rho1-G15V, but the penetrance is significantly lower, and overexpression of the dominant-negative allele rho5-T20N causes lysis like that of rho1-T20N. Importantly, overexpression of rho5+ but no other rho genes is able to rescue the lethality of rho1Δ cells. Shutoff experiments indicated that Rho5p can replace Rho1p, but it is not as effective in maintaining cell wall integrity or actin organization. rho5+ expression is hardly detected during log-phase growth but is induced under nutritional starvation conditions. rho5Δ cells are viable and do not display any defects during logarithmic growth. However, when rho1+ expression is repressed during stationary phase, rho5Δ cells display reduced viability. Ascospores lacking Rho5p are less resistant to heat or lytic enzymes than wild-type spores. Moreover, h90 mutant strains carrying the hyperactive rho5-G15V or the dominant-negative rho5-T20N alleles display severe ascospore formation defects. These results suggest that Rho5p functions in a way similar to, but less efficient than, Rho1p, plays a nonessential role during stationary phase, and participates in the spore wall formation.
Morphogenesis studies are important for understanding the eukaryotic cell. Yeasts have been used as model systems because they undergo morphogenic changes that require asymmetric cell growth and actin cytoskeleton reorganization during their life cycle (30). Schizosaccharomyces pombe cells are rod shaped, grow mainly by elongation of their ends, and divide by binary fission after forming a centrally placed division septum. Under nutritional deprivation, cells enter a nondividing resting state known as the stationary phase. This entry is accompanied by a series of biological changes such as decrease in growth, increased resistance to environmental stresses, or thickening of the cell wall (14, 15).
Under nutritional deprivation, haploid cells of opposite mating types mate and form a diploid zygote, which subsequently undergoes meiosis and sporulation forming an ascus with four ascospores. These ascospores are resistant to unfavorable conditions due to their specialized cell wall.
In all eukaryotic organisms, Rho GTPases are key molecules in polarity processes (reviewed in references 12, 21, and 38). These small GTPases act as molecular switches that are turned on and off by binding to GTP or GDP, respectively. The GTP-bound form interacts with its effector molecules to perform its cellular functions. In yeast, Rho GTPases are responsible for the coordinated regulation of cell wall biosynthesis and actin cytoskeleton organization required to maintain cell integrity and polarized growth (4, 9).
The S. pombe genome contains six Rho GTPases, Cdc42p, and Rho1p-Rho5p. cdc42+ and rho1+ are essential genes involved in polarity processes. The Rho1p GTPase is required for maintenance of cell integrity and polarization of the actin cytoskeleton (2). Rho1p localizes to sites of polarized growth, the cell poles, and the septum (3). Cells lacking Rho1p lyse, but this defect is not rescued by an osmotic stabilizer. Rho1p is a regulatory component of 1,3-β-d-glucan-synthase (2). Additionally, Rho1p interacts with and stabilizes the protein kinase C homologues Pck1p and Pck2p (5). rho1+ overexpression produces cells with thick walls and some multiseptated cells. Overexpression of the dominant-negative allele rho1-T20N causes lysis. rho2+ is not an essential gene, but its overexpression is lethal. Rho2p also localizes to sites of polarized growth (16) and interacts with Pck2p, acting as a positive regulator of Mok1p, an essential 1,3-α-d-glucan-synthase (10). Therefore, Rho1p and Rho2p regulate the synthesis of the two main cell wall polymers. Rho3p interacts with the For3p formin and is involved in polarity processes (27). Additionally, Rho3p has been implicated in the modulation of the exocyst complex, involved in exocytosis (39). Rho4p is a GTPase involved in cytokinesis and cell wall integrity. rho4Δ cells display defects in cell separation at high temperature (29, 32, 33).
Through this study and an independent study by Nakano and colleagues (28), it is clear that Rho5p functions in a way similar to, but less efficient than, that of Rho1p. In this article, we characterize S. pombe Rho5p. Our results indicate that rho5+ is a nonessential gene expressed preferentially upon nitrogen starvation during stationary phase, mating, and sporulation processes. Interestingly, rho5+ overexpression can rescue the rho1Δ lethality, but these cells have a weaker cell wall than wild-type cells. Additionally, rho1Δ rho5Δ cells, repressed for ectopic Rho1p expression, display reduced viability during stationary phase. We also show here that ascospores lacking Rho5p are less resistant to different treatments, such as heat or glusulase. Moreover, h90 strains carrying the mutant rho5-G15V or rho5-T20N allele display ascospore formation defects. rho5-G15V ascospores are abnormally shaped, and the rho5-T20N ascospores have a weak wall. All these data suggest that the Rho5p GTPase is not essential and does not have a role in vegetative growth but participates in critical situations contributing to the viability of stationary-phase cells, and it contributes to the formation of the spore cell wall.
MATERIALS AND METHODS
Strains, growth conditions, and genetic methods.
Fission yeast strains used in this study are listed in Table 1. Genetic methods and growth media were as described previously (24). Yeast transformations were performed by the lithium acetate method (17). Cell lysis was visualized by staining cells with 0.01% methylene blue.
TABLE 1.
List of strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| PPG101 | h− leu1+ ura4+ | 24 |
| PPG143 | h+ leu1+ ura4+ | This work |
| PPG371 | h− leu1-32 ura4-D18 | 24 |
| PPG1704 | h− leu1-32 ura4-D18 rho5::kanMX6 | This work |
| PPG1831 | h− leu1+ ura4+ rho5::kanMX6 | This work |
| PPG1833 | h+ leu1+ ura4+ rho5::kanMX6 | This work |
| PPG1863 | h− leu1-32 ura4-D18 rho5::kanMX6 leu1::HA-rho5+ | This work |
| PPG1867 | h90 leu1-32 ura4-D18 rho5::kanMX6 leu1::HA-rho5+ | This work |
| PPG3558 | h− leu1-32 ura4-D18 rho1::ura4+ pREP41XHA-rho1+ | 3 |
| PPG3559 | h− leu1-32 ura4-D18 rho1::ura4+ pREP41XHA-rho5+ | This work |
| PPG3560 | h− leu1-32 ura4-D18 rho1::ura+ 4 rho5::kanMX6 pREP41XHA-rho1+ | This work |
| PPG3570 | h90 leu1-32 | This work |
| PPG3571 | h90 rho5+::rho5-G15V:ura4+ leu1-32 ura4-D18 | This work |
| PPG3572 | h90 rho5+::rho5-T20N:ura4+ leu1-32 ura4-D18 | This work |
For overexpression experiments, cells were grown until mid-log phase in Edinburgh minimal media (EMM) containing 15 μM thiamine. Then cells were harvested, washed three times with water, and inoculated into fresh media without thiamine at an optical density at 600 nm (OD600) of 0.01. Samples were taken at the time points described for each experiment.
