Abstract
Blue light regulates many physiological and developmental processes in fungi. In Trichoderma atroviride the complex formed by the BLR-1 and BLR-2 proteins appears to play an essential role as a sensor and transcriptional regulator in photoconidiation. Here we demonstrate that the BLR proteins are necessary for carbon deprivation induced conidiation, even in the absence of light, pointing to the existence of an unprecedented cross talk between light and carbon sensing. Further, in contrast to what has been found in all other fungal systems, clear BLR-independent blue-light responses, including the activation of protein kinase A (PKA) and the regulation of gene expression, were found. Expression of an antisense version of the pkr-1 gene, encoding the regulatory subunit of PKA, resulted in a nonsporulating phenotype, whereas overexpression of the gene produced colonies that conidiate even in the dark. In addition, overexpression of pkr-1 blocked the induction of early light response genes. Thus, our data demonstrate that PKA plays an important role in the regulation of light responses in Trichoderma. Together, these observations suggest that the BLR complex plays a general role in sensing environmental cues that trigger conidiation and that such a role can be separated from its function as a transcription factor.
The influence of light on living organisms is critical, not only because of its importance as the main source of energy for the biosphere but also due to its capacity to induce changes in the behavior and morphology of nearly all forms of life. In particular, physiological responses to blue light have been studied in a wide variety of organisms from microbes to animals. Examples of such capacity include phototropism of coleoptiles in plants (27), induction of carotenoid synthesis in fungi (32), and entrainment of circadian rhythms in a diversity of organisms (12).
Blue light influences fungi in many ways. It can affect metabolism, growth, sexual and asexual development, pigment formation, and tropism, among other phenomena. The ascomycete Neurospora crassa is considered a paradigm for biochemical, genetic, and molecular studies of light responses. Several developmental and morphological processes of Neurospora are regulated by light. In all cases, light perception occurs in the UV-blue-light range. Light-mediated responses include induction of changes in membrane potential, gene expression, protein phosphorylation, induction of protoperithecia, phototropism of perithecial beaks, photocarotenogenesis, entrainment of the circadian clock, and conidiation (28). Photoinduced conidiation (asexual reproduction) of fungi provides an interesting model for biochemical, physiological, and morphological studies on differentiation since a relatively simple and natural external stimulus, such as light, is used to initiate a sequence of molecular events, which ultimately lead to conidiation (26).
The mycoparasite Trichoderma atroviride is used as a biological control agent due to its capacity to attack a broad range of important air- and soilborne phytopathogenic fungi (17). The main mechanism for survival and dispersal of Trichoderma is through the production of asexual spores (conidia). Conidiation in this organism is induced by environmental factors such as light and nutrient depletion. In the laboratory, exposure of a dark-grown colony of Trichoderma to a brief pulse of blue light results in the formation of a ring of dark green conidia at what had been the perimeter of the colony at the time of illumination (20). Another light response described in T. atroviride is the regulation of the expression of the photolyase gene phr-1. No phr-1 mRNA is detected in the dark, becoming detectable immediately after a blue-light pulse (4). Among the biochemical changes evoked by illumination in dark-grown colonies are shifts in membrane potential and ATP levels, a transient biphasic oscillation in intracellular cyclic AMP (cAMP) levels, activation of adenylyl cyclase, and phosphorylation of proteins (13, 14, 24). Exogenous cAMP promotes sporulation in the dark (30), whereas atropine, a compound known to inhibit adenylyl cyclase in Neurospora (37), prevents sporulation even after photoinduction (5). In addition, the inhibitor of cAMP-phosphodiesterase, 3-isobutylmethylxanthine, stimulates photoconidiation (42). Light regulation of phr-1, however, is indifferent to these effectors (5). Induction of photolyase expression occurs as a direct, rapid response to light, independent of the induction of sporulation (5). These data suggest that photoconidiation and light-induced expression of phr-1 either follow divergent signal transduction cascades or that there could be more than one blue-light receptor.
A complex formed by the proteins WC-1 and WC-2 controls all known light responses in N. crassa. It has been demonstrated that WC-1 is a blue-light receptor in this fungus, that WC-1 and WC-2 are subjected to light-dependent phosphorylation, and that WC-1 is phosphorylated in parallel with the transient increase in the transcript levels of light-regulated genes (16). A second photoreceptor (VVD), which enables Neurospora to perceive and respond to daily changes in light intensity, has recently been characterized (36). Furthermore, the analysis of the N. crassa genome has revealed the presence of genes that encode other potential photoreceptors, namely, cry-1, phy-1, and phy-2 (6).
Recently, we cloned two T. atroviride genes, named blue-light regulators 1 and 2 (blr-1 and blr-2), homologues of N. crassa wc-1 and wc-2, respectively. The deduced protein sequence of BLR-1 indicates that it has all of the characteristics of a blue-light receptor, whereas that of BLR-2 suggests that it could interact with BLR-1 through PAS domains to form a complex. Both BLR proteins were shown to be essential for photoconidiation in Trichoderma. It was also demonstrated that Trichoderma responds to mycelial injury producing conidia at the damaged area and that the BLR proteins were not necessary for this response (7).
