Abstract
Using mutational and proteomic approaches, we have demonstrated the importance of the glycosylphosphatidylinositol (GPI) anchor pathway for cell wall synthesis and integrity and for the overall morphology of the filamentous fungus Neurospora crassa. Mutants affected in the gpig-1, gpip-1, gpip-2, gpip-3, and gpit-1 genes, which encode components of the N. crassa GPI anchor biosynthetic pathway, have been characterized. GPI anchor mutants exhibit colonial morphologies, significantly reduced rates of growth, altered hyphal growth patterns, considerable cellular lysis, and an abnormal “cell-within-a-cell” phenotype. The mutants are deficient in the production of GPI-anchored proteins, verifying the requirement of each altered gene for the process of GPI-anchoring. The mutant cell walls are abnormally weak, contain reduced amounts of protein, and have an altered carbohydrate composition. The mutant cell walls lack a number of GPI-anchored proteins, putatively involved in cell wall biogenesis and remodeling. From these studies, we conclude that the GPI anchor pathway is critical for proper cell wall structure and function in N. crassa.
In eukaryotic cells, a number of proteins are anchored to the outer leaflet of the plasma membrane via glycosylphosphatidylinositol (GPI) anchors. The presence of the GPI anchor is thought to play an important role in the trafficking of these proteins and providing them with an attachment to the plasma membrane, and in the case of fungi, to the cell wall as well (24, 41). Proteins destined to receive a GPI anchor are directed into the lumen of the endoplasmic reticulum (ER) by a typical signal peptide. The carboxyl termini of these proteins have a sequence motif that is recognized by a protein complex located in the ER, known as the GPI transamidase. The GPI transamidase complex cleaves the substrate protein at a position within this motif, termed the omega site, and transfers the GPI anchor en bloc to the newly generated C terminus of the protein.
The structures of the GPI anchor in the trypanosome, yeast, and mammalian systems have been determined. Although there are differences in the various substituents present on the GPI anchors produced by these organisms, all GPI anchors appear to share a common core structure (15, 19, 20). This core structure consists of a phosphatidylinositide (or inositol-containing sphingolipid) with an attached oligosaccharide chain that is terminated with a phosphoethanolamine residue. The linkages between the sugar units within the carbohydrate chain are conserved, and the amino group of the phosphoethanolamine moiety is used to attach the GPI anchor to the C terminus of the target protein. The organization of this basic GPI anchor structure is as follows: protein—phosphoethanolamine—6Mannoseα1—2Mannoseα1—6Mannoseα1—4Glucos amineα1—6inositol—phospholipid.
The process of GPI anchor production and attachment is mediated by the concerted actions of approximately 20 proteins, which are organized into biosynthetic complexes in the ER membrane. Seven primary steps have been identified in the GPI anchor pathway, beginning with anchor biosynthesis and concluding with the final attachment of the completed anchor structure to the recipient protein (15, 36). Some of the proteins involved in the biosynthesis and attachment of the GPI anchor catalyze the steps in the pathway, while others function as auxiliary factors.
In our analysis of the GPI anchor pathway in the filamentous fungus Neurospora crassa, we have focused on the functions and components of the phosphoethanolamine transferase and GPI transamidase complexes. The phosphoethanolamine transferase complex has been shown to consist of at least 4 components in mammals and Saccharomyces cerevisiae. The PIG-N/Mcd4p, hGPI7/Gpi7p, and PIG-O/Gpi13p proteins are involved in the addition of phosphoethanolamine substituents to the first, second, and third mannose residues in the mammalian/S. cerevisiae GPI-anchors, respectively (4, 26, 30, 31, 64, 66). The PIG-F/Gpi11p proteins serve as an auxiliary factor within the phosphoethanolamine transferase complex (31, 64, 66). The GPI transamidase complex is minimally composed of 5 proteins in both the mammalian and Saccahromyces cerevisiae systems. The GPI8/Gpi8p proteins likely function as the proteolytic activity of the complex, which cleaves the target protein at the omega site (48). The PIG-T/Gpi16p proteins have been shown to be important for the formation and stabilization of the GPI transamidase complex (22, 48, 49). The PIG-U/Cdc91p proteins have been implicated in the recognition of either the GPI anchor attachment motif in the substrate protein or the long chain fatty acids of the GPI anchor (32). The functions of the remaining GPI transamidase subunits, PIG-S/Gpi17p and GAA1/Gaa1p, are somewhat unclear, but the mammalian GAA1 has been shown to be involved in the binding of the GPI anchor (68). Homologs of each of these mammalian and S. cerevisiae phosphoethanolamine transferase and GPI transamidase complex components exist in N. crassa.
We have identified and characterized N. crassa mutants affected in three genes encoding components of the phosphoethanolamine transferase complex and one gene encoding a component of the GPI transamidase complex. We have also further characterized a previously identified N. crassa mutant affected in the enzymatic activity of the N-acetylglucosamine transferase complex, which catalyzes the transfer of N-acetylglucosamine to phosphatidylinositol during the first step in the GPI anchor biosynthetic pathway. Mutants affected in these genes are unable to make normal hyphae and fail to produce many of the traditional cell types found in the N. crassa life cycle. The mutants have a vastly reduced rate of growth and grow in a tight colonial manner. Functional studies demonstrate that these mutants produce a weaker, altered cell wall. Electron micrographs of mutant cells illustrate an unusual “cell-within-a-cell” morphology, which we attribute to a defective cell wall. In addition, we show that the mutants generate a cell wall that differs extensively from the normal hyphal cell wall in carbohydrate and protein components. We conclude that GPI anchoring plays an important role in the biosynthesis, structure, and function of the N. crassa cell wall.
MATERIALS AND METHODS
Strains and culturing conditions.
The arg-12 (FGSC 1527) and GTH-16 strains of N. crassa were used as the wild-type parental strains for the isolation of mutants affected in the gpip-1, gpip-2, gpip-3, and gpit-1 genes, respectively. The arg-12 strain was obtained from the Fungal Genetics Stock Center (Kansas City, Kansas). The GTH-16 strain has an al-2 aro-9 inv qa-2 genotype (37). All cells were grown on supplemented Vogel's medium as described by Davis and DeSerres (12). Gene mapping experiments were conducted by mating strains on a corn meal agar medium supplemented with needed amino acids, vitamins, and 0.4% glucose and using standard mapping procedures (12). The gpig-1 mutant, a temperature-sensitive mutant affected in the catalytic subunit of the N-acetylglucosamine transferase complex, was obtained from Seiler and Plamann (60).
Isolation of the MSA-7 mutant.
The MSA-7 mutant, which contains a mutation in the gpip-1 gene, was isolated in a mutant screening experiment designed to identify mutants affected in the process of cell fusion. Conidia from the arg-12 strain were harvested into 10 ml of sterile water, transferred to petri dishes, and mutagenized by a 10-min exposure to a UV light source held at a distance of 10 cm above the petri dish. The UV mutagenesis resulted in a 99.7% killing of the conidia. The mutagenized conidia were plated onto sorbose agar medium supplemented with arginine. Individual isolates were picked into test tubes containing Vogel's medium and subsequently tested for their ability to participate in the process of cell fusion using a heterokaryon formation assay. To do so, hyphae and conidia from the arg-12 mutant isolates were transferred to test tubes containing unsupplemented medium and a second strain having a different type of auxotrophy (inl FGSC 1438). If cell fusion occurs, the heterokaryon formed from the two auxotrophs is capable of growth on the unsupplemented medium. The gpip-1 mutant was identified as a mutant with a colonial growth phenotype and as being impaired in the ability to form heterokaryons in the cell fusion test system. It was also unable to generate protoperithecia, the Neurospora female mating structure. Fine mapping of the gpip-1 mutation was done using the al-3 inl strain (FGSC 2308) as the female partner in a standard mating.
PCR analysis and sequencing of MSA-7 candidate genes.
All PCR experiments were carried out using primer oligonucleotides designed to amplify genes of interest in the short genomic region identified as containing the mutant gene. The sequences for the oligonucleotides were derived from the published genomic DNA sequence provided by the Neurospora genome project at the Broad Institute/MIT Center for Genome Research. Genomic DNA was isolated from all strains using the Trizol reagent as described in the manufacturer's instructions (Invitrogen, Carlsbad, CA). The amplified genomic DNA regions were sequenced at Retrogen, Inc. (San Diego, CA).
Use of RIP to isolate gpip-2, gpip-3, and gpit-1 mutants.
Mutants affected in the gpip-2, gpip-3, and gpit-1 genes were obtained using the Neurospora RIP (repeat-induced point mutation) phenomenon. RIP is a process in which multiple point mutations (C to T and G to A mutations) occur in DNA regions that are found in two or more copies in the haploid Neurospora genome during the premeiotic phase of the mating process (61). These mutations are generated in both copies of the duplicated DNA sequences. To produce mutations in the gpip-2, gpip-3, and gpit-1 genes, PCR-amplified sequences from each gene were subcloned into the pRAL1 vector (1) and the GPI gene/pRAL1 constructs were used to transform the N. crassa GTH-16 strain. The pRAL1 vector includes a copy of the N. crassa qa-2 gene and can be used to select for transformants (37). The resultant transformants were then mated with the inl strain (FGSC 1453), and mutant progeny were identified by virtue of the colonial growth phenotype. The mutations present in the gpip-2, gpip-3, and gpit-1 genes from these RIP-generated mutants were then identified by PCR amplification and sequencing of the gene as described above.
GPIP-3 and ACW-1 antibody production and Western blot analyses.
Peptides representing amino acid numbers 388 to 407 (NH2-PKPVFGRTKPEYVTPPATAK- COOH) of the predicted GPIP-3 protein and 186 to 208 (NH2-IQANGLDMEVGFPNLIWAMNMAI-COOH) of the predicted ACW-1 protein were synthesized and used to immunize rabbits (Proteintech Group, Inc., Chicago, IL).
