Abstract
Recently, the academic interest in the yeast Torulaspora delbrueckii has increased notably due to its high resistance to several types of stress, including salt and osmotic imbalance. However, the molecular mechanisms underlying these unusual properties are poorly understood. In Saccharomyces cerevisiae, the high-salt response is mediated by calcineurin, a conserved Ca2+/calmodulin-modulated protein phosphatase that regulates the transcriptional factor Crz1p. Here, we cloned the T. delbrueckii TdCRZ1 gene, which encodes a putative zinc finger transcription factor homologue to Crz1p. Consistent with this, overexpression of TdCRZ1 enhanced the salt tolerance of S. cerevisiae wild-type cells and suppressed the sensitivity phenotype of cnb1Δ and crz1Δ mutants to monovalent and divalent cations. However, T. delbrueckii cells lacking TdCrz1p showed phenotypes distinct from those previously observed in S. cerevisiae crz1Δ mutants. Quite remarkably, Tdcrz1-null cells were insensitive to high Na+ and were more Li+ tolerant than wild-type cells. Clearly, TdCrz1p was not required for the salt-induced transcriptional activation of the TdENA1 gene, encoding a putative P-type ATPase homologue to the main S. cerevisiae Na+ pump ENA1. Furthermore, T. delbrueckii cells were insensitive to the immunosuppressive agents FK506 and cyclosporine A, both in the presence and in the absence of NaCl. Signaling through the calcineurin/Crz1 pathway appeared to be essential only on high-Ca2+/Mn2+ media. Hence, T. delbrueckii and S. cerevisiae differ in the regulatory circuits and mechanisms that drive the adaptive response to salt stress.
Exposure of cells to saline stress implies both a specific cation toxicity and osmotic stress. Sodium and lithium ions are particularly toxic to the cells of most living organisms because of their ability to inhibit specific metabolic pathways. Therefore, regulation of intracellular ion content is a primary issue in the cellular reprogramming of almost all organisms subjected to salt stress (4, 58).
The high degree of evolutionary conservation of stress pathways between higher eukaryotes and Saccharomyces cerevisiae and the genetic advantages of budding yeast have made this organism a model system for studying stress responses (24). Yeast genes involved in salt tolerance have been identified by the ability to protect cells at increasing gene dosage or by the growth defects of yeast mutants at elevated ion concentrations (53). Thus, studies with yeast have covered basic mechanisms of ion homeostasis and have identified key genes in the maintenance of a high K+/Na+ ratio (48, 54). Protein kinases and signaling pathways involved in salt responses have been also identified and characterized (52). Despite these advances, we are far from completely understanding the mechanisms, the nature of signaling pathways, and the functions of gene targets that allow cells to adapt to salt stress. Moreover, there is evidence that signaling pathways and stress responses have evolved in different organisms, including yeasts, in a niche-dependent manner (8, 59). It is clear, for example, that S. cerevisiae is not the best model of a salt-resistant microorganism. Nonconventional yeasts such as Zygosaccharomyces rouxii, Debaryomyces hansenii, and Torulaspora delbrueckii are by far more resistant to the combined effects of ion toxicity and osmotic stress (11, 32). Thus, the identification and characterization of the cellular mechanisms regulating salt tolerance in these non-Saccharomyces species are of major interest.
In S. cerevisiae, toxic concentrations of Na+ and Li+ promote their extrusion by induction and activation of the specific ATP-driven ion pump Ena1p (40). ENA1 expression is regulated by two different signaling pathways, the HOG (for “high osmolarity glycerol”) pathway (64), one of the five known mitogen-activated protein kinase cascades in S. cerevisiae (16), and the calcineurin/Crz1p pathway (19). Calcineurin is a highly conserved Ca2+/calmodulin-dependent Ser/Thr protein phosphatase of type 2B (30, 49). In its native form, calcineurin is present as a heterodimer containing a catalytic subunit, encoded by the functionally redundant genes CNA1 and CNA2, complexed with a regulatory subunit, the product of CNB1. The phosphatase activity of calcineurin is dispensable for growth under standard conditions. However, cna1 cna2 or cnb1 mutants show decreased tolerance to Na+/Li+, Mn2+, and OH− ions (41, 43, 44). Calcineurin is also required for escape from cell cycle arrest after exposure to pheromone (20, 21) and plays an important role in regulating cell wall structure (22, 25).
When cells are exposed to salt stress, cytosolic Ca2+ levels rise, inducing its binding to calmodulin. This interaction promotes a conformational change in calmodulin, allowing it to bind and activate calcineurin (39), which in turn dephosphorylates the transcriptional factor Crz1p (41, 61). Dephosphorylation of Crz1p causes its nuclear import (62) and binding to a consensus DNA sequence (42), the calcineurin-dependent response element (CDRE) (61), found in the promoter of most salt-responsive genes (67). In consonance with this, cells lacking Crz1p display hypersensitivity to α-factor, Mn2+, or Li+ (41, 61). Nevertheless, crz1 and calcineurin mutant cells show opposite phenotypes under specific conditions, such as exposure to Ca2+ and OH− ions (41, 42). This observation strongly suggests that calcineurin regulates additional yeast proteins (19). It is also possible that Crz1p might respond to signals other than those driven by calmodulin-calcineurin. Whether this signaling pathway plays a similar role in other yeasts, particularly in highly osmotolerant species, remains unclear.
Recently, homologues to S. cerevisiae Crz1p have been identified in Schizosaccharomyces pombe (34) and Candida albicans (46). Proteins with some degree of similarity to Crz1p have also been found in Candida glabrata and Kluyveromyces lactis through the Génolevures sequencing project (available at http://cbi.labri.fr/Genolevures/index.php). Among these, only the prz1+ and CaCRZ1 genes from S. pombe and C. albicans, respectively, have been studied in detail (34, 46, 51). Like Crz1p, Prz1p and CaCrz1p act downstream of calcineurin and regulate Ca2+ homeostasis (34, 51). However, S. pombe prz1 and C. albicans crz1/crz1 defective strains show phenotypes distinct from those observed in S. cerevisiae crz1 mutants (34, 46). Hence, calcineurin and Crz1p homologues in fission yeast and C. albicans appear to play functional roles that are not shared by the S. cerevisiae pathway.
In this work we took advantage of the salt-sensitive phenotype of the S. cerevisiae strain CEN.PK2-1C to identify genes from T. delbrueckii that confer increased salt tolerance. Using this strategy, we cloned the TdENA1 and TdCRZ1 genes, which encode a putative Na+/Li+ P-type ATPase and a zinc finger protein homologue to S. cerevisiae Crz1p, respectively. As expected, T. delbrueckii cells lacking TdCrz1p showed some phenotypes similar to those reported for S. cerevisiae calcineurin and crz1Δ mutant strains. However, lack of the transcriptional factor in T. delbrueckii led to enhanced resistance to Li+, while no growth defects were observed at high Na+ concentrations. Furthermore, T. delbrueckii cells did not show the same calcineurin dependency in response to saline stress as that previously reported for S. cerevisiae. These results suggest that salt stress in T. delbrueckii is regulated differently, through uncovered regulators and molecular circuits.
MATERIALS AND METHODS
Strains, culture media, and general methods.
T. delbrueckii wild-type strain PYCC5321 (1) and S. cerevisiae strains (Table 1) were used throughout this work. The T. delbrueckii Tdcrz1Δ mutant strain (MJHY211) was constructed as described below. Cells were cultured at 30°C in defined medium, YPD (1% yeast extract, 2% peptone, 2% glucose) or SD (0.2% yeast nitrogen base without amino acids [Difco], 0.5%(NH4)2SO4, 2% glucose), supplemented with the appropriate auxotrophic requirements (55). Escherichia coli was grown in Luria-Bertani (LB) medium (1% peptone, 0.5% yeast extract, 0.5% NaCl) supplemented with ampicillin (50 mg/liter). Antibiotics were filter sterilized and added to autoclaved medium. Transformation of yeasts was performed by the lithium acetate method (37). T. delbrueckii transformants containing the nourseothricin resistance module (natMX4) were grown for 4 h in YPD at 30°C before being plated on YPD agar plates containing 10 mg/liter of nourseothricin (clonNAT; Werne Bioagents, Germany). E. coli was transformed by electroporation according to the manufacturer's instructions (Eppendorf).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| CEN.PK2-1C | MATα ura3-52 his3-Δ1 leu2-3,112 trp1-289 MAL2-8cSUC2 | 23 |
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | 57 |
| DD12 | Same as YPH499, except cnb1::hisG | 20 |
| ASY472 | Same as YPH499, except crz1::loxP-kanMX-loxP | 61 |
| ASY475 | Same as DD12, except crz1::loxP-kanMX-loxP | 61 |
| ASY832 | Same as YPH499, except ura3::TRP1-4x CDRE-lacZ | M. Cyert |
| ASY834 | Same as ASY472, except ura3::TRP1-4x CDRE-lacZ | M. Cyert |
| ASY835 | Same as ASY475, except ura3::TRP1-4x CDRE-lacZ | M. Cyert |
Stress sensitivity tests.
