Abstract
Ssy5p is a 77-kDa protein believed to be a component of the SPS amino acid sensor complex in the plasma membrane of Saccharomyces cerevisiae. Ssy5p has been suggested to be a chymotrypsin-like serine protease that activates the transcription factor Stp1p upon exposure of the yeast to extracellular amino acid. Here we overexpressed and partially purified Ssy5p to improve our understanding of its structure and function. Antibodies against Ssy5p expressed in Escherichia coli were isolated and used to detect Ssy5p processing in S. cerevisiae cells. Partial purification and N-terminal sequencing of processed Ssy5p revealed in vivo cleavage of Ssy5p between amino acids 381 and 382. We also isolated constitutively signaling SSY5 mutants and quantified target promoter activation and Stp1p processing. One mutant contained an amino acid substitution in the prodomain, whereas three others harbored amino acid substitutions in the protease domain. Dose-response analysis indicated that all four mutants exhibited increased basal levels of Stp1p processing. Interestingly, whereas the three constitutive mutants mapping to the protease domain of Ssy5p exhibited the decreased 50% effective concentration (EC50) characteristic of constitutive mutations previously found in Ssy1p, Ptr3p, and Ssy5p, the EC50 of the mutation that maps to the prodomain of Ssy5p remained essentially unchanged. In a model of Ssy5p derived from its similarities with α-lytic protease from Lysobacter enzymogenes, the sites corresponding to the mutations in the protease domain are clustered in a region facing the prodomain, suggesting that this region interacts with the prodomain and participates in the conformational dynamics of sensing.
As part of the regulatory machinery for nutrient uptake (9, 20), the yeast Saccharomyces cerevisiae is equipped with an amino acid sensor in the plasma membrane that initiates signal transduction when extracellular amino acids are available. The signaling results in proteolytic processing of downstream transcription factors and stimulation of transcription of various amino acid permease genes. The sensor consists of the Ssy1p integral membrane protein and two membrane-associated proteins, Ptr3p and Ssy5p (7, 13, 21, 23, 25), and has been designated SPS for the complex that its three components are suggested to form (15).
Ssy1p, which has high similarity to amino acid permeases, is believed to initiate the signal transduction by recognizing the inducing amino acids on the outside of the plasma membrane. Whereas little is known about the involvement of Ptr3p in amino acid signaling, the function of Ssy5p is now in the process of being unraveled. It has been determined that the C-terminal part of Ssy5p has similarity to chymotrypsin-like serine proteases, and mutational analysis is consistent with this function (1, 2). This suggests that Ssy5p is responsible for the proteolytic removal of the ∼10-kDa N-terminal fragment of each of the transcription factors Stp1p and Stp2p, resulting in their migration from the cytoplasm/plasma membrane to the nucleus (1, 2, 3, 4). Signaling has been measured by the activation of target promoters, such as the BAP2 promoter (12, 26) or the AGP1 promoter (21), and by quantifying the proteolytic processing of Stp1p processing (27, 28).
To initiate biochemical studies of the SPS sensor components we have overexpressed and partially purified Ssy5p from Escherichia coli and S. cerevisiae. We have produced antibodies directed against Ssy5p and have determined the site of an internal proteolytic cleavage of Ssy5p. To extend our knowledge of Ssy5p function, we isolated and characterized several constitutively signaling SSY5 mutants.
MATERIALS AND METHODS
Media.
The glucose-based media SD (synthetic minimal), SC (synthetic complete), and YPD (yeast extract-peptone-dextrose complex) were prepared as described (31). However, amino acid concentrations in SC were as specified elsewhere (19). Where indicated, the glucose in the SD and SC media was replaced with filter-sterilized 10% raffinose to give a final concentration of 2%.
Strains.
The microbial strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| S. cerevisiae | ||
| M3124 | MATaura3-52 leu2-3 leu2-112 his3-Δ200 prc1-Δ::HIS3 prb1-Δ::LEU2 pep4-d1137 | 35 |
| M5359 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ AGP1::PAGP1-lacZ | 28 |
| M5360 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ptr3Δ AGP1::PAGP1-lacZ | 28 |
| M5361 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy5Δ AGP1::PAGP1-lacZ | 28 |
| M5380 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy1Δ AGP1::PAGP1-lacZ | 28 |
| M5444 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy5Δ STP1::ZZ-kanMX | 28 |
| E. coli | ||
| BL21(DE3) Codon Plus RIL | B F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ(DE3) endA Hte(argU ileY leuW Camr) | Stratagene |
| XL10 | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15] Tn10(Tetr) Amy Camr | Stratagene |
Tet, tetracycline; Cam, chloramphenicol.
Plasmids.
Plasmid pSSY5 (23, 28) contains a 3-kb HindIII-SacII fragment with wild-type SSY5 inserted into the centromeric, URA3-based vector pRS316 (32).
Plasmid pPEP18 was constructed by insertion of SSY5, amplified from plasmid pSSY5 using primers SSY5-1 (5′ GTA CTG GTG TAA ACT CGA TAT ACC G 3′) and SSY5-16 (5′ TCC ATC TAG TTG TGG ATC AAT GTC 3′), into the pYES2.1/V5-His TOPO expression vector by TOPO TA cloning (Invitrogen). This placed the SSY5 open reading frame (ORF) behind a GAL1 promoter in frame with the His6 tag in the vector, adding 33 amino acid residues to the C terminus of Ssy5p.