For Western blot analyses of total cellular extracts, the gpip-3 mutant and the GTH-16 wild-type parental strain were grown on cellophane on standard agar medium. The cells were harvested, ground on liquid nitrogen to a fine powder, and resuspended in a solution of 50 mM Tris-Cl (pH 7.5) and 1% sodium dodecyl sulfate (SDS). Each extract was briefly sonicated, boiled, and centrifuged at 1,000 × g to pellet cell wall debris. The protein concentrations of the soluble fractions were determined using the Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, CA). Forty micrograms of total protein from each extract was separated on a 4 to 12% Bis-Tris NuPAGE gel (Invitrogen Life Technologies, Carlsbad, CA) and transferred to a polyvinylidene difluoride membrane. The GPIP-3 and ACW-1 proteins were detected using the GPIP-3 polyclonal antibody at a 1/25,000 dilution and the ACW-1 polyclonal antibody at a 1/5,000 dilution, respectively. Immunoreactive bands were visualized using an anti-rabbit alkaline phosphatase-conjugated secondary antibody (Sigma Aldrich, St. Louis, MO) at a 1/15,000 dilution.
For Western blot analysis of the GPIP-3 protein in a wild-type membrane preparation, the GTH-16 strain was grown, harvested, and ground as before. The frozen, powdered mycelia were then resuspended in an ice-cold solution containing 50 mM Tris-Cl (pH 7.5), 200 mM NaCl, 10 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (product number P8215 from Sigma Aldrich, St. Louis, MO). The extract was precleared by centrifugation at 1,000 × g and the supernatant was collected and centrifuged at 100,000 × g at 4°C for 1 h. The 100,000 × g supernatant was discarded, and the pellet containing the total cellular membrane was resuspended in 50 mM Tris-Cl (pH 7.5) and 1% SDS. The determination of the protein concentration, SDS-polyacrylamide gel electrophoresis (PAGE), and Western blot analysis of the sample were done as described above.
Assessment of gross and hyphal morphologies.
To assess the gross colony morphology of each strain, small inocula of the gpig-1, gpip-1, gpip-2, gpip-3, and gpit-1 mutants and the wild-type parents were made in the center of petri dishes containing standard agar growth medium. All strains, with the exception of the gpig-1 temperature-sensitive mutant, which was placed at 39°C, were allowed to grow at room temperature for the times indicated in Fig. 1. Images of the plates were then captured using a digital scanner.
FIG. 1.
GPI anchor mutants have altered gross and hyphal morphologies. Panels A to F are photographs of strains that were inoculated on agar growth medium in standard petri dishes. Panels G to L are pictures of the same strains that were inoculated between two cellophane sheets on agar growth medium, and the growing edge of each colony was photographed at a magnification of ×400. All cultures were incubated at room temperature, with the exception of the 34-15 (gpig-1) temperature-sensitive mutant, which was grown at 39°C to induce the mutant phenotype. Colonies of the wild-type (GTH-16) strain (A and G), MSA-7 (gpip-1) mutant (B and H), 34-15 (gpig-1) mutant (C and I), gpip-2 mutant (D and J), gpip-3 mutant (E and K), and gpit-1 mutant (F and L) are shown. The images in panels A to D, H, and J to L were captured at 48 h after inoculation. Panels E and F are shown at 10 days after inoculation. Panel G is shown at 24 h after inoculation. For the micrograph in panel I, the 34-15 (gpig-1) mutant was initially grown at room temperature for 24 h and then shifted to 39°C for an additional 6 h prior to examination. The scale bar in panel L represents a distance of 10 μm.
To analyze hyphal morphology, inocula of all mutant and wild-type strains were placed between two cellophane sheets on an agar medium in a petri dish. The two sheets of cellophane were cut from the same larger sheet of cellophane and oriented at 90° to one another as determined by their original orientation within the larger cellophane sheet. The gpig-1 mutant was initially grown at room temperature for 24 h before being shifted to 39°C for an additional 6 h to induce the mutant phenotype. All other strains were grown at room temperature for the times indicated in Fig. 1. A region of cellophane containing the growing edge of the colony was then cut from each dish and placed on a droplet of water, and the cells were photographed using a differential interference contrast microscope.
GTH-16 was chosen as the representative wild-type strain in Fig. 1, as it is the parental strain from which the majority of the mutants were derived (see “Use of RIP to isolate gpip-2, gpip-3, and gpit-1 mutants” above).
[3H]inositol labeling and incorporation into protein.
As a means to verify that the gpig-1, gpip-1, gpip-2, gpip-3, and gpit-1 mutants were impaired in the process of GPI anchoring, mutant and wild-type parental cells were labeled with [2-3H]inositol (Perkin Elmer) and the total 3H incorporated into protein was determined. For labeling experiments with the gpig-1 temperature-sensitive mutant, mutant and wild-type strains were grown to approximately mid-log phase in 50-ml liquid shaker cultures at room temperature (the permissive temperature) and then shifted to 39°C (the restrictive temperature) for an additional 4 h prior to labeling. While being retained at the restrictive temperature, each culture was labeled with 15 μCi of [3H]inositol for a 15-min time period. All cells were then harvested over a Buchner funnel, quickly washed with fresh medium that had been prewarmed to 39°C, frozen in liquid nitrogen, and stored at −20°C.
Due to the problems associated with growing the gpip-1, gpip-2, gpip-3, and gpit-1 mutant cells in liquid culture, these mutants were grown on cellophane sheets atop agar medium for labeling experiments. The mutants and wild-type parental strains were grown on cellophane to a point best estimated as a healthy and comparable stage of growth. All cells were then labeled with 1 μCi of [3H]inositol by removing the entire cellophane sheet from each original culture plate and placing it into a new petri dish containing 1 ml of labeled liquid medium. After 1 h, the cellophane sheets were removed from the labeling dishes and blotted dry on Whatman 3MM paper to remove excess medium. Cells were then scraped from the cellophane sheets using a razor blade, frozen in liquid nitrogen, and stored at −20°C.
All labeled, frozen mycelia were Dounce homogenized in 2 ml of an ice-cold extraction buffer containing 50 mM Tris-Cl (pH 7.5), 200 mM NaCl, 10 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. Each cellular extract was divided into 2 separate 1-ml aliquots, and SDS was added to a final concentration of 1% to one aliquot of each series. Those extracts containing 1% SDS were used for a determination of 3H-labeled protein. These extracts were precleared of cell wall debris by centrifugation at 3,000 × g for 5 min, and 150 μl of each supernatant was combined with 500 μl of chloroform and 500 μl of methanol (10:10:3 chloroform/methanol/extract) to precipitate and delipidate total protein for analysis. The samples were incubated at −20°C for 2 h, and the precipitated protein was collected by centrifugation at 17,000 × g for 10 min. The protein pellets were washed five times in a solution of chloroform/methanol/water (10:10:3), briefly dried, and resuspended by boiling in extraction buffer supplemented with 1% SDS. Total protein-associated counts were then determined using a scintillation counter.
The extracts devoid of SDS were used for a determination of 3H-labeled lipids as described by Hamburger et al. (29). The sum of the protein- and lipid-associated counts was taken as the total amount of [3H]inositol incorporated by each strain. The amount of [3H]inositol incorporated into precipitable protein was then expressed as a percentage of the total incorporation and compared among the mutant and wild-type strains.
Growth in the presence of cell wall perturbing reagents.
To assess the sensitivity of the GPI anchor mutants to reagents that affect osmotic pressure or cell wall biosynthesis, cells were grown on standard Vogel's medium supplemented with NaCl, calcofluor white, or Congo red (Sigma Aldrich, St. Louis, MO).
Cell lysis assays.
To study the cell lysis phenotype associated with a defect in GPI anchoring, the protein released into the medium by the temperature-sensitive gpig-1 mutant and its wild-type parent at the permissive and restrictive temperatures was quantitated. To do so, two flasks of mutant cells and two flasks of wild-type cells were placed on a shaker at 150 rpm at room temperature and grown to approximately mid-log phase. One flask of each strain was then shifted to a shaker at 39°C, while the second was left to shake at room temperature. After 6 h, the cultures were harvested over a Buchner funnel and the cells and medium from each were saved for protein analysis. Released proteins were precipitated from the culture media in 10% trichloroacetic acid (TCA) in acetone at −20°C. The precipitated proteins were pelleted by centrifugation at 5,000 × g, washed three times in ice-cold 100% acetone, briefly dried, and resuspended by boiling in 1% SDS. The cells were ground in a mortar and pestle under liquid nitrogen and resuspended in an extraction buffer containing 50 mM Tris-Cl (pH 7.5), 200 mM NaCl, and 1% SDS. The cell extracts were then centrifuged at 3,000 × g to pellet cell wall debris. The amounts of protein recovered from the media and cells were determined using the Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, CA). For each strain, the amount of protein released into the medium was expressed as a percentage of the total protein present in the system (the total amount of protein present in the medium and cells).
We also assessed the lysis of the gpip-3 and gpit-1 mutant cells when placed in a hypo-osmotic environment. For this experiment, mutant and wild-type cells were grown on cellophane sheets on supplemented agar medium. The cells were washed from the cellophane into a solution of 10 mM sodium acetate buffer (pH 5.0) and shaken at 150 rpm at 37°C for 1 h. The cells were collected by centrifugation at 3,000 × g for 10 min, and aliquots of the supernatants were saved to assay for released protein. The cells were resuspended in 10 mM sodium acetate buffer (pH 5.0) containing 1% SDS and sonicated in 30-s bursts for a total of 3 min. The extracts were then precleared of debris by a second centrifugation, and aliquots of the supernatants were saved to assay for cellular protein. As described above, protein concentrations of all samples were determined using the Bio-Rad DC protein assay kit, and the amount of protein released into the hypo-osmotic buffer was expressed as a percentage of the total protein present in the system (the sum of the protein released to the buffer by lysis and the cellular protein released by sonication).
Electron microscopy.
