Abstract
Assembly of active Fe-hydrogenase in the chloroplasts of the green alga Chlamydomonas reinhardtii requires auxiliary maturases, the S-adenosylmethionine-dependent enzymes HydG and HydE and the GTPase HydF. Genes encoding homologous maturases had been found in the genomes of all eubacteria that contain Fe-hydrogenase genes but not yet in any other eukaryote. By means of proteomic analysis, we identified a homologue of HydG in the hydrogenosomes, mitochondrion-related organelles that produce hydrogen under anaerobiosis by the activity of Fe-hydrogenase, in the pathogenic protist Trichomonas vaginalis. Genes encoding two other components of the Hyd system, HydE and HydF, were found in the T. vaginalis genome database. Overexpression of HydG, HydE, and HydF in trichomonads showed that all three proteins are specifically targeted to the hydrogenosomes, the site of Fe-hydrogenase maturation. The results of Neighbor-Net analyses of sequence similarities are consistent with a common eubacterial ancestor of HydG, HydE, and HydF in T. vaginalis and C. reinhardtii, supporting a monophyletic origin of Fe-hydrogenase maturases in the two eukaryotes. Although Fe-hydrogenases exist in only a few eukaryotes, related Narf proteins with different cellular functions are widely distributed. Thus, we propose that the acquisition of Fe-hydrogenases, together with Hyd maturases, occurred once in eukaryotic evolution, followed by the appearance of Narf through gene duplication of the Fe-hydrogenase gene and subsequent loss of the Hyd proteins in eukaryotes in which Fe-hydrogenase function was lost.
Metallocatalytic hydrogenases occur as two distinct enzymes, known as NiFe- and Fe-hydrogenases, according to the type of their catalytic active sites (33). Heterodimeric NiFe-hydrogenases contain a bimetallic NiFe cluster at the hydrogen-activating site of the large subunit, while the small subunit contains up to three iron-sulfur (FeS) centers transporting electrons to the physiological acceptors. NiFe-hydrogenases are predominantly involved in H2 uptake in archaebacteria and eubacteria (7). In contrast, the mostly monomeric Fe-hydrogenases usually catalyze H2 evolution (12). These enzymes are found in anaerobic eubacteria (33), green algae (11, 15), and various anaerobic eukaryotes, among them chytridiomycetes (34), anaerobic ciliates (26), and amitochondriate protists such as the trichomonads (27), diplomonads, and Entamoeba histolytica (24). The hydrogen-activating center of Fe-hydrogenases is known as the H cluster and consists of a 4Fe4S cluster that is connected via a cysteinyl bridge to a binuclear 2Fe center (33). The H cluster is the only metallic FeS cluster in green algal Fe-hydrogenases, while the N-terminal domains of Fe-hydrogenases in other eukaryotes contain various numbers of additional, ferredoxin-like 2Fe2S clusters and 4Fe4S clusters (14, 17). Like the small subunit of NiFe-hydrogenases, the N-terminal domains of Fe-hydrogenases conduct electrons between the donors and the H2-activating center.
Synthesis and insertion of the metal clusters at the active centers of NiFe-hydrogenase apoproteins and proteolytic processing after completion of the active centers in bacteria require the action of an independent system of numerous auxiliary proteins (7, 33). In Escherichia coli at least seven proteins, HypABCDEF and HycI, are involved (19); similar systems are active in Helicobacter pylori and Ralstonia eutropha (6, 25). In contrast, no homologues of Hyp proteins are encoded in the genomes of bacteria that contain Fe-hydrogenases only (33). As NiFe- and Fe-hydrogenases are phylogenetically independent proteins, it is conceivable that their maturation is achieved by independent auxiliary proteins. Maturation of the Fe-hydrogenase in the chloroplasts of the green alga Chlamydomonas reinhardtii was only recently shown to require HydG, an S-adenosylmethionine (SAM)-dependent enzyme, and the fusion protein HydEF, another SAM-dependent protein (HydE) fused with a GTPase (HydF) (28). Genes for homologues of all three Hyd proteins were detected, without exception, in the genomes of eubacteria with Fe-hydrogenases. In some prokaryotic genomes these hyd genes were organized in putative operons together with the Fe-hydrogenase structural genes (28). Coexistence of the HydE, HydF, and HydG proteins with Fe-hydrogenase in prokaryotes and C. reinhardtii strongly suggests that their presence is mandatory for the maturation of Fe-hydrogenase. However, no HydE, HydF, or HydG homologues other than those from C. reinhardtii had been characterized to date.
