Abstract
The unicellular flagellated green alga Chlamydomonas reinhardtii has emerged as a model organism for the study of a variety of cellular processes. Posttranslational control via protein phosphorylation plays a key role in signal transduction, regulation of gene expression, and control of metabolism. Thus, analysis of the phosphoproteome of C. reinhardtii can significantly enhance our understanding of various regulatory pathways. In this study, we have grown C. reinhardtii cultures in the presence of an inhibitor of Ser/Thr phosphatases to increase the phosphoprotein pool. Phosphopeptides from these cells were enriched by immobilized metal-ion affinity chromatography and analyzed by nano-liquid chromatography-electrospray ionization-mass spectrometry (MS) with MS-MS as well as neutral-loss-triggered MS-MS-MS spectra. In this way, we were able to identify 360 phosphopeptides from 328 different phosphoproteins of C. reinhardtii, thus providing new insights into a variety of cellular processes, including metabolic and signaling pathways. Comparative analysis of the phosphoproteome also yielded new functional information on proteins controlled by redox regulation (thioredoxin target proteins) and proteins of the chloroplast 70S ribosome, the centriole, and especially the flagella, for which 32 phosphoproteins were identified. The high yield of phosphoproteins of the latter correlates well with the presence of several flagellar kinases and indicates that phosphorylation/dephosphorylation represents one of the key regulatory mechanisms of eukaryotic cilia. Our data also provide new insights into certain cilium-related mammalian diseases.
The unicellular green alga Chlamydomonas reinhardtii serves as a model for a wide range of biological processes (5, 8). Some of them, such as photosynthesis and nitrogen metabolism, are common to plant cells, while others, such as the structure and composition of flagella and basal bodies, are similar to animal cells and highly relevant for the understanding of several human diseases. Still others, such as sensing and acclimation to environmental factors or the control of biological processes by the circadian clock, are fundamental to all eukaryotes. In recent years, specific cellular processes and cell “compartments” of C. reinhardtii have been investigated by applying proteomic strategies. This is possible for two main reasons: (i) the alga can be easily and quickly grown in large amounts, and therefore biochemical purification procedures for certain subproteomes can be well established; and (ii) genome sequences from all three genetic compartments of Chlamydomonas (nucleus, mitochondria, and chloroplast) are available, as well as more than 200,000 expressed sequence tags (ESTs), which have been assembled into 10,000 contigs representing unique cDNAs (6). In the meantime, proteomics was used in C. reinhardtii to investigate, for example, the chloroplast 70S ribosome (33, 34), light-harvesting proteins (31), new thioredoxin targets (18), novel components of the circadian clock (32), the flagella (24), and centrioles (13).
Our current approach is to apply proteomics to the investigation of posttranslational modifications in C. reinhardtii. One of the key modifications of proteins, which is crucial in the control of many regulatory pathways and can effect protein function, activity, stability, localization, and interactions, is phosphorylation. Therefore, information about the phosphoproteome of C. reinhardtii is extremely useful for understanding a variety of cellular processes in this alga, which may serve as a basis for examining these processes in other organisms. Several phosphoproteins of C. reinhardtii, such as the α heavy chain of outer arm dynein (16) and IC138, a WD repeat dynein intermediate chain (10, 15), have already been identified. Also, several radial spoke proteins of the flagella have been characterized as phosphoproteins via in vivo 32P labeling (26). Metabolic enzymes such as cytosolic glutamine synthetase (27) and proteins of the photosynthetic machinery (e.g., Cp29-like protein [12]) of C. reinhardtii have also been identified as phosphoproteins.
Proteome analysis of phosphoproteins is a challenging task (20, 28). Phosphoproteins can possess more than one phosphorylation site, and the phosphorylation status of these sites can fluctuate depending on the physiological conditions under which the cells are kept. This leads to a great variety of phosphoproteins. In addition, the ratio of the phosphorylated to nonphosphorylated form of a protein can be very low. Although proteins can be identified down to the femtomole, and even attomole, level with modern mass spectrometry (MS), many phosphoproteins within a crude extract (especially those of cell signaling pathways) are not abundant enough to be unambiguously identified by MS. For this reason, enrichment of such proteins is often a prerequisite for efficient phosphoproteome analysis.
Different methods that can be used for this purpose have been described in the literature (reviewed in references 11 and 28). One of them, immobilized metal-ion affinity chromatography (IMAC), is based on the presence of negatively charged phosphate groups and enriches for phosphorylated Ser, Thr, and Tyr. This method has been applied, for example, to the analysis of phosphoproteins from yeast (4, 7) and from a lymphoma cell line (30). The IMAC method relies on direct identification of phosphopeptides by MS, in contrast to other methods that chemically substitute for the phosphate residue (11, 28). However, in tandem MS (MS2), phosphopeptide precursor ions can exhibit a neutral loss of phosphoric acid (−98 Da). The reason for this loss is that phosphopeptides (phosphoserine and phosphothreonine) can undergo gas-phase β elimination when subjected to collision-induced fragmentation (20). Because m/z (mass/charge) values, and not absolute masses, are measured in a mass spectrometer, doubly and triply charged peptide ions show an apparent loss of 49 and 32.66, respectively. In the MS2 spectrum, the presence of a neutral loss therefore indicates phosphorylation and can be used as a selection parameter for phosphopeptides.
Recently, a new type of linear-ion-trap electrospray ionization (ESI)-MS was developed, permitting the acquisition of data-dependent neutral-loss experiments. With this instrument (Finnigan LTQ; Thermo Electron Corp., San Jose, CA), neutral-loss analysis can be executed during MS measurements (MS2 scan). If a pair of peaks of the most prominent ion of the MS2 spectrum, versus the full-scan MS spectrum, is found with a mass difference of 98, 49, or 32.66 (depending on the charge of the peptide ion), a phosphorylation-specific neutral loss is indicated. This prominent ion of the MS2 spectrum (representing the dephosphorylated peptide) is then selected for an automatically triggered MS3 scan. Thus, sufficient fragment ion information with a significant cross-correlation (Xcorr) factor (2) can be obtained. The Xcorr factor describes the cross-correlation between the experimentally measured MS2/MS3 spectrum and an in silico-generated MS2/MS3 spectrum of candidate peptides in the databases. The data-dependent neutral-loss-triggered MS3 mode of operation has been recently applied successfully to the study of phosphoproteins from the yeast signaling pathway (7).
So far, relatively little information about phosphoproteomes in plants is known. Among the few examples, phosphoproteins of Arabidopsis plasma membranes (23) and phosphoproteins in the moss Physcomitrella patens (9) have been characterized. Here, we have enriched phosphoproteins from C. reinhardtii by using IMAC in combination with Ga3+ as the metal component. For this purpose, cells were grown in presence of a strong inhibitor of Ser/Thr phosphatases (okadaic acid) to increase the phosphorylation status of the proteins. It was previously shown that the growth of cells in the presence of such an inhibitor can robustly increase phosphorylation on specific sites (21), and such inhibitors have already been used successfully for phosphoproteome analysis (30). Using the information from MS2 and neutral-loss-triggered MS3 spectra, we identified 328 phosphoproteins in C. reinhardtii with 360 phosphopeptides. These results provide insight not only into phosphorylation events within various metabolic and signaling pathways but also into tactic movements, based on comparative proteome analysis with proteins from the centriole and the flagella.
MATERIALS AND METHODS
Cell culture.
C. reinhardtii wild-type strain SAG 73.72 was grown in high-salt-acetate medium under a 12-h light/12-h dark cycle with a light intensity of 71 μE/m2/s at 24°C and was then put under constant conditions of dim light (15 μE/m2/s) before harvest (22). At the time point at which the cells were transferred to dim light, okadaic acid was added to the culture, in some cases, with a final concentration of 1.5 μM. Cells were harvested after they had been kept for 29 h under dim light at a cell density of 2 × 106 to 3 × 106 cells/ml.