Electron micrographs of mutant and wild-type cells were prepared similar to the method described by Lenhard et al. (38). The gpip-3 and gpit-1 mutants were grown on cellophane on standard agar medium for 7 days. The GTH-16 wild-type strain was grown on cellophane on the same medium for 2 days. Samples of the mutant colonies and wild-type mycelia were floated off the cellophane into a solution of 2% glutaraldehyde, 100 mM sodium cacodylate, 800 mM sorbitol, and 0.1% dimethyl sulfoxide and fixed for 24 h at 4°C. The cells were then embedded in 2% agar and postfixed in a solution containing 1% osmium tetroxide and 100 mM sodium cacodylate. Following dehydration in a graded ethanol and acetone series, the samples were embedded in epon-araldite and thin sectioned. The sections were then stained with 2% uranyl acetate and Reynold's lead citrate and viewed with the transmission electron microscope.
Western blot analysis of cell wall-associated proteins.
To assess how the loss of GPI anchoring affects the synthesis of cell wall proteins, we performed a Western blot analysis on cell extracts from the temperature-sensitive gpig-1 mutant and wild-type strains using an “anti-cell wall” antibody. The “anti-cell wall” antibody is a polyclonal rabbit antibody raised against a crude preparation of N. crassa cell wall. The temperature-sensitive gpig-1 mutant and wild-type strains were grown at both 22°C and 39°C and harvested, and cell extracts of each were prepared. The cell extracts were centrifuged at 10,000 × g, and 30 μg aliquots of protein from the soluble fractions were analyzed by SDS-PAGE and Western blotting. The “anti-cell wall” antiserum was used at a 1/5,000 dilution, detected with a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Promega, Madison, WI) at a 1/5,000 dilution, and subsequently visualized with chemiluminescence (Pierce, Rockford, IL).
Purification of cell walls.
Mutant and wild-type strains were grown on cellophane sheets on standard agar growth medium. Cells were harvested and ground to a fine powder in a mortar and pestle while being maintained in a frozen state with liquid nitrogen. The ground material was resuspended in an extraction buffer containing 50 mM Tris-Cl (pH 7.5), 200 mM NaCl, 1% SDS, and 10 mM DTT and boiled for 20 min. All extracts were then centrifuged at 3,000 × g to obtain cell wall material. The isolated cell walls were washed once in extraction buffer, followed by five additional washes in ice-cold distilled H2O. Purified cell wall material was then lyophilized to complete dryness and used for aniline blue dye binding assays or trifluoromethanesulfonic (TFMS) acid digestions as described below.
Aniline blue binding assay.
As a measure of the beta-1,3-glucan present in the mutant and wild-type cell walls, we assessed the ability of the cell walls to absorb aniline blue dye. Purified cell walls were prepared from the gpip-3 and gpit-1 mutants, the mnt-1 mutant, and their wild-type parental strain, GTH-16, as described above. The mnt-1 is a null mutant for the N. crassa alpha-1,2-mannosyltransferase gene and has been shown to contain reduced levels of galactomannan and elevated levels of glucose in its cell wall (6). All lyophilized cell wall preparations were resuspended in distilled H2O to a final concentration of 2 mg/ml. Aliquots containing increasing amounts of cell wall material from the mutant and wild-type cells were centrifuged at 3,000 × g for 10 min, and the supernatants were decanted. The collected cell walls were then resuspended in 1 ml of 0.002% aniline blue (Sigma Aldrich, St. Louis, MO) and agitated on a platform shaker at room temperature for 24 h. After pelleting the cell wall material by centrifugation, the amount of unabsorbed aniline blue dye was determined using a spectrophotometer (optical density at 595 nm [OD595]).
TFMS acid digestion of cell walls and identification of integral cell wall proteins.
To analyze the proteins covalently linked to the mutant and wild-type cell walls, we digested purified cell walls with TFMS acid using a procedure modified from that described by Edge (14) for deglycosylation of glycoproteins. Cell walls of the gpip-3 and gpit-1 mutant and wild-type strains were prepared as described above. TFMS acid, anisole, and pyridine were obtained from Sigma Aldrich chemical company (St. Louis, MO). Prior to performing the acid digestions, 20 mg of each cell wall preparation were relyophilized overnight to ensure complete dryness of the samples. To maintain anhydrous conditions during the digestions, all glass tubes and syringes used were dried under a vacuum and all steps throughout the procedure were performed in a chamber being continually purged with N2 gas. Initially, a solution of 16% anisole in TFMS acid was prepared and 1.25 ml of this mixture was added to each of the cell wall preparations. The samples were then purged with N2 gas, quickly covered with Parafilm, and placed in the N2-filled chamber at 4°C for 5 h. During the course of the digestion, the samples were periodically mixed with a Pasteur pipette, purged, and recovered with Parafilm. After 5 h, 3.75 ml of a solution of pyridine/methanol/H2O (3:1:1) were added in a dropwise fashion to each of the digests, which were continually swirled in a dry ice-ethanol bath. The samples were then placed on dry ice for 20 min, followed by an additional 20 min incubation at −20°C. All samples were removed from −20°C and allowed to thaw, and 1 ml of 5% ammonium bicarbonate solution was added to each. The released proteins were then precipitated in 12.5% TCA in acetone at −20°C for 24 h. The precipitated proteins were collected by centrifugation at 5,000 × g, washed three times in ice-cold 100% acetone, briefly dried, and resuspended by boiling in 1% SDS. Protein concentrations of all samples were determined using the Bio-Rad DC protein assay kit. The amount of protein released from 1 mg of starting cell wall material was separated by SDS-PAGE on a 4 to 12% Bis-Tris NuPAGE gel and visualized using the SilverQuest silver staining kit (Invitrogen Life Technologies, Carlsbad, CA) or with Coomassie blue. Selected protein bands were then excised from the stained gels and sent to Midwest Bio Services (Overland Park, KS) for nano-liquid chromatography/mass spectrometry/ mass spectrometry (LC/MS/MS)-based identification.
RESULTS
Isolation and identification of the MSA-7 mutant.
The MSA-7 mutant was originally isolated in a screening experiment designed to generate mutants affected in the process of vegetative cell fusion. In addition to its cell fusion defect, the MSA-7 mutant grows in an extremely slow, spreading colonial manner, markedly different from that of its wild-type parent (Fig. 1A). The mutant cells do not readily extend across the medium and have a significantly reduced rate of growth (Fig. 1B). Microscopic examination of the MSA-7 mutant shows that the mutant has a very different hyphal morphology and branching pattern than does the wild-type strain. The characteristic vegetative hyphae of the wild-type strain are elongated and branch at points behind the growing hyphal tip (Fig. 1G). In contrast, the mutant hyphae are more bulbous in shape and highly branched (Fig. 1H). Clear evidence of cell lysis is also often found in MSA-7 mutant colonies.
Classical genetic mapping experiments were done to locate the mutation responsible for the MSA-7 phenotype on the N. crassa genome. Following the segregation of the colonial morphology among 1,000 total progeny from a number of crosses, it was determined that the relevant mutation mapped to a location 0.6 centimorgans to the left of the inl gene on linkage group V. From an analysis of the annotated genes at this locus, five genes were selected as likely candidates to contain the mutation of interest. Oligonucleotide primers were designed to amplify and sequence these genes from the mutant and wild-type parent genomes. Four of the five genes from the mutant were identical in sequence to those of the wild-type parent. However, the NCU06663 gene was found to contain a mutation in the normal stop codon (TGA to CGA) that results in the addition of 57 extra amino acids to the protein product. The NCU06663 gene encodes a homolog of the mammalian PIG-F and S. cerevisiae Gpi11p proteins, which function as an auxiliary factor in the phosphoethanolamine transferase complex involved in GPI anchor biosynthesis (31, 64, 66). We have named the N. crassa gene gpip-1 (glycosylphosphatidylinositol anchor phosphoethanolamine transferase gene-1).
In addition to the colonial gpip-1 mutant, Seiler and Plamann (60) reported on the isolation of three temperature-sensitive N. crassa mutants (34-15, 33-15, and 31-5) that grew as tight colonial mutants at the restrictive temperature. These mutants were described as having a slow growing, bulbous, branched phenotype and some degree of cell lysis (Fig. 1C and 1I). They demonstrated that transformation of the mutants with the NCU09757 gene, which encodes a homolog of the mammalian PIG-A and S. cerevisiae Gpi3p proteins, rescued the mutant phenotype. The mammalian PIG-A and S. cerevisiae Gpi3p proteins putatively function as the N-acetylglucosamine transferase responsible for catalyzing the first step in the GPI anchor biosynthetic pathway (15, 59). In their study, Seiler and Plamann named this gene gpi-3 after its S. cerevisiae homolog. As the abbreviation of “gpi” has been previously used to designate N. crassa mutants lacking glucosephosphate isomerase and to develop a specific nomenclature for naming multiple components of the GPI anchor pathway, we have opted to refer to this gene as gpig-1 (glycosylphosphatidylinositol anchor N-acetylglucosamine transferase gene-1). We sequenced the 34-15 (gpig-1) mutant allele and found a single missense mutation (TCG to TTG), which changes amino acid number 176 of the GPIG-1 protein from a serine to a leucine residue.
Isolation of additional mutants in the GPI anchor pathway.
As a means to further demonstrate the importance of GPI anchoring to the overall morphology of N. crassa, mutations in other genes in the GPI anchor pathway were generated via the RIP phenomenon. RIP is a process in which genes found in duplicate copies within the N. crassa haploid genome are mutated during mating. To generate these RIP mutants, a BLAST search of the N. crassa genome was first conducted to identify those genes having high levels of sequence similarity with genes encoding known S. cerevisiae and mammalian GPI anchor pathway components. Several such genes were readily identified, and those encoding selected components of the phosphoethanolamine transferase and GPI transamidase complexes were targeted for disruption. Specifically, we attempted to create RIP mutations within the following N. crassa genes as defined by the Broad Institute/MIT Neurospora genome project (25): NCU07999, NCU06215, and NCU06508 (members of the phosphoethanolamine transferase complex involved in the addition of phosphoethanolamine to the first, second, and third mannose residues of the GPI anchor, respectively) and NCU05644 (a component of the GPI transamidase complex required for complex formation and stabilization). We also attempted to use the RIP technique to obtain null mutants for the gpip-1 (NCU06663) and gpig-1 genes (NCU09757). Each of the genes was PCR amplified and subcloned into the pRAL1 plasmid (1). The resultant constructs were then used to transform the N. crassa GTH-16 strain (37). A number of individual transformants for each construct were obtained and subsequently mated with the inl (FGSC 1453) strain to activate the RIP process. Multiple matings for three of the six chosen putative GPI anchor pathway genes yielded progeny with a colonial phenotype characteristic of the gpip-1 and gpig-1 mutants. Colonial mutants were isolated for the NCU07999, NCU06508, and NCU05644 genes, suggesting that the products of these genes function in the GPI anchor pathway.