Plasmid pPEP21 was made by insertion of SSY5 amplified by PCR using pSSY5 as the template and primers SSY5-13 (5′ GAG CTC ATG GTC AGA TTT TTT GGT TTA AAC 3′) and SSY5-14 (5′ AAG CTT AGT TAC AGT CAT GTA GTC 3′) between the SacI and HindIII sites of the pET44b expression vector (Novagen). This allows expression of Ssy5p in E. coli as a fusion protein with the 495-amino-acid-residue NusA tag at the N terminus of Ssy5p (11).
Site-directed mutagenesis of SSY5.
Site-specific mutations were introduced into the SSY5 ORF using pSSY5 as the template and the QuickChange II XL kit (Stratagene) as described by the manufacturer. The primers used for mutagenesis are listed in Table 2.
TABLE 2.
Primers used for introduction of site-specific mutations in SSY5
| Primer | DNA sequence (5′→3′) |
|---|---|
| E131K | TCT TGA GTC CTG TTA AGA AGG AGG AAT CTC AGG ATA C |
| Anti E131K | GTA TCC TGA GAT TCC TCC TTC TTA ACA GGA CTC AAG A |
| F575V | CAT TCC CAG ATC CAA CAT TAA GAG TTC AAA ATT TAC ATG TGA AAC GAA AAA TT |
| Anti F575V | AAT TTT TCG TTT CAC ATG TAA ATT TTG AAC TCT TAA TGT TGG ATC TGG GAA TG |
| Q576P | CCC AGA TCC AAC ATT AAG ATT TCC AAA TTT ACA TGT GAA ACG AAA AAT T |
| Anti Q576P | AAT TTT TCG TTT CAC ATG TAA ATT TGG AAA TCT TAA TGT TGG ATC TGG G |
| K581N | CAA CAT TAA GAT TTC AAA ATT TAC ATG TGA ATC GAA AAA TTT TTA AAA TGA AGC CTG G |
| Anti K581N | CCA GGC TTC ATT TTA AAA ATT TTT CGA TTC ACA TGT AAA TTT TGA AAT CTT AAT GTT G |
| P632H | GCG AGT TTG TCG TTG CAT CTC CAA CTC ATT TAT TTG CTA GTG CGG GGG ATT CAG |
| Anti P632H | GCC TGA ATC CCC CGC ACT AGC AAA TAA ATG AGT TGG AGA TGC AAC GAC AAA CTC |
Production of NusA-Ssy5p inclusion bodies in E. coli.
E. coli strain BL21(DE3) Codon Plus RIL/pPEP21 was grown at 37°C in 100 ml LB medium supplemented with 100 μg/ml ampicillin until the optical density at 600 nm (OD600) reached 0.8. Isopropylthiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and shaking was continued for 3 h. Cells were harvested by centrifugation at 4,000 rpm for 25 min in an Eppendorf 5810 centrifuge and suspended in 6 ml BugBuster (Novagen) supplemented with 6 μl Benzonase (25 units/μl, Novagen) and 1.5 μl rLysozyme (30 kilounits/μl, Novagen). The cells were lysed by incubation for 40 min at room temperature with shaking, and inclusion bodies were sedimented at 20,000 × g for 20 min. The pellet was suspended in 4 ml phosphate-buffered saline (pH 7.5; 80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl) containing 4 M urea and washed by whirlimixing and repeated centrifugation. Inclusion bodies were suspended in 2 ml phosphate-buffered saline and sonicated for 1 min with intervals of 1 s on and 1 s off, using a Vibra-Cell sonicator (Sonics & Materials, Inc.). The sample contained ca. 1.3 mg NusA-Ssy5 fusion protein and was used for immunization of rabbits (Pineda, Berlin, Germany).
Overexpression of Ssy5p in S. cerevisiae.
Strain M3124 transformed with plasmid pPEP18 was grown exponentially in 50 ml SD medium containing 2% raffinose. At an OD600 of 1.0, galactose was added to a final concentration of 2% to induce the GAL1 promoter. When indicated, leucine was added to a final concentration of 0.2 mM. At the indicated time intervals samples of 2 ml were withdrawn from the culture, and the cells from each sample were immediately harvested by centrifugation. The cells were suspended in 200 μl YeastBuster mix and shaken for 15 min at room temperature. YeastBuster mix was made by mixing 4 ml YeastBuster (Novagen), 4 μl Benzonase (25 units/μl, Novagen), 40 μl of the reducing agent Tris(hydroxypropyl)phosphine (Novagen) and 200 μl protease inhibitor cocktail set II (Novagen); 50 μl of the lysate was mixed with 20 μl 4× lithium dodecyl sulfate sample buffer (LDS; Invitrogen) and 8 μl 0.5 M dithiothreitol, incubated at 95°C for 5 min, and whirlimixed; 20 μl of the denatured sample was applied to a 4 to 12% gradient NuPAGE Bis-Tris gel (Invitrogen) and subjected to electrophoresis for 50 min at 200 V with morpholinepropanesulfonic acid (MOPS)-sodium dodecyl sulfate (SDS) running buffer (Invitrogen) and an XCell SureLock Mini-Cell apparatus (Invitrogen). Proteins were blotted onto a polyvinylidene difluoride membrane using an Xcell II blot module and NuPAGE transfer buffer according to Invitrogen's recommendations.
Purification of the Ssy5p chymotrypsin-like serine protease domain produced in S. cerevisiae.