For stress experiments, cells were grown at 30°C to mid-exponential phase, collected, and transferred to fresh medium containing the stressor to be tested at the indicated concentration. Plate phenotype experiments were made by diluting the cultures to an optical density at 600 nm (OD600) of 0.3 and spotting (3-μl) 10-fold serial dilutions onto SD or YPD agar solid medium containing NaCl, LiCl, MnCl2, or CaCl2. FK506 (Fujisawa GmbH) (20 mg/ml in 90% ethanol-10% Tween 20) was added at the indicated concentrations on solid and liquid media. Cyclosporine A (10 mg/ml in ethanol) was purchased from Sigma (St. Louis, MO). Unless otherwise indicated, colony growth was inspected after 2 to 4 days of incubation at 30°C.
Strain and plasmid construction.
Plasmids pAMS345 (61) and pJQ10 (9), containing the S. cerevisiae CRZ1 and ENA1 genes, respectively, were a gift from M. Cyert and A. Rodríguez-Navarro. Plasmids pMJH1 and pMJH14, carrying DNA fragments containing the ENA1 and CRZ1 genes from T. delbrueckii, TdENA1 and TdCRZ1, respectively, and flanking regions around these genes were isolated from a genomic library (31) by complementation in S. cerevisiae of the salt sensitivity phenotype of strain CEN.PK2-1C. To construct plasmid YEpTdENA1, the PstI/SpeI fragment released from plasmid YEpMJH1, containing the coding region of the TdENA1 gene and the 5′ and 3′ noncoding regions, was moved into vector YEplac195 (26). Plasmid YEpTdCRZ1, containing the isolated TdCRZ1 gene, was constructed by cloning a 1,980-bp ScaI/EcoRI fragment from plasmid YEpMJH14 into the vectorYEplac195 (26), previously digested with EcoRI/SmaI. The TdCRZ1 disruption cassette containing the nourseothricin-resistance module natMX4 (27) was constructed by restriction. First, a 3′-side fragment of TdCRZ1 (+1322 to +1810) was obtained by PCR using two specific primers, FR142 and FR141 (Table 2), and plasmid YEpMJH14 as a template. The PCR product was cloned into the pGEM-T Easy vector (Promega), released by restriction with SalI/EcoRI, and inserted into the pBS plasmid (Stratagene), previously digested with the same set of enzymes. The resulting plasmid, pBSCRZ, was treated with BamHI/EcoRI and used to accommodate the natMX4 module obtained from the BamHI- and EcoRI-digested plasmid pAG25 (27), creating plasmid pBSCRZ-natMX4. A PCR fragment was amplified from the 5′ region of TdCRZ1 (−270 to +448) using oligonucleotides FR140 and FR139 (Table 2) and plasmid YEpMJH14 as a template. The PCR product was inserted into the pGEM-T Easy vector, released with NotI and SpeI, and subcloned into plasmid pBSCRZ-natMX4, obtaining plasmid pBSCRZ-natMX4-CRZ. This was digested with EcoRV, releasing the TdCRZ1 disruption cassette, which contains the natMX4 module flanked by 718 and 488 bp (5′ and 3′ sides, respectively), homologues to the TdCRZ1 gene.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′) | Comment(s) |
|---|---|---|
| FR55 | ATGACCATGATTACGCCAA | TdENA1 and TdCRZ1 sequencing |
| FR77 | GTAAAACGACGGCCAGT | TdENA1 and TdCRZ1 sequencing |
| FR94 | AATCATCGGCAACCTTAG | TdCRZ1 sequencing |
| FR103 | GGCGACATGCTACGACTTC | TdCRZ1 sequencing |
| FR136 | GTGGTTAAATAGGACATCGC | TdCRZ1 sequencing |
| FR90 | TGCTACTGAAAAGACAAGG | TdENA1 sequencing |
| FR96 | GCTTACAGGCGAGGAATT | TdENA1 sequencing |
| FR135 | TGAGTCCTTGCCTATCGC | TdENA1 sequencing |
| FR110 | CCTCGTTCTGCTTTGACA | TdENA1 sequencing |
| FR117 | CAGGATATCAAGGGTAAGCT | TdENA1 sequencing |
| FR376 | AATGGTTCAGACGTCGC | TdENA1 sequencing |
| FR377 | CCAGCTGATCACTTCGG | TdENA1 sequencing |
| FR101 | CCTAAAGCCCAAACTATAACA | TdCRZ1 sequencing, verify correct targeting of the natMX4 module |
| FR142 | CGAAGTCGACAGCTCAATCA | PCR amplification of TdCRZ1 3′ side |
| FR141 | TGAATTCGGGTAAGAAAAGG | PCR amplification of TdCRZ1 3′ side |
| FR140 | CATTGAGCTCCTTGGAAGG | PCR amplification of TdCRZ1 5′ side |
| FR139 | ATTCGGATCCTAAGTCACTC | PCR amplification of TdCRZ1 5′ side |
| FR126 | TTCAGTGCCGAAGGGACTAC | Verify correct targeting of the natMX4 module |
| FR76 | GTCAAGGAGGGTATTCTGG | Verify correct targeting of the natMX4 module |
| FR75 | AGTTAAGTGCGCAGAAAG | Verify correct targeting of the natMX4 module |
| FR96 | GCTTACAGGCGAGGAATT | TdENA1 probe for Northern blotting |
| FR121 | GCTGCACCAACAGACAAAG | TdENA1 probe for Northern blotting |
| FR390 | GGTATGTTCTAGCGCTTG | ACT1 probe for Northern blotting |
| FR391 | TCTGGGGCTCTGAATCTT | ACT1 probe for Northern blotting |
Correct disruption of the TdCRZ1 gene was detected by diagnostic PCR using whole yeast cells (36) from isolated colonies and a set of oligonucleotides designed to bind outside or inside of the replaced TdCRZ1 sequence and within the marker module (Table 2).
β-Galactosidase assay.
Exponentially SD-growing cells (OD600, 0.6 to 0.8) were collected, resuspended in YPD (pH 5.5) or the same medium supplemented with 0.2 M CaCl2, and incubated at 30°C and 200 rpm for 45 min. Then, aliquots of the yeast suspension (15 OD600 units) were harvested, washed with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4), and centrifuged at 3,000 × g for 2 min (4°C), and the cell pellets were frozen at −20°C for further analysis. Cell extracts were prepared as previously described (14). Total protein was determined with the Bio-Rad Bradford assay kit and bovine serum albumin as the standard protein. β-Galactosidase activity was determined at room temperature by using the substrate ONPG (o-nitrophenyl-β-d-galactopyranoside) as previously described (47). One unit is defined as the amount of enzyme that is able to convert 1 nmol of ONPG per min under the assay conditions.
Northern blotting.
Total RNA from T. delbrueckii cells was prepared as described previously (55). Equal amounts of RNA (10 μg) were separated in 1% (wt/vol) agarose gels containing formaldehyde (2.5% vol/vol), transferred to a nylon membrane, and hybridized with a 32P-labeled probe of TdENA1 (+90 to +1003). A fragment of the S. cerevisiae ACT1 gene (+10 to +1066) was used as the loading control. Probes were generated by PCR and radiolabeled with the random primer Ready-to-Go kit (Amersham Biosciences, Chalfont-St. Giles, England) and [α-32P]dCTP (Amersham Biosciences). Hybridization was carried out under standard conditions (55), except for the ACT1 probe. Briefly, after hybridization overnight at 35°C, the filters were rinsed once with 50 ml of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate and once with 50 ml of 0.2× SSC-0.1% sodium dodecyl sulfate at room temperature for 20 and 10 min, respectively. Filters were exposed to a high-resolution BAS-MP 2040S imaging plate (Fuji, Kyoto, Japan) for 24 h and scanned in a phosphorimager (FLA-3000; Fuji). Spot intensities were quantified with Image Gauge software, version 3.12 (Fuji). Values of spot intensity were corrected with respect to the ACT1 mRNA level and represented as the relative mRNA level. The highest relative TdENA1 mRNA level for each sample analyzed was set to 100.