The NCU07999-encoded protein is a homolog of the mammalian PIG-N and S. cerevisiae Mcd4p proteins. The PIG-N and Mcd4p proteins function in the addition of phosphoethanolamine to the first mannose residue during GPI anchor biosynthesis (26, 30, 34). The protein encoded by NCU06508 is a homolog of the mammalian PIG-O and S. cerevisiae Gpi13p proteins. The PIG-O and Gpi13p proteins function in the addition of phosphoethanolamine to the third mannose residue within the GPI anchor (31, 66). We have named the NCU07999 and NCU06508 genes gpip-2 and gpip-3, respectively (glycosylphosphatidylinositol anchor phosphoethanolamine transferase gene-2 and -3). The NCU05644 gene encodes a mammalian PIG-T and S. cerevisiae Gpi16p homolog. The PIG-T and Gpi16p proteins are thought to be involved in the formation and stabilization of the GPI transamidase complex, which cleaves specified proteins at the omega site and attaches the GPI anchor to the newly generated C terminus (22, 48, 49). We have opted to call the NCU05644 gene gpit-1 (glycosylphosphatidylinositol anchor transamidase gene-1).
Genomic DNA was isolated from three mutant progeny for each of the successful RIP experiments and used for PCR amplification and sequencing of the mutant genes. In each instance, we found that the progeny having the RIP-induced phenotype contained multiple mutations within the endogenous copy of the transforming genes and that the majority of the progeny isolated contained nonsense mutations. One isolate that clearly represented a null mutant for each of the genes was selected for further phenotypic characterization. The mutations present in these representative null mutants were as follows: the gpip-2 mutant had a total of 147 mutations, including the introduction of 4 stop codons and the disruption of a putative 5′ splice site; the gpip-3 mutant contained 33 total mutations, 5 of which produced stop codons; and the gpit-1 mutant had 32 total mutations, including the introduction of 6 stop codons.
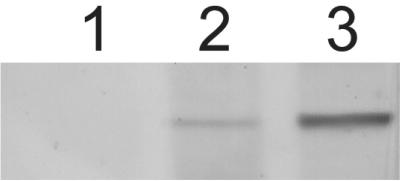
To verify the effectiveness of the mutations introduced by the RIP process and to demonstrate that the mutant lacked the relevant protein, antibodies were raised against a peptide representing amino acids 388 to 407 of the GPIP-3 protein and used to probe cell extracts of the representative gpip-3 null mutant and wild-type parental strain (Fig. 2). The GPIP-3 antisera detected an approximately 100-kDa protein in the wild-type cell extract (lane 2) that was absent in the gpip-3 null mutant (lane 1). The apparent size of the GPIP-3 protein is slightly smaller than that predicted for the translated gene product, most likely due to proteolytic processing. The same 100-kDa protein was found to be enriched in a wild-type membrane preparation (lane 3), consistent with the expected ER membrane localization of the GPIP-3 protein.
FIG. 2.
The GPIP-3 protein is absent in the gpip-3 mutant. Samples of gpip-3 null mutant (lane 1) and wild-type (GTH-16) (lane 2) total cellular extracts and a wild-type (GTH-16) membrane preparation (lane 3) were separated by SDS-PAGE and analyzed by Western blotting using an anti-GPIP-3 antibody.
We were unable to obtain RIP mutant progeny from matings of numerous transformants containing DNA sequences from the NCU06215 gene, or the gpip-1 and gpig-1 genes (for which we have the MSA-7 mutant with the 57-amino-acid extension and the 34-15 temperature-sensitive mutant, respectively). The NCU06215 gene is a homolog of the mammalian hGPI7 and S. cerevisiae GPI7, which encode a phosphoethanolamine transferase that functions in the addition of phosphoethanolamine to the second mannose residue of the GPI anchor (4, 64). One possible explanation for the lack of mutant progeny from RIP experiments using the NCU06215, gpip-1, and gpig-1 genes might be that their gene products are vital for the biosynthesis of the N. crassa GPI anchor and that null mutants for these genes are inviable. A second line of evidence also suggests that null mutations in the NCU06215 and gpip-1 genes may potentially result in a lethal phenotype. As part of the Neurospora genome project (Dartmouth College, Hanover, NH), gene knockouts are being generated via homologous recombination-mediated gene replacement and made available to the Neurospora community. Gene replacement experiments were done for the NCU06215 and gpip-1 genes, and in both instances, viable knockouts could not be recovered. Efforts to obtain knockout mutants for these genes are continuing at the Neurospora genome project (G. Park, H. V. Colot, L. Litvinkova, S. Curilla, C. Ringelberg, K. A. Borkovich, and J. C. Dunlap, personal communication).
As previously mentioned, each of the mutants was disrupted in a homolog of a known component of the S. cerevisiae and mammalian GPI anchor pathways. As a means of verifying that the mutants are defective in the process of GPI anchoring, we assessed the ability of each to synthesize GPI-anchored proteins. To do so, all mutants and the corresponding wild-type parental strains were pulse labeled with [3H]inositol, and the amount of [3H]inositol incorporated into the protein was measured as a percentage of the total [3H]inositol incorporation. In each instance, there was a three- to fourfold reduction in the amount of [3H]inositol-containing proteins produced by the mutants compared to the wild-type strains, demonstrating that the mutants are impaired in the production of GPI-anchored proteins. As presented below, an examination of the protein composition of mutant and wild-type cell walls provides additional evidence that the mutants are lacking in the ability to generate GPI-anchored proteins.
The GPI anchor pathway is required for cell wall integrity.
All of the null mutants obtained from the RIP experiments displayed a classical colonial growth phenotype, but the severity of the colonial phenotype differed among the individual mutants. The gpip-3 and gpit-1 mutants had extremely slow growing, tight, colonial growth patterns, while the gpip-2 mutant displayed a phenotype in which the colonial colonies showed some minor spreading. However, these GPI anchor pathway mutants, as well as the gpip-1 and gpig-1 mutants described earlier, all exhibited significantly reduced growth rates, altered gross and hyphal morphologies, and obvious points of cell swelling and lysis (Fig. 1). The mutant hyphae were extremely bulbous and apolar in shape and had many more septa than did the wild-type cells. Each of the mutants was impaired in the ability to undergo cell fusion events, as determined by a heterokaryon formation assay. In addition, the mutants were unable to produce either protoperithecia (the female mating structure) or conidia (asexual spores). These phenotypic defects were most apparent in the gpip-3 and gpit-1 mutants, which were seemingly identical in phenotype. Since the gpip-3 and gpit-1 mutants were the most severely affected, these two mutants were used for the majority of our GPI anchor mutant analyses. One likely explanation for the variation among the mutants is that the severity of the phenotype is dependent upon the role of the various gene products in the GPI anchor pathway and the relative amount of GPI anchoring that may remain in the mutant cells.
In addition to these GPI anchor pathway mutants, several other N. crassa mutants having a colonial phenotype have previously been isolated. The altered gene in a few of these colonial mutants has been identified and shown to be involved in cell wall biosynthesis, structure, or function (6, 17, 71). Studies of S. cerevisiae and Aspergillus have demonstrated the importance of the GPI anchor biosynthetic pathway and the role of certain GPI-anchored proteins for cell wall integrity (4, 21, 45, 56, 70). Based upon these studies, the putative functions of several N. crassa proteins predicted to be GPI anchored, and the significant degree of cell lysis observed within the mutant colonies, we hypothesized that the N. crassa GPI anchor mutants would have altered cell walls.
As a way of examining the cell wall, we cultured the mutant and wild-type cells in a variety of growth conditions. Salt sensitivity has been found to be associated with mutations affecting cell wall biosynthesis (11, 47). We tested the ability of the mutant and wild-type strains to grow on solid medium supplemented with elevated levels of salt and found that all of the mutants were impaired. There was a clear correlation between the tightness of the colonial growth pattern displayed by the individual mutants and the salt concentration that fully inhibited their growth. The wild-type parental strains were unable to survive on agar medium containing 12% NaCl. In contrast, the very slow-spreading colonial mutants, gpip-1, gpip-2, and gpig-1, were unable to grow on agar medium supplemented with 5% NaCl. The most severe, sickly mutants, gpip-3 and gpit-1, were unable to survive on agar medium containing 2% NaCl.
The GPI anchor mutants were unable to readily grow in liquid media. This fact, coupled with the cell lysis frequently seen within the mutant colonies, suggested that the mutants were osmotically sensitive. Sorbitol is often used to stabilize N. crassa spheroplasts and other osmotically sensitive cell types (69). We found that the addition of 1 M sorbitol to either liquid or solid media caused an apparent increase in the growth rate of the mutants.
We initially used the temperature-sensitive gpig-1 mutant to assess why the GPI anchor mutants might have difficulty growing in liquid medium. To assay for cell lysis, the gpig-1 mutant and its wild-type counterpart were grown at room temperature to approximately mid-log phase and then shifted to the restrictive temperature for an additional 6 h. The cells and their respective culture media were then harvested and assayed for total protein. The gpig-1 mutant was found to lose approximately 50% of its cellular protein to the medium at the restrictive temperature. Less than 5% of the protein from the wild-type parental strain is released under identical conditions. The extensive protein loss due to cell lysis is consistent with the slow, colonial growth pattern of the mutant and suggests that cell wall integrity is compromised when the GPI anchor pathway is disrupted.
