Abstract
Phylogenetic analyses show the single origin of a plastid metabolite translocator family in the Plantae from a gene encoding an existing endomembrane-derived protein. Red algal secondary endosymbiosis has spread a translocator gene into the ancestor of the “chromalveolate” protists, where it has diversified into a novel clade of proteins.
The photosynthetic organelle (plastid) in red, green, and glaucophyte algae (Plantae) likely had a single origin from a cyanobacterial endosymbiosis (8, 17). This ancient (ca. 1 to 1.5 billion years ago) (10, 24) event was followed by the transfer of genetic material from the endosymbiont to the nuclear genome of the host, the evolution of a protein import apparatus for the plastid and targeting sequences for nucleus-encoded plastid-targeted proteins, and the establishment of genome-plastome intracellular communication and regulation (4, 11, 23). Current hypotheses regarding plastid origin and evolution provide plausible explanations for the later stages of organelle establishment (e.g., gene transfer to the nucleus) but do not specifically address the initial formation of the endosymbiosis.
We hypothesize that the insertion of a metabolite antiporter into the ancestral plastid membrane was essential for establishing the primary endosymbiosis, allowing the ancestor of the Plantae to profit immediately from cyanobacterial carbon fixation. This antiporter likely evolved from an existing host metabolite translocator associated with either mitochondrial (5, 12) function or the endomembrane system. In contrast, autotrophic free-living cyanobacteria have no obvious need for such antiporters. Members of the nucleotide-sugar/triose phosphate translocator gene family (15) are likely ancestors to plastid translocators because (i) genes encoding these proteins are found in all sequenced eukaryotic genomes, (ii) orthologous proteins are absent from prokaryotes, and (iii) some gene family members are targeted to plant plastids.
To address plastid translocator origin, we gathered available sequences (genome and expressed sequence tag data) from the NCBI (http://www.ncbi.nlm.nih.gov/), DOE Joint Genome Institute (http://www.jgi.doe.gov/), Michigan State University Galdieria Database (http://genomics.msu.edu/galdieria/sequence_data.html), Porphyra yezoensis EXPRESSED SEQUENCE TAG Index (http://www.kazusa.or.jp/en/plant/porphyra/EST/), and the Cyanidioschyzon merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp/). Homologs were identified using BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) with an E-value cutoff of ≤10−4. A translocator phylogeny was inferred using the protein maximum-likelihood method (PHYML V2.4.3 [http://atgc.lirmm.fr/phyml/]), neighbor joining (NJ) (PHYLIP V3.63 [http://evolution.genetics.washington.edu/phylip.html]), maximum parsimony (MP) (PAUP*V4.0b10 [http://paup.csit.fsu.edu/]), and Bayesian (MrBayes V3.0b4 [http://mrbayes.csit.fsu.edu/index.php]) inference (for details of the phylogenetic approach, see reference 13). A total of 250 amino acids from 65 plastid phosphate translocators from red and “chromalveolate” (i.e., chlorophyll c-containing) algae and land plants and their nonplastid homologs were included in these analyses.
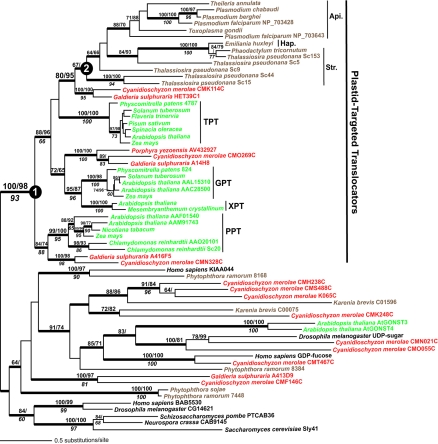
The resulting tree (Fig. 1) shows that plastid translocators are monophyletic (Fig. 1) and had a single origin from an existing endomembrane translocator (PHYML bootstrap support, 100%; NJ, 98%; MP, 93%), consistent with our model. Each major group of plant plastid translocators (i.e., triose phosphate translocator [TPT], glucose-6-phosphate translocator [GPT]/xylulose-5-phosphate translocator [XPT], and phosphoenolpyruvate translocator [PPT] [shown in green text in Fig. 1]) form robustly supported (ML and MP bootstrap values of >90%) lineages that are sisters to homologs in the red algae (Fig. 1, shown in red text). This result supports the monophyly of red and green algae and land plants (see, e.g., reference 19) and is consistent with the origin of the different plastid translocators in their common ancestor; i.e., all plastid translocators are monophyletic, and each is divided into sister groups (two of which are well supported, PPT and GPT/XPT) comprised of the red and green clades. Presumably, these Plantae translocators diversified because of the selective advantage they offer, i.e., to harvest the different fixed carbon products of the cyanobacterial primary endosymbiont. This phylogeny provides strong evidence for a single primary endosymbiosis in the studied Plantae, reflecting a critical and early step in plastid evolution in its topology. The monophyly of GPTs and XPTs has previously been reported (15) and likely reflects a more recent plant-specific gene duplication event.