Sequencing and sequence analysis.
DNA sequencing was performed on both strands by the dideoxy chain termination procedure (50). Analysis of sequence data was carried out with DNAMAN sequence analysis software (Lynnon BioSoft). Similarity searches were performed using BLAST software (3) at the Munich Information Center for Protein Sequences (http://mips.gsf.de/). TdEna1p and TdCrz1p domains were searched by scanning protein sequences in the ExPASy Molecular Biology Server (http://www.expasy.ch/) from the Swiss Institute of Bioinformatics (http://www.isb-sib.ch/) against the PROSITE database of protein families (56). Multiple sequence alignment was done with MultAlin software (15) at INRA (http://prodes.toulouse.inra.fr/).
Nucleotide sequence accession numbers.
The nucleotide sequences of TdCRZ1 and TdENA1 have been deposited in the GenBank database (available at http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession numbers DQ097180 and DQ097181, respectively.
RESULTS
Isolation of T. delbrueckii genes that confer increased salt tolerance in S. cerevisiae.
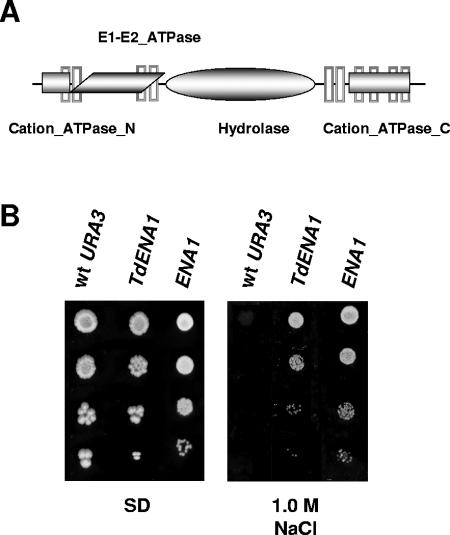
We transformed cells of the S. cerevisiae strain CEN.PK2-1C with a high-copy-number genomic library from T. delbrueckii (31). This Saccharomyces strain is very sensitive to saline stress and therefore is a good recipient to detect genes that could confer salt tolerance. After transformation, 18 yeast colonies were isolated, purified, and confirmed on SD medium plates containing NaCl at 0.5 M, a salt concentration that inhibits the growth of the host strain. Plasmid restriction analysis established four plasmid groups that were confirmed by dot blot analysis (data not shown). Two of them were studied in detail in this work. The first, named YEpMJH1, permitted identification of a 3,273-bp open reading frame (ORF) (GenBank accession number DQ097181) that encodes a putative polypeptide closely similar to Ena proteins isolated from other yeasts, such as D. hansenii (55% identity), Schwanniomyces occidentalis (56%), Z. rouxii (67%), and S. cerevisiae (69%). These proteins belong to the large P-type ATPase family, subfamily IID, whose members perform active ion transport across biological membranes (7, 10). Consistent with this, the putative protein identified in plasmid YEpMJH1 was found to contain the typical ATPase α chains involved in Na+ and K+ transport and responsible for ATP hydrolysis (13), the E1-E2 ATPase domain characteristic of the superfamily P-type ATPases (60), and one hydrolase and 10 transmembrane domains (Fig. 1A).
FIG. 1.
Multiple copies of TdENA1, encoding a P-type ATPase, enhance NaCl resistance in S. cerevisiae. (A) Schematic structure of TdEna1p showing the conserved domains for N-terminal cation transporting ATPase (Cation_ATPase_N), C-terminal cation transporting ATPase (Cation_ATPase_N), hydrolase, the E1-E2 ATPase-associated region, and transmembrane segments (shown as vertical boxes). (B) Mid-exponential-phase cultures of the S. cerevisiae strain CEN.PK2-1C transformed with plasmid YEpTdENA1 (TdENA1), plasmid pJQ10 (ENA1), or the empty plasmid YEplac195 (wt URA3) were adjusted to an OD600 of 0.3, diluted (1 to 10−3), and spotted (3 μl) onto SD plates or SD plates containing NaCl at the indicated concentration. Plates were inspected after 2 to 5 days at 30°C. A representative experiment is shown.
In S. cerevisiae, three isoforms of Ena proteins (encoded by the ENA1, ENA2, and ENA5 genes) have been characterized. Among them, ENA1 encodes the main ATPase involved in Na+ extrusion, whose function determines tolerance to NaCl (28). Therefore, we tried to further confirm the identity of the gene contained in plasmid YEpMJH1. A PstI/SpeI restriction fragment was subcloned into the vector YEplac195 (26) and used to transform the wild-type strain CEN.PK2-1C. As expected, overexpression of TdENA1 conferred an increased growth ability to yeast cells on NaCl medium (Fig. 1B). Moreover, the phenotype was similar to that observed in high-copy-number expression of the S. cerevisiae ENA1 gene. Thus, the ORF identified in plasmid YEpMJH1 was named TdENA1, the ENA1 gene from T. delbrueckii.
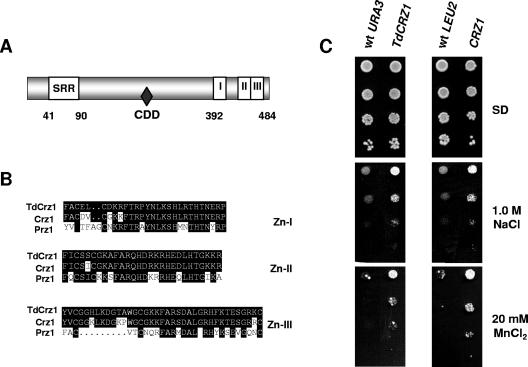
DNA sequencing of the second plasmid analyzed in this work, YEpMJH14, revealed a 1,518-bp ORF (GenBank accession number DQ097180) that encodes a protein of 506 amino acids with overall 36% identity to S. cerevisiae Crz1p (41, 61). Accordingly, the gene was designated TdCRZ1. As shown in Fig. 2A, the gene product of TdCRZ1 contains three C2H2-type zinc finger motifs at the carboxyl terminus that are highly homologous to those of Crz1p and Prz1p (Fig. 2B). Like S. cerevisiae Crz1p and S. pombe Prz1p, the protein from T. delbrueckii displayed a serine-rich region (residues 41 to 90) (Fig. 2A) essential for protein dephosphorylation by calcineurin (62). Inspection of the protein sequence also showed the presence of a PVISVQ sequence, similar to the calcineurin-docking domains (Fig. 2A) defined in Crz1p (12) and human nuclear factors of activated T cells (NFAT) (5).
FIG. 2.
TdCRZ1 encodes the homologue to the transcriptional factor Crz1p, and its overexpression in S. cerevisiae confers enhanced salt tolerance. (A) Schematic representation of TdCrz1p. Denoted are the serine-rich region (SRR), the calcineurin-docking domain (CDD) and three putative C2H2-type zinc finger motifs at the carboxyl terminus. (B) Sequence alignment of the three zinc finger motifs from TdCrz1, Crz1p, and Prz1p. Residues conserved in at least two sequences are boxed and highlighted. (C) Cells of the S. cerevisiae CEN.PK2-1C wild-type strain were transformed with plasmid YEpTdCRZ1 (TdCRZ1), plasmid pAMS354 (CRZ1), or the empty plasmids YEplac195 (wt URA3) and YEplac181 (wt LEU2). Mid-exponential-phase SD-grown cultures were adjusted to an OD600 of 0.3, diluted (1 to 10−3), and spotted (3 μl) onto SD plates or SD plates containing NaCl or MnCl2 at the indicated concentrations. Plates were inspected after 2 to 5 days at 30°C. A representative experiment is shown.