The bulk of Fe-hydrogenase activity in the green algae is located in the chloroplasts, while in T. vaginalis it has been detected in the hydrogenosomes (32). Hydrogenosomes are hydrogen-producing organelles found in several ciliate lineages, anaerobic chytridiomycetes, and parabasalid flagellates (11). Like mitochondria, the hydrogenosomes generate ATP using pyruvate or malate as substrates; however, ATP is synthesized exclusively by substrate-level phosphorylation. The key steps in this process are mediated by various FeS proteins, including pyruvate-ferredoxin oxidoreductase (PFOR), NADH dehydrogenase, and 2Fe2S ferredoxin (18, 23). The maturation of ferredoxin and probably other FeS proteins is dependent on hydrogenosomal FeS cluster (ISC) assembly machinery similar to that recognized in mitochondria (31). Unlike mitochondria, the anaerobic hydrogenosomes possess Fe-hydrogenases, which serve as the terminal electron acceptors, evolving molecular hydrogen (22). Virtually no information is available about formation of the metallic clusters in these enzymes, although Fe-hydrogenases play a fundamental role in hydrogenosomal metabolism. Here we report the identification of three putative Fe-hydrogenase maturases (HydG, HydE, and HydF) in the hydrogenosomes of the human parasite T. vaginalis with homology to those from C. reinhardtii and Fe-hydrogenase-possessing anaerobic eubacteria.
MATERIALS AND METHODS
Organisms.
Trichomonas vaginalis strain C1:NIH (ATCC 3001) and strain T1 (J.-H. Tai, Institute of Biomedical Sciences, Taipei, Taiwan) were grown as described previously (8).
Cell fractionation, 2D electrophoresis, and mass spectrometry.
Hydrogenosomes were isolated and hydrogenosomal proteins were identified by two-dimensional (2D) electrophoresis and mass spectrometry, as described previously (29).
Selectable transformation of T. vaginalis.
HydE, HydF, and HydG open reading frames (ORFs) were amplified and introduced into plasmid Tagvag2 (18) between an α-succinyl coenzyme A synthetase (α-SCS) 5′ untranslated region and a 3′ hemagglutinin (HA)2 tag to yield pTvhydE, pTvhydF, and pTvhydG. T. vaginalis T1 cells were electroporated with pTvhydE, pTvhydF, or pTvhydG and selected in Trypticase-yeast extract-maltose medium supplemented with G418 (Sigma).
Immunofluorescence microscopy.
Hyd proteins and malic enzyme (ME) were visualized in T. vaginalis cells with mouse anti-HA monoclonal antibody (Sigma) and rabbit anti-ME polyclonal antibody (9) as primary antibodies and secondary Alexa Fluor-488 donkey anti-mouse (green) and Alexa Fluor-594 donkey anti-rabbit (red) antibodies (Molecular Probes, Eugene, OR), as described previously (31).
RT-PCR.
Reverse transcriptase (RT) PCR was used to detect Hyd maturase transcripts in T. vaginalis. Total RNA was isolated with the QuickPrep total RNA extraction kit, and DNase-treated RNA was tested for the presence of genomic DNA contamination by PCR. cDNA was synthesized by a standard protocol with the SuperScriptII First-Strand Synthesis System for RT-PCR (Invitrogen). Amplification of cDNA by PCR was performed with primers specific for HydE (5′-TGCCATTATTGCGGTGTT-3′, 5′-GCGGCCCATCTTTGTATC-3′), HydF (5′-TCTACACCCGGAACTACA-3′, 5′-TCTTTTGCGCTTTGATAA-3′), and HydG (5′-CGTCGAAATCCCAACACT-3′, 5′-AACAGCGCAACGAAGAAC-3′).
Computational methods.
pI and molecular weight (MW) were calculated with the ExPASy pI/MW tool (http://au.expasy.org/tools/pi_tool.html). Neighbor-Net analyses of sequence similarities were carried out as described previously (13). The alignment of HydG and HydE protein sequences contained 696 sites, with 257 sites left in the analysis after removal of gaps. The alignment of HydF sequences contained 724 residues, of which 263 were left after removal of gaps. Accession numbers of sequences used in the analyses are available as supplemental information.
RESULTS
Homologues of the putative Fe-hydrogenase maturases HydG, HydE, and HydF in the hydrogenosomes of T. vaginalis.