In the present study, we show that BLR-1 and BLR-2 are also necessary for conidiation when induced by glucose deprivation, that in Trichoderma there are blue-light responses independent of BLR, and that PKA activity is required for light-induced gene expression. Blue-light regulation of growth and development in basidiomycetes is under the control of genes equivalent to the T. atroviride brl-1/2 and the N. crassa wc-1/2 (21, 29, 45). Thus, our current research on these interactions has potential implications restricted not just within a class of fungi (sordariomycetes) but across the fungal Kingdom.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
T. atroviride IMI 206040, Δblr-1, and Δblr-2 (7) strains were used in the present study. Escherichia coli TOP10F′ (Invitrogen) was used for plasmid transformation. The plasmids used were pCR2.1 (Invitrogen), pGFP-Hyg, and pB (carrying a fragment of human 28S rRNA gene). Plasmid pGFP-Hyg is a derivative of pZEGA (47) carrying a hygromycin resistance cassette that contains the E. coli hygromycin phosphotransferase gene under the control of the Aspergillus nidulans trpC promoter. T. atroviride cultures were routinely grown at 25°C on potato dextrose agar plates (Difco). For isolation of protoplasts, photoconidiation assays and light induction, mycelia were grown in PDYC medium (24 g of potato dextrose broth, 2 g of yeast extract, and 1.2 g of casein hydrolysate medium [all from Difco]/liter). For protoplast transformation, potato dextrose agar with 200 μg of hygromycin/ml was used as a selection medium, and potato dextrose agar containing 1% agarose and 200 μg of hygromycin/ml soft selection medium was used as an overlay.
Southern and Northern blot analysis.
Total RNA was extracted from mycelia as previously described (5) and used for Northern analysis according to standard techniques (33). For Northern blot analysis, a 1.35-kb EcoRV fragment of phr-1 or the c51, blu1, blu6, blu17, and blu8 cDNA fragments were labeled with 32P and used as probes. Southern blot analysis was performed according to standard techniques (33), using an EcoRI pkr-1 fragment, containing the complete cDNA labeled with 32P as a probe.
Isolation of pkr-1.
A DNA fragment of about 550 bp was amplified by PCR using T. atroviride genomic DNA as a template and the oligonucleotides PKArF (forward; 5′-GGTGATTATTTCTATGTGGTAGAG-3′) and PKArR (reverse; 5′-CTGGCCGCGCGAGGGGCATC-3′) as primers, designed based on conserved regions of N. crassa PKA regulatory subunit gene. The amplification program consisted of 30 cycles performed as follows: 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, with a final extension period of 7 min at 72°C. The main product was subcloned into plasmid pCR2.1 and sequenced (34); its sequence showed high similarity to many PKA regulatory subunit encoding genes. The fragment was 32P labeled and used to screen, under high-stringency conditions, a Trichoderma cDNA library from cultures grown under blue light. Sequencing data of a selected clone confirmed its identity and indicated that it corresponded to the full-length cDNA (GenBank accession no. DQ077817).
Constructs.
Plasmids for sense and antisense expression were constructed as follows. For the sense construct, a pair of primers was designed based on the pkr-1 cDNA sequence in which NsiI and XbaI restriction sites were added to the forward and reverse primers, respectively. The same strategy was used for the antisense construct by inverting the restriction sites. Using the pairs of primers and the cDNA clone as a template, the different clones were amplified. The fragments corresponding to the coding region of the pkr-1 cDNA were subcloned in pCR2.1. The subclones were double digested with the enzymes NsiI and XbaI. The pkr-1 fragment obtained was used to replace the gfp coding sequence in plasmid pGFP-Hyg. The two resulting plasmids pPKArAS and pPKArOE constituted the antisense and sense constructs. These constructs were used for transformation of T. atroviride, as previously described (3). Transformants were selected and allowed to sporulate, and serial dilutions of spore suspensions were plated for three cycles to obtain monosporic cultures.
Light responses.
Colonies were induced to conidiate as previously described (4) by exposure to a standard blue-light source consisting of light from cool-white fluorescent tubes filtered through a blue acrylic filter (LEE #183; fluence rate, 3 μmol m−2 s−1). For RNA extractions the colonies were inoculated over cellophane sheets overlying the filter paper. At various times, the mycelia were scraped from the surface of the cellophane under safelight (red light; 0.1 μmol m−2 s−1). Samples of mycelia exposed to light or kept in the dark were immediately frozen in liquid nitrogen and used for RNA extraction.
cAMP-dependent protein kinase activity.
Approximately 50 mg of mycelium was frozen in liquid nitrogen, ground to a fine powder, and resuspended in 200 μl of a buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA (pH 8.0), and a protease inhibitor cocktail consisting of 5 μg of antipain and leupeptin ml−1 and 1 mM phenylmethylsulfonyl fluoride. Samples were then vortexed three times for 10 s and kept at 4°C. The homogenate was centrifuged for 120 min at 13,000 rpm and 4°C in a microfuge. Supernatants were transferred to fresh Eppendorf tubes, and the protein content was determined with Coomassie blue (41) using bovine serum albumin fraction V as a standard. PKA activity was determined immediately by using the nonradioactive PepTag test method with dye-labeled Kemptide as a substrate according to the manufacturer's protocol (Promega). The incubation time for the enzymatic reaction was 30 min at 30°C. One unit of enzyme activity was defined as the amount of enzyme required to transfer one nanomole of phosphate from ATP to the substrate (Kemptide) per minute at 30°C. The catalytic subunit of bovine heart PKA (Promega) was used as a standard.
Stress-induced conidiation assays.
For nutritional stress-induced conidiation, Trichoderma colonies were cultivated in the dark on top of a double layer of filter paper consisting of an 8-cm Whatman 50 filter overlaying a Whatman 1 filter soaked in minimal medium (MM; 1.66 mM MgSO4, 5.16 mM K2HPO4, 2.68 mM KCl, 12.5 mM NH4NO3, 7.19 μM FeSO4, 6.95 μM ZnSO4, 10.1 μM MnCl2) with 111 mM glucose. After 48 h of cultivation in the dark at 25°C, the filter papers with the fungus were washed in sterile water, transferred to petri dishes containing fresh liquid MM with or without glucose, allowed to grow for a further 24 h in total darkness, and photographed. All manipulations were carried out under red safelight.
Effect of cAMP and dB-cAMP on the conidiation of wild-type, pkr-1 modified, and blr mutant strains.