A culture of yeast strain M3124/pPEP18 grown to an OD600 of 6.2 in raffinose-based SC without uracil was used to inoculate 750 ml of the same medium supplemented with 2% galactose. The culture was shaken overnight at 30°C. Cells were harvested by centrifugation, suspended in 35 ml buffer A (50 mM NaH2PO4, pH 7.4, 500 mM NaCl), and disrupted in a One Shot cell disrupter (Constant Systems, England) at 2.70 kBar. The crude extract was centrifuged at 20,000 × g for 10 min at 4°C. Sedimented debris was suspended in 4 ml buffer A for later Western analysis; 10 μl Benzonase (25 units/μl, Novagen) was added to the supernatant, which was incubated for 1 h on ice to remove DNA.
The extract was then passed through a 45-μm filter and a 20-μm filter (Minisart, Sartorius) and applied to a 5-ml HisTrap column (Amersham Biosciences) equilibrated in buffer A. After washing the column with 50 ml buffer A, proteins were eluted in steps with 35, 75, and 500 mM imidazole. A sample of 25 μl from each of the 5-ml fractions and 2-ml fractions collected during the wash and the elution, respectively, was mixed with 10 μl 4× LDS sample buffer (Invitrogen) and 5 μl 0.5 M dithiothreitol, incubated at 95°C for 5 min, and whirlimixed; 20 μl of each denatured sample was subjected to electrophoresis as described above. Gels were then either stained using GelCode blue stain reagent (Pierce) or blotted onto a polyvinylidene difluoride membrane using an Xcell II blot module and NuPAGE transfer buffer according to the supplier's recommendations (Invitrogen). BenchMark prestained protein ladder and Mark 12 (both Invitrogen) were used as protein molecular weight markers. The apparent molecular weights of the protein bands in the BenchMark prestained protein ladder were calibrated to the Mark 12 proteins by comparison of the mobility of the two ladders running in parallel in the NuPAGE Bis-Tris gel system using MOPS-SDS running buffer.
Western analysis.
Western blots were prepared for chemiluminescent immunodetection using blocking solution, primary antibody diluent and antibody wash from Invitrogen using the accompanying guidelines. Immunodetection was carried out using ECL Plus detection reagents from Amersham Pharmacia Biotech and a STORM 850 scanner.
Primary antibodies were diluted as follows: anti-Ssy5p antibodies (this study), 1:2,000, and His6 monoclonal antibody-horseradish peroxidase conjugate (BD Biosciences), 1:10,000. Peroxidase-conjugated goat anti-rabbit immunoglobulins (DakoCytomation, code no. P 0448) were used as secondary antibodies where indicated in a 1:2,000 dilution.
Stp1p processing assays.
SSY5 mutants were characterized for the dose-response relationships between amino acid exposure and Stp1p processing as described (27, 28).
β-Galactosidase assays were carried out as described (28).
Amino acid sequencing.
Protein was blotted onto a ProBlott membrane (Applied Biosystems) using the Xcell II blot module and NuPAGE transfer buffer as described above. Proteins were visualized by Coomassie blue R-250 (Sigma) staining, and the desired protein was cut out of the membrane for sequencing on a PROCISE apparatus (Applied Biosystems).
DNA sequencing.
Custom DNA sequencing of the isolated constitutive mutants and plasmid constructs was carried out by MWG Biotech (Germany).
RESULTS
Ssy5p is expressed in E. coli as inclusion bodies.
Ssy5p was expressed abundantly in E. coli strain BL21(DE3) Codon Plus RIL, although the majority of the protein was present in insoluble inclusion bodies (not shown). Because expression of SSY5 as a NusA-Ssy5p fusion protein in this strain only slightly improved the solubility of the protein it was decided to use these preparations for raising antibodies in rabbits and to overexpress Ssy5p in yeast.
Ssy5p is overexpressed in S. cerevisiae as a proenzyme cleaved into an N-terminal prodomain and a C-terminal protease domain.
To overexpress Ssy5p in S. cerevisiae the SSY5 ORF was inserted into expression vector pYES2.1/V5-His to allow galactose-inducible expression of Ssy5p carrying a His6 tag at its C terminus. Western analysis of an induction experiment with this construct is shown in Fig. 1. Using monoclonal antibody against the His tag (Fig. 1A), Ssy5 proteins of ∼80 kDa (Ssy5p) and ∼45 kDa (Ssy5p-C) were detected after induction with galactose. With polyclonal Ssy5p antibodies (Fig. 1C) Ssy5p species of ∼80 kDa (Ssy5p) and ∼50 kDa (Ssy5p-N) were observed.
FIG. 1.
Overexpression of SSY5 in S. cerevisiae results in full-length Ssy5p (∼80 kDa), an N-terminal Ssy5p fragment (∼50 kDa) and a C-terminal Ssy5p fragment (∼45 kDa). (A) M3124/pPEP18 cells were grown exponentially in SD medium containing raffinose as carbon source. At an OD600 of 1.0, galactose was added to 2% to induce expression of Ssy5p C-terminally tagged with His6. At the indicated times, samples were withdrawn from the culture for Western analysis with His6 antibodies (panels A and B) and Ssy5p antibodies (panel C). (B) Leucine (0.2 mM) was added at the same time as the galactose. Ssy5p, full-length Ssy5p; Ssy5p-N, ca. 50-kDa N-terminal Ssy5p fragment; Ssy5p-C, ca. 45-kDa C-terminal Ssy5p fragment; M, BenchMark protein molecular size markers.