For mating and sporulation assays, 107 cells of each mating type were mixed in 50 μl of sterile water and spotted on an SPA medium plate. The plate was incubated at 25°C for 2 or 3 days. For spore preparation, the mating mixtures were treated overnight at room temperature with glusulase (New England Nuclear). To check the spore resistance to heat, spores were incubated for 40 min at 55°C and plated onto rich medium. Resistance of the spores to glusulase was analyzed by incubating 107 spores with 100 μl of glusulase at room temperature, taking samples every 30 min, and staining them with methylene blue to visualize lysed cells.
Plasmids and recombinant DNA techniques.
All DNA and RNA manipulations were carried out by using established methods (6, 31). Determination of nucleotide sequences was done by automated sequencing at the IMB (CSIC/USAL) sequencing service. The nmt1 promoter-containing vector pREP3X (13) was used for overexpression of rho5+ and its different alleles.
The open reading frame of rho5+ was amplified by PCR from a cDNA library by using appropriate primers and cloned into the XhoI-BamHI sites of the pREP3X vector, creating the plasmid pREP3Xrho5+, which contains the rho5+ gene under the control of the nmt1 promoter. To generate specific alleles of rho5+, site-directed mutagenesis was performed using PCR and the appropriate primers. The entire coding region sequence was confirmed for each mutant. The rho5-G15V and rho5-T20N alleles were cloned into the pREP3X vector, as described above.
To construct the plasmid pREP41XGFPrho5+, rho5+ was obtained by PCR amplification and cloned into the NdeI-BamHI sites of the pREP41XEGFPN vector (11).
Deletion and tagging of the rho5+ gene.
A rho5::kanMX6 null mutant (PPG1704 strain) was generated using a PCR-based disruption procedure (7).
The strain with 3XHArho5+ integrated under its own promoter (PPG1863) was constructed as follows. First, 0.5 kb upstream of the initiation codon of rho5+ was PCR amplified using the appropriate primers containing the PstI (forward primer) and XhoI (backward primer) sites, respectively. The PstI-XhoI fragment corresponding to the rho5+ promoter region was cloned into pREP3X replacing the nmt1 promoter. rho5+ was cloned as a XhoI-BamHI fragment, and the epitope 3XHA was cloned as an NdeI-NdeI fragment at the NdeI site created at the rho5+ start codon. After subcloning as a HindIII-EcoRI fragment into a Bluescript KS vector, a SalI-EcoRI fragment from the created plasmid containing 3XHA-rho5+ under the rho5+ promoter was cloned into the integrative vector pJK148 (19). The resulting pJK148-3XHArho5+ plasmid was cut with NruI and integrated at the leu1 locus of the strain PPG1704, which contains a rho5Δ null allele, to generate the strain PPG1863.
For construction of the h90 rho5-G15V or h90 rho5-T20N strain, a cassette containing the rho5+ promoter followed by the rho5 allele, the ADH terminator, and the ura4+ gene as a marker was substituted for the rho5+ genomic locus.
Northern blot and reverse transcription (RT)-PCR techniques.
Total RNA was obtained isolated using the FastRNA Pro Red extraction kit (Q-Biogene). For Northern blot analysis, 4 μg of RNA was used. RNA was transferred to an Immobilon N membrane and probed with 32P-labeled DNA fragments obtained by PCR corresponding to the coding regions of rho5+, rho1+, and act1+. The blots were exposed to PhosphorImager screens and visualized with ImageQuant 5.2 on an Amersham Biosciences Typhoon 9200 scanner.
For RT-PCR analysis, the obtained RNA was treated with 1 unit/μg of RNase-free DNase (Promega) to eliminate the genomic DNA. RT-PCR was performed with the one-step kit (Invitrogen) using 200 ng of RNA and 1 μl of RT Taq enzyme in a 50 μl final volume. The actin gene act1+ was amplified using primers 5′-GTG ATGAAGCTCAAAGCAAGC-3′ and 5′-TTGAAGTTCTTGCTCAAAGTCC-3′. For rho1+ amplification, primers 5′-GTAAACAAATCTAGGGATGGCG-3′ and 5′-TGTGGTGACGGGATGCTGATTA-3′ were used. For rho5+ amplification, primers 5′-ATTCGACATTATGACTACTGAG-3′ and 5′-TCGTAAATCCAC TTTACACC-3′ were used.
Western blotting.
To detect the HA-Rho5p protein, 2 × 108 cells were harvested by centrifugation, washed once in lysis buffer (50 mM Tris-HCl, pH 7.6, 10 mM EDTA, 0.5% NP-40, or 1% Triton), resuspended in 150 μl of lysing buffer containing protease inhibitors (Sigma), and broken with glass beads in a Fast-Prep (Bio 101) using two 15-s intervals at 5.5 speed. Lysates were centrifuged for 10 min at 3,000 × g, and the supernatant protein concentration was determined by the Bio-Rad protein assay method (Bio-Rad). Total cell lysates were mixed with a 1/5 volume of sixfold-concentrated Laemmli buffer and boiled for 5 min before loading into 12.5% polyacrylamide gels containing sodium dodecyl sulfate. After electrophoresis, proteins were blotted onto Immobilon-P (Millipore) and probed with antihemagglutinin (anti-HA, 12CA5; Roche) or antitubulin (B-5-1-2; Sigma) monoclonal antibodies. The reactive bands were detected using anti-mouse horseradish peroxidase-conjugated antibodies (Bio-Rad) and the ECL detection kit (Amersham Pharmacia Biotech).
Measurement of (1,3)β-d-glucan synthase activity.
Cell extracts were prepared and (1,3)β-d-glucan synthase activity was assayed as described previously (3) with some modifications. Basically, 3 × 109 cells were harvested, washed with 1 mM EDTA, and resuspended in 300 μl of 50 mM Tris-HCl, pH 8.0, in 1 mM EDTA. Lysis was achieved in a Fast-Prep as described above. The resulting homogenates were collected by adding 30 ml of the same buffer. Cell walls were removed by centrifugation for 5 min at 1,000 × g. The supernatant was centrifuged for 30 min at 50,000 × g, and the membrane pellet was resuspended in 0.5 ml of buffer containing 50 mM Tris-HCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 30% glycerol. Protein concentration was determined by the Bio-Rad protein assay method. The amount that catalyzes the incorporation of 1 μmol of substrate (UDP-d-glucose) per minute at 30°C was considered a unit of activity.
Microscopy techniques.