Preparation of crude extracts of C. reinhardtii.
Cells from a 100-ml culture were harvested by centrifugation and stored in liquid nitrogen. Extract preparation was based on the protocols of Shu et al. (30) and Mittag (22) with some modifications. Cells were resuspended in TriPure isolation reagent (Roche Applied Science), and glass beads (diameter, 0.25 to 0.30 mm) that had been prewashed once with TriPure isolation reagent were added. Cells were disrupted by vortexing this mixture five times for 1 min each, and the tubes were placed on ice for 1 min between each vortexing step. After a centrifugation step (200 × g, 2 s), the supernatant was subjected to the TriPure procedure for removal of RNA and DNA, according to the Roche manual. Proteins were precipitated with isopropanol, and the pellet was washed three times with 0.3 M guanidine hydrochloride in 95% ethanol and finally with 100% ethanol. A 6 M guanidine hydrochloride solution (100 μl) was added to dissolve the protein pellet. For tryptic digestion, the solubilized proteins were diluted 10-fold in 100 mM ammonium bicarbonate and the protein concentration was determined with the Bio-Rad protein assay. The digest was performed with 20 μg of trypsin (Promega) and 1 mg of solubilized protein overnight at 37°C. Then, acetonitrile and formic acid were added to the sample to reach final concentrations of 2% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid. The tryptic peptide solution was centrifuged (10,000 × g, 4°C, 5 min), and the supernatant was put on a fast protein liquid chromatography (LC) system (Amersham Biosciences)-driven 1-ml column (SOURCE RPC; Amersham Biosciences [now part of GE Healthcare]) for desalting. Peptides were eluted with 500 μl of 80% (vol/vol) acetonitrile-0.1% (vol/vol) formic acid. The desalted sample was subjected to the IMAC procedure.
Enrichment of proteins by IMAC.
The IMAC procedure was performed according to Shu et al. (30) with some modifications. For IMAC, we used homemade microcolumns prepared according to the procedure of Erdjument-Bromage et al. (3). Fifty microliters of POROS 20 MC (catalog no. 1-5428-02; Applied Biosystems) metal-chelating resin (66% [wt/wt] slurry) was pipetted into a microtip column (Eppendorf gel loading tip). Then, two 50-μl volumes of 0.1% (vol/vol) acetic acid were passed through the column. The charging of the metal-chelating resin was carried out by applying 150 μl of 100 mM GaCl3. To remove excess GaCl3, the column was washed with 50 μl of 0.1% (vol/vol) acetic acid. Afterward, the tryptically digested and desalted peptide solution (see above) was loaded onto the activated IMAC column. The column was washed successively with 50 μl of 0.1% (vol/vol) acetic acid, 50 μl of 50% (vol/vol) acetonitrile-0.1% (vol/vol) acetic acid, 50 μl of 50% (vol/vol) acetonitrile-0.1% (vol/vol) acetic acid-100 mM sodium chloride, and finally 50 μl of 0.1% (vol/vol) acetic acid. Then, phosphopeptides were eluted with three 20-μl volumes of 200 mM Na2HPO4 and desalted with a ZipTip microtip (Perseptive Biosystems) according to the method of Stauber et al. (31). The ZipTip contained C18 reversed-phase chromatographic medium. In addition, a 50-μl mixture of POROS R2 (Applied Biosystems) and methanol (1:5) was loaded, and the column was washed two times with 5% (vol/vol) methanol-5% (vol/vol) formic acid. To improve the binding of the phosphopeptides to the ZipTip microtip, formic acid was added to the sample to a final concentration of 5% (vol/vol) prior to loading. Then, the column was again washed two times with 50 μl of 5% (vol/vol) methanol-5% (vol/vol) formic acid. Peptides were eluted with two 50-μl volumes of 60% (vol/vol) methanol-5% (vol/vol) formic acid. The resulting 100-μl sample solution was dried in a SpeedVac concentrator (about 2 to 3 h), and the pellet was then stored at −70°C.
Peptide identification by nano-LC-ESI-MS (MS2 and neutral-loss-triggered MS3).
After the dried pellet was dissolved in 5 μl of 5% (vol/vol) acetonitrile-0.1% (vol/vol) formic acid, the phosphopeptides were subjected to LC-ESI-MS using a nanoscale C18 column (flow rate, 600 nl/min) coupled online with a linear-ion-trap mass spectrometer (Finnigan LTQ; Thermo Electron Corp., San Jose, CA). The LC system consisted of a Surveyor high-performance liquid chromatograph (Thermo Electron), including a Surveyor autosampler and MS pump. A gradient was used to elute peptides from the reversed-phase column (Picofrit) (length, 10 cm; inner diameter, 100 μm; Magic-C18 particle size, 5 μm [Spectronex]). The successive steps of the applied gradient were as follows: 10 min, 100% (vol/vol) A; 10 min, gradually shifting to 70% (vol/vol) A-30% (vol/vol) B; 20 min, gradually shifting to 40% (vol/vol) A-60% (vol/vol) B; 10 min, gradually shifting to 20% (vol/vol) A-80% (vol/vol) B; 10 min, 20% (vol/vol) A-80% (vol/vol) B; and 1 min, gradually shifting to 100% (vol/vol) A, where A consists of 0.1% (vol/vol) formic acid in water and B consists of 0.1% (vol/vol) formic acid in acetonitrile. The instrument was run by the data-dependent neutral-loss method, cycling between one full MS scan and MS2 scans of the four most-abundant ions. The detection of a neutral-loss fragment (98, 49, or 32.66 Da) in the MS2 scans triggered immediately an MS3 scan of the precursor ion representing the dephosphorylated peptide. The MS2 and MS3 data were used to search the available Chlamydomonas reinhardtii databases (see below) using Bioworks software (version 3.1; Thermo Electron Corp., San Jose, CA) including the SEQUEST algorithm (19). The software parameters were set to detect a modification of 79.96 Da in Ser, Thr, or Tyr in MS2 and MS3 spectra. When phosphoserine and phosphothreonine undergo gas-phase β elimination, dehydroalanine (Dha) and 2-aminodehydrobuturic acid (methyldehydroalanine [MeDha]), respectively, are produced. Thus, modifications of −18.00 Da in Ser and Thr residues were additionally used for database searches with MS3 data. Searches were done for tryptic peptides, allowing two missed cleavages. Mass tolerance was set to 1.5 Da for the peptide precursor ion in MS mode. For fragment ions (MS2 and MS3 modes), mass tolerance was set to 0.5 Da. It should be noted that Bioworks software (version 3.1) includes masses of fragment ions up to 1 Da when this setting is applied. Scores for the Xcorr factor (2) were set to the following limits: Xcorr of >1.5 if the charge of the peptide was 1, Xcorr of >2 if the charge of the peptide was 2, and Xcorr of >2.5 if the charge of the peptide was 3. Peptide searches were done with the following databases: genomic databases, including their predicted proteins (version 2), available from the Joint Genome Institute (JGI) facility (http://genome.jgi-psf.org/chlre2/chlre2.home.html); the Kazusa EST database (http://www.kazusa.or.jp/en/plant/chlamy/EST); the Chlamy Center EST database (http://www.chlamy.org) (31); the NCBI mitochondrion database (gi|11467088|ref|NC_001638.1|); and the chloroplast database (http://www.chlamy.org/chloro/default.html). As Bioworks software (version 3.1) ignores stop codons from translated DNA sequences of genome and EST databases, an in-house program was developed to automatically delete such false positives.
Phosphothreonine immunoblotting.