The finding that the anti-Ssy5p polyclonal antibodies did not detect the 45-kDa Ssy5-C protein indicates that these antibodies recognize an epitope(s) in the N-terminal part of Ssy5p. The size of the slow-migrating protein detected in the Western analysis is in agreement with the expected 699-amino-acid full-length Ssy5 protein (Mr = 77,633) (28) fused to the 33-amino-acid V5/His6 C-terminal tag. The apparent sizes of the two fast-migrating Ssy5p species suggest that these fragments originated from a single cleavage of full-length Ssy5p. These results suggest that overexpressed Ssy5p is synthesized as a proenzyme which is proteolytically cleaved into two fragments of nearly equal size: an ∼45-kDa C-terminal fragment harboring the protease domain of Ssy5p and an ∼50-kDa N-terminal prodomain reported previously (1, 2). The same migration patterns were observed throughout the 17 h of induction, indicating that the three forms of Ssy5p are stable when overexpressed overnight.
To test whether the extraction procedure used in these experiments influences Ssy5p cleavage, a similar induction experiment was carried out in which culture aliquots were added directly to ice-cold NaOH and β-mercaptoethanol and then precipitated with trichloroacetic acid on ice (10). The same migration pattern was observed (data not shown), indicating that the cleavage of Ssy5p takes place within the intact yeast cells and not during extraction.
To determine whether amino acid signaling conditions affect the proposed proteolytic cleavage of Ssy5p, the described experiment (Fig. 1A) was carried out in parallel with an experiment in which 0.2 mM leucine was added simultaneously with the addition of galactose (Fig. 1B). The same migration pattern of Ssy5p was observed, indicating that the presence of leucine did not influence the extent of proteolytic cleavage of Ssy5p.
Purification of the Ssy5p chymotrypsin-like serine protease domain.
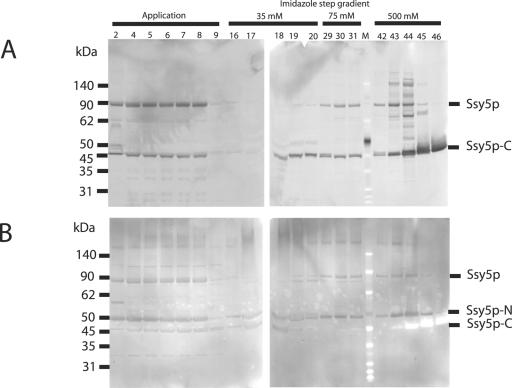
Overexpressed, C-terminally His-tagged Ssy5p was partially purified from yeast strain M3124/pPEP18 as described in Materials and Methods. Western analysis of the fractions representing the protein peaks collected during the elution from the HisTrap column was carried out using the antibody directed against the His6 C-terminal tag (Fig. 2A) and the polyclonal Ssy5p antibodies (Fig. 2B), allowing visualization of all three forms of Ssy5p (full-length Ssy5p, Ssy5p-N, and Ssy5p-C) throughout the purification. The three forms of Ssy5p were detected in different relative amounts during the purification: the application step (fractions 2 through 9), the 35 mM imidazole elution step (fractions 16 through 20), the 75 mM imidazole elution step (fractions 29 through 31), and the 500 mM imidazole elution step (fractions 42 through 46). Although a significant amount of Ssy5p was not bound to the column, microgram quantities of Ssy5p-C were eluted from the Ni-Sepharose resin into fractions 43 to 46 (Fig. 2A and Fig. 3).
FIG. 2.
Purification of Ssy5p C-terminally fused to His6 using Ni-Sepharose chromatography. An extract prepared from M3124/pPEP18 cells induced with galactose for 20 h was applied to a HisTrap column, and proteins bound to the Ni-Sepharose were eluted using a three-step gradient with 35, 75, and 500 mM imidazole. Samples from the fractions collected during application of the yeast extract and the three elution steps were analyzed by SDS-PAGE on a 4 to 12% gradient NuPAGE Bis-Tris gel and subsequent immunoblotting using His6 antibodies (panel A) and Ssy5p antibodies (panel B). Ssy5p, full-length Ssy5p; Ssy5p-N, ca. 50-kDa N-terminal Ssy5p fragment; Ssy5p-C, ca. 45-kDa C-terminal Ssy5p fragment; M, BenchMark protein molecular size markers.
FIG. 3.
Ssy5p 45-kDa C-terminal fragment can be isolated from fractions eluted with imidazole. Samples from the fractions collected during the 500 mM imidazole elution step (fractions 42 to 52) were analyzed by Coomassie staining of the proteins separated on a 4 to 12% gradient NuPAGE Bis-Tris gel. Ssy5p-C, ca. 45-kDa C-terminal Ssy5p protease fragment; M, BenchMark protein molecular size markers.
Remarkably, the Ssy5p-N and Ssy5p-C components coeluted in some of the fractions (e.g., fractions 43 to 46 in Fig. 2A and B). Since only Ssy5p and Ssy5p-C are expected to bind to the Ni-Sepharose, this observation suggests either that Ssy5p-N and Ssy5p-C remain associated during purification or that Ssy5p self-processing takes place on the resin or after elution from the resin.
Ssy5p processing results in a 381-amino-acid prodomain and a 318-amino-acid protease domain.