In order to confirm that the gene present in plasmid YEpMJH14 was responsible of the enhanced salt resistance of the S. cerevisiae strain CEN.PK2-1C, we constructed plasmid YEpTdCRZ1 by subcloning a 1,980-bp fragment containing the whole ORF plus 349 bp of the promoter region and 107 bp corresponding to the 3′ untranslated region into plasmid YEplac195 (26). As shown in Fig. 2C, overexpression of TdCRZ1 in the S. cerevisiae recipient strain produced a moderate increase in Na+ tolerance compared to the strain harboring an empty plasmid. These effects were more pronounced when Mn2+ tolerance was tested. Indeed, in SD medium, the CEN.PK2-1C strain transformed with YEpTdCRZ1 grew to 20 mM MnCl2, whereas the control strain showed only residual growth (Fig. 2C). Similar results were observed in transformant cells harboring plasmid pAMS345 (61), which affords high-copy-number expression of the S. cerevisiae CRZ1 gene (Fig. 2C). Hence, our results indicate that plasmid YEpMJH14 indeed contains the T. delbrueckii CRZ1 gene, the homologue to the transcriptional factor Crz1p, whose overexpression in S. cerevisiae confers enhanced tolerance to Na+ and Mn2+ ions.
TdCRZ1 suppresses the ion sensitivity of S. cerevisiae cnb1Δ and crz1Δ mutants.
Overexpression of S. cerevisiae CRZ1 compensates for the enhanced ion sensitivity of yeast calcineurin mutants, specifically to Na+, Li+, and Mn2+ (41, 61). Therefore, we were interested in determining whether TdCrz1p could affect these calcineurin phenotypes. For this, cnb1Δ and crz1Δ mutant cells of the YPH499 strain (20, 61) were transformed with plasmid YEpTdCRZ1 and examined for growth on NaCl-, MnCl2-, and LiCl-containing medium. The wild-type strain YPH499 shows higher MnCl2 sensitivity than the CEN.PK2-1C strain. Because of this, growth on this salt was inspected at 2.5 mM (final concentration). As shown in Fig. 3, production of the recombinant TdCrz1p increased Mn2+ tolerance of a cnb1Δ mutant to wild-type levels. Similar effects were observed in the presence of 0.4 M LiCl or 1.0 M NaCl. Moreover, overexpression of TdCRZ1 compensated for the ion sensitivity produced by the lack of Crz1p (Fig. 3). Like calcineurin mutants, crz1-null cells show retarded growth at high concentrations of Li+/Na+ and Mn2+ cations (41, 61).
FIG. 3.
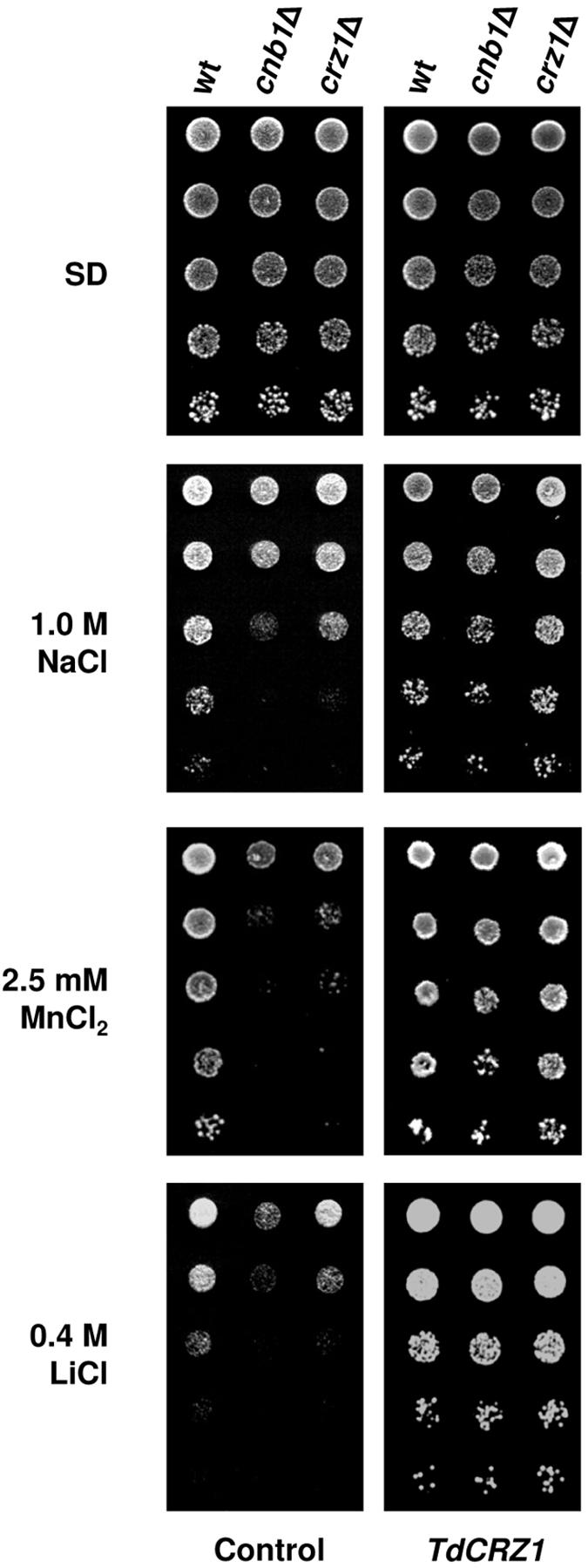
TdCrz1p restores growth of S. cerevisiae cnb1Δ and crz1Δ mutants in high-salt media. Mid-exponential-phase cultures of the S. cerevisiae strains YPH499 (wild type [wt]), DD12 (cnb1Δ), and ASY472 (crz1Δ), transformed with plasmid YEpTdCRZ1 or YEplac195 (empty plasmid control), were examined for growth on solid SD (SD) or SD containing NaCl, MnCl2, or LiCl at the indicated concentrations. SD-pregrown cells were diluted, spotted, and incubated as described in the legend to Fig. 1B. A representative experiment is shown.
We also tested whether the production of TdCrz1p could activate the expression of a 4× CDRE::lacZ reporter, which contains four tandem copies of the 24-bp CDRE (38). Multiple copies of the CDRE increase the calcineurin-dependent transcriptional activation of the reporter gene (61). Thus, ninefold inductions were observed in wild-type cells carrying an integrated copy of the heterologous construct after 45 min of exposure to 0.2 M CaCl2 (Table 3). Consistent with previous reports (61), CDRE-driven expression was completely dependent on the function of the calcineurin-Crz1p pathway. Thus, no significant β-galactosidase activity could be detected in S. cerevisiae cells lacking Crz1p or in the cnb1Δ crz1Δ double mutant transformed with an empty plasmid, YEplac195 (Table 3). In contrast, overproduction of TdCrz1p in a crz1-null background restored the CDRE-dependent transcriptional activation, although the induction level of β-galactosidase activity at high Ca2+ was lower, around fivefold, than that observed in the wild type. Nevertheless, the crz1Δ mutant transformed with plasmid pAMS435, which contains the S. cerevisiae CRZ1 gene (61), also showed lower induction, around threefold (Table 3). As could be expected, overexpression of TdCRZ1 in the strain lacking both Cnb1p and Crz1p was unable to restore the CDRE-mediated expression level observed in the crz1Δ single mutant (Table 3). Again, similar results were obtained by overexpression of multiple copies of CRZ1. Hence, TdCrz1p is able to mediate CDRE-driven expression and appears to function downstream of calcineurin in S. cerevisiae. In order to confirm this, we analyzed the level of β-galactosidase activity in YEpTdCRZ1 transformants of the crz1Δ strain treated with both Ca2+ and the drug FK506. The immunosuppressant FK506 is a potent inhibitor of calcineurin (6, 65) and has been used extensively for molecular studies in lower and higher eukaryotes (18, 29, 45). As shown in Table 3, addition of FK506 at 1 μg/ml decreased the Ca2+-stimulated induction of β-galactosidase activity observed in the absence of the immunosuppressant, whereas at doses of 5 μg/ml, induction was eliminated altogether (Table 3). As expected, the addition of FK506 also impaired the β-galactosidase activity in Ca2+-treated crz1Δ cells transformed with the S. cerevisiae CRZ1 gene (data not shown).
TABLE 3.