Two hundred fifty randomly chosen hydrogenosomal proteins from T. vaginalis were sequenced by mass spectrometry (29). Sequences of tryptic peptides from protein S29 corresponded to ORF 88811.m00081 in the T. vaginalis genome (www.tigr.org), encoding a protein of an estimated 58 kDa and pI 8.8. A second, incomplete, ORF (96484.m00069) encoded an almost identical protein but lacked 71 amino acids at the C terminus, including the peptide EVQNITDEK (Fig. 1). The protein encoded by ORF 88811.m00081 predicted a significantly larger MW and lower pI than those determined for S29 on 2D sodium dodecyl sulfate-polyacrylamide gels (data not shown). As the peptide sequences acquired by mass spectrometry all corresponded to the C-terminal half of ORF 88811.m00081 (Fig. 1), S29 on the 2D gel probably contained only a C-terminal fragment of the complete protein. When the N-terminal 150 amino acids were removed from the sequence encoded by ORF 88811.m00081 to generate a protein with a calculated size of 41 kDa, approximately matching the apparent size of S29, the predicted pI of that C-terminal fragment shifted to 9.2, corresponding to the observed pI of spot S29 on the 2D gels. The recombinant HydG protein with a C-terminal HA tag in overexpressing T. vaginalis cells had the expected size in a Western blot, and a minor band of the size of protein spot S29 was also recognized (not shown). Thus, the protein seems to be fragile, and the C-terminal fragment on the 2D gel most likely is an artifact caused by the isoelectric-focusing sample preparation procedure.
FIG. 1.
Alignment of HydG sequences from T. vaginalis (Triva), C. reinhardtii (Chlre), Clostridium thermocellum (Cloth), and Bacteroides thetaiotaomicron (Bacth). A putative hydrogenosomal targeting sequence is shown in bold, and peptide sequences from hydrogenosomal protein spot S29 are shaded in gray. ▿, cysteine residues of the CXXXCXXC motif conserved in SAM-dependent proteins (30); *, conserved sites.
A BLAST search (blastp) of the GenBank nonredundant database with the conceptual translation of ORF 88811.m00081 as a query retrieved HydG from C. reinhardtii as the best hit, with 54% amino acid identity, as well as multiple prokaryotic sequences annotated as ThiH, an enzyme involved in bacterial thiamine biosynthesis (4). The prokaryotic sequences were up to 49% identical to the hydrogenosomal protein. A BLAST search of the T. vaginalis genome database (www.tigr.org) for homologues of HydE and HydF with the HydEF fusion protein from C. reinhardtii as a query identified independent ORFs for both HydE (81202.m00098, 91566.m00125, and 93793.m00242, all encoding proteins of about 44 kDa) and HydF (87122.m00057, encoding a protein of 48 kDa). All genes in the genome encoding HydG, HydE, and HydF homologues are expressed, as their transcripts were either represented in the expressed sequence tag database (Petrus Tang, Chang Gung University, Taiwan) or detected by RT-PCR (HydE, 81202.m00098; HydF, 87122.m00057) (Fig. 2). These data confirm the presence of the complete set of proteins in T. vaginalis that had previously been characterized as maturases of Fe-hydrogenase in C. reinhardtii.
FIG. 2.
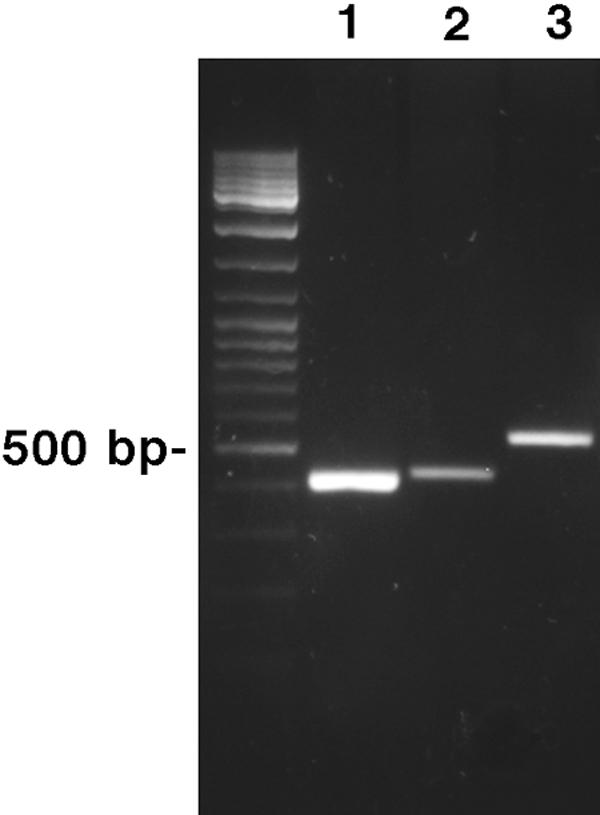
Detection of Hyd gene transcripts by RT-PCR. 1, HydG (88811.m00081); 2, HydF (87122.m00057); 3, HydE (81202.m00098).