For calcofluor staining, exponentially growing S. pombe cells were harvested, washed once with water, and resuspended for 5 min at room temperature in water with calcofluor at a final concentration of 50 μg/ml. After washing twice with water, cells were observed in a Leica DMRXA microscope. Green fluorescent protein (GFP)-Rho5p was observed with the same microscope using the appropriate filter. Images were captured with a Photometrics Sensys charge-coupled device camera. F-actin staining with Alexa Fluor 488-phalloidin (Molecular Probes) was basically performed as described previously (1). For 4′,6′-diamidino-2-phenylindole (DAPI) staining, cells were fixed with methanol and stained with 1 μg/ml of DAPI.
RESULTS
Rho5p is a GTPase closely related to Rho1p.
S. pombe possesses six members of the Rho family of GTPases (http://www.genedb.org/genedb/pombe); the SPAC20H4.11c-predicted open reading frame (ORF) is called rho5+. This ORF encodes a protein of 200 amino acids with significant homology to GTPases of the Rho family. Like all Rho proteins, Rho5p contains consensus amino acid sequences responsible for specific interaction with GDP and GTP and for GTPase activity. Additionally, Rho5p contains the CAAL sequence at its C terminus, which, in other Rho proteins, is posttranslationally modified by geranylgeranyl transferase type I, allowing them to be targeted to the membranes where they function (36). Using sequence comparison analysis, it is clear that Rho5p is more closely related to Rho1p than to the other Rho GTPases of the family (Fig. 1A). Interestingly, Rho5p shares 86% identity and 95% similarity with Rho1p (Fig. 1B). However, rho1+ is an essential gene, suggesting that Rho5p might function in a different way or at a different time than Rho1p.
FIG. 1.
Rho5p GTPase is similar to Rho1p. (A) S. pombe Rho GTPase tree based on similarities analyzed using the Clustal program. (B) Amino acid sequence comparison between Rho1p and Rho5p. Identical (*), conservative (:), and semiconservative (.) residues are indicated.
Overexpression of a rho5 dominant-negative allele causes cell lysis.
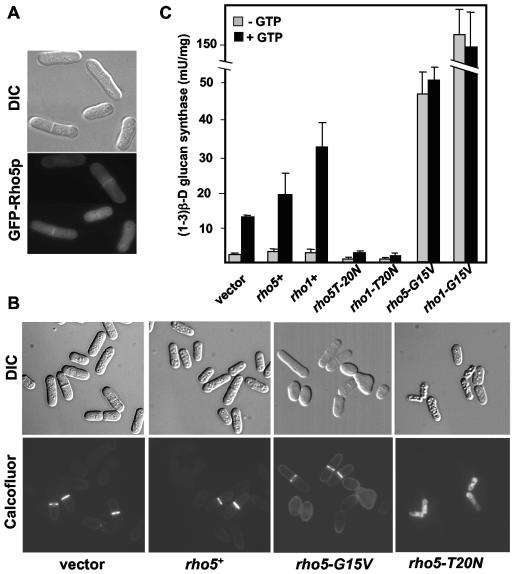
It has been shown that rho1+ overexpression in S. pombe causes a loss of polarity (2); cells become round and have a very thick wall. To study the role of Rho5p, we overproduced Rho5p and analyzed whether the effect was similar to rho1+ overexpression. First, GFP-rho5+ was cloned under the nmt41X promoter, and we checked the localization of this fusion protein expressed in logarithmic-phase cells. Cells do not display any morphological defects, and the overexpressed GFP-Rho5p was mainly localized to the septum (Fig. 2A). This suggests that, although Rho5p is not normally expressed when cells are dividing, the protein is able to localize to the septum. We could not detect any specific Rho5-GFP signal in the growth poles, as opposite to Rho1p (3, 26).
FIG. 2.
Overexpression of different rho5 alleles during logarithmic-phase growth. (A) GFP-Rho5p overproduced from pREP41X-GFPrho5+ during logarithmic phase localizes to the septum. (B) Morphological phenotype of S. pombe cells (PPG371) transformed with pREP3X, pREP3X-rho5+, pREP3X-rho5G15V, and pREP3X-rho5T20N. Upper panels, differential interference contrast (DIC) images; lower panels, calcofluor staining. (C) (1-3)β-d-glucan synthase activity from the wild type and cells overexpressing different rho5 and rho1 alleles. All extracts were prepared from strains grown at 32°C in minimal medium without thiamine for 14 h. The specific activities are the mean values of results from three independent experiments expressed as milliunits/mg of protein. Error bars represent standard deviations.
rho5+ was also cloned under the stronger nmt1 promoter into the pREP3X vector (see Materials and Methods). After 20 h of induction, overexpression of rho5+ caused no morphological effect (Fig. 2B), as opposite to rho1+ (3, 26). It is known that Rho proteins are tightly regulated; they are active when bound to GTP and inactive when bound to GDP. To further analyze Rho5p function, we generated two site-specific mutations described in other GTPases (8). The G15V mutation should generate a Rho5p permanently bound to GTP (constitutively active Rho5p), whereas the T20N mutation should generate a Rho5p permanently bound to GDP (constitutively inactive Rho5p). Some of the cells overexpressing rho5-G15V from the nmt1 promoter showed identical phenotypes to those of cells with high levels of Rho1G15Vp; they were lemon shaped with very thick walls, suggesting that Rho5p might function in a similar way to Rho1p, but with less activity (Fig. 2B). Interestingly, overexpression of the rho5-T20N dominant-negative allele caused cell lysis mainly in separating cells (Fig. 2B). The level of lysis determined in three independent experiments after 18 h of rho5-T20N overexpression (47.9 ± 0.3%) was similar to that caused by rho1-T20N (49.5 ± 0.3%), corroborating the idea that Rho5p and Rho1p might function similarly.
To further compare Rho5p and Rho1p function, we analyzed the (1,3)β-d-glucan synthase activity in all the strains overexpressing rho5 and rho1 alleles (Fig. 2 C). The (1,3)β-d-glucan synthase levels of cells overexpressing rho5+ were higher than those of wild-type cells, but they were always around 50% lower than those from cells overexpressing rho1+. On the other hand, the overexpression of the rho5-T20N dominant-negative allele caused a (1,3)β-d-glucan synthase decrease similar to that observed in the rho1-T20N overexpression. This result could explain the similar lysis phenotype caused by the rho5-T20N and rho1-T20N alleles. Additionally, cells overexpressing rho5-G15V had more (1,3)β-d-glucan synthase activity than wild-type cells, and this activity is GTP independent, similar to that of cells overexpressing rho1-G15V (Fig. 2C) (2). Together, these results suggest that Rho5p, like Rho1p, could function as a direct activator of the (1,3)β-d-glucan synthase. The main difference between both GTPases is that Rho1p is more efficient.
Overexpression of rho5+ rescues rho1Δ lethality.