Based on our previous microscopic analyses of these mutants and our direct examination of the cell lysis associated with the temperature-sensitive gpig-1 mutant, we expected the gpip-3 and gpit-1 mutants would be susceptible to hypo-osmotic conditions. To test this hypothesis, mutant and wild-type cells were grown atop cellophane sheets, transferred to a hypo-osmotic solution of 10 mM sodium acetate (pH 5.0), and shaken at 150 rpm at 37°C. After 1 h of agitation, the cells and hypo-osmotic buffer were collected and assayed for total protein as described in Materials and Methods. It was determined that approximately 45% of the total cellular protein was released or secreted from the gpip-3 and gpit-1 mutant cells into the buffer. In contrast, the wild-type parent released or secreted less than 5% of its total cellular protein under the same conditions. These findings are consistent with the degree of lysis experienced by the gpig-1 mutant at the restrictive temperature and further demonstrate the importance of the GPI anchor pathway for cell wall strength and stability.
The sensitivity of the gpip-3 and gpit-1 mutants to various cell wall-perturbing agents provides additional evidence for these mutants having fragile cell walls. We found that the mutants were hypersensitive to calcofluor white and Congo red, two reagents that bind to and affect the synthesis of the chitin component of the cell wall (57). Specifically, the gpip-3 and gpit-1 mutants were unable to grow on solid media containing calcofluor white or Congo red at a concentration of 1 mg/ml. In contrast, the wild-type parental strain was able to grow in the presence of either reagent up to a concentration of 30 mg/ml. Similar results have been reported for S. cerevisiae mutants defective in the GPI anchor and cell wall biosynthetic pathways (4, 5, 21, 54, 66). It has been shown that when cell wall integrity is compromised, fungal cells respond by increasing cell wall biosynthesis (35, 67). We would interpret our results to mean that when chitin synthesis is compromised by the presence of calcofluor white or Congo red, the gpip-3 and gpit-1 mutant cells are less able to compensate for its loss than are wild-type cells.
Transmission electron micrographs further illustrate the importance of GPI anchoring in the synthesis of the N. crassa cell wall. In approximately 20% of the gpip-3 and gpit-1 mutant micrographs, we find cells with an abnormal “cell-within-a-cell” organization. These mutant cells have clearly defined cytosolic, plasma membrane, and cell wall constituents that are enclosed within what appears to be another cell or second region of surrounding cytosol, plasma membrane, and cell wall (Fig. 3B and C). This unusual cellular morphology was never found in micrographs of the wild-type parental cells (Fig. 3A). The “cell-within-a-cell” morphology has some apparent similarity to the previously described phenomenon of intrahyphal hyphae. Intrahyphal hyphae have been reported in several fungi and are predominantly thought to occur under certain conditions of growth or in response to cellular damage or various genetic mutations (18, 33, 39, 40, 43). In each of these instances, the intrahyphal hyphae, or “invading hyphae,” are thought to reside within the remnants of older, empty, or degenerating host hyphae. The host hyphae either lack cytosol or contain cytosol that is highly vacuolated and disorganized. This is distinct from the “cell-within-a-cell” organization observed in the GPI anchor mutant colonies, where we find no evidence indicating that either the inner or outer cell is degenerating. Given the tight, colonial growth phenotype of the mutants, we have not yet been able to determine the mechanism by which the “cell-within-a-cell” structures are formed. However, we attribute the phenomenon to alterations in cell wall structure and function.
FIG. 3.
The gpip-3 and gpit-1 mutants have an abnormal “cell-within-a-cell” morphology. The gpip-3 mutant, gpit-1 mutant, and wild-type (GTH-16) strains were grown atop cellophane sheets on standard agar growth medium and prepared for electron microscopy as described in Materials and Methods. Representative electron micrographs of a wild-type cell (A), the gpip-3 mutant (B), and the gpit-1 mutant (C) are shown. Note the “cell-within-a-cell” morphology characteristic of the mutant cells. The scale bar in panel C represents a distance of 1 μm.
GPI anchor mutants have alterations in cell wall carbohydrate composition.
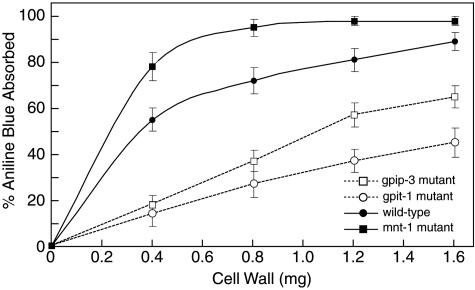
Several GPI-anchored proteins have been shown to be involved in the biosynthesis and remodeling of the glucan layer of the fungal cell wall. The GPI-anchored S. cerevisiae Gas1p and the Aspergillus Gel1p and Gel2p have been shown to function as endoglucanases/glucanosyltransferases that cleave and rejoin molecules of beta-1,3-glucan (44, 45, 46, 53). The N. crassa genome contains three Gas1p/Gel1p/Gel2p homologs, which are predicted to be GPI anchored (16). If N. crassa GPI-anchored proteins were critical for the synthesis of beta-1,3-glucan, we would expect that disruptions of the GPI anchor pathway would result in alterations of the beta-1,3-glucan component of the cell wall. To assay for differences in cell wall carbohydrate composition, we used the beta-1,3-glucan-specific dye, aniline blue (63). Cell walls from the gpip-3 and gpit-1 mutant and wild-type cells were purified, and the amount of aniline blue absorbed by each was determined. We found that the mutant cell walls bound less aniline blue than the wild-type cell wall on a per milligram of cell wall basis (Fig. 4). At low cell wall concentrations, the mutant cell walls bound only between 25% and 33% as much aniline blue as the wild-type cell wall. The data strongly suggest that the GPI anchor mutants have reduced levels of beta-1,3-glucan in their cell walls.
FIG. 4.
Cell wall absorption of aniline blue dye. Purified cell walls were prepared from the gpip-3 and gpit-1 mutants, the mnt-1 mutant, and the wild-type (GTH-16) parental strain as described in Materials and Methods. Increasing amounts of the mutant and wild-type cell wall preparations were incubated with a solution of 0.002% aniline blue dye, and the amount of aniline blue dye absorbed by each was determined as described in Materials and Methods. Graphed values are the means ± standard deviations of the results from three independent determinations.
As a control for the aniline blue binding experiment, we used cell wall from the colonial mnt-1 mutant, which is affected in the production of the galactomannan component of the N. crassa cell wall (6). The mnt-1 mutant cell wall bound much more aniline blue than either the GPI anchor mutant or wild-type cell walls. This is not unexpected, given the fact that the mnt-1 mutant cell wall has a higher percentage of glucose than that of the wild type. It is possible that the mnt-1 mutant compensates for the loss of galactomannan by increasing the amount of beta-1,3-glucan in its cell wall.
GPI anchor mutants have alterations in integral cell wall proteins.
Based upon the importance of GPI anchoring in targeting proteins to the cell surface, we decided to look for differences in the proteins associated with the mutant and wild-type cell walls. Polyclonal antibodies directed against a crude cell wall fraction were used to look at the pattern of cell wall-associated proteins in the temperature-sensitive gpig-1 mutant and wild-type cells at both the permissive and restrictive temperatures. The mutant and wild-type parent were grown at the permissive and restrictive temperatures, and cellular extracts of each were subjected to SDS-PAGE and Western blot analysis with the polyclonal antibody (Fig. 5). As is clear from the Western blot, at the restrictive temperature, the gpig-1 mutant extract lacked some larger-sized proteins (≥83 kDa) that were found in extracts from the mutant at the permissive temperature and from wild-type cells at both temperatures. We would conclude that when GPI anchoring is impaired, some cell wall-associated proteins are lost. Because the polyclonal antibody is directed against a constellation of cell wall proteins, we were unable to identify specific GPI-anchored proteins from this analysis.
FIG. 5.
The gpig-1 mutant lacks a number of “cell wall-associated” proteins at the restrictive temperature. The gpig-1 temperature-sensitive mutant and wild-type strain were grown at the permissive (22°C) and restrictive (39°C) temperatures and harvested, and cell extracts of each were prepared. Aliquots of the SDS-soluble material from each cell extract were separated by SDS-PAGE and analyzed by Western blotting using an “anti-cell wall” antibody. Samples of the gpig-1 mutant at 22°C (lane 1) and 39°C (lane 2) and the wild-type strain at 22°C (lane 3) and 39°C (lane 4) are shown. The molecular masses indicated at the right are in kilodaltons.
The proteins detected in Fig. 5 represent either detergent extractable “cell wall-associated” proteins or “integral cell wall” proteins that were captured in transit to the cell surface. Those proteins which are lost in the mutant at the restrictive temperature are likely to be GPI anchored. Given the importance of GPI-anchored proteins to cell wall formation and stability, we decided to examine the “integral cell wall” protein content and composition of the mutant and wild-type cell walls. Among the “integral cell wall” proteins, some may have GPI anchors, while others may be transmembrane proteins or secreted nonanchored proteins. We expected these proteins to be glycoproteins, with long carbohydrate chains that are often covalently cross-linked into the beta-1,3-glucan of the cell wall (7).