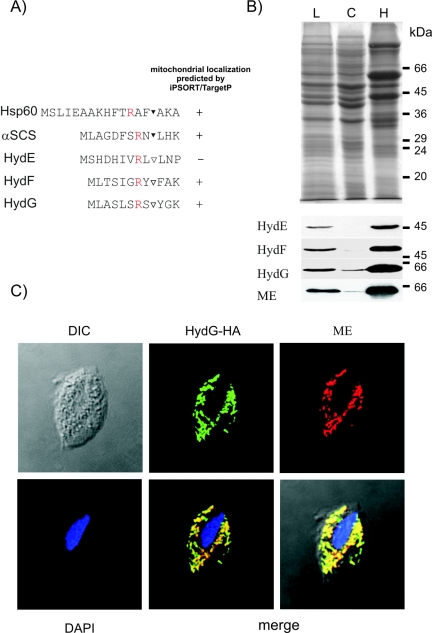
Targeting and translocation of proteins into hydrogenosomes is dependent on the presence of N-terminal presequences, which are cleaved off during protein maturation in the organellar matrix (5). Indeed, the deduced N termini of the HydG homologue that was identified in the hydrogenosomal proteome, as well as of HydE and HydF homologues retrieved from the T. vaginalis genome database, strongly resembled the targeting sequences of proteins known to reside in the hydrogenosomes of T. vaginalis (Fig. 3A). Although usually much shorter, hydrogenosomal targeting sequences resemble those of their relatives, the mitochondria, in several respects (5). Based on this similarity we analyzed the N termini of the Hyd proteins as well as of two hydrogenosomal proteins, Hsp60 and α-SCS, with the protein localization prediction programs TargetP (www.cbs.dtu.dk/services/TargetP/) and iPSORT (http://psort.nibb.ac.jp/). Although these programs were designed to detect targeting signals for import into mitochondria rather than the related hydrogenosomes, they both predicted the N termini of hydrogenosomal Hsp60 and α-SCS, as well as HydG and HydF, to be mitochondrial import signals, thus recognizing these proteins as organelle targeted (Fig. 3A). The third putative maturase, HydE, was not recognized as an organellar protein by either program. To prove localization of HydE, HydF, and HydG in the hydrogenosomes, HA-tagged recombinants of the proteins were expressed in T. vaginalis T1 cells. Western blots of subcellular fractions from transfected cells probed with monoclonal anti-HA antibody detected all three proteins in the hydrogenosomal fractions (Fig. 3B). In addition, immunofluorescence microscopy of T. vaginalis cells expressing recombinant HydG proved colocalization of HydG with ME, the biochemical marker of hydrogenosomes (Fig. 3C).
FIG. 3.
Intracellular localization of Hyd proteins in T. vaginalis. (A) Comparison of the N-terminal targeting sequences of the hydrogenosomal proteins Hsp60 and α-SCS with the N-terminal sequences of Hyd proteins. ▾, protease processing site; ▿, putative protease processing site. The typical arginine residue (R) at position −2 of the protease processing site of hydrogenosomal targeting sequences is shown in red. (B) Localization of HA-tagged HydE, HydF, and HydG overexpressed in T. vaginalis. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of HydF-overexpressing cells (top) and Western blots of cellular fractions from cells expressing tagged HydE, HydF, or HydG (bottom) are shown. ME was used as a hydrogenosomal marker protein. L, cell lysate; C, cytosolic fraction; H, hydrogenosomal fraction. (C) Immunolocalization of HydG in transfected cells shows colocalization of HA-tagged HydG with hydrogenosomal ME. DIC, differential interference contrast image; DAPI, 4′,6′-diamidino-2-phenylindole-stained image.
Phylogenetic relationships of Hyd proteins.