FIG. 1.
Maximum-likelihood phylogeny of endomembrane and plastid translocators. PHYML bootstrap values (200 replications) are shown above the branches on the left of the slash mark, whereas the values to the right are from an NJ analysis (100 replications). The bootstrap values shown below the branches in italics are from an unweighted MP analysis (2,000 replications). Only bootstrap values ≥60% are shown. The red algae, plants, and chromalveolates are shown in red, green, and brown text, respectively. The different plastid translocators are GPT, PPT, TPT, and XPT. Api. is apicomplexans, Hap. is haptophytes, and Str. is stramenopiles. The numbers in the filled circles indicate translocators to the right that resulted from primary (1) and secondary (2) endosymbiosis.
The chromalveolates are a taxonomically diverse assemblage of protists comprised of alveolates (dinoflagellate algae, ciliates, and parasitic apicomplexans) and chromists (cryptophytes, haptophytes, and stramenopiles) that are thought to have ancestrally contained a plastid of red algal origin (7). This group is yet to be substantiated in a global eukaryotic phylogeny. It is therefore of interest that the available (i.e., apicomplexan, haptophyte, and stramenopile) chromalveolate sequences are monophyletic in our tree (Fig. 1) and solidly associated (ML, 80%; NJ, 95%) with one clade of red algal proteins (i.e., Cyanidioschyzon merolae CMK114C and Galdieria sulfuraria HET39C12). The addition of cryptophyte and dinoflagellate plastid translocators is needed to verify this result. Interestingly, preliminary biochemical analysis of the G. sulfuraria HET39C12 translocator suggests that it is a TPT (A. P. M. Weber and M. Linka, unpublished data); therefore, the ancestral chromalveolate plastid translocator was likely of this type.
The topology of the chromalveolate subtree supports a single origin of the plastid translocator gene in the common ancestor of these species (i.e., supporting their monophyly) from the nucleus of the red algal endosymbiont. It is noteworthy that PPT and GPT/XPT from Plantae are absent from the chromalveolates. In organisms containing plastids of secondary endosymbiotic origin with three or four plastid envelope membranes, transfer of a phosphate translocator-related gene to the host nucleus and retargeting to the inner plastid membrane alone are not sufficient for the export of photosynthates to the host. Additional translocators with identical transport properties would be required in the third (i.e., the remnant of the endosymbiont plasma membrane) and the fourth (i.e., the plastid endoplasmic reticulum) membranes to connect the metabolism of the host and the endosymbiont. This essential metabolic connection has been accomplished in the ancestor of the chromalveolates in our tree with the red algal gene undergoing diversification through duplication and divergence giving rise to the complex branching pattern shown in Fig. 1. The specific functions and the detailed membrane localizations of the different chromalveolate translocators remain to be determined.