Fractions 42 through 52 from the elution described above were analyzed in more detail by Coomassie staining (Fig. 3) and Western analysis (data not shown). The greatest amount of Ssy5p-C, comprising the catalytically active region of Ssy5p, was found in fraction 44 (Fig. 3), which contains approximately 100 μg Ssy5p-C. Part of this fraction was therefore subjected to preparative SDS-polyacrylamide gel electrophoresis (PAGE), the proteins were blotted onto a membrane, and the Ssy5p-C band was cut out of the membrane and used for N-terminal sequencing. The analysis yielded the 18-residue sequence ASAVGSIPSHTAATIDTI. This sequence is identical to the Ssy5p sequence from residues 382 to 399, showing that the Ssy5p-C fragment analyzed originated from cleavage between residues 381 and 382, producing a 381-amino-acid prodomain and a 318-amino-acid protease domain.
Identification of single-site SSY5 mutations that activate a target promoter in the absence of extracellular amino acids.
To gain further insight into the functional roles of the Ssy5p domains, five constitutive SSY5 mutants (SSY5-1, SSY5-2, SSY5-3, SSY5-10, and SSY5-12) were isolated as described previously (28), using the PAGP1-KAT1 potassium channel reporter system for amino acid sensing (18). The mutant SSY5 alleles were expressed from a centromeric plasmid and were identified by their ability to activate the AGP1 promoter in M5361 (ssy5Δ AGP1::PAGP1-lacZ-kanMX) in medium lacking amino acids (Table 3). Tests for dominance were performed by introducing the plasmids expressing mutant SSY5 alleles into strain M5359, which carries the wild-type SSY5 gene at its normal chromosomal location and the AGP1 promoter-lacZ construct integrated at the AGP1 locus. β-Galactosidase activity in these transformants grown in the absence of leucine was increased over that of control cells expressing a plasmid-borne wild-type SSY5 allele. These results (not shown) are similar to those found for the previously characterized constitutive mutant SSY5-6 (28). Like this mutant, the new mutants were also dependent on the presence of Ssy1p and Ptr3p for constitutive signaling (data not shown).
TABLE 3.
Characterization of constitutive SSY5 mutants
| SSY5 allele | Amino acid substitution(s) | AGP1 activation (Miller units)a | % Basal Stp1p processingb (SEM) | EC50c (μM [SEM]) |
|---|---|---|---|---|
| SSY5 wild type | None | 0.121 | 5.5 (1.2) | 8.1 (1.9) |
| SSY5-1 | E131K, L288V | 1.461 | ND | ND |
| SSY5-2 | S430N, F575V, L645M | 1.916 | ND | ND |
| SSY5-3 | V412A, K581N, P632H | 1.297 | ND | ND |
| SSY5-10 | K304R, Q576P | 2.187 | ND | ND |
| SSY5-12 | K268M, G562C, F575V | 2.158 | ND | ND |
| SSY5-13 | E131K | 1.015 | 15.5 (2.1) | 10.3 (2.2) |
| SSY5-14 | F575V | 1.201 | 29.5 (5.5) | 1.4 (0.2) |
| SSY5-15 | Q576P | 0.810 | 32.1 (2.7) | 0.8 (0.2) |
| SSY5-16 | K581N | 0.145 | ND | ND |
| SSY5-17 | P632H | 0.087 | ND | ND |
| SSY5-18 | K581N, P632H | 0.730 | 9.3 (1.2) | 2.1 (0.4) |
| SSY5-6d | E512K | 1.646 | 36.9 (3.6) | 3.2 (0.6) |
β-Galactosidase activity in M5361 (ssy5Δ AGP1::PAGP1-lacZ-kanMX) cells transformed with the centromere-based vector pRS316 carrying the SSY5 mutant gene and grown in SD medium without amino acids.
Stp1p processing in M5444 (ssy5Δ STP1::ZZ-kanMX) cells transformed with the centromeric vector pRS316 carrying the SSY5 mutant gene and grown in SD medium without amino acids. The average of at least two individual determinations is shown. ND, not determined.
EC50 values (apparent Kd) were determined as described (27, 28). The averages of at least two individual determinations are shown.
Data are from reference 28.
Because each of the new mutants contained two or more nucleotide substitutions in the SSY5 open reading frame, each of the different single mutants was generated by site-directed mutagenesis of the original plasmid expressing wild-type SSY5 (Table 3). The single-amino-acid substitutions E131K, F575V, and Q576P of SSY5-13, SSY5-14, and SSY5-15, respectively, were found to activate the AGP1 promoter in cells growing in SD medium without amino acids (Table 3) and thus confer constitutive signaling. The analysis of SSY5-3, which contained the three amino acid substitutions V412A, K581N, and P632H, revealed that the single substitutions K581N and P632H (SSY5-16 and SSY5-17, respectively) are unable to cause constitutive signaling. However, when the K581N and P632H substitutions were combined, resulting in the allele SSY5-18, a constitutive phenotype was obtained. In this case, two substitutions are required to cause Ssy5p to signal in the absence of extracellular amino acid.
Effect of constitutive SSY5 mutants on basal levels of Stp1p processing and the EC50 for induction by leucine.
An ssy5Δ STP1::ZZ recipient (M5444) was transformed with plasmids expressing wild-type SSY5 or SSY5-13, SSY5-14, SSY5-15, or SSY5-18 constitutive allele to determine their effects on Stp1p processing in response to different concentrations of leucine. The recipient strain expressed the ZZ tag fused to Stp1p, enabling us to perform quantitative Western analysis to estimate Stp1p processing in the absence of extracellular amino acids (basal level) as described (27, 28).