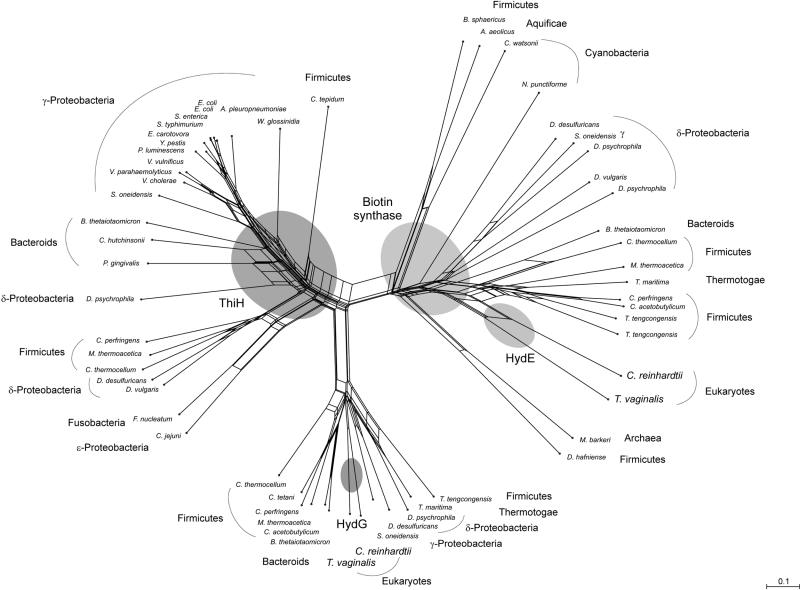
Relationships of HydG, HydE, and HydF from T. vaginalis and C. reinhardtii with similar proteins from numerous prokaryotes were assessed in Neighbor-Net analyses. HydG and HydE were related to different members of the SAM-dependent enzyme family (Fig. 4). HydG from T. vaginalis and C. reinhardtii were most similar to eubacterial sequences annotated as ThiH, a protein involved in thiamine biosynthesis in prokaryotes. HydE appeared in a cluster of eubacterial sequences annotated as biotin synthase. The eukaryotic HydG sequences belonged to a divergent subgroup of ThiH sequences with an overall amino acid identity of 47%. This cluster was separated by a strong split from all other ThiH and biotin synthase homologues in the analysis. Amino acid identity was 35% with the main ThiH cluster and 26% with the biotin synthase cluster. Only HydG from C. reinhardtii was functionally characterized in heterologous coexpression experiments (28), while all prokaryotic sequences of the cluster were annotated as “putative ThiH” or “similar to ThiH” solely on the basis of sequence similarity. HydG from C. reinhardtii and its homologue from T. vaginalis were separated from all other sequences by a common split and showed no specific affinity to any of the neighboring clusters of gram-positive bacteria or γ- and δ-proteobacteria in the subgroup.
FIG. 4.
Neighbor-Net analysis of sequence similarities of HydG and HydE and related proteins from prokaryotes. HydG sequences from T. vaginalis and C. reinhardtii form a monophyletic cluster within a subset of prokaryotic ThiH sequences. HydE sequences from both eukaryotes appear on a common branch among prokaryotic biotin synthases.
HydE from T. vaginalis and the N-terminal half of C. reinhardtii HydEF, corresponding to HydE, formed a cluster with prokaryotic sequences annotated as biotin synthases with an overall amino acid identity of 29%. Again, the two eukaryotic sequences were grouped together, with a mixed group of gram-positive bacteria and γ- and δ-proteobacteria as their closest neighbors.
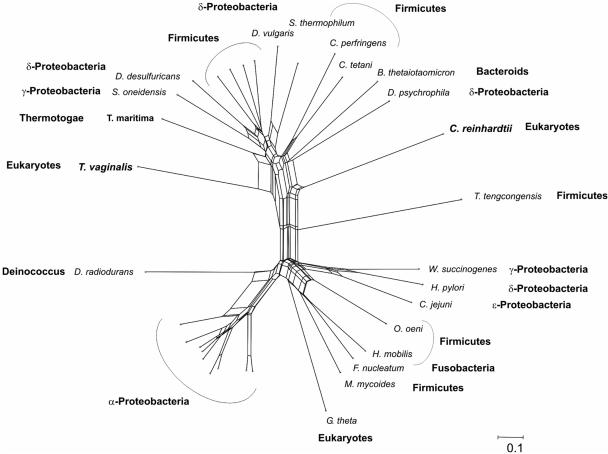
HydF from C. reinhardtii and its homologue from T. vaginalis belong to a family of putative GTPases (Fig. 5). In the sequence similarity network, both eukaryotic sequences are members of an unresolved subgroup of eubacterial sequences that is separated by a strong split from another subgroup of prokaryotic GTPases containing a third eukaryotic sequence from the nucleomorph of the cryptomonad Guillardia theta.
FIG. 5.
Neighbor-Net analysis of HydF and related GTPases. Sequences from T. vaginalis and C. reinhardtii appear in the same subset of GTPases but do not form a monophyletic group.