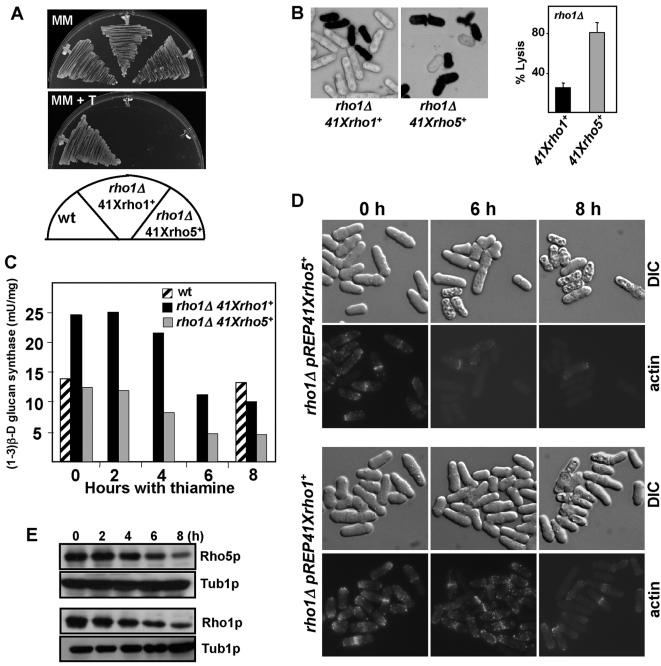
To test that Rho5p could functionally substitute for Rho1p, we analyzed whether overexpression of rho5+ could bypass the essential requirement of Rho1p. To do that, a heterozygous rho1+/rho1::ura4+ diploid was transformed with plasmid pREP41XHArho5+ or pREP41XHArho1+ and sporulated. rho1::ura4+ haploid segregants containing either plasmid grew in medium without thiamine and exhibited wild-type morphology (Fig. 3A and D). As expected, repression of rho5+ expression by adding thiamine resulted in lethality, as happens after repression of rho1+ (Fig. 3A). These results clearly demonstrated that Rho5p overproduction can rescue the lethality caused by rho1+ deletion. Importantly, this effect is specific for rho5+ because overexpression of the other GTPases (rho2+, rho3+, or rho4+) cannot rescue the lethality of rho1Δ cells (data not shown).
FIG. 3.
Overexpression of rho5+ can rescue rho1Δ lethality. (A) Wild-type (wt) and rho1::ura4+ cells transformed with either pREP41XHA-rho5+ (PPG3559) or pREP41XHA-rho1+ (PPG3558) were grown on plates with minimal medium without thiamine (MM) or with thiamine (MM + T). (B) The same cells were grown without thiamine and switched to medium with thiamine for 8 h. Lysis was assessed by staining with methylene blue. Quantification is shown in the right panel. (C) (1-3)β-d-glucan synthase activity of the cells described above. All extracts were prepared from cells grown at 32°C in minimal medium without thiamine and switched to medium with thiamine for the indicated times. The specific activities, expressed as milliunits/mg of protein, are the mean values of results from three independent experiments. Wild-type activity is shown at 0 and 8 h because it does not change throughout the experiment. (D) Actin staining of the PPG3559 or PPG3558 cells grown with thiamine. At the indicated times after switching to medium with thiamine, cells were fixed and stained with Alexa Fluor 488-phalloidin. (E) HA-Rho5p or HA-Rho1p levels in the same cells after switching to medium with thiamine during the indicated times to repress rho5+ or rho1+ expression. Cell extracts were immunoanalyzed using anti-HA or antitubulin monoclonal antibodies. DIC, differential interference contrast.
As described before (3), rho1+ repression caused lysis at any time during the cycle. Repression of rho5+ in the rho1Δ strain also resulted in cell lysis, but this lysis occurred mainly during cytokinesis (Fig. 3B). Quantification of cell lysis by methylene blue staining showed that 80% (n = 420) of rho1Δ pREP41XHArho5+ cells were lysed after 8 h of repression, whereas only 25% (n = 510) of rho1Δ pREP41XHArho1+ cells were lysed (Fig. 3B). At 12 h, most cells were lysed in both strains (data not shown).
Comparing 1,3-β-d-glucan synthase activity of rho1Δ cells that had been maintained alive with either pREP41XHArho1+ or pREP41XHArho5+, in vitro measurements showed a higher level of activity in rho1Δ cells overexpressing rho1+ (Fig. 3C) (3). Additionally, during the first 6 h after adding thiamine, there was a 50% reduction in the enzyme activity of the rho1Δ cells overexpressing rho5+ compared with wild-type cells, whereas the cells overexpressing rho1+ still had a high level of activity (Fig. 3C). These results suggest that Rho5p is not as effective as Rho1p in activating the β-glucan synthase, and this could cause the earlier lysis of the rho1Δ cells overexpressing rho5+ during the repression of the genes.
It has been described that, during rho1+ shutoff, the actin cytoskeleton disappears after 8 h of repression and before cell lysis occurs (Fig. 3D) (3). When rho5+ is overexpressed in rho1Δ cells, actin is perfectly localized (Fig. 3D, 0 h), but after 6 h of repression, when most cells have not yet undergone lysis, the actin patches had largely disappeared (Fig. 3D). Western blot analysis showed that both Rho1p and Rho5p proteins disappear with the same dynamics in the shutoff experiments (Fig. 3E). Therefore, Rho5p can also perform the function of Rho1p in maintaining actin polymerization, although less efficiently.
Taken together, all these results indicate that Rho5p is capable of acting on the same targets as Rho1p but is less effective; thus, an excess of Rho5p allows the survival of the cells in the absence of Rho1p.
Rho5p is expressed during stationary phase.
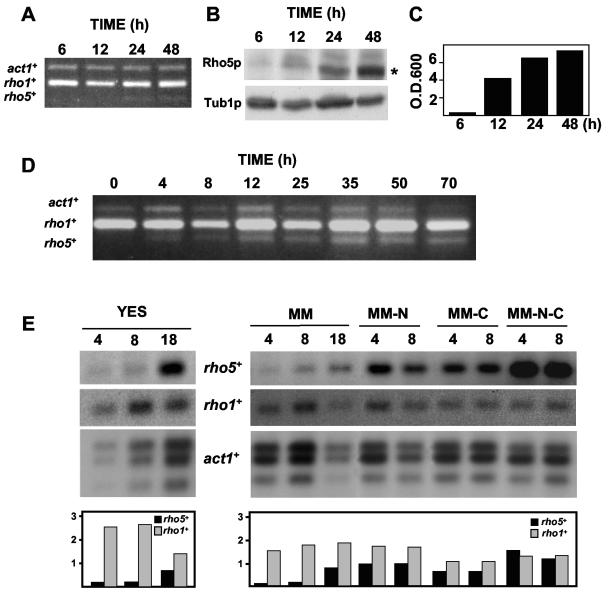
To investigate the function of rho5+, the gene was deleted. However, rho5Δ cells were viable at all temperatures and did not show any morphological defects (data not shown). Being so similar to Rho1p, one possibility was that rho5+ would be a pseudogene. To know if the gene rho5+ was expressed, we analyzed the amount of rho5+ mRNAs during vegetative growth using the RT-PCR technique. As shown in Fig. 4A, rho5+ is expressed at very low levels when cells are actively growing (6 and 12 h) but is clearly induced when the cells are entering stationary phase (24 and 48 h). On the other hand, rho1+ is constantly expressed, like act1+ used as a control (Fig. 4A). These results indicate that rho5+ is a gene preferentially expressed in stationary phase.