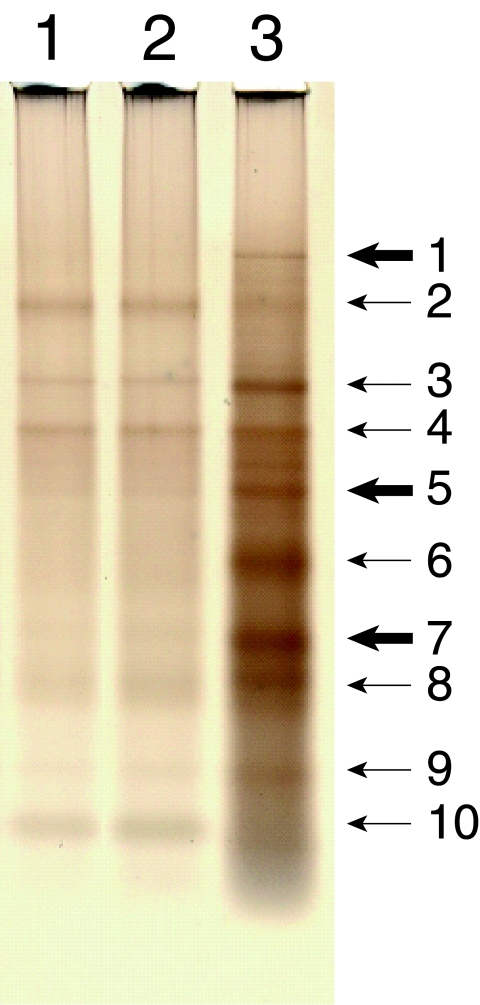
To characterize “integral cell wall” proteins, they have to be released by enzymatic or chemical hydrolysis of the cell wall carbohydrates. We opted to use a chemical digestion to free these proteins from the cell wall. TFMS acid has been previously used to remove sugars from purified glycoproteins and is able to hydrolyze glycosidic linkages without hydrolyzing peptide bonds if used in a temperature-controlled, water-free environment (14). Cell walls from the gpip-3 and gpit-1 mutant and wild-type cells were purified and treated with the TFMS acid as described in Materials and Methods. The released proteins were then recovered by TCA precipitation, and the relative amount of protein released from each cell wall was determined. On a mass basis, we found that the gpip-3 and gpit-1 cell walls contained 2.4% and 3.8% protein, respectively. This is to be compared with the wild-type cell wall, which we found to be 15.3% protein by mass. Clearly, the loss of GPI anchoring has a dramatic effect on the incorporation of protein into the cell wall. The protein released from 1 mg of purified cell wall material was subjected to SDS-PAGE and visualized by silver staining (Fig. 6). As can be seen from the figure, the total amount of protein recovered from equivalent amounts of mutant and wild-type cell walls reflects the levels of protein previously determined for each of the samples. In addition, there were obvious differences in the number and relative intensities of various protein bands from the mutant (lanes 1 and 2) and wild-type (lane 3) cell walls. The 10 major protein bands were excised from the wild-type cell wall lane of the gel. The identities of the proteins within these bands were determined by a comparison of the masses and amino acid sequences of tryptic fragments obtained from the gel slices by a nano-LC/MS/MS analysis (Midwest Bio Sciences, Overland Park, KS) with the proteins encoded in the N. crassa genome (25). The analysis showed that some of the bands contained more than a single protein. The major proteins present in each of the bands were considered to be those that yielded the greatest number of identified tryptic fragments in the nano-LC/MS/MS analyses. These major proteins are indicated in Table 1.
FIG. 6.
The gpip-3 and gpit-1 mutants have altered “integral cell wall” protein compositions. Purified cell walls from the gpip-3 and gpit-1 mutants and the wild-type (GTH-16) strain were digested with TFMS acid as described in Materials and Methods. The total protein released from 1 mg of starting cell wall material from the gpip-3 mutant (lane 1), gpit-1 mutant (lane 2), and wild-type strain (lane 3) were separated by SDS-PAGE and visualized by silver staining. The 10 major protein bands from the wild-type cell wall are indicated with arrows. Those bands containing GPI-anchored proteins are highlighted with thick arrows. See Table 1 for a listing of both the GPI-anchored and nonanchored proteins detected in the major protein bands.
TABLE 1.
“Integral cell wall” proteins from wild-type Neurospora crassa hyphae
| Protein band no. (predicted molecular mass [kDa]) | Identified GPI-anchored protein(s) (gene) | Identified nonanchored protein(s) (gene) |
|---|---|---|
| 1 (135) | Endochitinase (NCU02184) | None |
| 2 (80) | Mixed-linked glucanase (NCU01353) | Catalase-3 (NCU00355) |
| 3 (74) | None | NCW-1 (NCU05137) |
| 4 (63) | None | Beta-glucosidase (NCU09326), NCW-2 (NCU01752) |
| 5 (49) | ACW-1 (NCU08936), glucan/beta-glucanase (NCU09175) | None |
| 6 (37) | None | None |
| 7 (28) | ACW-2 (NCU00957), ACW-3 (NCU05667) | None |
| 8 (25) | None | NCW-3 (NCU07817) |
| 9 (17) | None | Cellular proteins |
| 10 (11) | None | Cellular proteins |
Of the cell wall proteins identified, six are predicted to be GPI anchored based upon a computer analysis, which examines their carboxyl termini for a GPI anchor motif (16), and their sequence similarity with other known fungal GPI-anchored proteins. Figure 6 indicates in which of the major protein bands these GPI-anchored proteins were found (bands 1, 5, and 7). An examination of the cell wall proteins from the gpip-3 and gpit-1 mutants (lanes 1 and 2) shows that these bands are either absent or greatly reduced in the mutant cell walls, providing additional evidence that the mutants are defective in the process of GPI anchoring. The most striking distinction was the complete absence of a putative GPI-anchored endochitinase from the cell walls of the mutants (band 1). The endochitinase-2 is a homolog of the Aspergillus nidulans ChiA protein and was the only protein detected in band 1. The ChiA protein has been shown to be required for proper growth and morphogenesis of A. nidulans (65). ChiA contains a probable GPI anchor addition site at its C terminus. Bands 5 and 7 each contained two major proteins, which are also predicted to be GPI anchored. Band 5 contained a glucan/beta-glucanase and a protein which we have designated ACW-1 (anchored cell wall protein-1). The ACW-1 is a homolog of the S. cerevisiae GPI-anchored Sps2p/Ecm33p (50, 51), which has been shown to be important for the integrity of the S. cerevisiae cell wall. The glucan/beta-glucanase protein and the ACW-1 (band 5) were reduced in the gpip-3 and gpit-1 mutant cell walls compared to that of the wild type. We have designated the two major proteins detected in band 7 ACW-2 and ACW-3. ACW-2 and ACW-3 are putative GPI-anchored proteins, which are also expressed at reduced levels in the mutant cell walls. The ACW-2 is a homolog of the Fusarium oxysporum FEM1p. FEM1p has been shown to be a GPI-anchored protein covalently linked to the cell wall of F. oxysporum (58). The ACW-3 is a serine- and threonine-rich protein without homology to other previously identified cell wall proteins. The final putative N. crassa GPI-anchored protein identified was a mixed-linked glucanase. This mixed-linked glucanase is a homolog of the Mlg1a and Mlg1b proteins from Cochliobolus carbonum, which serve as bifunctional beta-1,3-1,4/beta-1,3-glucanases (28). The mixed-linked glucanase was detected as a minor component of band 2. The major protein present in band 2 was catalase-3, a nonanchored protein. Thus, it is not unexpected that the intensity of band 2 was not diminished in the gpip-3 and gpit-1 mutant lanes.
To analyze the expression of a single GPI-anchored protein, antibodies were raised against a peptide representing amino acids 186 to 208 of ACW-1. The ACW-1 protein, the product of the NCU08936 gene, was chosen as the representative GPI-anchored protein because its sequence is highly similar to that of a known GPI-anchored protein (the S. cerevisiae Sps2p/Ecm33p) and it is highly expressed (based upon its abundance in the vegetative hyphal cell wall and the high number of known expressed sequence tags for the gene). The ACW-1 antisera detected three major bands at 59, 50, and 45 kDa in the wild-type cell extract (Fig. 7, lane 2) that were greatly reduced or absent in the gpip-3 mutant (lane 1). This, along with the data showing that ACW-1 is lost from the mutant cell wall (Fig. 6), suggests that ACW-1 is rapidly degraded in the absence of GPI anchoring. The 45-kDa band is consistent with the predicted molecular mass of the ACW-1 proprotein. The upper two bands most likely represent different species of the protein, which might be expected to differ from the proprotein in GPI anchor and/or glycosylation status. The detection of such species is not unexpected, since the ACW-1 protein seen in Fig. 7 is presumably in transit to the cell surface and may be found in multiple modification states. The small amount of ACW-1 protein detected in the gpip-3 null mutant might indicate that a minimal level of GPI anchoring persists in the mutant cells or might simply represent the steady-state level of the ACW-1 protein that is being rapidly degraded in the absence of the GPI anchor.
FIG. 7.
The gpip-3 mutant has reduced levels of ACW-1. Samples of gpip-3 null mutant (lane 1) and wild-type (GTH-16) (lane 2) total cellular extracts were separated by SDS-PAGE and analyzed by Western blotting using an anti-ACW-1 antibody.
As shown in Table 1, we also identified five cell wall proteins whose amino acid sequences indicate they are secreted without a GPI anchor. These nonanchored cell wall proteins were represented in bands 2, 3, 4, and 8 of Fig. 6. As mentioned, catalase-3 (CAT-3) was the major protein comprising band 2. The N. crassa CAT-3 is a secreted protein which has been identified as the major catalase within vegetative hyphae and is induced in response to different conditions of environmental stress (10, 42). It is interesting that the CAT-3 protein appeared in relatively equal amounts in the gpip-3 and gpit-1 mutant and wild-type cell walls. We determined that band 4 included two cell wall proteins, a beta-glucosidase and a protein that we have designated NCW-2 (nonanchored cell wall protein-2). The beta-glucosidase is a homolog of the S. cerevisiae Scw11p. Scw11p is a member of a family of cell wall glucanases involved in the mating process of S. cerevisiae (9, 62). NCW-2, as well as NCW-1 and NCW-3 found in bands 3 and 8, respectively, are serine- and threonine-rich proteins without known homologs. Such serine- and threonine-rich regions are often characteristic of cell wall proteins and may serve as O-linked glycosylation sites that might function to direct and covalently link them to the fungal cell wall (23, 24).
The TFMS acid digestion leaves a single sugar from the N-linked and O-linked oligosaccharides attached to the protein. The presence of these sugars interferes with the identification of peptide fragments containing them in the analysis. For this reason, it is possible we may have been unable to identify some of the smaller cell wall proteins in our analysis (for example, from band 6).
It should also be noted that, in our proteomic analyses, we detected a number of small cytosolic, ribosomal, and mitochondrial proteins in bands 5 through 10. These proteins, which we have referred to as “cellular proteins” in Table 1, were the primary constituents of bands 9 and 10. Such small “intracellular” proteins are often found in fungal cell wall preparations, and there is some evidence suggesting they are cross-linked into the cell wall (2, 3, 27).