Overexpression of TdCRZ1 permits the calcineurin-dependent induction of a 4x CDRE-lacZ gene fusion in response to Ca2+
| Relevant genotype (+ growth condition) of S. cerevisiae strain useda | β-Galactosidase activity (U/mg of protein)b
|
|
|---|---|---|
| Control | Ca2+ | |
| Wild type | 152.7 ± 43.2 | 1,336.6 ± 98.1 |
| crz1Δ | 0.9 ± 0.3 | 1.8 ± 0.4 |
| crz1Δ TdCRZ1 | 57.0 ± 4.0 | 322.4 ± 19.8 |
| crz1Δ CRZ1 | 85.0 ± 7.0 | 246.1 ± 23.2 |
| cnb1Δ crz1Δ | 1.7 ± 0.2 | 4.2 ± 1.3 |
| cnb1Δ crz1Δ TdCRZ1 | 121.0 ± 14.2 | 184.6 ± 12.2 |
| cnb1Δ crz1Δ CRZ1 | 67.8 ± 8.4 | 110.5 ± 17.1 |
| crz1Δ TdCRZ1 + FK506 (1 μg/ml) | 52.0 ± 12.0 | 157.4 ± 31.0 |
| crz1Δ TdCRZ1 + FK506 (5 μg/ml) | 57.0 ± 21.0 | 60.4 ± 18.0 |
The S. cerevisiae strains used were YPH499 (wild type), ASY834 (crz1Δ), and ASY835 (cnb1Δ crz1Δ) transformed with plasmid YEplac195 (empty plasmid), YEpTdCRZ1 (TdCRZ1), or pAMS435 (CRZ1).
Data are means ± standard errors for three independent experiments. β-Galactosidase activities are shown for the indicated strains treated with 0.2 M CaCl2 in the presence or absence of FK506.
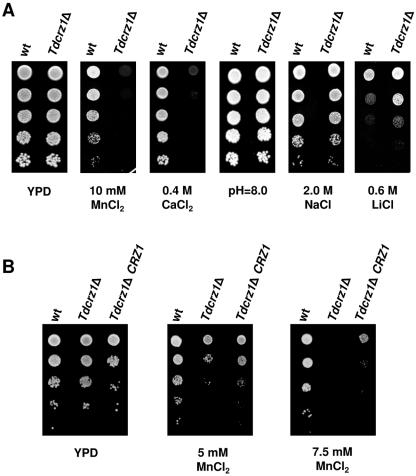
Tdcrz1Δ shows conserved phenotypes distinct from those of S. cerevisiae crz1Δ mutants.
To clarify the function of TdCRZ1 in T. delbrueckii, we constructed a Tdcrz1-null mutant (MJH211 strain) and analyzed cells for phenotypes previously reported for S. cerevisiae calcineurin and crz1Δ mutants (41, 44, 61, 66). On YPD plates containing 10 mM MnCl2 or 0.4 M CaCl2, Tdcrz1Δ cells displayed a clear growth defect (Fig. 4A). Unlike S. cerevisiae cnb1Δ mutants, cells lacking TdCrz1p did not exhibit sensitivity to high pH. These phenotypes coincide completely with those reported for crz1Δ mutants (61). However, T. delbrueckii mutant cells were indifferent to the presence of 2.0 M NaCl and, remarkably, TdCrz1p deficiency increased Li+ tolerance (Fig. 4A). Both calcineurin and crz1Δ mutant cells of S. cerevisiae have been reported to be sensitive to high concentrations of monovalent Na+ and Li+ cations (41, 44, 61).
FIG. 4.
Tdcrz1Δ cells exhibit specific phenotypes in response to diverse ionic stresses. (A) Exponentially growing cultures of the T. delbrueckii strains PYCC5321 (wild type [wt]) and MJH211 (Tdcrz1Δ) were adjusted to an OD600 of 0.3, diluted (1 to 10−4), and spotted (3 μl) onto YPD agar medium, YPD adjusted to pH 8.0, or YPD supplemented with MnCl2, CaCl2, NaCl, or LiCl at the indicated concentrations. Plates were incubated at 30°C for 2 to 5 days. A representative experiment is shown. (B) Tdcrz1Δ mutant cells were transformed with plasmid pAMS354, which contains the S. cerevisiae CRZ1 gene, and transformants (Tdcrz1Δ CRZ1) were examined for growth on MnCl2 medium. The wild-type strain PYCC5321 (wt) and the mutant strain MJH211 (Tdcrz1Δ) were used as controls. Cells were grown in liquid YPD containing 0.2 M CaCl2, diluted as described for panel A, and spotted onto YPD agar medium lacking or containing MnCl2 at the indicated concentrations. In all cases, a representative experiment is shown.
We also examined whether overexpression of the S. cerevisiae CRZ1 gene might suppress the salt-dependent phenotypes observed in Tdcrz1Δ mutant cells. Since the T. delbrueckii strains used in our work are prototrophic, pAMS435 (CRZ1) transformants were selected on solid YPD medium containing 0.4 M CaCl2. Under these conditions, the recipient mutant strain displays residual growth (Fig. 4A). Transformants were verified by plasmid isolation and G-418 resistance prior to use (data not shown). As can be seen in Fig. 4B, the MnCl2 sensitivity phenotype observed in Tdcrz1Δ mutant cells was partially suppressed by expression of multiple copies of CRZ1. Similar results were obtained in CaCl2- or LiCl-containing medium, while no phenotype could be detected at pH 8.0 or in the presence of NaCl (data not shown). Therefore, CRZ1 partially complements phenotypes associated with deletion of the T. delbrueckii CRZ1 gene.
The T. delbrueckii calcineurin-Crz1p pathway plays no role in Na+ tolerance.
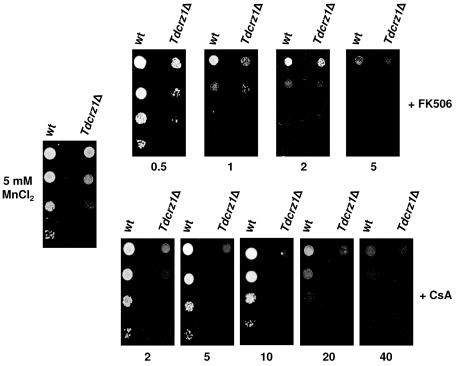
The results reported above led us to investigate whether calcineurin was involved in the phenotypes exhibited by Tdcrz1Δ cells. Since the T. delbrueckii genes for the calcineurin catalytic and regulatory subunits are unknown, it is not possible to delete these genes selectively to assess their biological function. As an alternative, we analyzed the ability of FK506 and cyclosporine A to enhance the salt sensitivity of wild-type and Tdcrz1Δ mutant cells. Like FK506, cyclosporine A is one of the most potent, specific, and well-known inhibitors of calcineurin (49). As shown in Fig. 5, the MnCl2 sensitivity phenotype associated with loss of TdCRZ1 was not suppressed in the presence of a range of cyclosporine A concentrations. Moreover, the Tdcrz1Δ mutant strain was clearly more sensitive to the immunosuppressant than was the wild-type strain. Indeed, mutant cells displayed impaired growth in response to low doses of cyclosporine A (>2 μg/ml). On the contrary, much higher levels of the drug, about 20 μg/ml, were required for enhanced sensitivity to MnCl2 in wild-type cells (Fig. 5). Similar results were obtained when phenotypes were tested in the presence of 0.5 to 5 μg/ml FK506 (Fig. 5).
FIG. 5.
The MnCl2 sensitivity phenotype associated with loss of TdCRZ1 is not suppressed by exposure to FK506 or cyclosporine A. YPD-grown cultures of the T. delbrueckii strains PYCC5321 (wild type [wt]) and MJH211 (Tdcrz1Δ) were adjusted to an OD600 of 0.3, diluted (1 to 10−3), and spotted (3 μl) onto 5 mM MnCl2-YPD agar medium with or without FK506 or cyclosporine A (CsA) at the indicated concentration (in micrograms per milliliter). Plates were incubated at 30°C for 2 to 5 days. A representative experiment is shown.
The physiological role of calcineurin was also examined on CaCl2 medium. In this case, single FK506 (1 μg/ml) and cyclosporine A (10 μg/ml) concentrations were tested. As shown in Fig. 6, addition of FK506 had dramatic inhibitory effects on the growth of wild-type cells treated with 0.2 M CaCl2. This result was surprising since S. cerevisiae calcineurin mutants are more Ca2+ tolerant than wild-type cells (66). Moreover, we found that the growth defect of the Tdcrz1Δ mutant strain was again more pronounced than that observed in the wild-type strain on Ca2+/FK506-containing medium. Again, similar results were observed when cyclosporine A was used instead of FK506 (Fig. 6). Thus, our results indicate that Mn2+/Ca2+ tolerance in T. delbrueckii requires a functional calcineurin-Crz1p pathway. However, T. delbrueckii calcineurin and TdCrz1p appear to function independently.