To observe the effect of cAMP on conidiation in the Δblr1, Δblr2, pkr-1 AS, pkr-1 OE, and wild-type strains, Trichoderma was cultivated in the dark by using a double layer of filter paper as described for the nutrient deprivation experiments, but colonies were transferred to medium with or without 10 mM cAMP as indicated. dB-cAMP was used to test its effect on conidiation in rich medium. In this case, Trichoderma was cultivated in the dark but the double layer of filter paper soaked in PDYC medium. After 48 h of cultivation at 25°C, the filter papers with the fungus were washed in sterile water and transferred to petri dishes containing fresh medium with or without the addition of 200 μM dB-cAMP. When the colonies were exposed to light, the dB-cAMP was added to the PDYC medium 30 min before the light pulse. The colonies were then incubated in the dark for 24 h and photographed. All other manipulations were carried out under red safelight.
RESULTS
blr-1 and blr-2 are necessary for conidiation triggered by carbon deprivation.
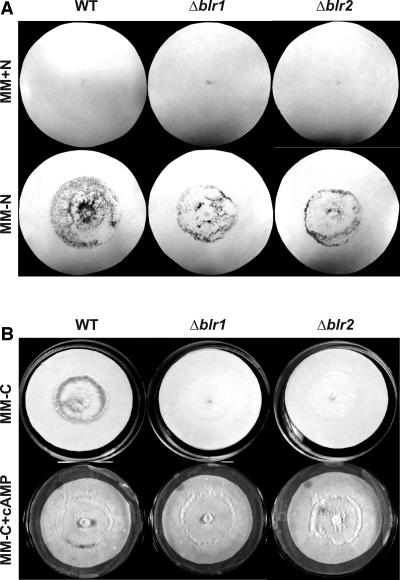
Because nutrient deprivation is considered a universal inducer of sporulation in fungi and it has previously been shown that in Trichoderma a sudden carbon deprivation mimics the effect of light by triggering the development of a discrete ring of conidia at the growing edge of a colony (20), we decided to verify whether the BLR proteins played any role in the induction of conidiation by nutrient deprivation by testing this response in the corresponding mutants. Briefly, the wild type, and the Δblr-1 and Δblr-2 mutant strains were grown for 48 h in MM and then transferred to either nitrogen- or glucose-free MM medium and allowed to grow for a further 24 h in complete darkness (Fig. 1) . As expected, nitrogen deprivation triggered the production of conidia in the wild-type strain. Similarly, the Δblr-1 and Δblr-2 mutant strains produced abundant conidia in response to the sudden lack of nitrogen (Fig. 1A). Surprisingly, upon glucose deprivation, no conidiation was observed in the mutant strains, whereas a ring of conidia was clearly visible in the wild-type strain (Fig. 1B). Controls transferred to medium containing glucose and nitrogen did not conidiate (data not shown). These results suggested that the BLR-1 and BLR-2 proteins play a major role in carbon deprivation-induced conidiation. cAMP has been linked to the control of a number of functions in fungi, including utilization of endogenous and exogenous carbon sources and conidiation (25). In order to test whether the phenotype observed upon glucose deprivation could be linked to cAMP, the strains were transferred to medium containing this compound and no carbon source. As seen on Fig. 1B, all strains responded to the addition of exogenous cAMP producing a ring of conidia at the growing edge of a colony, clearly resembling that observed in the wild type in response to the sudden lack of a carbon source.
FIG. 1.
Effect of glucose and nitrogen deprivation on the conidiation phenotype. Pictures show dark-grown colonies of the wild-type and blr mutant strains 24 h after a change of growth medium from 2% glucose to medium without nitrogen (A) or without glucose (B). A 10 mM concentration of cAMP was added to the medium as indicated. The hyaline mycelium is not visible on the white filter paper.
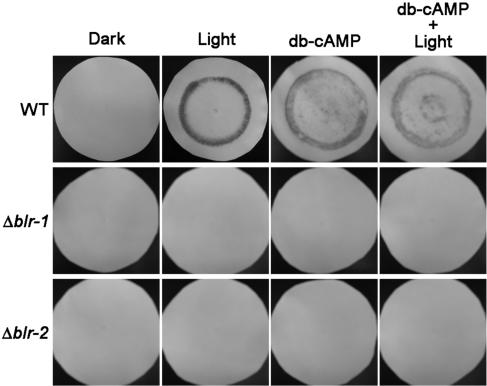
dB-cAMP does not restore photoconidiation in the blr mutants.
Data available in the literature suggest that cAMP could be involved in blue light-induced conidiation in Trichoderma (13, 14, 24). In addition, we have previously shown that application of exogenous dB-cAMP to Trichoderma cultures growing on rich medium triggers conidiation even in the dark (5). In order to test whether exogenous cAMP could restore the nonsporulating phenotype of the mutants upon light exposure, the blr-null mutants and the wild-type strain were transferred to rich medium containing 200 μM dB-cAMP before exposure to light. A ring of green conidia in the growing front of the colony was observed in the absence of light in the wild-type strain, whereas the blr mutants were unable to conidiate under these conditions (Fig. 2). The addition of dB-cAMP to colonies exposed to light did not induce conidiation in the blr mutants.
FIG. 2.
Effect of dB-cAMP on the conidiation of the blr mutants and wild-type strains. The pictures show dark-grown colonies 24 h after exposure to 200 μM dB-cAMP, a 5-min blue-light pulse (Light), or a combination of both. Control colonies were treated identically except for the fact that the light pulse was not applied (Dark) or no dB-cAMP was added.