Each of the constitutively signaling mutants exhibited an increased basal level of Stp1p processing (Table 3). Cells expressing SSY5-6, SSY5-14, and SSY5-15 showed an Stp1p processing level in the range of 30%, whereas cells expressing SSY5-13 and SSY5-18 exhibited 9.3 and 15.5% processing, respectively. As previously noted (27, 28), a modest increase in Stp1p processing is sufficient to cause high AGP1 promoter activation. In addition, we used the Stp1p-ZZ processing assay to estimate the median effective concentration (EC50; i.e., the concentration of leucine in the medium that results in 50% processing of Stp1p) as reported (27, 28). As shown in Table 3, the EC50 values for the SSY5-6, SSY5-14, SSY5-15, and SSY5-18 mutants, which have amino acid substitutions in the protease domain, were 4- to 10-fold lower than that obtained with the wild-type gene. In other words, these mutants require less extracellular leucine to give strong signaling. This behavior is similar to that of the previously characterized constitutive SSY1 and PTR3 mutants (18, 28). Remarkably, however, the SSY5-13 mutation, which maps in the prodomain of Ssy5p, exhibited an EC50 value similar to that of the wild type.
Ssy5p is structurally similar to α-lytic protease from Lysobacter enzymogenes.
Because of the lack of direct structural data, the Ssy5p sequence was modeled using PHYRE (http://www.sbg.bio.ic.ac.uk/phyre [24]). The highest scoring structure found by PHYRE was that of the trypsin-like serine protease α-lytic protease from Lysobacter enzymogenes (Protein Data Bank code 1SSX) (17). A precision value (PHYRE) of 70% was obtained.
The PHYRE alignment covers residues 544 to 689 of Ssy5p, amounting to most of the protease domain. This region was predicted to have predominantly β-structure, while the N-terminal proregion has a high α-helical content. The sequence identity between Ssy5p and the α-lytic protease over the aligned region was 13% and includes the regions around Ser and Asp of the catalytic triad. We therefore used the similarity with the α-lytic protease as a structural framework to interpret the effects of the constitutive SSY5 mutations.
Three of the constitutive mutations (F575V, Q576P, and K581N + P632H) alter side chains in two loops between secondary-structure elements conserved between Ssy5p and the α-lytic protease. These loops are longer in Ssy5p and are indicated in Fig. 4B by broken red lines. The loops are in the region of the protease domain that in the α-lytic protease interacts with the N-terminal proregion (30). It is notable that the constitutive mutations alter sites that are spatially clustered according to this model, and we interpret this to mean that this part of Ssy5p is subject to conformational changes in relation to signaling, perhaps affecting interactions with the prodomain.
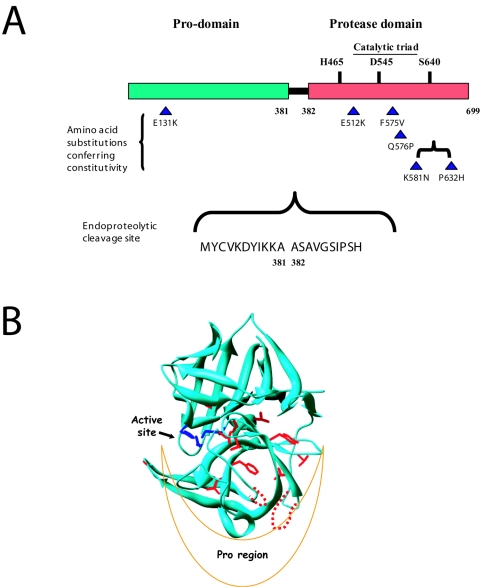
FIG. 4.
Structural features of the Ssy5 protease. (A) The positions of amino acid substitutions in the constitutive SSY5 mutants constructed in the present work are indicated by triangles together with the position of the E512K substitution in SSY5-6 reported previously (28). The proposed (1, 2) catalytic triad, as conserved in chymotrypsin-like serine proteases, is also indicated. The site of endoproteolytic cleavage, between amino acid residue positions 381 and 382, is indicated with flanking residues. (B) The structure of α-lytic protease from Lysobacter enzymogenes (PDB code 1SSX) is shown in ribbon representation, with the catalytic triad (in blue) and identical residues aligned by PHYRE (in red) shown as sticks. The loops predicted to be longer in Ssy5p and containing four of the substitutions encoded by the constitutive mutations are shown as dashed lines in red.
DISCUSSION
Ssy5p, together with the Ssy1p integral membrane protein and Ptr3p, is believed to be part of the SPS amino acid sensor complex (8, 15, 28). It was recently proposed that Ssy5p is the protease that activates the transcription factors Stp1p and Stp2p, which then stimulate transcription of the genes for several amino acid permeases (1, 2). In support of this, InterProScan analysis revealed that the C-terminal 230 amino acids of Ssy5p have weak similarity to chymotrypsin-like serine proteases, and expression analysis showed that Ssy5p is endoproteolytically processed into an N-terminal prodomain and a C-terminal protease domain (Fig. 4A). Mutational alteration of the proposed catalytic triad (H465, D545, and S640) led to complete loss of signaling (1, 2).
To initiate biochemical studies of Ssy5p, we expressed Ssy5p in E. coli and overexpressed it in S. cerevisiae. Expression of Ssy5p in E. coli resulted in inclusion bodies, and these were successfully used to raise polyclonal antibodies that proved valuable in the analysis of Ssy5p processing in yeast. Using these antibodies and a monoclonal antibody that recognizes the His6 tag we found that the overexpression of Ssy5p-His6 in yeast results in the production of full-length Ssy5p-His6 protein and two cleavage products (Fig. 1). All three species of Ssy5p accumulated when overexpressed, whether or not leucine was present in the growth medium (Fig. 1 and 2). These results confirm and extend similar findings by Abdel-Sater et al. (1).