FIG. 6.
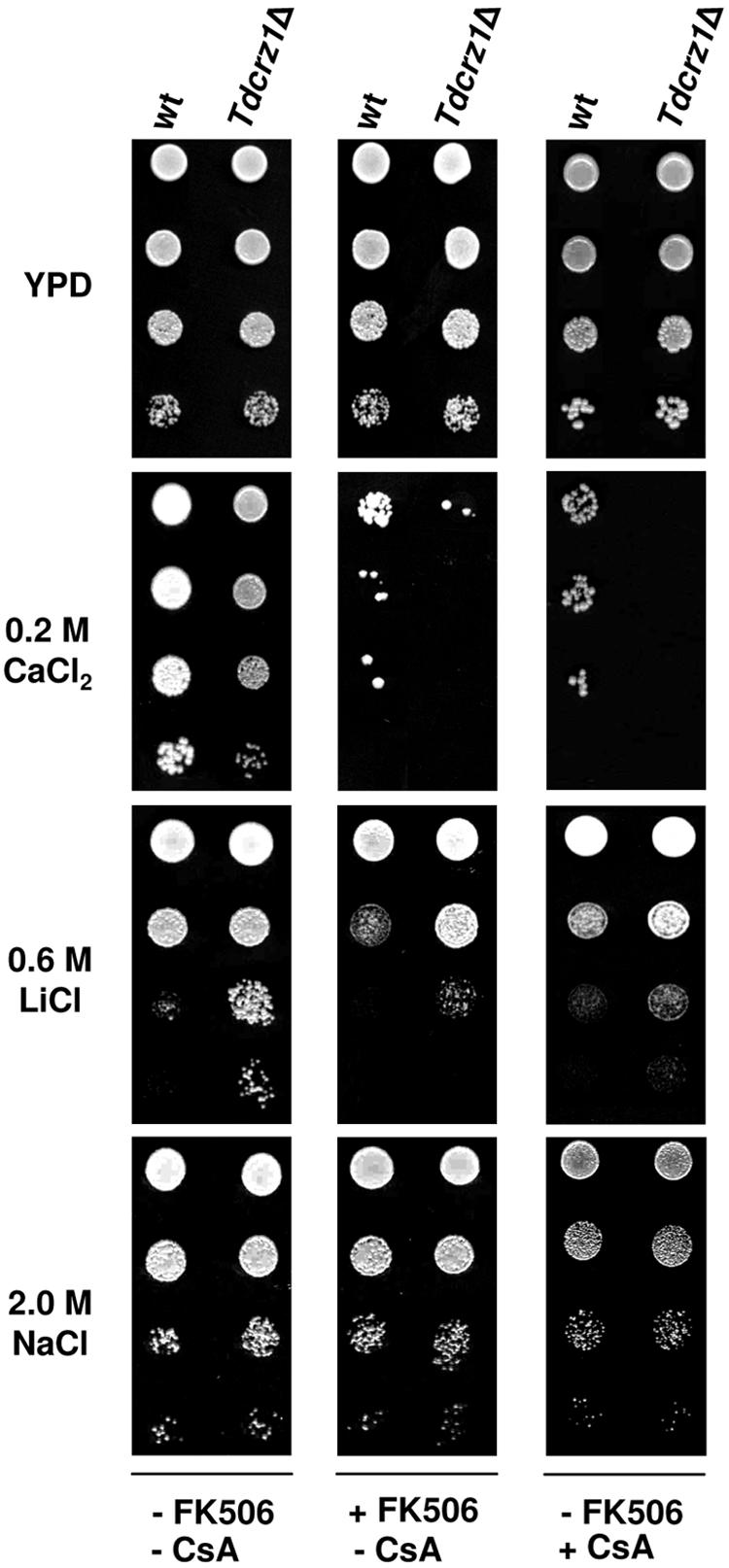
The T. delbrueckii calcineurin-Crz1p pathway plays no role in Na+ tolerance. Exponentially growing cultures of the T. delbrueckii strains PYCC5321 (wild type [wt]) and MJH211 (Tdcrz1Δ) were adjusted to an OD600 of 0.3, diluted (1 to 10−3), and spotted (3 μl) onto YPD agar medium containing CaCl2, NaCl, or LiCl at the indicated concentrations, in the presence or absence of 1 μg/ml FK506 or 10 μg/ml cyclosporine A (CsA). Plates were incubated at 30°C for 2 to 5 days. A representative experiment is shown.
We also characterized the properties of wild-type and Tdcrz1Δ cells exposed to NaCl or LiCl in the presence or absence of FK506 or cyclosporine A. Unlike S. cerevisiae, neither the lack of TdCrz1p nor exposure to the immunosuppressive agents affected the Na+ tolerance of this organism (Fig. 6). In contrast, addition of FK506 or cyclosporine A impaired the growth of cells exposed to Li+ (Fig. 6). Nevertheless, their presence had weak effects on Li+ sensitivity compared to that observed for Ca2+ and did not affect the phenotype of Tdcrz1Δ mutant cells (Fig. 6). Hence, the T. delbrueckii calcineurin-Crz1p pathway plays conserved roles distinct from those reported in S. cerevisiae and has positive or negative effects in an ion-dependent manner.
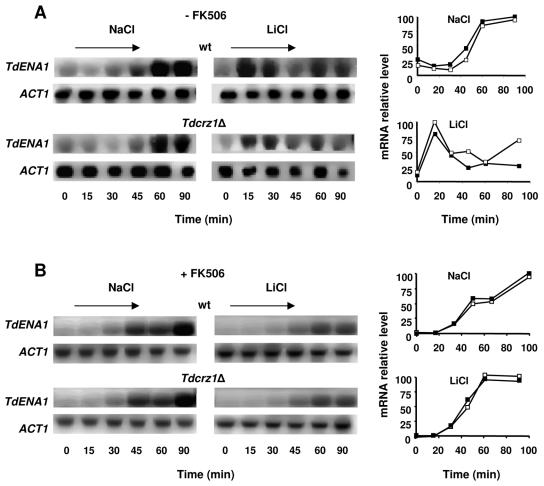
TdCrz1p is not required for salt-induced transcriptional activation of TdENA1.
In S. cerevisiae, adaptation to salinity is primarily based on the Na+/Li+-extruding ATPase encoded by the gene ENA1. Consequently, we were interested in discovering the effects of TdCRZ1 deletion on the levels of TdENA1 mRNA in cells exposed to NaCl or LiCl. Figure 7A shows a Northern blot analysis of total RNA from wild-type and Tdcrz1 mutant cells probed with a 913-bp fragment of TdENA1. As expected, expression of the P-type ATPase was induced in response to either Na+ or Li+, suggesting a functional role of the T. delbrueckii pump in cation homeostasis. TdENA1 mRNA accumulation was induced rapidly after the addition of 0.4 M LiCl. Then, the TdENA1 mRNA levels fell and shifted back to high at the end of the period assayed. When 1.4 M NaCl was used, the response was delayed and only one peak, at 60 min, could be detected. However, TdCrz1p was not required for the Na+- or Li+-induced expression of TdENA1 (Fig. 7A).
FIG. 7.
Activity of TdCrz1p is not required for the salt-induced transcriptional activation of the TdENA1 gene. YPD-grown cells of the T. delbrueckii strains PYCC5321 (wild type [wt], ▪) and MJH211 (Tdcrz1Δ, □) were transferred to 1.4 M NaCl-YPD or 0.5 M LiCl-YPD lacking (A) or containing (B) 5 μg/ml FK506. Samples were taken at the indicated times and analyzed by Northern blotting, as described in Materials and Methods. Filters were probed for TdENA1 mRNA. The graphs represent quantification of the mRNA levels of TdENA1 relative to those of the S. cerevisiae ACT1 gene. Results from a representative experiment are shown.
Then, we analyzed the induction profile of TdENA1 in the presence of FK506. As shown in Fig. 7B, addition of the immunosuppressant had no effect on the NaCl-induced expression of TdENA1 observed in either wild-type or Tdcrz1Δ mutant cells. In contrast, addition of FK506 affected the temporal pattern of expression of TdENA1 in cells exposed to Li+. Thus, the response was clearly delayed compared to that observed in the absence of the drug (Fig. 7A), and only a late peak of mRNA accumulation could be detected (Fig. 7B). However, similar results were found in both wild-type and Tdcrz1Δ mutant cells.