Lysates from control and okadaic acid-treated cells were immunoblotted with an antiphosphothreonine antibody. C. reinhardtii cells were either left untreated or treated with 1.5 μM okadaic acid for 29 h under dim light. Crude extracts were prepared as described previously (22), and proteins were denatured in sodium dodecyl sulfate (SDS) sample buffer (30). The proteins (50 and 100 μg, respectively) were resolved by 9% SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and immunoblotted with antiphosphothreonine antibody, according to the manufacturer's protocol (catalog no. 9381; Cell Signaling Technologies) with one modification: the commercially available soya-based powder Slimfast was used instead of nonfat dried milk powder in the blocking buffer.
RESULTS
Treatment of C. reinhardtii with okadaic acid, a potent inhibitor of Ser/Thr phosphatases, leads to a significant increase in the level of phosphoproteins.
Incubation of growing cells with an inhibitor of Ser/Thr phosphatases (e.g., calyculin) can robustly increase the phosphoprotein pool (30). Another inhibitor, okadaic acid, was shown to change specifically the ratio of nonphosphorylated versus phosphorylated sites within a given protein (21). To check whether treatment of C. reinhardtii with such an inhibitor would significantly increase the yield of phosphoproteins, cells were grown in the presence or absence of okadaic acid. We checked different incubation times to determine how long cells can be incubated with okadaic acid without influencing their viability. On the second (sample 1), fourth (sample 2), and fifth (sample 3) days of incubation with okadaic acid, a small amount of cells was removed and the motility of the cells was examined under the microscope. On the second and fourth days (samples 1 and 2), nearly all cells were fully motile. On day five (sample 3), only 10 to 15% of the cells were still motile and about 10% appeared dead; 75 to 80% of the cells had flagella that were still beating, but the cells were not able to move forward. Thus, we selected an incubation period of 29 h with okadaic acid for further experiments.
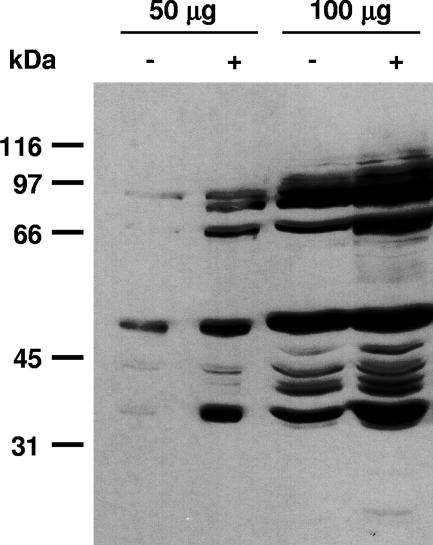
To analyze whether treatment with okadaic acid for 29 h would significantly increase the levels of phosphoproteins in C. reinhardtii, we prepared crude extracts from okadaic acid-treated and untreated cells and analyzed proteins by immunoblotting with antiphosphothreonine antibodies (for details, see Materials and Methods). Immunoblotting showed that okadaic acid significantly increases Thr phosphorylation in C. reinhardtii (Fig. 1). An increase in the level of phosphoproteins in okadaic acid-treated versus untreated cells was also observed when the yield of phosphoproteins within these two phosphoproteomes was compared (see “Identification of phosphopeptides from C. reinhardtii by nano-LC-ESI-MS,” below).
FIG. 1.
Enrichment of phosphorylated proteins by okadaic acid treatment of C. reinhardtii. Cells were either left untreated (−) or treated with 1.5 μM okadaic acid (+), as described in Materials and Methods, harvested, and used for preparation of a crude extract. Proteins (50 and 100 μg) were denatured in SDS sample buffer, resolved on a 9% SDS gel, transferred to a nitrocellulose membrane, and immunoblotted with antiphosphothreonine polyclonal antibody (Cell Signaling Technologies).
Purification procedure for enrichment of phosphopeptides and efficiency of MS analysis with complex peptide mixtures tested with internal-standard phosphopeptides.
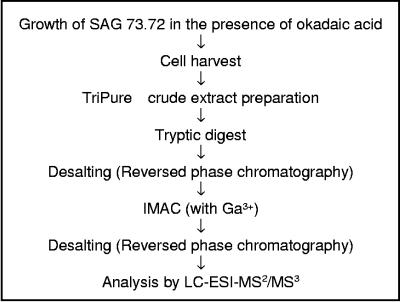
Considering the low expression levels and phosphorylation stoichiometries of many proteins regulated by phosphorylation, an efficient enrichment strategy for phosphopeptides is a prerequisite for analysis of the phosphoproteome. We used a crude extract preparation based on TriPure agent, digestion of the proteins with trypsin, and subsequently IMAC in combination with Ga3+ as the metal component for this enrichment procedure (for details, see Materials and Methods). A short outline of the procedure is shown in Fig. 2.
FIG. 2.
Schema for the enrichment and analysis of phosphopeptides from C. reinhardtii. The details are described in Materials and Methods. Nano-LC-ESI-MS (MS2 and MS3) analysis was carried out in a mass spectrometer with a linear ion trap, permitting the acquisition of data-dependent neutral loss (Finnigan LTQ; Thermo Electron Corp., San Jose, CA).
We examined the correct assignment of phosphopeptides and their phosphorylation sites by using an internal phosphopeptide standard. Thereby, phosphopeptides from 250 μg of beta-casein (Bos taurus) were isolated by IMAC from the trypsin-digested peptides of beta-casein by using the procedure described above. These phosphopeptides were then mixed with each phosphopeptide sample from C. reinhardtii before MS analysis to a concentration of 10 μM (under the premise that 100% of the phosphopeptides from beta-casein would have been enriched by the IMAC procedure). The complete phosphopeptide mixture was submitted to LC-ESI-MS using the data-dependent neutral-loss experiment, which selectively triggers MS3 scans only of those MS2 fragment ions for which a prominent neutral loss is detected. The experimentally measured MS2 and MS3 spectra were then compared with in silico-generated spectra of candidate peptides from beta-casein. Beta-casein is known to contain two phosphopeptides. One of them contains one phosphorylation site (FQSpEEQQQTEDELQDK), while the other has four (RELEELNVPGEIVESpLSpSpSpEESITR). Detection of the tetraphosphopeptide is known to be problematic for LC/MS (14). We were able to detect both phosphopeptides from beta-casein within the phosphopeptide mixture of C. reinhardtii (Table 1). The monophosphopeptide was identified with significant Xcorr factors from MS2 (see Fig. S1A in the supplemental material), as well as from the neutral-loss-triggered MS3 spectra (see Fig. S1B in the supplemental material). The tetraphosphopeptide could not be identified based on the MS2 spectra. However, it could be found in the neutral-loss-triggered MS3 spectra with a significant Xcorr factor (see Fig. S2 in the supplemental material). For both peptides, Dha residues were detected in the MS3 spectra, showing at which particular Ser the neutral loss took place. Notably, not all four phosphoserines showed a neutral loss, and the site of the neutral-loss event was subject to change in the tetraphosphopeptide. These data show the advantage of neutral-loss-triggered MS3 spectra. They also reveal that neutral-loss events of a specific phosphoserine/phosphothreonine can be subject to variation within a given phosphopeptide and that phosphoserines/phosphothreonines do not necessarily show a neutral loss.
TABLE 1.
Identification of phosphopeptides from beta-casein, which were added to the total phosphopeptide sample from C. reinhardtii, by LC-ESI-MS2 and neutral-loss-triggered MS3
| Phosphopeptide sequencea | NCBI accession no. | MS type | zb | Xcorr |
|---|---|---|---|---|
| FQS*EEQQQTEDELQDK | gi|30794310 | |||
| FQSpEEQQQTEDELQDK | MS2 | 2 | 5.20 | |
| FQ(Dha)EEQQQTEDELQDK | MS3 | 2 | 5.44 | |
| RELEELNVPGEIVES*LS*S*S*EESITR | gi|30794310 | |||
| RELEELNVPGEIVESpLSp(Dha)(Dha)EESITR | MS3 | 3 | 5.60 | |
| RELEELNVPGEIVESpL(Dha)(Dha)SpEESITR | MS3 | 3 | 5.55 |
*, known phosphorylation sites within the beta-casein phosphopeptides (15); p, phosphorylated residue; Dha, dehydroalanine.
z, charge.