DISCUSSION
Here we report the characterization of five mutants, each affected in a different gene in the GPI anchor pathway of Neurospora crassa. All of the mutants have a colonial morphology and altered hyphal growth pattern and frequently undergo cell lysis. We attribute these abnormalities to structural and functional defects in the cell wall that result from the absence of a number of GPI-anchored proteins.
Although the GPI anchor mutants shared a general phenotype, the severity of the phenotype varied among the mutants. The gpig-1 mutant (containing an amino acid substitution in the N-acetylglucosamine transferase), gpip-1 mutant (containing a 57-amino-acid extension to an auxiliary factor of the phosphoethanolamine transferase complex), and gpip-2 mutant (a null mutant for the phosphoethanolamine transferase involved in the modification of the first mannose) most closely resembled one another and were less impaired than the gpip-3 mutant (a null mutant for the phosphoethanolamine transferase involved in the modification of the third mannose) and gpit-1 mutant (a null mutant for a subunit of the GPI transamidase complex involved in complex stabilization), which were phenotypically indistinguishable. Such variation could be explained by the type of mutation(s) present in each mutant gene, the potential role of the mutant gene product in the process of GPI anchoring, and the relative amount of anchoring that may persist with the alteration or abolition of that gene product.
Null mutants were obtained for the gpip-2, gpip-3, and gpit-1 genes yet could not be recovered for the gpig-1, gpip-1, or NCU06215 genes, despite repeated attempts. One likely explanation for this finding is that the gpig-1, gpip-1, and/or NCU06215 gene products may be absolutely essential for biosynthesis of the GPI anchor and survival of the cell. This explanation presumes that the mutants we have isolated experience a greatly reduced amount of GPI anchoring but are not completely devoid of the process. The gpig-1 temperature-sensitive (34-15) and gpip-1 (MSA-7) mutants were generated by the introduction of missense mutations in their respective GPI genes, which perhaps allows for a minimal level of product function and subsequent GPI anchoring.
Microscopic examination and functional assays indicated that all of the N. crassa GPI anchor mutants had altered hyphal morphologies and experienced a significant degree of cell lysis. Subsequent analyses demonstrated that each of the GPI anchor mutants was defective in the production of GPI-anchored proteins. We focused on the gpip-3 and gpit-1 mutants because they had the most severely impaired phenotype and sought to associate this phenotype with their defect in the process of GPI anchoring. The gpip-3 and gpit-1 mutants had defects in cell wall strength, as determined by their hypersensitivity to hypo-osmotic conditions and the cell wall-perturbing agents calcofluor white and Congo red. Aniline blue binding assays and TFMS acid digestions illustrated obvious alterations in both carbohydrate and protein composition of the gpip-3 and gpit-1 mutant cell walls. Proteomic analyses of “integral cell wall” proteins demonstrated the abolition or reduction of a number of putative GPI-anchored proteins from the gpip-3 and gpit-1 mutant cell walls. Western blot analysis of an individual GPI-anchored protein, ACW-1, showed that its synthesis was affected in the gpip-3 mutant. In completing these studies, we have demonstrated clear defects in the gpip-3 and gpit-1 mutant cell walls and established a connection between these cell wall alterations and the deficiency of GPI-anchored proteins.
Another interesting observation was the unusual “cell-within- a-cell” organization found in electron micrographs of the gpip-3 and gpit-1 mutants. The biogenesis of this abnormal cellular morphology remains to be elucidated.
Consistent with the findings for other fungi (8, 72), we have shown that GPI-anchored proteins represent a significant fraction of the N. crassa cell wall protein. Specifically, we have identified 6 “integral cell wall” proteins from vegetative hyphae which are predicted to contain a GPI anchor based upon their primary sequence and sequence similarity to other known GPI-anchored proteins. The N. crassa GPI-anchored proteins were likely integrated into the cell wall via covalent cross-linking of N- and O-linked glycosylation and/or the GPI anchor present on the proteins to the carbohydrate component of the cell wall. These proteins, as well as other “nonanchored integral cell wall” proteins, were released from the cell wall by chemical hydrolysis with TFMS acid. To the best of our knowledge, this is the first report of the use of TFMS acid to digest a fungal cell wall. Using this procedure, we were able to isolate and identify a number of N. crassa cell wall proteins.
A consideration of the potential roles of the GPI-anchored proteins detected in the wild-type cell wall (Table 1) provides a likely explanation for the phenotype of the GPI anchor mutants. Some of these GPI-anchored proteins might be expected to have important roles in cell wall biogenesis and remodeling (the identified endochitinase, beta-glucanase, and mixed-linked glucanase might clip cell wall polymers to allow for wall remodeling during growth or function as cross-linking enzymes), while others may serve as major structural components of the wall (ACW-1, ACW-2, and ACW-3). The loss of such proteins would not only contribute to the decreased protein content of the mutant cell wall but would also lead to an overall decline in cell wall stability and function. Including those identified here, the N. crassa genome encodes approximately 90 proteins predicted to be GPI anchored (13, 16). Many of these proteins putatively serve as glycosyl hydrolases, glycosyl transferases, or peptidases, which would also be required for proper cell wall synthesis, structure, and function. Among the predicted N. crassa GPI-anchored proteins are homologs of the S. cerevisiae Gas1p and Aspergillus Gel1p and Gel2p. Gas1p, Gel1p, and Gel2p function in the remodeling of cell wall glucans (44, 45, 46). The absence of these N. crassa homologs could account for the reduction of beta-1,3-glucan in the cell walls of the GPI anchor mutants.
The GPI anchor mutants had decreased amounts of two major cell wall components, protein (Fig. 6) and beta-1,3-glucan (Fig. 4). It is unclear what component(s) are found in increased levels in the mutant cell walls as measured on a per-mg-cell wall basis. One likely possibility is that the mutant cell walls could have a higher percentage of chitin than the wild-type cell wall. Elevated levels of chitin have been associated with defects in cell wall integrity and result from the activation of the cell wall salvage pathway (52, 54, 55). In addition, our calcofluor white and Congo red sensitivity assays demonstrate that the mutants are extremely sensitive to the loss of chitin synthesis. It is also possible that the mutant cells compensate for the lack of beta-1,3-glucan by producing other polymers, such as beta-1,6-glucan. Although the exact composition of the mutant cell wall remains a subject for further study, it is clear that, when GPI anchoring is lost, the cell wall lacks key components and has an abnormal organization.
Acknowledgments
We thank Alan Siegel for technical assistance with the differential interference contrast and electron microscopy and James Stamos for helping in the preparation of illustrations for the manuscript.
This work was supported by funds from the UB Foundation.
REFERENCES
- 1.Akins, R. A., and A. M. Lambowitz. 1985. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol. Cell. Biol. 5:2272-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alloush, H. M., J. L. Lopez-Ribot, B. J. Masten, and W. L. Chaffin. 1997. 3-Phosphoglycerate kinase: a glycolytic enzyme present in the cell wall of Candida albicans. Microbiology 143:321-330. [DOI] [PubMed] [Google Scholar]
- 3.Angiolella, L., M. Facchin, A. Stringaro, B. Maras, N. Simonetti, and A. Cassone. 1996. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173:684-690. [DOI] [PubMed] [Google Scholar]
- 4.Benachour, A., G. Sipos, I. Flury, F. Reggiori, E. Canivenc-Gansel, C. Vionnet, A. Conzelmann, and M. Benghezal. 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274:15251-15261. [DOI] [PubMed] [Google Scholar]
- 5.Benghezal, M., P. N. Lipke, and A. Conzelmann. 1995. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J. Cell Biol. 130:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, S. M., A. Piwowar, M. Ciocca, and S. J. Free. 2005. Mannosyltransferase is required for cell wall biosynthesis, morphology, and control of asexual development in Neurospora crassa. Mycologia 97:872-879. [DOI] [PubMed] [Google Scholar]
- 7.Brul, S., A. King, J. M. van der Vaart, J. Chapman, F. Klis, and C. T. Verrips. 1997. The incorporation of mannoproteins in the cell wall of S. cerevisiae and filamentous Ascomycetes. Antonie Leeuwenhoek 72:229-237. [DOI] [PubMed] [Google Scholar]
- 8.Bruneau, J.-M., T. Magnin, E. Tagat, R. Legrand, M. Bernard, M. Diaquin, C. Fudali, and J.-P. Latge. 2001. Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22:2812-2823. [DOI] [PubMed] [Google Scholar]
- 9.Cappellaro, C., V. Mrsa, and W. Tanner. 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180:5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chary, P., and D. O. Natvig. 1989. Evidence for three differentially regulated catalase genes in Neurospora crassa: effects of oxidative stress, heat shock, and development. J. Bacteriol. 171:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da-Silva, M. M., M. L. Polizeli, J. A. Jorge, and H. F. Terenzi. 1994. Cell wall deficiency in “slime” strains of Neurospora crassa: osmotic inhibition of cell wall synthesis and beta-D-glucan synthase activity. Braz. J. Med. Biol. Res. 12:2843-2857. [PubMed] [Google Scholar]
- 12.Davis, R. H., and F. J. DeSerres. 1970. Genetic and microbiological techniques for Neurospora crassa. Methods Enzymol. 17:79-143. [Google Scholar]
- 13.de Groot, P. W. J., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 14.Edge, A. S. 2003. Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem. J. 3:339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhaber, B., S. Maurer-Stroh, M. Novatchkova, G. Schneider, and F. Eisenhaber. 2003. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays 25:367-385. [DOI] [PubMed] [Google Scholar]