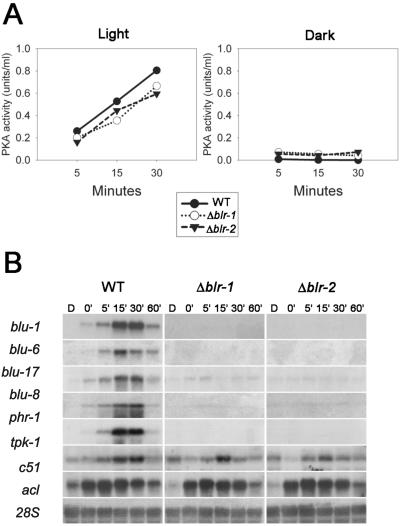
PKA activity and gene expression are induced by blue light through a novel receptor.
To better understand the absence of sporulation in the blr mutants upon application of dB-cAMP, PKA activities were measured in cell extracts of the blr mutants exposed to light or kept in total darkness. A clear increase in the PKA activity levels was detected upon light exposure of the wild-type strain. Surprisingly, both blr mutants showed exactly the same response (Fig. 3A), suggesting the existence of an alternative blue light receptor. To verify the possible existence of an additional blue light receptor, we used a subset of light-regulated genes from a set previously identified by using microarrays, where gene expression in T. atroviride exposed to a pulse of white light was compared to that observed when the fungus was grown in the dark. The expression pattern of this subset of genes (blu1, blu6, blu17, and blu8) was analyzed by using the phr-1 gene as a control (Fig. 3B). This subset of genes is induced by blue light in a blr-dependent manner, showing maximum levels of expression 30 min after illumination and decreasing to basal levels after 120 min. Two more genes were selected from Trichoderma subtractive libraries (blue-light versus dark): tpk-1, encoding a protein kinase and a clone (c51) with no significant similarity to any sequence available in public databases. As shown in Fig. 3B, these two genes accumulated in response to a pulse of blue-light with a time course similar to that observed for phr-1, except that basal transcript levels of these genes could be detected even in the dark. This response was maintained in the blr mutants, where the level of tpk-1 and c51 transcripts increase very rapidly, reaching their maximum between 5 and 15 min after the light pulse (Fig. 3B). Red light had no effect on the mRNA abundance of these genes (data not shown).
FIG. 3.
(A) PKA activity in the wild type and in blr mutants. Total protein was prepared from different strains from dark grown colonies and exposed to a 5-min pulse of blue light, and activities were determined subsequently at the indicated times. Experiments were made in triplicate and the figure shows the results of a representative experiment. (B) Northern blot analysis of total RNA extracted from blr mutants and wild-type colonies, hybridized with the blu1, blu6, blu17, blu8, phr-1, tpk-1, and c51 cDNAs. 28S was included as loading control. RNA was extracted at the indicated times (in minutes) after a 5-min light pulse. D, dark control.
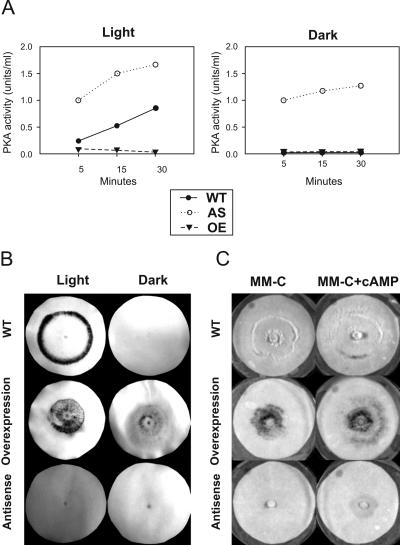
cAMP-dependent protein kinases (PKAs) are involved in light-induced conidiation.
To test the possible role of cAMP-dependent protein kinases in conidiation, we cloned a gene encoding the PKA regulatory subunit (pkr-1) from T. atroviride and generated transformants that express either an antisense (AS) or overexpress the sense version of the gene (OE). In both constructs the pkr-1 gene was under the control of the constitutive pki promoter and the cbh2 terminator of Trichoderma reesei. In order to prove the functionality of the AS and OE constructs in the transformants, PKA activities were measured in total cell extracts of the wild-type strain and AS and OE representative transformants (Fig. 4A). Elevated levels of PKA activity were found in the AS strains both under dark and light conditions. In contrast, the OE strains had nearly undetectable levels of PKA activity in both conditions. Photoconidiation assays were carried out to evaluate the effect of the constructs on the blue-light response of the generated strains, showing that the AS transformants were unable to respond to light while the OE transformants hypersporulated in response to light and conidiated even in the dark (Fig. 4B). Furthermore, mechanical injury did not trigger sporulation in AS strains, whereas the OE transformants sporulated at the damaged area, thereby suggesting that PKA is involved in the response to injury in Trichoderma (data not shown). To further investigate the cross talk between conidiation and the cAMP pathway, colonies of the AS, OE, and WT strains were grown on minimal medium and then transferred to fresh medium with or without glucose, containing or not containing cAMP. As shown in Fig. 1, the wild-type strain responded to the lack of glucose producing a ring of conidia, regardless of the presence of cAMP. The OE transformants produced conidia both in the presence (data not shown) and in the absence of glucose (Fig. 4C). However, more conidia were observed, especially toward the edge of the colony, when the medium contained cAMP (Fig. 4C), whereas the AS transformants did not conidiate under any condition, except in the presence of cAMP (Fig. 4C; see the dark area). These data suggest that the observed conidiation, triggered by the presence of cAMP, is at least in part independent of PKA activity.
FIG. 4.
(A) PKA activity in wild type and pkr-1 AS and OE transformants. Total protein was prepared from different strains from dark-grown colonies and exposed to a 5-min pulse of blue light. Activities were subsequently determined at the indicated times. Experiments were made in triplicate, and the figure shows the results of a representative experiment. (B) Photoconidiation assay for the wild-type, AS, and OE strains. The pictures show dark-grown colonies 24 h after a 5-min blue-light pulse (Light) and colonies treated identically except for the absence of the light pulse (Dark). (C) Effect of cAMP on the conidiation phenotypes of the AS, OE, and wild-type strains. The pictures show dark-grown colonies 24 h after transfer to medium with no glucose with or without 10 mM cAMP, as indicated.