DISCUSSION
This study is the first to report the identification and characterization of a putative C2H2 zinc finger transcriptional factor, TdCrz1p, from the osmotolerant yeast T. delbrueckii; TdCrz1p is the homologue of S. cerevisiae Crz1p (41, 61), S. pombe Prz1p (34), and C. albicans Crz1p (46, 51). The budding yeast transcriptional factor Crz1p mediates the Ca2+/calcineurin-dependent induction of genes in response to salt stress and is necessary for survival under these conditions (19). Similar to its yeast homologues (12) and mammalian NFATc transcription factors (5, 63), the primary structure of TdCrz1p exhibits motifs that are characteristic of calcineurin-regulated proteins. In our study, overexpression of TdCRZ1 in S. cerevisiae wild-type cells led to improved growth on media containing NaCl or MnCl2. Furthermore, production of TdCrz1p suppressed the growth defect at high Na+/Li+ or Mn2+ concentrations in calcineurin and crz1Δ mutants and mediated the calcinuerin/Ca2+-dependent activation of a CDRE-containing reporter gene. Hence, TdCrz1p appears to be a calcineurin target and is able to compensate for the lack of a functional calcineurin-Crz1p pathway in S. cerevisiae and provide tolerance to salt stress.
These results led us to postulate that TdCrz1p and by extension calcineurin might play a role similar to that of their S. cerevisiae counterparts in regulating the salt stress response in T. delbrueckii. In contrast, however, our results demonstrated that this signaling pathway has conserved roles that are different from those described for the S. cerevisiae pathway. As shown, Tdcrz1-null phenotypes differ from those associated with Crz1p deficiency. S. cerevisiae crz1Δ mutants are sensitive to extracellular Ca2+ and Mn2+ and monovalent Na+ and Li+ cations (61). However, growth of Tdcrz1Δ cells was diminished only upon exposure to divalent cations. In S. cerevisiae, adaptation to Ca2+ requires the calcineurin/Crz1p-dependent induction of genes by the vacuolar and secretory Ca2+ pumps Pmc1p and Pmr1p (17, 61). Mn2+ tolerance has also been related to the function of Pmr1p, the Golgi-localized Ca2+ pump (41). A similar Prz1p-dependent regulation of PMC1 expression in S. pombe has also been reported (34). Thus, a common regulatory mechanism involving Crz1p/Prz1p homologues appears to control Ca2+/Mn2+ homeostasis in different yeasts. However, calcineurin-null cells of S. cerevisiae and S. pombe show opposite phenotypes in response to high concentrations of Ca2+. Whereas S. cerevisiae cnb1Δ strains are resistant to Ca2+ (17), calcineurin mutants in fission yeast display decreased Ca2+ tolerance, a phenotype shared with prz1-null cells (34). Similarly, our results indicate that T. delbrueckii wild-type cells treated with FK506 or cyclosporine A are hypersensitive to Ca2+. Hence, unlike in S. cerevisiae, calcineurin activation is required for both Ca2+ and Mn2+ tolerance in T. delbrueckii. Moreover, FK506- or cyclosporine A-treated cells of the Tdcrz1Δ strain exhibited a greater degree of sensitivity to Ca2+/Mn2+ than did wild-type cells. Hence, TdCrz1p must carry out functions in tolerance to divalent cations that are independent of calcineurin signaling.
The differences between the S. cerevisiae and T. delbrueckii calcineurin-Crz1p pathways were further demonstrated by analysis of their respective phenotypes in response to monovalent Na+/Li+ cations. In sharp contrast to the situation in S. cerevisiae, Tdcrz1Δ mutants were insensitive to high external NaCl levels. Growth of wild-type cells was also unaffected by combined exposure to Na+/FK506 or Na+/cyclosporine A. In consonance with these phenotypes, TdCrz1p was not required to activate the NaCl-induced expression of TdENA1 either in the presence or absence of FK506. Hence, the T. delbrueckii calcineurin-Crz1p pathway has no apparent role in Na+ homeostasis. On the other hand, Tdcrz1-null cells are more tolerant to high levels of external Li+ than are wild-type cells. This fact suggests that Li+ and Na+ extrusion in T. delbrueckii is regulated, at least in part, by independent mechanisms. Again, this is in striking contrast to the situation in S. cerevisiae, where both ions are extruded trough the same calcineurin-regulated Na+/Li+ ATPase, ENA1 (40). Our results indicate that calcineurin mediates the early induction of TdENA1 in LiCl medium. However, the accumulation of TdENA1 mRNA at the late stage after a shift to high Li+ was indifferent to the presence of FK506. Consistent with this, exposure of yeast cells to calcineurin inhibitors had a weak effect on Li+ tolerance. Interestingly, the enhanced Li+ tolerance observed in the Tdcrz1Δ strain indicates that TdCrz1p might function as a repressor and not as an activator of the Li+ extrusion system. Therefore, the calcineurin-Crz1p pathway has evolved to carry out different cellular roles in T. delbrueckii. Moreover, Na+ and Li+ signals appear to be transduced by unknown regulatory mechanisms and might activate distinct gene targets.
In recent years, the major biological role played by calcineurin in Ca2+-dependent eukaryotic signal transduction pathways has been demonstrated (49). Some of the most prominent research has been devoted to deciphering the function of the calcineurin-Crz1p pathway in the adaptation of the model yeast S. cerevisiae to salt stress (19, 35). However, this signaling pathway appears to have different functions in other yeasts and fungi (33, 34, 45). Moreover, stress responses and stress response mechanisms appear to have diverged among different yeasts in a niche-dependent manner. A clear example of this is the specialization of the HOG pathway toward virulence in pathogenic fungi (2, 8, 59). In conclusion, the differences in the biological functions of the calcineurin-Crz1p pathway highlighted by our results may explain the high resistance to salt stress in T. delbrueckii compared to S. cerevisiae. However, further experimentation is required to confirm this possibility and to clarify the regulatory mechanisms operating in this salt-tolerant, unconventional yeast.
Acknowledgments
We thank M. Cyert (Stanford University) and A. Rodríguez-Navarro (Universidad Politécnica de Madrid) for plasmids and strains used in this work. We also thank M. Cyert for critical reading of the manuscript, A. Blasco for technical assistance, and J. Aguilera for helpful discussions.
This research was funded by the Comisión Interministerial de Ciencia y Tecnología project (AGL2001-1203) from the Ministry of Science and Technology of Spain. M.J.H.-L. and J.P. were supported by CSIC-EPO and FPI fellowships, respectively.