Identification of phosphopeptides from C. reinhardtii by nano-LC-ESI-MS (MS2 and neutral-loss-triggered MS3).
The above-mentioned results with okadaic acid (Fig. 1) suggested that treatment of the cells with okadaic acid would significantly increase the yield of the phosphoproteome. We analyzed once the number of phosphoproteins using cells that were untreated and compared the yield to that of cells that were incubated for 29 h with okadaic acid. For comparison of the yields, only phosphopeptides that (i) had a significant Xcorr factor (see Materials and Methods) and (ii) could be found within the open reading frame (ORF) of any predicted gene model were counted. There was a significant increase visible in the yield of phosphoproteins when the cells were grown in the presence of the inhibitor. In this case, 38% more phosphoproteins were obtained. Therefore, we chose incubation of cells with okadaic acid for performance of a large-scale analysis of the phosphoproteome of C. reinhardtii.
In four independent experiments, cells were grown in the presence of 1.5 μM okadaic acid for 29 h and harvested, and the phosphopeptides were obtained by the procedure described in Fig. 2. To increase the confidence of the phosphopeptide assignment, we chose only phosphopeptides that fulfilled three criteria for further analysis. (i) They appeared at least in two of the four independent experiments. (ii) They showed a significant Xcorr factor (see Materials and Methods). (iii) They could be identified (a) within the protein sequence of a predicted gene model, (b) within the ORF of an EST assembly, or (c) within a potential part of an ORF (potential exon) predicted after translation of the genome sequence itself. The latter (c) was included only if this potential exon had significant homology (a BLAST E score of ≤1 × 10−5) to an already known protein from any other organism. Thus, the yield of the phosphopeptides correlates to a high degree with the current number of gene models predicted by JGI from the available genome sequence (version 2) and the available EST data.
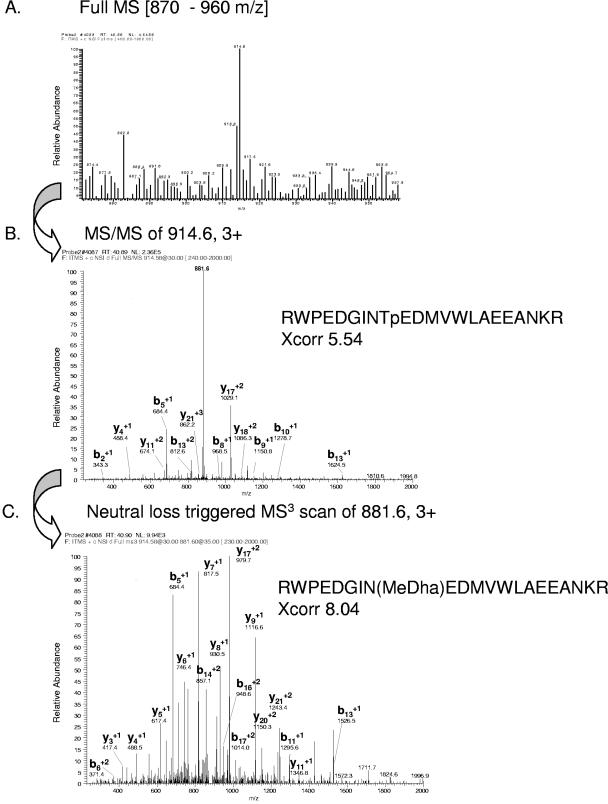
Using these criteria, we were able to identify 186 phosphoproteins of known and putative function (see Table S1 in the supplemental material) and 142 novel phosphoproteins of as-yet-unknown function (see Table S2 in the supplemental material). Certain phosphopeptides could be identified solely by their MS2 spectra with a significant Xcorr factor. These include almost all phosphotyrosine-containing peptides but also Ser/Thr phosphorylated peptides. Some phosphopeptides were identified by MS2 spectra and additionally by the corresponding MS3 spectra of their neutral-loss peptide (indicated in bold in Tables S1 and S2 in the supplemental material). An example is depicted in Fig. 3. A detailed view of the full MS scan with the precursor ion m/z 914.6 is shown in Fig. 3A. The MS2 spectrum of the peptide ion showing the neutral loss led to identification of the phosphopeptide RWPEDGINTpEDMVWLAEEANKR with an Xcorr factor of 5.54 (Fig. 3B). Analysis of the MS3 fragmentation spectrum from the peptide ion m/z 881.6 revealed the same peptide with the 2-aminodehydrobuturic acid residue (MeDha) as a result of the neutral-loss event with an Xcorr factor of 8.0, showing nearly a complete set of the corresponding y- and b-ion series (Fig. 3C).
FIG. 3.
Identification of a phosphopeptide from a MORN (for “membrane occupation and recognition nexus”) repeat protein (gene model C_190173) by nano-LC-ESI-MS2 and by neutral-loss-triggered MS3. (A) Full MS scan in the m/z range of 870 to 960 with the prominent peptide ion 914.6. (B) Identification of the phosphopeptide RWPEDGINTpEDMVWLAEEANKR by the MS2 fragmentation pattern of peptide ion 914.6. “p” indicates the phosphorylation site on Thr. (C) Identification of the peptide RWPEDGIN(MeDha)EDMVWLAEEANKR by the MS3 fragmentation pattern of the detected neutral-loss fragment 881.6. “MeDha” indicates the site of the neutral loss of phosphoric acid from the phosphothreonine. Only prominent y- and b-fragment ions have been labeled.
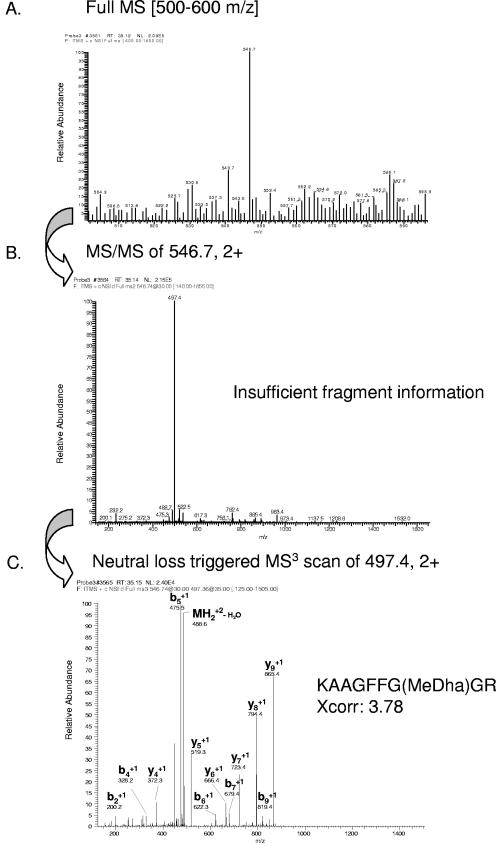
In some cases, the peptide sequence could not be unambiguously identified with the MS2 spectra but only with the MS3 spectra (indicated by parallel bold and italic text in Tables S1 and S2 in the supplemental material). Again, an example is depicted (Fig. 4). Figure 4A shows the full MS scan. The corresponding MS2 spectra of the peptide ion m/z 546.7 revealed insufficient fragment information, with only one major peak from the neutral-loss peptide ion (Fig. 4B). However, the MS3 spectra from the neutral-loss-derived peptide ion m/z 497.4 led to identification of the peptide KAAGFFG(MeDha)GR, which reveals phosphorylation of the Thr residue (Fig. 4C).
FIG.4.