FIG. 4.
Analysis of rho5+ expression. (A) RT-PCR of rho5+, rho1+, and act1+ from cells grown in rich medium (YES) at 32°C for different periods of time. (B) Western blot analysis of HA-Rho5p levels in the same cultures used in panel A. Total cell extracts from PPG1863 cells were prepared and immunoblotted with anti-HA antibody. The antitubulin blot was performed as a loading control. Rho5p is marked with an asterisk. (C) OD600 of cultures used in panels A and B. (D) RT-PCR of rho5+, rho1+, and act1+ from h90 HA-rho5+ cells (strain PPG1867) incubated in SPA medium for different periods of time. (E) rho5+ expression is increased in starving cells. Samples from wild-type cultures (PPG101) in different media were taken at the indicated time points (hours) for RNA extraction. RNA was hybridized with specific probes for rho5+, rho1+, and act1+. The media used were rich medium (YES), minimal medium (MM), minimal medium without nitrogen (MM-N), minimal medium with 0.1% glucose (MM-C), and minimal medium without nitrogen and with 0.1% glucose (MM-N-C). Lower panels show the amount of rho5+ and rho1+ mRNAs relative to act1+ mRNA.
To further corroborate this data, we also checked Rho5p levels during vegetative growth by Western blot analysis using the HA-rho5+ strain (PPG1863). HA-Rho5p is clearly produced at the end of the logarithmic phase (postdiauxic phase), when nutrients in the culture are getting low and the cells divide very slowly, and it is retained during stationary phase (Fig. 4B and C).
rho5+ is expressed during the sexual cycle.
Since Rho5p does not seem to have a relevant role during vegetative growth, we considered the possibility that Rho5p could be functioning during mating or sporulation, two processes in which cell wall remodeling and actin organization are required.
To check this hypothesis, we first analyzed the levels of rho5+ mRNAs and of the Rho5 protein in an h90 HA-rho5+ strain during the sexual cycle. Conjugation and sporulation were induced in SPA medium at 25°C. In these conditions, zygotes began to appear at 9 to 12 h and asci at 22 to 24 h. rho5+ expression is clearly induced at 4 h and the expression levels are maintained even after 70 h (Fig. 4D). rho1+ expression is constant during the whole process. In previous studies of the meiotic transcription program using DNA microarrays, rho5+ was classified as a middle meiotic gene (23). However, using RT-PCR, we observed that rho5+ expression was induced in early stationary phase and during mating (Fig. 4A and D), suggesting that rho5+ might be induced by nutrient starvation. In fact, the protein was clearly detected after 4 h in SPA medium, indicating that rho5+ responds to nutrient starvation (data not shown).
To further corroborate the RT-PCR data, we used Northern blotting to analyze rho5+ expression in different nutrient conditions: rich medium (YES), minimal medium (MM), minimal medium without nitrogen (MM-N), minimal medium with 0.1% glucose, and minimal medium without nitrogen with 0.1% glucose. The cells were grown in rich medium to an OD600 of 0.6, washed, and resuspended in the different media at the same OD. rho5+, rho1+, and act1+ expression were analyzed at different times. There was a sevenfold increase of rho5+ RNA levels in poor media, whereas rho1+ RNA did not vary significantly (Fig. 4E). In YES or MM, the maximum increase was observed at 18 h, whereas in MM-N, the increase was clear at 4 h. That could explain the rho5+ RNA induction during mating, since the medium used (SPA) is limited for nitrogen.
Rho5p plays a role in stationary-phase cell integrity.
Since rho5+ expression is induced during stationary phase, we analyzed if Rho5p, by analogy with the Rho1p, could be required in the synthesis of the cell wall during the stationary phase. The viability of wild-type and rho5Δ cells after 4 days of growth in liquid culture was checked, and no differences were observed (data not shown). Similarly, there were no differences in the survival of wild-type and rho5Δ stationary cells to treatments with zymolyase or novozyme, suggesting that no drastic cell wall changes occur in cells lacking rho5+ (data not shown). Additionally, rho5Δ cells have no defect in reentry into the cell cycle after maintenance during 4 days in liquid culture (data not shown).
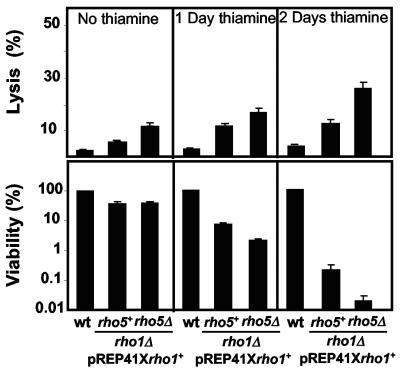
In the experiments described above, we did not find any phenotype in rho5Δ stationary-phase cells. If Rho5p is redundant with Rho1p, the role of Rho5p would be difficult to examine when Rho1p is present in the cells. Thus, we analyzed the effect of the lack of Rho5p on the viability of stationary-phase cells in the absence of Rho1p. Wild-type, and rho1Δ or rho1Δ rho5Δ cells transformed with pREP41Xrho1+ were grown during 2 days in medium without thiamine; then thiamine was added to the medium to repress the expression of rho1+. Cells were cultured for 1 or 2 days more, and the lysis and the viability were checked. The percentage of lysed cells during stationary phase was higher in rho1Δ than in wild-type cells, and there was always an increase in the rho1Δ rho5Δ double mutant (Fig. 5, upper panels). Furthermore, the viability of rho1Δ cells was very low compared with the wild type, but there is a 10-fold further reduction in the viability of the rho1Δ rho5Δ cells (Fig. 5, lower panels). These results suggest that Rho5p plays a role, additional to that of Rho1p, in the maintenance of cell viability during stationary phase.
FIG. 5.