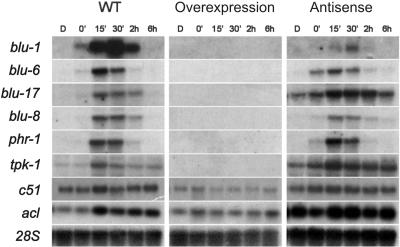
Altered expression of blue light induced genes in the AS and OE transformants of pkr-1 gene.
As shown here, conidiation is affected by both expression of an antisense version of pkr-1 and its overexpression. Thus, assuming that at least part of the blue-light-induced genes are directly related to the early stages of conidiation in Trichoderma, it could be expected that their expression pattern in response to light could be altered in the AS and OE strains. Accordingly, we decided to determine whether the expression of the light-regulated genes tested in the present study was altered in these strains. Transcription of all blr-dependent genes and that of tpk-1 was completely blocked in the OE transformants, whereas in the AS strains the corresponding mRNAs were clearly induced by light (Fig. 5). In addition, the bli-3 and tpk-1 genes showed an induction, which persisted for a longer period of time than that observed for the wild type. The expression of the acl gene, included as a control because its expression is not controlled by light, remained unaltered. These data provide evidence for the existence of a cross talk between the PKA and blr gene-controlled signal transduction pathways.
FIG. 5.
Northern blot analysis of total RNA extracted from AS and OE transformants and the wild-type strains, hybridized to the blu1, blu6, blu17, blu8, phr-1, tpk-1, c51, and acl cDNAs. The acl gene was included as a light-independent transcription control, and 28S was included as a loading control. RNA was extracted at the indicated times after a 5-min light pulse. D, dark control.
DISCUSSION
Nutrient deprivation is considered a universal inducer of sporulation in fungi. Trichoderma is no exception, and nutrient deprivation dutifully triggers the conidiation process in this organism. A major finding in the present study was that carbon deprivation did not induce conidiation in photoreceptor deletion mutants (blr mutants), indicating that the BLR proteins are involved in sensing different signals that allow the fungus to sporulate. In contrast, conidiation in response to nutrient deprivation of mutants in the orthologous genes in N. crassa was not altered (2, 40). However, it has been shown in several other organisms that blue light generates changes in carbon metabolism. Exposure of Aspergillus ornatus to a pulse of light leads to protein phosphorylation and low glucose uptake (18). Similarly, when microplasmodia of Physarum polycephalum were exposed to light, glucose consumption was reversibly inhibited and the production of glucan and trehalose decreased (35). In addition, a starvation state was observed in the chytrid Blastocladiella britannica after a light stimulus (19). A transporter-based mechanism for sensing nutrients in fungi is thought to be universal. Accordingly, in yeast, glucose sensing is mediated by the receptors/sensors Snf3 and Rgt2, which act with hexokinase as an important component of this pathway (10). In plants, light responses and sugar sensing are clearly linked, and hexokinase appears to play an important role in this cross talk (9). However, the molecular bases linking glucose and light sensing have not yet been elucidated.
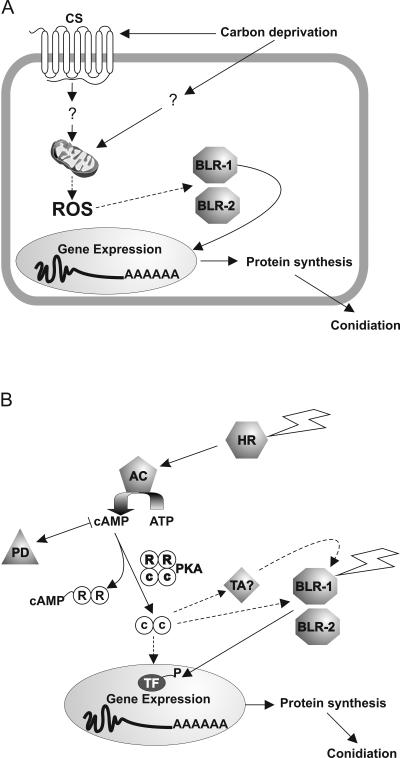
Various hypotheses could be postulated to explain the fact that BLR-1 and BLR-2 are required for conidiation in response to carbon deprivation. We favor one that postulates that the BLRs themselves perceive or transduce the signal originated from the lack of glucose (see Fig. 6A). In this sense, PAS domains could be important since they have been shown to be involved in several responses to environmental and internal stimuli, i.e., redox and oxygen sensing (44). PAS domains are also known to mediate protein-protein interactions and to bind various cofactors depending on their functional properties (15). The requirement for the BLR-1 and BLR-2 proteins for the responses analyzed here could imply that one or more of their PAS or LOV domains are required for this process, indicating that they may be involved in binding specific signals derived from carbon metabolism in T. atroviride. A model involving light modulated changes in the binding affinity between the LOV domain and its partner domain(s), in which the dynamic state of the LOV domain is the main determinant of its interactions with partner domains, has been proposed (8). The human aryl hydrocarbon receptor AhR, another PAS/LOV domain protein, binds different ligands to regulate the expression of several genes in a ligand-dependent manner. Besides its capacity to bind flavin, which is suggested by its structure (7), BLR-1 could have a function comparable to that of AhR, binding to one or several different ligands and thus responding to the presence or absence of certain carbon sources in the medium. Alternatively, any of the PAS domains found in either BLR-1 or BLR-2 could have dual-sensing capabilities that integrate both redox (carbon deprivation) and light signals. The fact that the BLRs are involved in controlling carbon deprivation responses in Trichoderma represents a novel link between light and carbon source sensing.
FIG. 6.