Identification of a phosphopeptide from a novel protein of unknown function (gene model C_50124) by neutral-loss-triggered nano-LC-ESI-MS3. (A) Full MS scan in the m/z range of 500 to 600 detects the prominent peptide ion 546.7. (B) The MS2 fragmentation spectrum of peptide ion 546.7 reveals the neutral loss (fragment ion 497.4) but no fragment information sufficient for peptide identification. (C) Identification of the peptide KAAGFFG(MeDha)GR by neutral-loss-triggered MS3 of peptide ion 479.4. “MeDha” indicates the site of the neutral loss of phosphoric acid from the phosphothreonine. Only prominent y- and b-fragment ions have been labeled.
Most of the phosphoproteins were identified by one peptide bearing one or more phosphorylated residues. However, we also detected phosphoproteins (32 in all) that had more than one phosphorylated peptide or a different number of phosphorylation sites within a given peptide. In certain cases, we could not specify the exact site(s) of phosphorylation within a peptide with certainty. These cases are all marked in Tables S1 and S2 (in the supplemental material) by addition of an “n” at the end of the peptide sequence.
Functional analysis of the phosphoproteome of C. reinhardtii.
We searched for the functions of all phosphoproteins by using the information on the JGI website, including annotation notes as well as information about conserved domains. Further, we did an NCBI homology search (NCBI protein BLAST). An overview of the output of the functional analysis is shown in Table 2, and a detailed view is presented in Tables S1 and S2 in the supplemental material. Thereby, proteins were labeled as novel proteins of as-yet-unknown function (see Table S2 in the supplemental material) if (i) the protein had not yet been characterized for C. reinhardtii, (ii) it had either no or weak homology (a BLAST E score of ≤1 × 10−5) to a protein from any other organism, or (iii) it had significant homology to a protein from any other organism but the function of the protein in the other organism was unknown and no conserved domain could be found indicating any putative function.
TABLE 2.
Functional analysis of identified phosphoproteins
| Functional categories | No. of proteinsa |
|---|---|
| Proteins of the flagella and the centriole | 36 |
| Nucleic acid-binding and ribosomal proteins | 31 |
| Kinases and phosphatases | 26 |
| Enzymes | 23 |
| Transport/membrane proteins and receptors | 17 |
| Proteins with protein-protein interaction domains | 8 |
| ATPases | 6 |
| Proteins with other functions as listed above | 39 |
| Novel proteins of as-yet-unknown function | 142 |
Proteins that appear in the flagella/centriole and can be grouped at the same time under some other category have been counted only once (under flagella/centriole).
In Table S1 in the supplemental material, all information about functional implications is listed, indicating whether (i) the protein has already been characterized for C. reinhardtii or (ii) its function either is predicted based on homology to proteins from other organisms or is putative if only conserved domains are present. Within the phosphoproteome of C. reinhardtii, we were able to identify proteins from a variety of cellular processes, including, e.g., enzymes from different metabolic pathways, DNA- and RNA-binding proteins, and several kinases, transporters, and receptors as well as proteins that are located in the flagella or the centriole. Still, the functions of a great percentage of phosphoproteins cannot yet be predicted (see Table S2 in the supplemental material) since they are unique and unexamined in C. reinhardtii or since their homologs in other organisms have also not been investigated up to now.
We also performed a comparative proteome analysis with already known subproteomes from the chloroplast 70S ribosome (33, 34), thioredoxin interaction partners (18), the flagella (24), and the centriole (13). In this way, we were able to identify several proteins of these subproteomes that are subject to phosphorylation (Table 3). A few of them even appeared in more than one subproteome; these are indicated in Table 3. In summary, 1 protein of the chloroplast 70S ribosome, 7 from the thioredoxin targets, 4 from the centriole, and 32 from the flagella were detected. The relatively high yield of phosphoproteins from the flagella is reflected by higher phosphopeptide coverage (41 phosphopeptides in all). Some of the phosphoproteins of the flagella are related to certain human diseases, which are discussed below.
TABLE 3.
Comparative proteome analysis with the phosphoproteome of C. reinhardtii
| Gene model (JGI, version 2)a | Function and/or homologies of depicted proteins to proteins of other organisms, their conserved domains, and their NCBI accession no.b |
|---|---|
| Flagella (24) | |
| C_20170 | gi|23128981|ref|ZP_00110817.1| COG2319: FOG: WD40 repeat (Nostoc punctiforme PCC 73102) |
| C_20337 (C)* | gi|1353876|gb|AAB01817.1| glutamine synthetase (Chlamydomonas reinhardtii); u.a., GS1, glutamine synthetase, cytosolic isozyme, CO2-responsive gene |
| C_50072 | gi|23128003|ref|ZP_00109860.1| COG2453: predicted protein, tyrosine phosphatase (N. punctiforme PCC73102); KOG: protein tyrosine phosphatase (CDC14) |
| C_50129 | gi|12620209|gb|AAG60619.1| adenylate cyclase (Cryptococcus neoformans var. grubii); gnl|CDD|37 smart00044, CYCc, Adenylyl/guanylyl cyclase, catalytic domain |
| C_60100 | gi|23479871|gb|EAA16587.1| R27-2 protein (Plasmodium yoelii yoelii); gnl|CDD|10914 COG1196, Smc, chromosome segregation ATPases |
| C_90101 | gi|21039486|gb|AAM33652.1| mastigoneme-like protein (C. reinhardtii); KOG: fibrillins and related proteins containing Ca2+-binding EGF-like domains |
| C_110095 | u.a., coiled-coil protein |
| C_150121 | gi|50906913|ref|XP_464945.1| putative PKG-lb (Oryza sativa japonica cultivar group); gnl|CDD|17776 cd00180, S_TKc, serine/threonine protein kinases, catalytic domain; gnl|CDD|16536 cd00143, PP2Cc, serine/threonine phosphatases, family 2C, catalytic domain; gnl|CDD|14778 cd00038, CAP_ED, effector domain of the CAP family of transcription factors |
| C_270111 | gi|62650163|ref|XP_575971.1| similar to hypothetical protein A530045M11 (Rattus norvegicus); u.a., FAP74, conserved uncharacterized flagellum-associated protein |
| C_290071 | gi|68353830|ref|XP_690098.1| similar to cold autoinflammatory syndrome 1 protein (cryopyrin) (NACHT-, LRR-, and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like) (Danio rerio); c.d., leucine-rich repeats, ribonuclease inhibitor-like subfamily |
| C_360006 | gi|27529748|dbj|BAA76788.2| KIAA0944 protein (Homo sapiens); u.a., DHC6, dynein heavy chain 6 (putative flagellar inner-arm dynein heavy chain) |
| C_410060 | gi|57087161|ref|XP_536793.1| similar to hydrocephalus-inducing protein (Canis familiaris); u.a., similar to mouse hydrocephaly protein hydin HY3 |
| C_420012 | gi|29135339|ref|NP_619615.2| polycystic kidney and hepatic disease 1-like 1 (Mus musculus); gnl|CDD|27705 cd00603, IPT_PCSR, IPT domain of plexins and cell surface receptors (PCSR) and related proteins |
| C_550085 | No significant similarity found; u.a., FAP1 |
| C_570035 | gi|50057451|emb|CAH03435.1| hypothetical transmembrane protein (Paramecium tetraurelia); u.a., FAP113 |
| C_590094 | No significant similarity found |
| C_590099 | gi|68371158|ref|XP_695404.1| similar to SI:PACKTRZ.2 (novel protein similar to human polycystic kidney disease 2-like 1 [PKD2L1]), partial (Danio rerio); u.a., PKD2, weakly similar to polycystin-2; KOG: Ca2+-modulated nonselective cation channel polycystin |