DISCUSSION
We have identified three proteins with high similarity to the Fe-hydrogenase maturases HydG and HydEF from C. reinhardtii in the hydrogenosomes of T. vaginalis, the cellular compartments which also harbor the target of their putative function, Fe-hydrogenase (22). Colocalization of HydG, HydE, and HydF with Fe-hydrogenase in the hydrogenosomes strongly supports our hypothesis that these three proteins are required for the synthesis of active Fe-hydrogenase, as demonstrated for C. reinhardtii homologues. Fe-hydrogenase of C. reinhardtii contains no FeS clusters other than the catalytic H cluster. Thus, the Hyd proteins must be involved in the synthesis and/or insertion of this specific cluster. In contrast, at least four different types of Fe-hydrogenases are encoded in the genome of T. vaginalis; they contain, in addition to the H cluster, various numbers of additional, but less-complex, FeS clusters in their N-terminal domains (references 14, 16, and 17 and our unpublished data). This raises the question of whether the Hyd proteins are responsible for the generation of all FeS clusters of Fe-hydrogenase in T. vaginalis or whether they are required only for assembly of the H cluster, while the additional FeS clusters are synthesized by a distinct mitochondrial-type ISC system. In mitochondria, formation of FeS clusters is mediated by an ISC assembly machinery that consists of at least 10 distinct proteins (20). The main components of this machinery were recently found in T. vaginalis hydrogenosomes, and their involvement in the formation of the 2Fe2S clusters of ferredoxin was demonstrated (31). Thus, it is likely that both the ISC and Hyd systems contribute to FeS cluster formation in Fe-hydrogenases in T. vaginalis, although the particular roles of both systems remain to be elucidated.
The presence of Hyd maturases can also be expected in other anaerobic eukaryotes with Fe-hydrogenase activity. Likely candidates are the anaerobic, hydrogenosome-bearing ciliates and chytridiomycetes, as they have previously been demonstrated to generate H2 in hydrogenosomes with the activity of Fe-hydrogenases (1, 34). Interestingly, two other anaerobic eukaryotes, E. histolytica (24) and Giardia intestinalis (21), have recently been reported to exhibit low levels of H2 production, although they both lack hydrogenosomes. The completely sequenced genomes of both E. histolytica and G. intestinalis contain several genes with strong similarity to Fe-hydrogenases, but contrary to our expectations we were not able to discover homologues of either HydG, HydE, or HydF in the respective genome databases. Notably, while C. reinhardtii Fe-hydrogenase was obtained in its active form only when coexpressed in E. coli with both HydG and HydEF (28), the Fe-hydrogenase from E. histolytica was reportedly active in the heterologous system without coexpression of auxiliary proteins (24). Thus, Fe-hydrogenase maturation in E. histolytica, and most likely in G. intestinalis, may be achieved by a mode different from that in green algae and possibly in T. vaginalis.
Neighbor-Net analysis to determine the origins of eukaryotic HydG and HydE proteins shows that the proteins belong to different subfamilies of SAM-dependent proteins (Fig. 4). C. reinhardtii HydG and the homologue from T. vaginalis form a cluster with prokaryotic sequences annotated as ThiH, which is highly divergent from the other major cluster of prokaryotic ThiH sequences in the network. As C. reinhardtii HydG is the only functionally characterized protein in this cluster (28), and all organisms in the cluster contain Fe-hydrogenases, it is likely that these proteins are all active as HydG rather than ThiH. HydE from C. reinhardtii and its homologue from T. vaginalis cluster with prokaryotic sequences mostly annotated as putative biotin synthases or biosynthase-like proteins, but in this case no apparent separation between putative HydE proteins and biotin synthases was observed. The eukaryotic HydG and HydE sequences from T. vaginalis and C. reinhardtii are most similar to their respective orthologues in the other eukaryote, to the exclusion of all prokaryotic sequences in the data set, a topology that is consistent with a common origin of the HydG and HydE proteins in T. vaginalis and C. reinhardtii. Due to the low resolution of prokaryotic groups in the network, a specific prokaryotic source of the eukaryotic homologues cannot be identified. Although the independent proteins HydG and HydE behaved similarly in our analysis, we cannot exclude the possibility that the observed monophyly of HydG and HydE from Trichomonas and Chlamydomonas is an artifact caused by the small sample size until more eukaryotic sequences become available.