Rho5p is important in stationary phase. rho1Δ or rho1Δ rho5Δ cells, maintained alive with pREP41X-rho1+, were grown in minimal medium without thiamine to stationary phase (2 days), and then thiamine was added for 1 or 2 additional days. Corresponding wild-type (wt) cultures served as controls. Aliquots of each culture were removed at the indicated times, and cells were counted in the presence of methylene blue to evaluate lysis (n = 200; top panels) and plated on minimal medium without thiamine. The percentage of viable cells in each culture was determined (lower panels). Values are the means of results from three independent experiments. Error bars represent standard deviations.
Rho5p plays a role in spore wall integrity.
Since rho5+ expression is induced and maintained during the sexual cycle, we investigated whether rho5Δ cells display defects in the mating and/or meiotic processes. rho5Δ zygotes do not display any morphological defects, and they are able to complete meiosis and form asci with four ascospores (data not shown). To study whether Rho5p could be involved in the process of cell wall synthesis required for the formation and maturation of the ascospores, the integrity of wild-type and rho5Δ spores was analyzed in two ways: first, incubating the spores at 55°C for up to 40 min and evaluating the viability (Fig. 6A); second, by treating the spores with high concentrations of glusulase and counting those that lysed (Fig. 6B). In both cases, rho5Δ spores were more sensitive to these treatments than wild-type spores, suggesting that the cell wall integrity in rho5Δ spores is mildly compromised.
FIG. 6.
Rho5p participates in spore integrity. (A) Wild-type (PPG101 × PPG143) and rho5Δ (PPG1831 × PPG1833) spores were incubated at 55°C for the indicated times, and viability was assessed by counting colony formation after plating on rich medium. (B) Wild-type and rho5Δ spores were incubated with glusulase for the indicated times, and lysis was evaluated by counting methylene blue-stained spores.
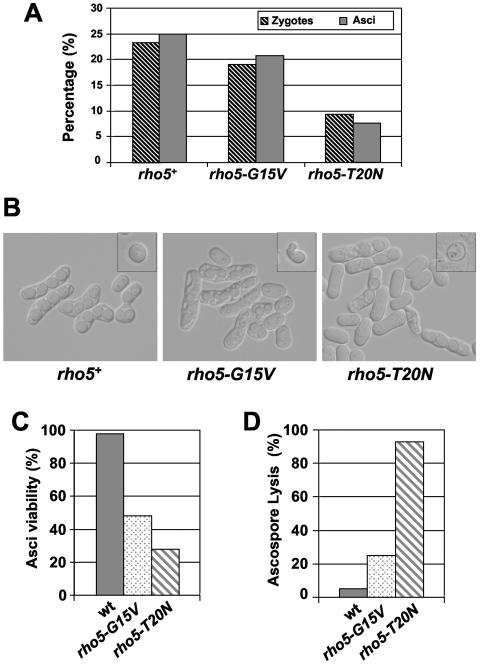
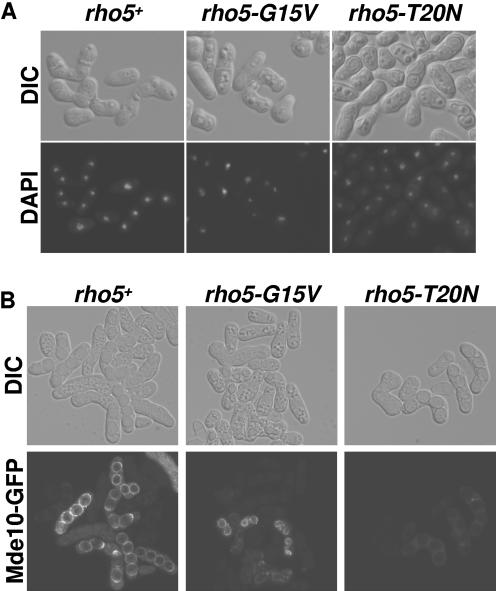
To study further the possible role of Rho5p in the spore formation process, we decided to construct two h90 strains in which the genomic copy of the rho5+ gene was replaced by the rho5-G15V or rho5-T20N allele. These strains did not show any defects during vegetative growth. They were also able to sporulate, but the efficiency was considerably reduced compared to the wild-type h90 strain, mainly in the h90 rho5-T20N strain (Fig. 7A). Microscopic analysis indicated that the rho5-G15V ascospores were always four in number, but some of them displayed elongated or aberrant morphologies. Additionally, the rho5-T20N ascospores have normal shape, but they were much less refringent than wild-type spores (Fig. 7B). Moreover, we analyzed the viability of complete asci, and it is clear that there is reduced viability in both rho5-G15V and rho5-T20N strains compared to the wild-type strain (Fig. 7C). To study the sporulation defect in more detail, we monitored the nuclear divisions in the mutant strains. The cells were fixed and stained with the DNA-specific dye DAPI. Both mutants progressed into meiosis I and II and completed meiotic divisions similar to the wild-type strain (Fig. 8A). We could not find spores without or with more than one nucleus. One possibility was that Rho5p could be involved in spore wall construction. To test this hypothesis, we used the protein Mde10-GFP as a fluorescent marker to visualize the spore walls under the microscope. Mde10p is an ADAM family metalloprotease that localizes to the spore walls (25). In the wild-type strain, the Mde10-GFP protein was uniformly distributed in the four spore walls and displayed four rings. However, in the rho5-G15V strain, the spore walls contained Mde10-GFP, but it was distributed irregularly and the Mde10-GFP signals were visualized as deformed rings. In the rho5-T20N spores, Mde10-GFP is localized to the walls, but the signal is much weaker than the wild-type spores, suggesting that these cells could contain a defective spore wall (Fig. 8B). Ascospores with mature functional spore walls are resistant to glusulase treatment. To asses whether the mutant spores have functional spore walls, we examined their sensitivity to glusulase. The rho5-G15V ascospores were slightly sensitive to glusulase, but the treatment of the rho5-T20N ascospores with glusulase produced the lysis of almost all the spores (Fig. 7D). All of these data indicated that Rho5p has a role in spore wall formation or maturation.
FIG. 7.
Changes in active Rho5p levels affect the sporulation process. (A) Percentages of zygotes (striped bars) and asci (solid bars) formed in h90 wild-type (PPG3570), rho5-G15V (PPG3571), and rho5-T20N (PPG3572) strains, grown for 36 h on malt extract plates at 28°C. (B) Differential interference contrast images of the asci formed in the strains detailed above using similar conditions. In the upper right corner, a representative ascospore of each strain after treatment with glusulase is shown. (C) Several asci of the strains detailed above were separated, and growth in rich medium was analyzed. Viability of the asci is shown as a percentage. (D) Sporulated cultures were treated with glusulase, and the lysed ascospores were quantified by microscopic observation. The percentage of ascospore lysis is shown. wt, wild type.
FIG. 8.