| C_700061 | gi|67475672|ref|XP_653525.1| plasma membrane calcium-transporting ATPase, putative (Entamoeba histolytica HM-1:IMSS) |
| C_750005 (C)* | gi|10441431|gb|AAG17036.1|S-adenosylmethionine synthetase (Pinus contorta) |
| C_750038 | gi|71755035|ref|XP_828432.1| hypothetical protein Tb11.02.0990 (Trypanosoma brucei); u.a., FAP75, flagellum-associated P-loop-containing protein |
| C_760066 | No significant similarity found |
| C_850039 | No significant similarity found |
| C_870044 | No significant similarity found |
| C_910056 | gi|29827935|ref|NP_822569.1| putative ABC transporter solute-binding protein (Streptomyces avermitilis MA-4680) |
| C_970003 | gi|50750584|ref|XP_422048.1| similar to tubulin alpha-5 chain, chicken (Gallus gallus); u.a., FAP248 |
| C_1120028 (B)* | gi|16974385|gb|AAL31118.1| gene info AT5g19940/F28l16_90 (Arabidopsis thaliana); gnl|CDD|16052 pfam04755, PAP_fibrillin |
| C_1150005 | gi|7441384|pir||T08164 dynein α heavy chain (C. reinhardtii) (fragment); u.a., DHC1, flagellar inner-arm dynein 1 heavy chain α |
| C_1340006 | gi|18164|emb|CAA32061.1| OEE3 precursor protein (C. reinhardtii); u.a., oxygen-evolving enhancer protein 3 (PsbQ) |
| C_1460023 (C)* | gi|619932|gb|AAB61446.1| isocitrate lyase (C. reinhardtii) |
| C_1580045 | gi|130264|sp|P18068|PLAS_CHLRE plastocyanin, chloroplast precursor (PC6-2); c.d., gnl|CDD|15127 pfam00127, copper-binding proteins, plastocyanin/azurin family |
| C_1710010 | gi|2494209|sp|Q39575|DYHG_CHLRE dynein γ chain, flagellar outer arm; u.a., ODA2 flagellar outer dynein arm heavy chain γ (PF28); gnl|CDD|26007 pfam03028 dynein heavy chain |
| C_2350009 | u.a., FAP157, conserved uncharacterized flagellum-associated protein |
| Centriole (13) | |
| C_20130 | gi|62860034|ref|NP_001016608.1| hypothetical protein LOC549362 (Xenopus tropicalis); u.a., novel protein (WD repeat domain), described as protocilium-class FABP protein in human centrosome |
| C_200025 | c.d., coiled-coil protein |
| C_200196 | gi|70879596|gb|EAN92756.1| hypothetical protein, conserved (Trypanosoma cruzi); u.a., coiled-coil protein with homology to a putative transcript, CG15792-PC, isoform C (Drosophila melanogaster), a hypothetical protein CE2713 (Corynebacterium efficiens YS-314), and to ciliary rootlet coiled-coil rootletin NP_055490.2 (H. sapiens) |
| C_1120028 (A)* | gi|16974385|gb|AAL31118.1| AT5g19940/F28l16_90 (A. thaliana); gnl|CDD|16052 pfam04755, PAP_fibrillin, PAP_fibrillin |
| Thioredoxin-interacting partners (18) | |
| C_20337 (A)* | gi|1353876|gb|AAB01817.1| glutamine synthetase (C. reinhardtii); u.a., GS1, cytosolic glutamine synthetase, CO2-responsive gene |
| C_30202 | gi|515618|emb|CAA52439.1| sedoheptulose-bisphosphatase (C. reinhardtii) |
| C_120103 | gi|24030434|gb|AAN41372.1| dihydroxy-acid dehydratase (A. thaliana); CDD|25654 pfam00920, ILVD_EDD, dehydratase family |
| C_200083 | gi|39936346|ref|NP_948622.1| elongation factor Tu (Rhodopseudomonas palustris CGA009); u.a., EEF1, putative mitochondrial translation factor Tu; gnl|CDD|9925 COG0050, TufB, GTPases, translation elongation factors |
| C_750005 (A)* | gi|10441431|gb|AAG17036.1|S-adenosylmethionine synthetase (Pinus contorta) |
| C_1460023 (A)* | gi|619932|gb|AAB61446.1| isocitrate lyase |
| C_1600004 | gi|50660327|gb|AAT80888.1| chloroplast chaperonin 21 (Vitis vinifera) |
| Chloroplast 70S ribosome (33, 34) | |
| C_150160 | gi|34398358|gbAAO22241.1| 41-kDa ribosome-associated protein precursor (C. reinhardtii); u.a., chloroplast RNA-binding protein, chloroplast ribosome-associated RAP41 stem-loop binding protein |
*, proteins that appear in different subproteomes (A, flagella; B, centriole; C, thioredoxin target).
u.a., user annotation notes, which can be found on the JGI website under the gene model number; c.d., conserved domains.
DISCUSSION
By the current approach we were able for the first time to obtain insights into the phosphoproteome of C. reinhardtii, which serves as a model organism for plants as well as for animals. We believe that the information about proteins that are subject to phosphorylation can be extremely useful in the study of their specific roles within certain pathways. As mentioned above, phosphorylation can affect protein function, activity, stability, localization, and interactions.
Addition of okadaic acid, an inhibitor of Ser/Thr phosphatases, was provided to increase the phosphorylation status of proteins that contain phosphoserine or phosphothreonine. Treatment of cells with the Ser/Thr phosphatase inhibitor calyculin had been already successfully applied for determining the phosphoproteome of a lymphoma cell line (30). In this case, it was shown that incubation of cells with the inhibitor led to a robust increase in the levels of phosphothreonine-containing proteins in comparison to untreated cells. By use of the inhibitor, the authors were able to identify a total of 107 phosphoproteins in a WEHI-231 B lymphoma cell line. Incubation of cells with okadaic acid had also been already studied, though with regard to a specific protein (human p53). It was shown thereby that the ratio of nonphosphorylated to specific phosphorylated amino acids from p53 was significantly shifted toward the phosphorylated form, permitting efficient determination of phosphorylation sites by LC-MS (21). For example, Ser315 of p53 had a ratio of 0.95 (nonphosphorylated form) to 0.05 (phosphorylated form) in wild-type cells and a ratio of 0 (nonphosphorylated form) to 1 (phosphorylated form) in okadaic acid-treated cells. This shows clearly the potential impact of inhibitor treatment in specifically increasing phosphorylation sites. Additionally, in the case of C. reinhardtii, the yield of phosphoproteins was significantly enhanced in the presence of okadaic acid in comparison to untreated cells. This was tested (i) by immunoblotting with antiphosphothreonine antibody and (ii) by comparing the yield of the phosphoproteins from untreated versus treated cells, in which it increased by 38%. Although okadaic acid enhanced the yield, phosphatase inhibition is not without biological consequences. In our case, treatment for 5 days caused defects in motility for about 75 to 80% of the cells and presumable cell death for about 10% of the cells. For this reason, we used an incubation time of 29 h, producing cell viability comparable to that of untreated controls.
To enrich the phosphopeptides in a complex trypsin-digested peptide mixture from C. reinhardtii, we used IMAC, which had already been successfully used to investigate, e.g., phosphoproteins from yeast (4, 7) and a lymphoma cell line (30). Since it had been reported (28, 30) that a change in the metal component from Fe3+ to Ga3+ could significantly increase the yield of phosphopeptides versus nonphosphorylated peptides, we used IMAC in combination with Ga3+ as the metal component. The internal phosphopeptide standard from beta-casein showed that MS2 spectra are sufficient to identify phosphopeptides, as was the case for the monophosphopeptide of beta-casein. However, it also became clear that in some cases the neutral-loss-triggered MS3 event can be crucial for the identification of a phosphorylation site, as was found for the tetraphosphopeptide of beta-casein. A similar situation was found for the identification of phosphopeptides from C. reinhardtii. Many of them could be identified based on the MS2 spectra alone or in combination with MS3 spectra. Notably, some phosphopeptides could be identified based only on the MS3 spectra, showing the advantage of the data-dependent neutral-loss method that automatically triggers MS3 scan events.