(A) Hypothetical model showing the function of the BLR complex in carbon sensing. The BLR-1 and/or BLR-2 could perceive or transduce the signal originated from the lack of glucose detected by a sensor/receptor (CS) or due the lack of reducing power in the cell, via reactive oxygen species (ROS) originated from the mitochondria (M), resulting in the induction of gene expression. (B) Hypothetical model integrating the distinct elements that participate in blue-light perception in Trichoderma atroviride. Two independent light inputs are necessary induce conidiation. The BLR independent pathway could activate adenylyl cyclase (AC), leading to the production cAMP, which in turn binds to the PKA regulatory subunit (R), resulting in the activation of the catalytic subunit (C). Phosphodiesterase (PD) would regulate the levels of cAMP, exerting a negative control on photoconidiation. The increase in PKA activity would activate the BLR complex, triggering the expression of the blue-light-responsive genes; such a function may involve direct phosphorylation of either of the BLR proteins or phosphorylation of an as-yet-unidentified regulatory partner (TA). Alternatively, PKA may phosphorylate a putative transcription factor (TF), whose modification is necessary for gene activation. Noncontinuous lines indicate hypothetical steps.
As mentioned before, the addition of dB-cAMP to Trichoderma cultures promotes sporulation in the dark (5, 30), while atropine prevents sporulation, even after a blue-light pulse (5). The addition of cAMP triggered conidiation in the blr mutants grown under carbon limitation conditions, resembling the response of the wild type to carbon deprivation. Surprisingly, in rich medium the addition of dB-cAMP did not trigger sporulation in the blr mutants as it does in the wild-type strain, not even when dB-cAMP was added to colonies before exposure to light. These data demonstrate that the BLR proteins have functions even in the dark and that they are necessary for this response. Thus, the BLR proteins could be part of a particular cAMP pathway that drives the fungus into the conidiation process, which is clearly influenced by the nutritional status of the cells.
Based on biochemical data (13, 14, 24), it has been suggested that a cAMP pathway, and consequently PKA, participates in light-induced conidiation in Trichoderma. Measurements of PKA activity in the wild type and in blr mutants revealed that it increased rapidly in the wild-type fungus after a pulse of blue light and that such induction was blr independent. Thus, two independent light inputs may induce the fungus to conidiate, with both inputs being necessary and with the blr-independent pathway triggering an increase of PKA activity. This notion is further supported by the fact that blue-light induction of at least two genes (c51 and tpk-1) was clearly maintained in the blr mutants, indicating that this response is independent of BLR-1 and BLR-2. In contrast, all known responses to blue light in N. crassa are initiated by the WC-1 and WC-2 proteins and modulated by Vivid. Mutations in either of the corresponding genes result in total blindness (28). Even though genes showing similarity to other types of photoreceptors have been found in the N. crassa genome, WC-1 remains as the only known functional, primary blue-light receptor (16). Similar to what has been shown for N. crassa, we demonstrated that another set of genes (blu1, blu6, blu17, and blu8) responded to blue light in a blr-dependent fashion, providing further evidence that supports the key role of the BLR proteins in blue light perception in T. atroviride.
To further investigate the role of PKA in photoconidiation and its possible interaction with the light perception pathway controlled by the BLR proteins, we generated T. atroviride transformants expressing sense and antisense versions of the PKA regulatory subunit-encoding gene. The antisense transformants showed high PKA activities and did not sporulate, whereas the overexpressing strains showed low activities and sporulated even in the dark. In agreement with our data, S. cerevisiae mutants affected in the regulatory PKA encoding gene BCY1 showed high PKA activity, and diploid homozygote strains affected in BCY1 did not sporulate (46). It should be noted, however, that sporulation in the case of S. cerevisiae is meiotic, whereas conidia in Trichoderma are produced through a mitotic process. Moreover, low PKA activities in Dictyostelium discoideum resulted in an uncontrollable sporulation phenotype (39). In A. nidulans, sporulation was partially inhibited in strains that overexpressed the catalytic subunit of PKA, whereas the sporulation process increased drastically in gene-disruptant strains (38). Interestingly, in our case the overexpressing strains continued responding to light by forming a ring of green conidia at what had been the colony perimeter at the time of the light pulse, even when the PKA activity detected was low. Based on these observations, we conclude that sustained low PKA activity provokes sporulation of Trichoderma, as it happens in other organisms, perhaps simulating starvation conditions.
Our data are apparently contradictory since it could be expected that strains expressing an antisense version of the PKA regulatory subunit would mimic the effect of the addition of dB-cAMP extracellularly. In theory, both conditions would result in high PKA activity by releasing the catalytic subunit from its inactive complex with the regulatory subunit. Our current model explains this apparent contradiction by considering that these two phenomena in fact use separate signaling pathways: (i) the classical pathway in which cAMP activates PKA and (ii) an alternate pathway that uses a membrane receptor for exogenous cAMP. Thus, if the blr mutants do not express this receptor or are blocked in any other downstream element of the pathway, they will not be able to respond to the stimulus. In support of this proposal, Scott and Solomon (37) reported in Neurospora a phosphodiesterase-like activity in the culture medium, concluding that N. crassa could secrete cAMP. Ivey et al. (22) demonstrated that the Gα protein mutant strains Δgna-1 were insensitive to cAMP supplementation. Later, these authors showed that the wild-type and Δgna-1 strains secreted similar amounts of cAMP into the medium and proposed that cAMP might function as an extracellular signal in N. crassa (23). Recently, with the conclusion of the Neurospora genome project, a putative cAMP receptor has been described (6). Our data on the exposure of AS and OE pkr-1 transformants to cAMP support this hypothesis, since both of them appear to conidiate in response to this stimulus against what would be expected. In fact, the effect could clearly be observed only under carbon deprivation conditions.