The third putative set of members of the maturase system, the HydF GTPases from T. vaginalis and C. reinhardtii, again appear in the same subgroup of GTPases but are not clustered together as a monophyletic group. Another eukaryotic sequence, from the nucleomorph of the red algal endosymbiont of the cryptomonad G. theta, an organism that does not contain Fe-hydrogenase, appears in a different subgroup of the GTPase network and probably is involved in processes different from those of HydF.
Whether the unique anaerobic enzymes of hydrogenosomal metabolism, PFOR and Fe-hydrogenase, are remnants of the eubacterial ancestor of mitochondria and hydrogenosomes or are independent acquisitions in adaptation to an anaerobic lifestyle is an unanswered question. Previous analyses did not reject a common origin for PFOR and Fe-hydrogenase in eukaryotes, but due to poor sampling and low resolution of the gene trees, the eubacterial sources of these genes could not be identified (10, 14, 17). HydG, HydE, and HydF, like the Fe-hydrogenases, are clearly of eubacterial origin, as virtually no archaebacterial homologues, with the exception of M. barkeri biotin synthase, were detected in the databases. Only one α-proteobacterial Fe-hydrogenase, from Rhodospseudomonas palustris, which does not show a close relationship to eukaryotic homologues, is available to date (10), but no α-proteobacterial sequences similar to HydG and HydE, not even in R. palustris, have been found. Accordingly, Fe-hydrogenase and HydG, HydE, and HydF in eukaryotes are clearly of eubacterial origin, but there is currently no evidence that this eubacterium was the α-proteobacterial ancestor of mitochondria. The present data support monophyly of HydG and HydE in the two eukaryotes. As Fe-hydrogenase function is dependent on the activity of the maturases, it is likely that hydrogenase and maturases were acquired together in a single event. Homologues of HydG, HydE, and HydF in eubacteria are often organized in operons together with their target enzyme, Fe-hydrogenase (28), so that acquisition of an entire operon might have ensured activity of Fe-hydrogenase in the eukaryotic lineage in that event. Although Fe-hydrogenases exist in only a few distant eukaryotic lineages, closely related proteins called Narf, which are involved in different cellular functions, such as prelamin binding in the nucleus (3) and FeS cluster synthesis in the cytosol (2), are widely distributed among eukaryotes. Based on these observations we propose that the acquisition of Fe-hydrogenases together with Hyd maturases occurred once in eukaryotic evolution, followed by the appearance of Narf through gene duplication of the Fe-hydrogenase gene and subsequent loss of the Hyd proteins in eukaryotes in which Fe-hydrogenase function was lost.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to K.H. (He2951/3-2) and from the Grant Agency of the Czech Republic to J.T. (204/04/0435).
Genomic sequences of HydG, HydE, and HydF were acquired from the T. vaginalis genome database provided by TIGR. We thank Miklos Müller for helpful comments.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akhmanova, A. S., F. G. J. Voncken, T. A. van Alen, A. H. A. M. van Hoek, B. Boxma, F. G. Volgels, M. Veenhuis, and J. H. P. Hackstein. 1998. A hydrogenosome with a genome. Nature 396:527-528. [DOI] [PubMed] [Google Scholar]
- 2.Balk, J., A. J. Pierik, D. J. Aguilar Netz, U. Mühlenhoff, and R. Lill. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, R. M., and H. J. Worman. 1999. Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 42:30008-30018. [DOI] [PubMed] [Google Scholar]
- 4.Begley, T. P., D. M. Downs, S. E. Ealick, F. W. McLafferty, A. P. G. M. Van Loon, S. Taylo, N. Campobasso, H.-J. Chiu, C. Kinsland, J. J. Reddick, and J. Xi. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171:293-300. [DOI] [PubMed] [Google Scholar]
- 5.Bradley, P. J., C. J. Lahti, E. Plumper, and P. J. Johnson. 1997. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 16:3484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhrke, T., B. Bleijlevens, S. P. J. Albracht, and B. Friedrich. 2001. Involvement of hyp gene products in maturation of the H2-sensing [NiFe] hydrogenase of Ralstonia eutropha. J. Bacteriol. 183:7087-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe]-hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. G., and L. S. Diamond. 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 15:329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drmota, T., P. Proost, M. van Ranst, F. Weyda, J. Kulda, and J. Tachezy. 1996. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol. Biochem. Parasitol. 83:221-234. [DOI] [PubMed] [Google Scholar]