In all, 328 phosphoproteins, along with 360 phosphopeptides, from C. reinhardtii were identified. The yield of phosphoproteins was correlated directly with the current number of correctly predicted gene models (based on the genome sequence, version 2) and available EST data. Gene model predictions for Arabidopsis thaliana were reported by Peltier et al. (25) to contain significant errors for about 30% of the analyzed models, varying from incorrect N and C terminus predictions to errors in intron/exon boundary predictions and missing exons. In our case, 31% of the analyzed models were supported by EST data covering at least part of the exons of each gene model. However, 13% of the positive hits were based purely on EST data, since no gene models were available in these cases. This is due possibly to incorrect gene model predictions and certainly to the fact that parts of the genome of C. reinhardtii still contain regions with undetermined nucleotides (due to the high GC content of its DNA, which affects sequencing). It can be predicted that the yield of positive hits will increase after the final version of the C. reinhardtii genome has been released.
Our selection model provided rigorous criteria. We selected only phosphopeptides that occurred in at least two of the four independent experiments and had significant Xcorr factors (see Materials and Methods). Further hits that covered parts of potential exons within the genomic sequence of C. reinhardtii but showed no or only weak homology to currently known proteins and were not supported by gene models or EST data were neglected. It is possible that they have been omitted from the gene models due to errors in gene model prediction, but they may also represent false positives.
It is estimated that about 30% of all proteins of a eukaryotic cell (with regard to vertebrates) are subject to phosphorylation (20). Even if the possibility that the genome still has nondetermined nucleotides and that some gene models may be not correct is taken into account, one can predict that the current data do not cover all phosphoproteins from C. reinhardtii. Currently, there are about 19,000 predicted gene models (5), accounting for the same number of proteins in C. reinhardtii. If one assumes that about 30% of all proteins from C. reinhardtii are phosphorylated, a potential yield of 5,700 phosphoproteins would have to be considered. Taking into account the possibility that about 30% of the gene models might be wrong, as found previously (25), one would still expect 3,990 phosphoproteins. Thus, the yield of phosphoproteins identified in this study would cover about 8.2% of the total. Of course, the level of phosphoproteins in C. reinhardtii could be less than 30%, and indeed there is some indication, for example, that it differs in comparison to lymphoma cells (30). When the levels of proteins labeled with the antiphosphothreonine antibody (Cell Signaling Technologies) in the lymphoma cell line (20 μg of protein per lane [see Fig. 2 in reference 30]) are compared with those in C. reinhardtii (50 μg of protein per lane [Fig. 1]), one can detect more labeled phosphoproteins in the lymphoma cell line even though the amount of protein loaded per lane was less.
There are several reasons why the yield of phosphoproteins can be decreased: (i) very low abundance phosphopeptides are not enriched sufficiently by IMAC and thus have been missed; (ii) multiply phosphorylated peptides are more enriched by IMAC, as reported previously (20), and thus monophosphopeptides may be more easily missed; and (iii) phosphorylation of many proteins requires specific stimuli, and therefore any particular growth condition for the organism reflects only some of these stimuli. Thus, it was shown previously that calcium can repress the phosphorylation of an 85-kDa axonemal protein of C. reinhardtii at a concentration of ≥10−6 M, while the phosphorylation of a 95-kDa protein (designated b4) was increased at a similar concentration (29). These proteins seem not to be present in the current phosphoproteome. In this context, it could be of interest that a flagellar, putative plasma membrane calcium-transporting ATPase (C_700061) was identified among the phosphoproteins. Another example of stimulus-dependent phosphorylation is the light-harvesting complex II protein CP29. It contains four phosphorylated amino acids when the cells are exposed to so-called State 2 conditions, while in State 1 condition-exposed cells only two phosphorylated amino acids could be found (12). The State 1 to State 2 transition in the photosynthetic membranes involves the functional coupling of phosphorylated light-harvesting complexes of photosystem II to photosystem I. Thus, a set of different phosphoproteins will be present in the cell depending on the growth conditions (e.g., light, temperature, nutritional supplements, time of harvesting). Under our conditions, two phosphopeptides from CP29 (gene model C_10030) that cover the same amino acids were found. In the first phosphopeptide, one threonine (Thr6) was phosphorylated, while in the other, two threonines were phosphorylated within the same peptide. Only Thr6, not Thr10, of CP29 was reported to be phosphorylated under specific State conditions (12).
Our phosphoproteome data show that not only rather abundant phosphoproteins (e.g., metabolic enzymes) but also proteins that might be assumed to be present at low concentrations in the cell were detected. These include proteins from signaling pathways, such as several transporters, receptors, kinases, and phosphatases. Some of them, such as protein kinases, which can be subjected to positive and negative regulation by phosphorylation (17), could be expected to appear in the phosphoproteome. Further, putative transcription factors (e.g., plant homeodomain zinc fingers or SBP domain-containing proteins, which function as transcription factors in early flower development) and putative RNA-binding proteins containing, e.g., RRM domains, as well as DNA/RNA modifying proteins were detected. Also, proteins having domains that communicate protein-protein interactions (e.g., PAS, ankyrin repeats, TPR, Leu-rich repeats) were found.
Some of the proteins that were identified (e.g., CP29) had been already known to represent phosphoproteins of C. reinhardtii. Another example, the α dynein heavy chain (DHC) from the outer arm of the flagellum, was shown to have multiple sites of phosphorylation (16). From quantitative analysis it was concluded that DHC is phosphorylated at a minimum of six sites. In our study, we were able to detect four phosphopeptides from DHC with a total of eight phosphorylation sites. A further example is cytosolic glutamine synthetase, which was described previously as a phosphoprotein (27). One phosphopeptide with two phosphorylation sites was found in this case.
Comparative proteome analysis (Table 3) revealed that 32 proteins (with a total of 41 phosphopeptides) of the flagella and 4 proteins of the centriole are phosphorylated. A large number of phosphoproteins within the flagella could be expected, since several kinases and phosphatases have been shown to be located directly in the flagella (24, 35, 36), suggesting that dephosphorylation/phosphorylation plays a key role within the eukaryotic cilia. In addition to the above-mentioned DHC, which has multiple phosphorylation sites, a protein (gene model C_410060) from C. reinhardtii that is similar to the mouse hydrocephaly protein hydin (HY3) also contains numerous phosphorylation sites (eight sites situated in three phosphopeptides). HY3 is an example of a protein that is conserved in mammals and is involved in mammalian diseases. Thus, a mutation in mouse HY3 results in lethal communicating hydrocephalus with perinatal onset (1). Other phosphoproteins from the flagella (Table 3) are also related to diseases, such as, for example, predicted proteins from gene models C_290071 (similar to cold autoinflammatory syndrome 1 protein), C_420012 (polycystic kidney and hepatic disease 1-like protein), and C_590099 (a novel protein similar to human polycystic kidney disease 2-like protein 1). These findings open the possibility of specifically studying the mechanisms of posttranslational control for these proteins. Furthermore, some phosphoproteins from the flagella having a putative or unknown function can now be analyzed in a more focused way. However, the phosphoproteins identified within the flagella also reveal the limits of our analysis. For example, the phosphoprotein IC138, a WD repeat dynein intermediate chain that is hyperphosphorylated in paralyzed flagellar mutants (10, 15), was not found in the current phosphoproteome. Also, a significant number of radial spoke proteins of the flagella, which were identified as phosphoproteins by in vivo pulse labeling with 32P (26), are missing in the analyzed phosphoproteome.
Comparison of the proteome from the 70S ribosome (33, 34) with the phosphoproteome (Table 3) revealed only one hit, namely, RAP41. RAP41 represents one of two proteins that are specific for the complete 70S ribosome and could not be found in its 50S and 30S subunits (33). No components of the 50S and 30S subunits of the chloroplast ribosome could be found in the phosphoproteome. In contrast, proteins with significant homology to those from the cytosolic 80S ribosome (60S and 40S subunits) were found in the phosphoproteome.