REFERENCES
- 1.Almeida, M. J., and C. Pais. 1996. Leavening ability and freeze tolerance of yeasts isolated from traditional corn and rye bread doughs. Appl. Environ. Microbiol. 62:4401-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., F. Navarro-Garcia, G. Molero, R. Diez-Orejas, M. Gustin, J. Pla, M. Sanchez, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apse, M. P., and E. Blumwald. 2002. Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13:146-150. [DOI] [PubMed] [Google Scholar]
- 5.Aramburu, J., F. Garcia-Cozar, A. Raghavan, H. Okamura, A. Rao, and P. G. Hogan. 1998. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 5:627-637. [DOI] [PubMed] [Google Scholar]
- 6.Arndt, C., M. C. Cruz, M. E. Cardenas, and J. Heitman. 1999. Secretion of FK506/FK520 and rapamycin by Streptomyces inhibits the growth of competing Saccharomyces cerevisiae and Cryptococcus neoformans. Microbiology 145:1989-2000. [DOI] [PubMed] [Google Scholar]
- 7.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 8.Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benito, B., F. J. Quintero, and A. Rodríguez-Navarro. 1997. Overexpression of the sodium ATPase of Saccharomyces cerevisiae: conditions for phosphorylation from ATP and Pi. Biochim. Biophys. Acta 1328:214-225. [DOI] [PubMed] [Google Scholar]
- 10.Benito, B., B. Garciadeblás, and A. Rodríguez-Navarro. 2002. Potassium- or sodium-efflux ATPase, a key enzyme in the evolution of fungi. Microbiology 148:933-941. [DOI] [PubMed] [Google Scholar]
- 11.Blomberg, A., and L. Adler. 1992. Physiology of osmotolerance in fungi. Adv. Microb. Physiol. 33:145-212. [DOI] [PubMed] [Google Scholar]
- 12.Boustany, L. M., and M. S. Cyert. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 16:608-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broude, N. E., N. N. Modyanov, G. S. Monastyrskaya, and E. D. Sverdlov. 1989. Advances in Na+,K+-ATPase studies: from protein to gene and back to protein. FEBS Lett. 257:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Ciriacy, M. 1975. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol. Gen. Genet. 138:157-164. [DOI] [PubMed] [Google Scholar]
- 15.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan, K. J., and K. B. Storey. 2003. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 206:1107-1115. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham, K. W., and G. R. Fink. 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cyert, M. S. 1993. The function of Ca+2/calmodulin-regulated phosphatase in yeast. Adv. Protein Phosphatases 7:429-443. [Google Scholar]
- 19.Cyert, M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143-1150. [DOI] [PubMed] [Google Scholar]
- 20.Cyert, M. S., and J. Thorner. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cyert, M. S., R. Kunisawa, D. Kaim, and J. Thorner. 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 88:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng, W. K., L. Faucette, M. M. McLaughlin, R. Cafferkey, Y. Koltin, R. A. Morris, P. R. Young, R. K. Johnson, and G. P. Livi. 1994. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene 151:61-71. [DOI] [PubMed] [Google Scholar]
- 23.Entian, K. D., and P. Kötter. 1998. Yeast mutant and plasmid collections, p. 431-449. In A. Brown and M. Tuite (ed.), Yeast gene analysis. Methods in microbiology, vol. 26. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 24.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24:469-486. [DOI] [PubMed] [Google Scholar]
- 25.Garrett-Engele, P., B. Moilanen, and M. S. Cyert. 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+-ATPase. Mol. Cell. Biol. 15:4103-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 28.Haro, R., B. Garciadeblas, and A. Rodriguez-Navarro. 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291:189-191. [DOI] [PubMed] [Google Scholar]
- 29.Heitman, J., A. Koller, J. Kunz, R. Henriquez, A. Schmidt, N. R. Movva, and M. N. Hall. 1993. The immunosuppressant FK506 inhibits amino acid import in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemenway, C. S., and J. Heitman. 1999. Calcineurin. Structure, function, and inhibition. Cell Biochem. Biophys. 30:115-151. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez-Lopez, M. J., J. A. Prieto, and F. Randez-Gil. 2002. Isolation and characterization of the gene URA3 encoding the orotidine-5′-phosphate decarboxylase from Torulaspora delbrueckii. Yeast 19:1431-1435. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez-Lopez, M. J., J. A. Prieto, and F. Randez-Gil. 2003. Osmotolerance and leavening ability in sweet and frozen sweet dough. Comparative analysis between Torulaspora delbrueckii and Saccharomyces cerevisiae baker's yeast strains. Antonie Leeuwenhoek 84:125-134. [DOI] [PubMed] [Google Scholar]
- 33.Higuchi, S., J. Tamura, P. R. Giri, J. W. Polli, and R. L. Kincaid. 1991. Calmodulin-dependent protein phosphatase from Neurospora crassa. Molecular cloning and expression of recombinant catalytic subunit. J. Biol. Chem. 266:18104-18112. [PubMed] [Google Scholar]
- 34.Hirayama, S., R. Sugiura, Y. Lu, T. Maeda, K. Kawagishi, M. Yokoyama, H. Tohda, Y. Giga-Hama, H. Shuntoh, and T. Kuno. 2003. Zinc finger protein Prz1 regulates Ca2+ but not Cl− homeostasis in fission yeast. Identification of distinct branches of calcineurin signaling pathway in fission yeast. J. Biol. Chem. 278:18078-18084. [DOI] [PubMed] [Google Scholar]
- 35.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huxley, C., E. D. Green, and I. Dunham. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6:236. [DOI] [PubMed] [Google Scholar]
- 37.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang, B., and M. S. Cyert. 1999. Identification of a novel region critical for calcineurin function in vivo and in vitro. J. Biol. Chem. 274:18543-18551. [DOI] [PubMed] [Google Scholar]
- 39.Klee, C. B., and P. Cohen. 1988. The calmodulin-regulated protein phosphatase, p. 225-248. In P. Cohen and C. B. Klee (ed.), Molecular aspects of cellular regulation: calmodulin, vol. 5. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 40.Marquez, J. A., and R. Serrano. 1996. Multiple transduction pathways regulate the sodium extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 382:89-92. [DOI] [PubMed] [Google Scholar]
- 41.Matheos, D. P., T. J. Kingsbury, U. S. Ahsan, and K. W. Cunningham. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11:3445-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425:323-328. [DOI] [PubMed] [Google Scholar]
- 43.Mendoza, I., F. Rubio, A. Rodriguez-Navarro, and J. M. Pardo. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol. Chem. 269:8792-8796. [PubMed] [Google Scholar]
- 44.Nakamura, T., Y. Liu, D. Hirata, H. Namba, S. Harada, T. Hirokawa, and T. Miyakawa. 1993. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 12:4063-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanthakumar, N. N., J. S. Dayton, and A. R. Means. 1996. Role of Ca++/calmodulin binding proteins in Aspergillus nidulans cell cycle regulation. Prog. Cell Cycle Res. 2:217-228. [DOI] [PubMed] [Google Scholar]
- 46.Onyewu, C., F. L. Wormley, Jr., J. R. Perfect, and J. Heitman. 2004. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect. Immun. 72:7330-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platt, T., B. Muller-Hill, and J. H. Miller. 1972. Assay of β-galactosidase, p. 351-355. In J. H. Miller (ed.), Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Rodriguez-Navarro, A. 2000. Potasium transport in fungi and plants. Biochim. Biophys. Acta 1469:1-30. [DOI] [PubMed] [Google Scholar]
- 49.Rusnak, F., and P. Mertz. 2000. Calcineurin: form and function. Physiol. Rev. 4:1483-1521. [Google Scholar]
- 50.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos, M., and I. F. de Larrinoa. 2005. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr. Genet. 48:88-100. [DOI] [PubMed] [Google Scholar]
- 52.Serrano, R., and A. Rodriguez-Navarro. 2001. Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13:399-404. [DOI] [PubMed] [Google Scholar]
- 53.Serrano, R., J. A. Márquez, and G. Ríos. 1997. Crucial factors in salt stress tolerance, p. 147-169. In S. Hohmann and W. H. Mager (ed.), Yeast stress responses. Springer-Verlag, Heidelberg, Germany.
- 54.Serrano, R., J. Mulet, G. Rios, J. Marquez, I. de Larrinoa, M. Leube, I. Mendizabal, A. Pascual-Ahuir, M. Proft, R. Ros, and C. Montesinos. 1999. A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 50:1023-1036. [Google Scholar]
- 55.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Sigrist, C. J., L. Cerutti, N. Hulo, A. Gattiker, L. Falquet, M. Pagni, A. Bairoch, and P. Bucher. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinformatics 3:265-274. [DOI] [PubMed] [Google Scholar]
- 57.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sleator, R. D., and C. Hill. 2001. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 59.Smith, D. A., S. Nicholls, B. A. Morgan, A. J. Brown, and J. Quinn. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, D. L., T. Tao, and M. E. Maguire. 1993. Membrane topology of a P-type ATPase. The MgtB magnesium transport protein of Salmonella typhimurium. J. Biol. Chem. 268:22469-22479. [PubMed] [Google Scholar]
- 61.Stathopoulos, A. M., and M. S. Cyert. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stathopoulos-Gerontides, A., J. J. Guo, and M. S. Cyert. 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 13:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugiura, R., S. O. Sio, H. Shuntoh, and T. Kuno. 2001. Molecular genetic analysis of the calcineurin signaling pathways. Cell. Mol. Life Sci. 58:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westfall, P. J., D. R. Ballon, and J. Thorner. 2004. When the stress of your environment makes you go HOG wild. Science 5701:1511-1512. [DOI] [PubMed] [Google Scholar]
- 65.Wiederrecht, G., E. Lam, S. Hung, M. Martin, and N. Sigal. 1993. The mechanism of action of FK-506 and cyclosporin A. Ann. N. Y. Acad. Sci. 696:9-19. [DOI] [PubMed] [Google Scholar]
- 66.Withee, J. L., J. Mulholland, R. Jeng, and M. S. Cyert. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshimoto, H., K. Saltsman, A. P. Gasch, H. X. Li, N. Ogawa, D. Botstein, P. O. Brown, and M. S. Cyert. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079-31088. [DOI] [PubMed] [Google Scholar]