- 10.Embley, T. M., M. van der Giezen, D. S. Horner, P. L. Dyal, S. Bell, and P. G. Foster. 2003. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life 55:387-395. [DOI] [PubMed] [Google Scholar]
- 11.Florin, L., A. Tsokoglou, and T. Happe. 2001. A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J. Biol. Chem. 276:6125-6132. [DOI] [PubMed] [Google Scholar]
- 12.Frey, M. 2002. Hydrogenases: hydrogen-activating enzymes. Chembiochem 3:153-160. [DOI] [PubMed] [Google Scholar]
- 13.Gelius-Dietrich, G., and K. Henze. 2004. Pyruvate formate lyase (PFL) and PFL activating enzyme in the chytrid fungus Neocallimastix frontalis: a free-radical enzyme system conserved across divergent eukaryotic lineages. J. Eukaryot. Microbiol. 51:456-463. [DOI] [PubMed] [Google Scholar]
- 14.Hackstein, J. H. P. 2005. Eukaryotic Fe-hydrogenases—old eukaryotic heritage or adaptive acquisitions. Biochem. Soc. Trans. 33:47-50. [DOI] [PubMed] [Google Scholar]
- 15.Happe, T., B. Mosler, and D. Naber. 1994. Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222:769-774. [DOI] [PubMed] [Google Scholar]
- 16.Horner, D. S., P. G. Foster, and T. M. Embley. 2000. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 17:1695-1709. [DOI] [PubMed] [Google Scholar]
- 17.Horner, D. S., B. Heil, T. Happe, and T. M. Embley. 2002. Iron hydrogenases—ancient enzymes in modern eukaryotes. Trends Biochem. Sci. 27:148-153. [DOI] [PubMed] [Google Scholar]
- 18.Hrdy, I., R. P. Hirt, P. Dolezal, L. Bardonová, P. G. Foster, J. Tachezy, and T. M. Embley. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618-622. [DOI] [PubMed] [Google Scholar]
- 19.Hube, M., M. Blokesch, and A. Böck. 2002. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J. Bacteriol. 184:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill, R., and U. Mühlenhoff. 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30:133-141. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd, D., J. R. Ralphs, and J. C. Harris. 2002. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiology 48:727-733. [DOI] [PubMed] [Google Scholar]
- 22.Müller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879-2889. [DOI] [PubMed] [Google Scholar]
- 23.Müller, M. 2003. Energy metabolism. Part I: anaerobic protozoa, p. 125-139. In J. J. Marr, T. W. Nilsen, and R. W. Komuniecki (ed.), Molecular medical parasitology. Academic Press, New York, N.Y.
- 24.Nixon, J. E. J., J. Field, A. G. McArthur, M. L. Sogin, N. Yarlett, B. J. Loftus, and J. Samuelson. 2003. Iron-dependent hydrogenases of Entamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer. Biol. Bull. 204:1-9. [DOI] [PubMed] [Google Scholar]
- 25.Olson, J., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 26.Paul, R. G., A. G. Williams, and R. D. Butler. 1990. Hydrogenosomes in the rumen entodiniomorphid ciliate Polyplastron multivesiculatum. J. Gen. Microbiol. 136:1981-1989. [DOI] [PubMed] [Google Scholar]
- 27.Payne, M. J., A. Chapman, and R. Cammack. 1993. Evidence for an Fe-type hydrogenase in parasitic protozoan Trichomonas vaginalis. FEBS Lett. 317:101-104. [DOI] [PubMed] [Google Scholar]
- 28.Posewitz, M. C., P. W. King, S. L. Smolinski, L. Zhang, M. Seibert, and M. L. Ghirardi. 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active Fe hydrogenase. J. Biol. Chem. 279:25711-25720. [DOI] [PubMed] [Google Scholar]
- 29.Pütz, S., G. Gelius-Dietrich, M. Piotrowski, and K. Henze. 2005. Rubrerythrin and peroxiredoxin: two novel putative peroxidases in the hydrogenosomes of the microaerophilic protozoon Trichomonas vaginalis. Mol. Biochem. Parasitol. 142:212-223. [DOI] [PubMed] [Google Scholar]
- 30.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutak, R., P. Dolezal, H. L. Fiumera, I. Hrdy, A. Dancis, M. Delgadillo-Correa, P. J. Johnson, M. Müller, and J. Tachezy. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 101:10368-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Giezen, M., and J. Tovar. 2005. Degenerate mitochondria. EMBO Rep. 6:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vignais, P. V., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 34.Voncken, F. G. J., B. Boxma, A. Akhmanova, F. G. Vogels, M. Huynen, M. Veenhuis, and J. H. P. Hackstein. 2002. A hydrogenosomal Fe-hydrogenase from the anaerobic chytrid Neocallimastix sp. L2. Gene 284:103-112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.