Seven thioredoxin target proteins in C. reinhardtii (18) turned out to be phosphoproteins. This finding suggests that they are controlled in a complex manner by phosphorylation and redox regulation.
The present analysis has depicted a significant number of phosphoproteins of C. reinhardtii within the whole cell. It aimed to get a first insight into the variety of phosphoproteins within C. reinhardtii. Nevertheless, many phosphoproteins from this alga are still missing, based on prediction data for the phosphoprotein content within a eukaryotic cell (20). An efficient approach for the future to increase the yield of phosphoproteins may be to concentrate on the phosphoproteome of subcellular fractions. For example, flagella that bear many phosphoproteins could be isolated and used specifically for analysis. However, one must also point out that certain phosphoproteins may be ultimately missed by subjecting only subcellular fractions to analysis. Another approach may be to grow and harvest cells in the presence of different stimuli that might up-regulate phosphorylation of certain proteins. Thus, knowledge of stimulus-dependent phosphorylation would become available at the same time.
Supplementary Material
Acknowledgments
We thank several students (Jens Bösger, René Rainer Nötzold, Hendrik Rohn, and Sarah Werner) and Frank Meißner for help with bioinformatics analysis, and we thank Einar Stauber for critical reading of the manuscript. We appreciate very much the free delivery of information by the U.S. (DOE) and Japanese C. reinhardtii genome projects.
Our work was supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Davy, B. E., and M. L. Robinson. 2003. Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in hydin, a large novel gene. Hum. Mol. Genet. 12:1163-1170. [DOI] [PubMed] [Google Scholar]
- 2.Eng, J., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 3.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. McNulty, S. A. Carr, and P. Tempst. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826:167-181. [DOI] [PubMed] [Google Scholar]
- 4.Ficarro, S. B., M. L. McCleland, P. T. Stukenberg, D. J. Burke, M. M. Ross, J. Shabanowitz, D. F. Hunt, and F. M. White. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301-305. [DOI] [PubMed] [Google Scholar]
- 5.Grossman, A. R. 2005. Paths toward algal genomics. Plant Physiol. 137:410-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez, D. Rokhsar, J. Shrager, C. D. Silflow, D. Stern, O. Vallon, and Z. Zhang. 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2:1137-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruhler, A., J. V. Olsen, S. Mohammed, P. Mortensen, N. J. Faergeman, M. Mann, and O. N. Jensen. 2005. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4:310-327. [DOI] [PubMed] [Google Scholar]
- 8.Harris, E. H. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363-406. [DOI] [PubMed] [Google Scholar]
- 9.Heintz, D., V. Wurtz, A. A. High, A. Van Dorsselaer, R. Reski, and E. Sarnighausen. 2004. An efficient protocol for the identification of protein phosphorylation in a seedless plant, sensitive enough to detect members of signalling cascades. Electrophoresis 25:1149-1159. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson, T. W., C. A. Perrone, P. Griffin, K. Wuichet, J. Mueller, P. Yang, M. E. Porter, and W. S. Sale. 2004. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell 15:5431-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalume, D. E., H. Molina, and A. Pandey. 2003. Tackling the phosphoproteome: tools and strategies. Curr. Opin. Chem. Biol. 7:64-69. [DOI] [PubMed] [Google Scholar]
- 12.Kargul, J., M. V. Turkina, J. Nield, S. Benson, A. V. Vener, and J. Barber. 2005. Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J. 272:4797-4806. [DOI] [PubMed] [Google Scholar]
- 13.Keller, L. C., E. P. Romijn, I. Zamora, J. R. Yates III, and W. F. Marshall. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090-1098. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J., D. G. Camp, and R. D. Smith. 2004. Improved detection of multi-phosphorylated peptides in the presence of phosphoric acid in liquid chromatography/mass spectrometry. J. Mass Spectrom. 39:208-215. [DOI] [PubMed] [Google Scholar]
- 15.King, S. J., and S. K. Dutcher. 1997. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J. Cell Biol. 136:177-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, S. M., and G. B. Witman. 1994. Multiple sites of phosphorylation within the alpha heavy chain of Chlamydomonas outer arm dynein. J. Biol. Chem. 269:5452-5457. [PubMed] [Google Scholar]
- 17.Krupa, A., G. Preethi, and N. Srinivasan. 2004. Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J. Mol. Biol. 339:1025-1039. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire, S. D., B. Guillon, P. LeMarechal, E. Keryer, M. Miginiac-Maslow, and P. Decottignies. 2004. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 101:7475-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 20.Mann, M., S. E. Ong, M. Grønborg, H. Stehen, O. N. Jensen, and A. Pandey. 2002. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 20:261-268. [DOI] [PubMed] [Google Scholar]
- 21.Merrick, B. A., W. Zhou, J. Martin, S. Jeyarajah, C. E. Parker, J. K. Selkirk, K. B. Tomer, and C. H. Borchers. 2001. Site-specific phosphorylation of human p53 protein determined by mass spectrometry. Biochemistry 40:4053-4066. [DOI] [PubMed] [Google Scholar]
- 22.Mittag, M. 1996. Conserved circadian elements in phylogenetically diverse algae. Proc. Natl. Acad. Sci. USA 93:14401-14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nühse, T. S., A. Stensballe, O. N. Jensen, and S. C. Peck. 2003. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2:1234-1243. [DOI] [PubMed] [Google Scholar]
- 24.Pazour, G. J., N. Agrin, J. Leszyk, and G. B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltier, J. B., O. Emanuelsson, D. E. Kalume, J. Ytterberg, G. Friso, A. Rudella, D. A. Liberles, L. Soderberg, P. Roepstorff, G. von Heijne, and K. J. van Wijk. 2002. Central functions of the luminal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14:211-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piperno, G., B. Huang, Z. Ramanis, and D. J. L. Luck. 1981. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88:73-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pozuelo, M., C. MacKintosh, A. Galvan, and E. Fernandez. 2001. Cytosolic glutamine synthetase and not nitrate reductase from the green alga Chlamydomonas reinhardtii is phosphorylated and binds 14-3-3 proteins. Planta 212:264-269. [DOI] [PubMed] [Google Scholar]
- 28.Reinders, J., and A. Sickmann. 2005. State-of-the-art in phosphoproteomics. Proteomics 5:4052-4061. [DOI] [PubMed] [Google Scholar]
- 29.Segal, R. A., and D. J. Luck. 1985. Phosphorylation in isolated Chlamydomonas axonemes: a phosphoprotein may mediate the Ca2+-dependent photophobic response. J. Cell Biol. 101:1702-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu, H., S. Chen, Q. Bi, M. Mumby, and D. L. Brekken. 2004. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell. Proteomics 3:279-286. [DOI] [PubMed] [Google Scholar]
- 31.Stauber, E. J., A. Fink, C. Markert, O. Kruse, U. Johanningmeier, and M. Hippler. 2003. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2:978-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, V., M. Fiedler, C. Markert, M. Hippler, and M. Mittag. 2004. Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett. 559:129-135. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi, K., M. V. Beligni, S. Prieto, P. A. Haynes, W. H. McDonald, J. R. Yates III, and S. P. Mayfield. 2003. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of the proteins unique to the 70S ribosome. J. Biol. Chem. 278:33774-33785. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi, K., S. Prieto, M. V. Beligni, P. A. Haynes, W. H. McDonald, J. R. Yates III, and S. P. Mayfield. 2002. Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14:2957-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, P., L. Fox, R. J. Colbran, and W. S. Sale. 2000. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J. Cell Sci. 113:91-102. [DOI] [PubMed] [Google Scholar]
- 36.Yang, P., and W. S. Sale. 2000. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J. Biol. Chem. 275:18905-18912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.