When Forsberg and Ljungdahl (15) examined myc-Ssy5p expressed at presumably normal levels from the SSY5 promoter in a pRS316 low-copy construct, they detected a single ca. 67-kDa protein that disappeared rapidly upon induction with leucine. In contrast, Abdel-Sater et al. (1) found that N-terminally hemagglutinin-tagged Ssy5p expressed from its own promoter resulted in the production of full-length Ssy5p and the Ssy5p prodomain after 30 min of induction. The different tagging techniques and expression levels in the two studies cited may influence not only the molecular behavior of Ssy5p but also the signaling phenotype. Indeed, the N-terminal hemagglutinin tagging gave a constitutive signaling phenotype (2), and it could be that insertion of the c-myc epitope results in fast removal of the N terminus of Ssy5p. Our results show unambiguously that overexpression of Ssy5p results in the stable accumulation of all three Ssy5p forms. In future experiments it will be interesting to analyze native Ssy5p forms in plasma membrane fractions using polyclonal Ssy5p antibodies.
Ni-Sepharose chromatography allowed isolation of the C-terminal fragment of Ssy5p-His6 from yeast cells overexpressing the protein overnight. N-terminal sequencing of this fragment revealed that Ssy5p is cleaved between amino acid residues 381 and 382, resulting in a prodomain of 381 amino acid residues and a protease domain of 318 residues (Fig. 4). This result contradicts the proposal of an endoproteolytic cleavage site in the region around residue 420 (1).
The coelution of the prodomain and the protease domain from Ni-Sepharose is intriguing, since the prodomain had no His tag. Whether this reflects a complex between the two components or Ssy5p self-processing during purification is an important issue that must be addressed in the future.
We also isolated and characterized constitutive gain-of-function SSY5 mutants that map in the prodomain (Ssy5pE131K) and the protease domain (Ssy5pF575V, Ssy5pQ576P and Ssy5pK581N, P632H) (Fig. 4A). The mutations confer amino acid-independent activation of the AGP1 promoter through an increased basal level of processing of the transcription factor Stp1p. We analyzed these mutants by quantifying their dose-response relationships for signaling by extracellular leucine and found that the EC50 values for the three mutants mapping in the protease domain were 4- to 10-fold lower than that of the wild type. In other words, the three protease domain mutants are hyperresponsive, behaving similarly to the previously isolated SSY1 (18), PTR3, and SSY5 (28) constitutive and hyperresponsive mutants.
We interpret the increase of the basal level of Stp1p processing concomitant with the decrease of EC50 in this type of mutant to mean that the amino acid substitution influences the equilibrium between a signaling and a nonsignaling conformation of the whole sensor complex (18, 28). The finding of this type of mutation in SSY1, PTR3, and SSY5 mutants suggests that all three components of the SPS sensor have coordinated dynamics of these conformational shifts. Strikingly, a mutation mapping to the Ssy5p prodomain does not change the apparent affinity for leucine, indicating a qualitatively different effect of this mutation.
Many intracellular and extracellular proteases of both prokaryotic and eukaryotic origin are synthesized as inactive proenzymes (zymogens), from which a proregion can be cleaved, releasing the catalytically active protease. Proregions have roles in folding of the associated protease domains, intracellular protein transport, and inhibition of the protease activity. In some cases the cleaved proregion has high binding affinity to the protease (5, 6); in other cases this affinity is low, although transient binding can be sufficient for the proregion to function as a folding catalyst for the protease domain (29, 34, 35).
Similarly, the prodomain of Ssy5p may function as a catalyst for the folding of the protease domain and have an inhibitory effect on protease activity. In this view, a signaling conformation of the SPS complex could relieve the inhibitory effect of the prodomain, e.g., by promoting release or breakdown of the prodomain. The Ssy5pE131K prodomain mutation may result in less inhibition of the enzymatic activity of the protease by the prodomain in a way that is independent of the putative conformational shift of the SPS sensor, e.g., by increased release or degradation of the prodomain.
The predicted structure of the Ssy5p protease domain obtained by an analysis using PHYRE suggests that the Ssy5pF575V, Ssy5pQ576P and Ssy5pK581N, P632H substitutions are located in a region close to the prodomain (dashed loops in Fig. 4B). This location is striking, and we find it very significant, since it supports the possibility that the prodomain has an important, presumably negative, role in signaling and that these mutations may decrease effective prodomain-protease interaction.
On the basis of our findings it is likely that the release of an inhibitory effect of the prodomain is a key event in signaling. We suggest the following steps for the amino acid-sensing signal transduction mechanism: when an extracellular amino acid molecule binds to Ssy1p, a signaling conformation of the SPS sensor complex is stabilized, resulting in alteration or perhaps disruption of binding between the prodomain and the protease domain and subsequent cleavage of Stp1p (and Stp2p), which is then translocated to the nucleus to activate target promoters.
Recent investigations have revealed many components needed for proper amino acid signaling, including the three SPS sensor components (7, 13, 21, 23, 25), the transcription factors Stp1p and Stp2p (1, 3, 22), the Grr1p component of the SCFGrr1 ubiquitin ligase (1, 4, 33), the Yck1p and Yck2p casein kinases (1, 33), and the Rts1p regulatory subunit of protein phosphatase 2A (14). In addition, several SPS sensor-independent genes (ASI) that suppress ssy1 and ptr3 null mutants have been isolated (16).