Changes in active Rho5p levels affect spore wall formation. (A) h90 wild-type (PPG3570), rho5-G15V (PPG3571), and rho5-T20N (PPG3572) strains in sporulation conditions stained with DAPI (lower panels). (B) The same strains detailed above expressing Mde10-GFP and observed by fluorescence microscopy (lower panels). In both figures the corresponding differential interference contrast (DIC) images are presented in the upper panels.
DISCUSSION
The Rho family of GTPases is present in all eukaryotic cells, acting as regulators in signaling pathways that control actin organization and morphogenetic processes. The fission yeast S. pombe possesses six Rho GTPases, Rho1p to Rho5p and Cdc42p. Rho1p is an essential protein required for maintenance of cell integrity and polarization of the actin cytoskeleton. Rho1p acts as regulatory component of the 1,3-β-d-glucan synthase and stabilizes Pck1p and Pck2p.
In this report, we characterized the S. pombe Rho GTPase named Rho5p (SPAC20H14.11C). It has been reported that budding yeast Rho5p is a negative regulator of the cell integrity pathway (34). In S. pombe, Rho5p is very similar to Rho1p, suggesting a possible role of Rho5p in cell wall biosynthesis and morphogenesis, as it is also the case for Rho2p which regulates the biosynthesis of the α-d-glucan (10). However, rho5Δ cells have no morphological or growth defects, and their cell walls are not affected (data not shown).
Overexpression of rho1+ produces round and multiseptated cells with thick walls (2). Nakano and colleagues (28) have recently published that rho5+ overexpression caused depolarization of F-actin and aberrant cell wall formation. However, in our study, overproduction of Rho5p has no growth or morphological effects. Additionally, overexpression of rho1-G15V is lethal and the cell walls are extremely thick (2), whereas overexpression of Rho5-G15Vp only caused morphological alteration and cell wall thickening in a small percentage of the cells. This suggests that Rho5p is not as efficient as Rho1p in activating the glucan synthesis. Interestingly, overexpression of rho5-T20N caused levels of cell lysis overexpression similar to those of rho1-T20N. It is likely that Rho5-T20Np could be sequestering proteins that positively regulate Rho1p, such as those with GEF (GTPase exchange factor) activity. In fact, it has been described that Rgf3p, a Rho1 GEF expressed during cytokinesis, interacts with both Rho1T20Np and Rho5T20Np, and deletion of rgf3+ is lethal due to cell lysis (35). When the fusion protein GFP-Rho5p was overproduced during logarithmic phase, it was primarily localized to the septum. The fact that lysis caused by overproduction of Rho5T20Np mainly occurs during septation might be due to the preferential localization of Rho5p to the septum. In the recent study by Nakano and colleagues (28), Rho5p is shown to also localize to the cell poles. However, in both studies (ours and that of Nakano et al. [28]), the localization experiments in vegetative growth have been performed using rho5+ gene expressed under an exogenous promoter, repressible by thiamine (nmt1) in our studies or constitutive (ADH) in the experiments of Nakano et al. We have found that Rho5p is not present in the logarithmic phase; therefore, these localizations might not be significant.
We have demonstrated that overexpressed rho5+ is able to rescue the rho1Δ lethality. This effect is specific for rho5+, overexpression of other GTPases has no effect. This result implies that Rho5p could perform, at least partially, the essential functions of Rho1p in cell wall synthesis, actin cytoskeleton dynamics, cell cycle progression, or secretion. However, several lines of evidence suggest that rho5+ is not as active as rho1+ in some of these functions. First, glucan synthase levels of rho1Δ cells mildly overexpressing rho5+ are reduced compared to rho1Δ cells mildly overexpressing rho1+. Second, during the rho5+ shutoff experiments, rho1Δ cells die earlier, and actin patches disappear more rapidly than during the rho1+ shutoff. All of these results indicate that cell integrity is compromised when rho1+ is replaced by rho5+. We also tried to replace directly the rho1+ ORF for that of rho5+ to express rho5+ at the same level as rho1+. However, those cells were not viable, suggesting that Rho5p can only replace Rho1p function when Rho5p is overexpressed (data not shown). Based on overexpression studies, Nakano and colleagues (28) have described that Rho5p controls cell shape and septation in fission yeast. However, we have found that Rho5p is not present in logarithmic phase and is preferentially expressed upon nutrient starvation and during sexual differentiation, when septation does not occur. Therefore, Rho5p must function as Rho1p but in different processes. Curiously, rho5+ levels are always reduced compared with the rho1+ levels. However, in the conditions when rho5+ is expressed, the levels of rho1+ are decreased compared to the rho1+ expression levels in active growing cells, and the transcription levels of both GTPase RNA are similar. So, one possibility is that Rho5p is required to counterbalance this decrease of Rho1p in stress conditions.
Using DNA microarray studies, it was also found that rho5+ was induced during sexual differentiation, belonging to the class of “middle” meiotic genes (23). According to our results, rho5+ expression is induced early during mating and could correspond to the starvation-induced genes that are up-regulated upon nitrogen removal; after that, the expression is continuous during the process. However, we did not use the pat1 mutant cells to get a synchronous meiosis, as Mata et al. did, and therefore it might be possible that the continuous expression of rho5+ reached a peak similar to other “middle” meiotic genes. Indeed, Rho5p might participate in the spore cell wall formation because rho5Δ spores are more sensitive to heat and lytic enzymes than wild-type spores and rho5-G15V ascospores are aberrant in shape. This function is consistent with a “middle” or “late” meiotic expression (23). It is known that Bgs2p, a 1,3-β-d-glucan synthase subunit, is required for the spore cell wall synthesis (20, 22); perhaps, Rho5p is a regulator of Bgs2p, redundant with Rho1p. In summary, we have studied the function of Rho5p in S. pombe and have found that it is preferentially produced during stationary phase and sexual development process. During stationary phase, S. pombe cells are more rounded, losing their typical cylindrical shape, and undergoing several changes including increased resistance to environmental stresses, thickening of the cell wall, and increased ability to survive starvation. We have discarded the hypothesis that Rho5p functions exclusively during stationary phase and Rho1p exclusively during active growth because rho5Δ stationary cells are viable and have no defects in reentering the cell cycle. Moreover, rho1+ is also expressed during stationary phase. This possible redundant role of Rho5p in stationary phase might not have been uncovered in rho5Δ cells because Rho1p was present. Indeed, experiments where rho1+ was repressed during stationary phase have shown that the strain without Rho5p is significantly less viable than the one containing Rho5p. This role might be essential in natural habitats with more stringent life conditions than a laboratory.