Using Northern blot analysis of RNA from blue-light-induced mycelium of the AS and OE strains, we detected an altered expression pattern of a set of light-induced genes. For the AS strains the expression of two of the genes tested was prolonged, whereas expression of all genes in the OE transformants was undetectable. The gene expression data in the OE transformants, together with their phenotype, suggest that the rapid expression of the set of light-induced genes studied here is not essential for photoconidiation. In N. crassa, there are pharmacological and genetic data that suggest that PKC is involved in desensitization and adaptation to light (1, 10). WC-1 becomes phosphorylated upon light exposure, probably a primary effect of light reception (36, 43). The light-dependent phosphorylation of WC-1 is transient, but phosphorylation of WC-1 is also observed in the dark. PKC interacts in vivo with WC-1 and phosphorylates in vitro the Zn finger domain. This interaction, however, is observed only in dark-grown mycelia or after 2 h of illumination, when gene photoactivation has ceased (11). Regulatory mutations in PKC result in changes in the amount of WC-1, with corresponding changes in gene photoactivation, albeit without affecting the photophosphorylation pattern of WC-1, confirming that PKC is a negative regulator of WC-1 (11). These results suggest that other kinase(s) must be responsible for the observed photophosphorylation of WC-1. In addition, the authors of that study demonstrated the phosphorylation of the WC-1 protein by PKC (1, 11). The data obtained in the present study show that the blue-light-induced genes in the AS strain have a behavior similar to that reported for the N. crassa blue-light response when PKC activity is abolished. The opposite results were observed in the overexpressing strains, where the transcription of light-induced genes was blocked, similar to what was observed in Neurospora when PKC activity was stimulated. In addition to these observations, the analysis of the BLR proteins revealed putative PKA and PKC phosphorylation sites (data not shown). The fact that the BLR proteins are necessary for conidiation triggered by dB-cAMP allowed us to suggest that they play an important role even in the dark. Further, since strains overexpressing the PKA regulatory subunit still conidiated in response to light and genes that respond rapidly to this stimulus were switched off in these strains, we conclude that the roles of the BLR complex, as a sensor and transcription factor, may be separated.
Considering the molecular evidence shown in the present study and the similarity with findings reported for Neurospora, we hypothesize that PKA has a function in regulating the expression of blue-light-responsive genes in T. atroviride. Such a function may involve direct phosphorylation by PKA of either of the BLR proteins or phosphorylation of an as-yet-unidentified regulatory partner (see model in Fig. 6B). In this sense, the activation of the aryl hydrocarbon receptor by cAMP has recently been demonstrated. In that case, cAMP activates the AhR, moving the receptor to the nucleus (31). According to our model, PKA activity would be necessary to activate the function of the BLR complex by promoting its entry into the nucleus, by modifying its affinity for target promoter sequences, or by allowing the complex to bind to a third partner that allows the activation of transcription. It is worth mentioning that, in contrast to N. crassa WC-1, BLR-1 does not contain typical activation domains. Alternatively, the phosphorylation by PKA may activate an additional transcription factor necessary for the activation of the light-responsive genes or inactivate a repressor that may be blocking their expression in the absence of light. Consistent with our data, there would be transcription of BLR-dependent genes in the pkr-1 AS transformants and no transcription in the overexpressors, and the blr mutants would not be able to respond to dB-cAMP. However, our model cannot explain why the pkr-1 OE transformants still conidiate in response to light, unless very subtle or fast, transient increases in PKA activity could still take place in our transformants. Such changes in PKA activity would then be sufficient to trigger conidiation but not transcription of early light response genes. Thus, we propose that the BLR complex plays a general role in sensing signals that lead to conidiation and that such a role can be independent of its function as a transcription factor in the control of at least a subset of genes.
Acknowledgments
This study was supported by grant 37601-N from Conacyt to A.H.-E.
We are grateful to John Delahno and Hayley Ridgway for critical reading of the manuscript. S.C.-F. and M.R.-M. are indebted to Conacyt/Concyteg and SRE, respectively, for doctoral fellowships.
REFERENCES
- 1.Arpaia, G., F. Cerri, S. Baima, and G. Macino. 1999. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 262:314-322. [DOI] [PubMed] [Google Scholar]
- 2.Arpaia, G., J. Loros, J. Dunlap, G. Morelli, and G. Macino. 1995. Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol. Gen. Genet. 247:157-163. [DOI] [PubMed] [Google Scholar]
- 3.Baek, J. M., and C. M. Kenerley. 1998. The arg2 gene of Trichoderma virens: cloning and development of a homologous transformation system. Fungal Genet. Biol. 23:34-44. [DOI] [PubMed] [Google Scholar]
- 4.Berrocal-Tito, G., L. Sametz-Baron, K. Eichenberg, B. A. Horwitz, and A. Herrera-Estrella. 1999. Rapid blue light regulation of a Trichoderma harzianum photolyase gene. J. Biol. Chem. 274:14288-14294. [DOI] [PubMed] [Google Scholar]
- 5.Berrocal-Tito, G. M., T. Rosales-Saavedra, A. Herrera-Estrella, and B. A. Horwitz. 2000. Characterization of blue-light and developmental regulation of the photolyase gene phr1 in Trichoderma harzianum. Photochem. Photobiol. 71:662-668. [DOI] [PubMed] [Google Scholar]
- 6.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casas-Flores, S., M. Rios-Momberg, M. Bibbins, P. Ponce-Noyola, and A. Herrera-Estrella. 2004. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150:3561-3569. [DOI] [PubMed] [Google Scholar]
- 8.Crosson, S., S. Rajagopal, and K. Moffat. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42:2-10. [DOI] [PubMed] [Google Scholar]
- 9.Dijkwel, P. P., C. Huijser, P. J. Weisbeek, N. H. Chua, and S. C. Smeekens. 1997. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9:583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebbole, D. J. 1998. Carbon catabolite repression of gene expression and conidiation in Neurospora crassa. Fungal Genet. Biol. 25:15-21. [DOI] [PubMed] [Google Scholar]
- 11.Franchi, L., V. Fulci, and G. Macino. 2005. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 56:334-345. [DOI] [PubMed] [Google Scholar]
- 12.Gehring, W., and M. Rosbash. 2003. The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol. 57:286-289. [DOI] [PubMed] [Google Scholar]
- 13.Gresik, M., N. Kolarova, and V. Farkas. 1988. Membrane potential, ATP, and cyclic AMP changes induced by light in Trichoderma viride. Exp. Mycol. 12:295-301. [Google Scholar]
- 14.Gresik, M., N. Kolarova, and V. Farkas. 1989. Light-stimulated phosphorylation of proteins in cell-free extracts from Trichoderma viride. FEBS Lett. 248:185-187. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y. Z., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 16.He, Q., P. Cheng, Y. Yang, L. Wang, K. H. Gardner, and Y. Liu. 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297:840-843. [DOI] [PubMed] [Google Scholar]
- 17.Herrera-Estrella, A., and I. Chet. 1998. Biocontrol of bacteria and phytopathogenic fungi, p. 263-282. In A. Altman (ed.), Agricultural bio/technology. Marcel Dekker, Inc., New York, N.Y.