The constitutive mutants described here and in previous publications (18, 28) may help to further unravel the complexity of interactions of the components of the pathway. When detailed structural information on some of these components becomes available, the constitutive mutants may prove particularly informative in elucidating the roles of specific amino acid residues in signaling.
Acknowledgments
We thank Lisbeth F. Petersen and Annette Kure for excellent technical assistance and Clive Phipps Walter (Danisco Innovation, Copenhagen) for carrying out protein sequencing. We also thank Richard F. Gaber for carefully reading the manuscript and Per O. Ljungdahl for access to unpublished results and inspiring discussions.
The work was partly supported by a grant from the Danish Natural Science Research Council to Leila Lo Leggio.
REFERENCES
- 1.Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. André. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C. 2004. Ph.D. thesis. Karolinska University Press, Stockholm, Sweden.
- 3.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andréasson, C., and P. O. Ljungdahl. 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full SPS sensor control. Mol. Cell. Biol. 24:7503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, D., J. L. Silen, and D. A. Agard. 1992. Protease pro region required for folding is a potent inhibitor of the mature enzyme. Proteins 12:339-344. [DOI] [PubMed] [Google Scholar]
- 6.Baker, D., A. K. Shiau, and D. A. Agard. 1993. The role of pro regions in protein folding. Curr. Opin. Cell Biol. 5:966-970. [DOI] [PubMed] [Google Scholar]
- 7.Barnes, D., W. Lai, M. Breslav, F. Naider, and J. M. Becker. 1998. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 29:297-310. [DOI] [PubMed] [Google Scholar]
- 8.Bernard, F., and B. André. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41:489-502. [DOI] [PubMed] [Google Scholar]
- 9.Boles, E., and B. André. 2004. Role of transporter-like sensors in glucose and amino acid signalling in yeast. Curr. Top. Genet. 9:121-153. [Google Scholar]
- 10.Brandt, A. 1991. Pulse labeling of yeast cells as a tool to study mitochondrial protein import, p. 369-376. In A. M. Tartakoff (ed.), Methods in cell biology, vol. 34. Vectorial transport of proteins into and across membranes. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 11.Davis, G. D., C. Elisee, D. M. Newham, and R. G. Harrison. 1999. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65:382-388. [PubMed] [Google Scholar]
- 12.Didion, T., M. Grauslund, M. C. Kielland-Brandt, and H. A. Andersen. 1996. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178:2025-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didion, T., B. Regenberg, M. U. Jørgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 14.Eckert-Boulet, N., K. Larsson, B. Wu, P. Poulsen, B. Regenberg, J. Nielsen, and M. C. Kielland-Brandt. 2006. Deletion of RTS1, encoding a regulatory subunit of protein phosphatase 2A, results in constitutive amino acid signaling via increased Stp1p processing. Eukaryot. Cell 5:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg, H., M. Hammar, C. Andréasson, A. Molinér, and P. O. Ljungdahl. 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158:973-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrmann, C. N., B. A. Kelch, N. Ota, and D. A. Agard. 2004. The 0.83 Å resolution crystal structure of α-lytic protease reveals the detailed structure of the active site and identifies a source of conformational strain. J. Mol. Biol. 338:999-1013. [DOI] [PubMed] [Google Scholar]
- 18.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grauslund, M., T. Didion, M. C. Kielland-Brandt, and H. A. Andersen. 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269:275-280. [DOI] [PubMed] [Google Scholar]
- 20.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van de Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29:556-563. [DOI] [PubMed] [Google Scholar]
- 21.Iraqui, I., S. Vissers, F. Bernard, J.-O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen, M. U., C. Gjermansen, H. A. Andersen, and M. C. Kielland-Brandt. 1997. STP1, a gene involved in pre-tRNA processing in yeast, is important for amino acid uptake and transcription of the permease gene BAP2. Curr. Genet. 31:241-247. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, L. A., R. M. MacCallum, and M. J. E. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 25.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 27.Poulsen, P., B. Wu, R. F. Gaber, K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2005. Amino acid sensing by Ssy1. Biochem. Soc. Trans. 33:261-264. [DOI] [PubMed] [Google Scholar]
- 28.Poulsen, P., B. Wu, R. F. Gaber, and M. C. Kielland-Brandt. 2005. Constitutive signal transduction by mutant Ssy5p and Ptr3p components of the SPS amino acid sensor system in Saccharomyces cerevisiae. Eukaryot. Cell 4:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos, C., J. R. Winther, and M. C. Kielland-Brandt. 1994. Requirement of the propeptide for in vivo formation of active yeast carboxypeptidase Y. J. Biol. Chem. 269:7006-7012. [PubMed] [Google Scholar]
- 30.Sauter, N. K., T. Mau, S. D. Rader, and D. A. Agard. 1998. Structure of alpha-lytic protease complexed with its pro region. Nat. Struct. Biol. 5:945-950. [DOI] [PubMed] [Google Scholar]
- 31.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 32.Sikorsky, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Hazel, H. B., M. C. Kielland-Brandt, and J. R. Winther. 1993. The propeptide is required for in vivo formation of stable active yeast proteinase A and can function even when not covalently linked to the mature region. J. Biol. Chem. 268:18002-18007. [PubMed] [Google Scholar]
- 35.Winther, J. R., P. Sørensen, and M. C. Kielland-Brandt. 1994. Refolding of a carboxypeptidase Y folding intermediate in vitro by low-affinity binding of the proregion. J. Biol. Chem. 269:22007-22013. [PubMed] [Google Scholar]