Additionally, we have demonstrated that changes in active Rho5p levels in the cells cause defects in the sporulation process. The strain containing the constitutively active Rho5p produced aberrant ascospores that are irregularly stained with a spore wall marker. It has been described that the deletion of other genes such as meu10+, omt1+, or omt2+ caused similar phenotypes due to an aberrant thickness of the spore wall (18, 37). Oppositely, the mutant strain containing the constitutively inactive Rho5p produced ascospores with weak walls that lyse easily in the presence of glusulase. These phenotypes are in agreement with a role of Rho5p in spore wall integrity. The use of rho5+ dominant alleles allowed us to see the function of Rho5p that was not obvious in rho5Δ cells, probably due to the redundancy with Rho1p function and regulation. Based in these results, we conclude that Rho5p reinforce the Rho1p GTPase activity during spore wall formation, and the stationary phase changes.
Acknowledgments
J. C. Ribas, H. Valdivieso, and P. San Segundo provided critical comments on the manuscript. We thank D. Posner for English reviewing. We thank Elvira Portales for technical help. Y. Hiraoka provided plasmid A799 containing Mde10-GFP.
S.A.R. was supported by a fellowship from the Spanish Ministerio de Educación. This work was supported by grants BIO-2001-1531 and BIO-2004-00384 from the Comision Interministerial de Ciencia y Tecnología, Spain.
REFERENCES
- 1.Alfa, C. P., J. Fantes, M. Hyams, E. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Arellano, M., A. Durán, and P. Pérez. 1996. Rho1 GTPase activates the (1-3)β-d-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 15:4584-4591. [PMC free article] [PubMed] [Google Scholar]
- 3.Arellano, M., A. Durán, and P. Pérez. 1997. Localization of the Schizosaccharomyces pombe rho1 GTPase and its involvement in the organization of the actin cytoskeleton. J. Cell Sci. 110:2547-2555. [DOI] [PubMed] [Google Scholar]
- 4.Arellano, M., P. M. Coll, and P. Pérez. 1999. Rho GTPases in the control of cell morphology, cell polarity, and actin localization in fission yeast. Microsc. Res. Tech. 47:51-60. [DOI] [PubMed] [Google Scholar]
- 5.Arellano, M., M. H. Valdivieso, T. M. Calonge, P. M. Coll, A. Durán, and P. Pérez. 1999. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J. Cell Sci. 112:3569-3578. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 7.Bähler, J., J.-Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 8.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 9.Cabib, E., J. Drgonova, and T. Drgon. 1998. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67:307-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calonge, T. M., K. Nakano, M. Arellano, R. Arai, S. Katayama, T. Toda, I. Mabuchi, and P. Perez. 2000. Schizosaccharomyces pombe Rho2 GTPase regulates the cell wall α-glucan biosynthesis, through the protein kinase Pck2p. Mol. Biol. Cell 11:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 1:59-68. [DOI] [PubMed] [Google Scholar]
- 12.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 13.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman, P. K. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5:602-607. [DOI] [PubMed] [Google Scholar]
- 16.Hirata, D., K. Nakano, M. Fukui, H. Takenaka, T. Miyakawa, and I. Mabuchi. 1998. Genes that cause aberrant cell morphology by overexpression in fission yeast: a role for a small GTP-binding protein Rho2 in cell morphogenesis. J. Cell Sci. 111:149-159. [DOI] [PubMed] [Google Scholar]
- 17.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakihara, Y., K. Nabeshima, A. Hirata, and H. Nojima. 2003. Overlapping omt1+ and omt2+ genes are required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 8:547-558. [DOI] [PubMed] [Google Scholar]
- 19.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, J., X. Tang, H. Wang, and M. Balasubramanian. 2000. Bgs2p, a 1,3-b-glucan synthase subunit, is essential for maturation of ascospore wall in Schizosaccharomyces pombe. FEBS Lett. 478:105-108. [DOI] [PubMed] [Google Scholar]
- 21.Mackay, D. J., and A. Hall. 1998. Rho GTPases. J. Biol. Chem. 273:20685-20687. [DOI] [PubMed] [Google Scholar]
- 22.Martin, V., J. C. Ribas, E. Carnero, A. Durán, and Y. Sánchez. 2000. bgs2+, a sporulation-specific glucan synthase homologue is required for proper ascospore wall maturation in fission yeast. Mol. Microbiol. 38:308-321. [DOI] [PubMed] [Google Scholar]
- 23.Mata, J., R. Lyne, G. Burns, and J. Bahler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 24.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, T., H. Abe, A. Hirata, and C. Shimoda. 2004. ADAM family protein Mde10 is essential for development of spore envelopes in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano, K., R. Arai, and I. Mabuchi. 1997. The small GTP-binding protein Rho1 is a multifunctional protein that regulates actin localization, cell polarity, and septum formation in the fission yeast Schizosaccharomyces pombe. Genes Cells 2:679-694. [DOI] [PubMed] [Google Scholar]
- 27.Nakano, K., J. Imai, R. Arai, E. A. Toh, Y. Matsui, and I. Mabuchi. 2002. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 115:4629-4639. [DOI] [PubMed] [Google Scholar]
- 28.Nakano, K., R. Arai, and I. Mabuchi. 2005. Small GTPase Rho5 is a functional homologue of Rho1, which controls cell shape and septation in fission yeast. FEBS Lett. 579:5181-5186. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, K., T. Mutoh, R. Arai, and I. Mabuchi. 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells 8:357-370. [DOI] [PubMed] [Google Scholar]
- 30.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, N.Y.
- 32.Santos, B., J. Gutiérrez, T. M. Calonge, and P. Pérez. 2003. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos, B., A. B. Martin-Cuadrado, C. R. Vazquez de Aldana, F. del Rey, and P. Perez. 2005. Rho4 GTPase is involved in the secretion of glucanases during fission yeast cytokinesis. Eukaryot. Cell 4:1639-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz, H. P., S. Huppert, A. Lorberg, and J. J. Heinisch. 2002. Rho5p downregulates the yeast cell integrity pathway. J. Cell Sci. 115:3139-3148. [DOI] [PubMed] [Google Scholar]
- 35.Tajadura, V., B. García, I. García, P. García, and Y. Sánchez. 2004. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J. Cell Sci. 117:6163-6174. [DOI] [PubMed] [Google Scholar]
- 36.Takai, Y., T. Sasaki, Tanaka, and T. Matozaki. 2001. Small GTP-binding proteins. Phys. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 37.Tougan, T., Y. Chiba, Y. Kakihara, A. Hirata, and H. Nojima. 2002. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 7:217-231. [DOI] [PubMed] [Google Scholar]
- 38.Van-Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signalling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 39.Wang, H., X. Tang, and M. K. Balasubramanian. 2003. Rho3p regulates cell separation by modulating exocyst function in Schizosaccharomyces pombe. Genetics 164:1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]