- 16.Eisenhaber, B., G. Schneider, M. Wildpaner, and F. Eisenhaber. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. J. Mol. Biol. 337:243-253. [DOI] [PubMed] [Google Scholar]
- 17.Enderlin, C. S., and C. P. Selitrennikoff. 1994. Cloning and characterization of a Neurospora crassa gene required for (1,3)β-glucan synthase activity and cell wall formation. Proc. Natl. Acad. Sci. USA 91:9500-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farley, J. F., R. A. Jersild, and D. J. Niederpruem. 1975. Origin and ultrastructure of intra-hyphal hyphae in Trichophyton terrestre and T. rubrum. Arch. Microbiol. 106:195-200. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson, M. A. 1999. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112:2799-2809. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson, M. A., J. S. Brimacombe, J. R. Brown, A. Crossman, A. Dix, R. A. Field, M. L. Guther, K. G. Milne, D. K. Sharma, and T. K. Smith. 1999. The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim. Biophys. Acta 1455:327-340. [DOI] [PubMed] [Google Scholar]
- 21.Flury, I., A. Benachour, and A. Conzelmann. 2000. YLL031c belongs to a novel family of membrane proteins involved in the transfer of ethanolaminephosphate onto the core structure of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 275:24458-24465. [DOI] [PubMed] [Google Scholar]
- 22.Fraering, P., I. Imhof, U. Meyer, J.-M. Strub, A. Dorsselaer, C. Vionnet, and A. Conzelmann. 2001. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 12:3295-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frieman, M. B., J. M. McCaffery, and B. P. Cormack. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479-492. [DOI] [PubMed] [Google Scholar]
- 24.Frieman, M. B., and B. P. Cormack. 2004. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae. Microbiology 150:3105-3114. [DOI] [PubMed] [Google Scholar]
- 25.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 26.Gaynor, E. C., G. Mondesert, S. J. Grimme, S. I. Reed, P. Orlean, and S. D. Emr. 1999. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 10:627-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gil-Navarro, I., M. L. Gil, M. Casanova, J. E. O'Connor, J. P. Martinez, and D. Gozalbo. 1997. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorlach, J. M., E. Van der Knaap, and J. D. Walton. 1998. Cloning and targeted disruption of MLG1, a gene encoding two of three extracellular mixed-linked glucanases of Cochliobolus carbonum. Appl. Environ. Microbiol. 64:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamburger, D., M. Egerton, and H. Riezman. 1995. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 129:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong, Y., Y. Maeda, R. Watanabe, K. Ohishi, M. Mishkind, H. Riezman, and T. Kinoshita. 1999. Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 274:35099-35106. [DOI] [PubMed] [Google Scholar]
- 31.Hong, Y., Y. Maeda, R. Watanabe, N. Inoue, K. Ohishi, and T. Kinoshita. 2000. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275:20911-20919. [DOI] [PubMed] [Google Scholar]
- 32.Hong, Y., K. Ohishi, J. Y. Kang, S. Tanaka, N. Inoue, J. Nishimura, Y. Maeda, and T. Kinoshita. 2003. Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell 5:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imhof, I., I. Flury, C. Vionnet, C. Roubaty, D. Egger, and A. Conzelmann. 2004. Glycosylphosphatidylinositol (GPI) proteins of Saccharomyces cerevisiae contain ethanolamine phosphate groups on the α1,4-linked mannose of the GPI anchor. J. Biol. Chem. 279:19614-19627. [DOI] [PubMed] [Google Scholar]
- 35.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita, T., and N. Inoue. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632-638. [DOI] [PubMed] [Google Scholar]
- 37.Kothe, G. O., and S. J. Free. 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149:117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenhard, J. M., E. Kasperek, B. R. Moore, and S. J. Free. 1989. Developing Dictyostelium discoideum cells contain two distinct acid hydrolase-containing vesicles. Exp. Cell Res. 182:242-255. [DOI] [PubMed] [Google Scholar]
- 39.Lim, L. L., B. A. Fineran, and A. L. J. Cole. 1983. Ultrastructure of intrahyphal hyphae of Glomus fasciculatum (thaxter) Gerdemann and Trappe in roots of white clover (Trifolium repens L.). New Phytol. 95:231-239. [Google Scholar]
- 40.Lowry, R. J., and A. S. Sussman. 1966. Intra-hyphal hyphae in “clock” mutants of Neurospora. Mycologia 58:541-548. [PubMed] [Google Scholar]
- 41.Mayor, S., and H. Riezman. 2004. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5:110-120. [DOI] [PubMed] [Google Scholar]
- 42.Michan, S., F. Lledias, J. D. Baldwin, D. O. Natvig, and W. Hansberg. 2002. Regulation and oxidation of two large monofunctional catalases. Free Radic. Biol. Med. 33:521-532. [DOI] [PubMed] [Google Scholar]
- 43.Miller, C. V., and N. A. Anderson. 1961. Proliferation of conidiophores and intrahyphal hyphae in Aspergillus niger. Mycologia 53:433-436. [Google Scholar]
- 44.Mouyna, I., M. Monod, T. Fontaine, B. Henrissat, B. Lechenne, and J.-P. Latge. 2000. Identification of the catalytic residues of the first family of beta(1-3)glucanosyl-transferases identified in fungi. Biochem. J. 347:741-747. [PMC free article] [PubMed] [Google Scholar]
- 45.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J.-P. Latge. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882-14889. [DOI] [PubMed] [Google Scholar]
- 46.Mouyna, I., W. Morelle, M. Vai, M. Monod, B. Lechenne, T. Fontaine, A. Beauvais, J. Sarfati, M.-C. Prevost, C. Henry, and J.-P. Latge. 2005. Deletion of GEL2 encoding for a β(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 56:1675-1688. [DOI] [PubMed] [Google Scholar]
- 47.Noventa, M. A., J. A. Jorge, and H. F. Terenzi. 1991. Role of the cell wall on the expression of osmotic-sensitive (os-1) and temperature-sensitive (cot-1) phenotypes of N. crassa. A comparative study on mycelial and wall-less phenotypes of the “slime” variant. Fungal Genet. Newsl. 38:80-82. [Google Scholar]
- 48.Ohishi, K., N. Inoue, Y. Maeda, J. Takeda, H. Riezman, and T. Kinoshita. 2000. Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell 11:1523-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohishi, K., N. Inoue, and T. Kinoshita. 2001. PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 20:4088-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pardo, M., L. Monteoliva, P. Vazquez, R. Martinez, G. Molero, C. Nombela, and G. Concha. 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150:4157-4170. [DOI] [PubMed] [Google Scholar]
- 51.Percival-Smith, A., and J. Segall. 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Sacharomyces cerevisiae. Mol. Cell. Biol. 6:2443-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popolo, L., D. Gilardelli, P. Bonfante, and M. Vai. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popolo, L., and M. Vai. 1999. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1426:385-400. [DOI] [PubMed] [Google Scholar]
- 54.Ram, A. F. J., A. Wolters, R. T. Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 55.Ram, A. F. J., M. Arentshorst, R. A. Damveld, P. A. vanKuyk, F. M. Klis, and C. A. M. J. J. van den Hondel. 2004. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine:fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 150:3315-3326. [DOI] [PubMed] [Google Scholar]
- 56.Richard, M., P. de Groot, O. Courtin, D. Poulain, F. Klis, and C. Gaillardin. 2002. GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans. Microbiology 148:2125-2133. [DOI] [PubMed] [Google Scholar]
- 57.Roncero, C., and A. Duran. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoffelmeer, E. A. M., J. H. Vossen, A. A. van Doorn, B. J. C. Cornelissen, and M. A. Haring. 2001. FEM-1, a Fusarium oxysporum glycoprotein that is covalently linked to the cell wall matrix and is conserved in filamentous fungi. Mol. Genet. Genomics 265:143-152. [DOI] [PubMed] [Google Scholar]
- 59.Schonbachler, M., A. Horvath, J. Fassler, and H. Riezman. 1995. The yeast spt14 gene is homologous to the human PIG-A gene and is required for GPI anchor synthesis. EMBO J. 14:1637-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seiler, S., and M. Plamann. 2003. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14:4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selker, E. U. 1999. Gene silencing: repeats that count. Cell 97:157-160. [DOI] [PubMed] [Google Scholar]
- 62.Sestak, S., I. Hagen, W. Tanner, and S. Strahl. 2004. Scw10p, a cell wall glucanase/transglucosidase important for cell wall stability in Saccharomyces cerevisiae. Microbiology 150:3197-3208. [DOI] [PubMed] [Google Scholar]
- 63.Shedletzky, E., C. Unger, and D. P. Delmer. 1997. A microtiter-based fluorescence assay for (1,3)-B-glucan synthases. Anal. Biochem. 249:88-93. [DOI] [PubMed] [Google Scholar]
- 64.Shishioh, N., Y. Hong, K. Ohishi, H. Ashida, Y. Maeda, and T. Kinoshita. 2005. GPI7 is the second partner of PIG-F and involved in modification of glycosylphosphatidylinositol. J. Biol. Chem. 280:9728-9734. [DOI] [PubMed] [Google Scholar]
- 65.Takaya, N., D. Yamazaki, H. Horiuchi, A. Ohta, and M. Takagi. 1998. Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci. Biotechnol. Biochem. 62:60-65. [DOI] [PubMed] [Google Scholar]
- 66.Taron, C. H., J. M. Wiedman, S. J. Grimme, and P. Orlean. 2000. Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate Gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell 11:1611-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terashima, H., N. Yabuki, M. Arisawa, K. Hamada, and K. Kitada. 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:64-74. [DOI] [PubMed] [Google Scholar]
- 68.Vainauskas, S., and A. K. Menon. 2004. A conserved proline in the last transmembrane segment of Gaa1 is required for glycosylphosphatidylinositol (GPI) recognition by GPI transamidase. J. Biol. Chem. 279:6540-6545. [DOI] [PubMed] [Google Scholar]
- 69.Vollmer, S. J., and C. Yanofsky. 1986. Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. USA 83:4869-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vossen, J. H., W. H. Muller, P. N. Lipke, and F. M. Klis. 1997. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 179:2202- 2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yarden, O., and C. Yanofsky. 1991. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 5:2420-2430. [DOI] [PubMed] [Google Scholar]
- 72.Yin, Q. Y., P. W. J. de Groot, H. L. Dekker, L. de Jong, F. M. Klis, and C. G. de Koster. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls. J. Biol. Chem. 280:20894-20901. [DOI] [PubMed] [Google Scholar]