- 18.Hill, E. P. 1976. Effect of light on growth and sporulation of Aspergillus ornatus. J. Gen. Microbiol. 95:39-44. [DOI] [PubMed] [Google Scholar]
- 19.Horenstein, E. A., and E. C. Cantino. 1964. An effect of light on glucose uptake by the fungus Blastocladiella britannica. J. Gen. Microbiol. 37:59-65. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz, B. A., S. Malkin, and J. Gressel. 1984. Roseoflavin inhibition of photoconidiation in a Trichoderma riboflavin auxotroph: indirect evidence for flavin requirement for photoreactions. Photochem. Photobiol. 40:763-769. [DOI] [PubMed] [Google Scholar]
- 21.Idnurm, A., and J. Heitman. 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. Plos. Biol. 3:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivey, F. D., Q. Yang, and K. A. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a galpha i homolog in Neurospora crassa. Fungal Genet. Biol. 26:48-61. [DOI] [PubMed] [Google Scholar]
- 24.Kolarova, N., J. Haplova, and M. Gresik. 1992. Light-activated adenyl cyclase from Trichoderma viride. FEMS Microbiol. Lett. 72:275-278. [DOI] [PubMed] [Google Scholar]
- 25.Kronstad, J., A. D. De Maria, D. Funnell, R. D. Laidlaw, N. Lee, M. M. de Sa, and M. Ramesh. 1998. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch. Microbiol. 170:395-404. [DOI] [PubMed] [Google Scholar]
- 26.Lauter, F. 1996. Molecular genetics of fungal photobiology. J. Genet. 75:375-386. [Google Scholar]
- 27.Lin, C. 2002. Blue light receptors and signal transduction. Plant Cell 14:207-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linden, H., P. Ballario, and G. Macino. 1997. Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 22:141-150. [DOI] [PubMed] [Google Scholar]
- 29.Lu, Y. K., K. H. Sun, and W. C. Shen. 2005. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol. Microbiol. 56:480-491. [DOI] [PubMed] [Google Scholar]
- 30.Nemcovic, M., and V. Farkas. 1998. Stimulation of conidiation by derivatives of cAMP in Trichoderma viride. Folia Microbiol. 43:399-402. [Google Scholar]
- 31.Oesch-Bartlomowicz, B., A. Huelster, O. Wiss, P. Antoniou-Lipfert, C. Dietrich, M. Arand, C. Weiss, E. Bockamp, and F. Oesch. 2005. Aryl hydrocarbon receptor activation by cAMP versus dioxin: divergent signaling pathways. Proc. Natl. Acad. Sci. USA 102:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rau, W., and Mitzka-Schnabel, U. 1985. Carotenoid synthesis in Neurospora crassa. Methods Enzymol. 110:253-267. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, N.Y.
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreckenbach, T., B. Walckhoff, and C. Verfuerth. 1981. Blue-light receptor in a white mutant of Physarum polycephalum mediates inhibition of spherulation and regulation of glucose metabolism. Proc. Natl. Acad. Sci. USA 78:1009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwerdtfeger, C., and H. Linden. 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22:4846-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott, W. A., and B. Solomon. 1975. Adenosine 3′,5′-cyclic monophosphate and morphology in Neurospora crassa: drug-induced alterations. J. Bacteriol. 122:454-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, M. N., O. Pelegrini, M. Veron, and R. R. Kay. 1992. Mutation of protein kinase A causes heterochronic development of Dictyostelium. Nature 356:171-172. [DOI] [PubMed] [Google Scholar]
- 40.Sokolovsky, V., F. Lauter, B. Mueller-Roeber, M. Ricci, T. Shmidhauser, and V. Russo. 1992. Nitrogen regulation of blue light-inducible genes in Neurospora crassa. J. Gen. Microbiol. 138:2045-2049. [Google Scholar]
- 41.Spector, T. 1978. Refinement of the Coomassie blue method of protein quantitation: a simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal. Biochem. 86:142-146. [DOI] [PubMed] [Google Scholar]
- 42.Sulová, Z., and Farkás, V. 1991. Photoinduced conidiation in Trichoderma viride: a study with inhibitors. Folia Microbiol. 36:267-270. [Google Scholar]
- 43.Talora, C., L. Franchi, H. Linden, P. Ballario, and G. Macino. 1999. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 18:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terashima, K., K. Yuki, H. Muraguchi, M. Akiyama, and T. Kamada. 2005. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics 171:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeilinger, S., C. Galhaup, K. Payer, S. Woo, R. Mach, C. Fekete, M. Lorito, and C. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]