We analyzed chromalveolate translocators with significant N-terminal extensions to determine if they are putatively plastid (or apicoplast) targeted. Using PATS (http://gecco.org.chemie.uni-frankfurt.de/pats/pats-index.php) and PlasmoAP (http://www.plasmodb.org/restricted/PlasmoAPcgi.shtml), apicoplast targeting is supported for the apicomplexan Plasmodium spp. (e.g., Plasmodium falciparum, GenBank accession number NP_703643 [PATS probability {prob}, 0.954; PlasmoAP, 5/5 tests positive). Similarly, the N-terminal extension in the diatom (stramenopile) Thalassiosira pseudonana Sc15 (i.e., gene located on scaffold 15) sequence contains a typical bipartite plastid-targeting sequence (16) comprised of a signal sequence (using SignalP [http://www.cbs.dtu.dk/services/SignalP/]; prob, 0.997; predicted length, 18 amino acids) followed by a transit peptide (using TargetP; prob, 0.740). These results suggest that the chromalveolate clade in our tree includes plastid-targeted translocators, although it is possible that others in this group may have lost this function. For example, the translocator from Theileria annulata apparently does not encode an N-terminal extension but is clearly related to the other apicomplexan sequences.
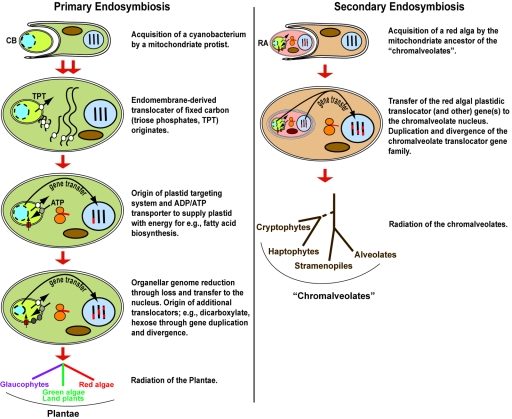
Although we do not have glaucophyte translocators in the tree, the signature activity of plastidic phosphate translocators has been detected in isolated cyanelles of Cyanophora paradoxa, indicating the presence of plastidic phosphate translocators (21). We therefore hypothesize that the insertion of a phosphate translocator into the plasma membrane of the endosymbiont occurred before the split of the Plantae and was probably a critical step in rendering the association between the cyanobacterium and the mitochondriate eukaryote irreversible. The proposed sequence of events for plastid establishment (Fig. 2) does not involve major evolutionary innovations. The basic components of a plastid envelope protein import apparatus were present in the cyanobacterium (see, e.g., reference 18), and it is reasonable to assume that this machinery was capable of importing proteins from the host cell cytosol, albeit with low efficiency; i.e., the outer leaflet of the plastid outer envelope membrane consists mainly of endoplasmic reticulum-derived phospholipids (6, 9), indicating a close interaction between plastid envelope membranes and the host endomembrane system. It is likely that the cyanobacterium was engulfed as a prey item (as often occurs today) through phagocytosis, an event that likely occurred countless times, and in some of these cells, the prey was not digested in the food vacuole but rather was maintained as an endosymbiont. A descendant of these cells gave rise to the Plantae. The origin (or replacement) of plastids through cell (or organelle) engulfment has occurred several times in evolution and can be found in taxa such as the filose amoeba Paulinella chromatophora (2) and in several dinoflagellate lineages (13, 20, 22). The position of the Plantae in the tree of life (see, e.g., references 1 and 19) suggests that its common ancestor was a highly developed flagellate (or had flagellated stages) that most likely had the capacity for phagocytosis and endomembrane formation (for a discussion, see reference 6).
FIG. 2.
Model for plastid and translocator origin in the common ancestor of the Plantae and the chromalveolates. CB is cyanobacterium, and RA is red alga. The chromalveolate tree reflects the present understanding of the phylogeny of this group (see, e.g., references 3 and 14).
Acknowledgments
This work was supported by NSF awards EF 03-32882 to A.P.M.W. and MCB 02-36631 and EF 04-31117 to D.B.
REFERENCES
- 1.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya, D., T. Helmchen, C. Bibeau, and M. Melkonian. 1995. Comparisons of nuclear-encoded small-subunit ribosomal RNAs reveal the evolutionary position of the Glaucocystophyta. Mol. Biol. Evol. 12:415-420. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, D., H. S. Yoon, and J. D. Hackett. 2004. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26:50-60. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E. C., A. Somanchi, and S. P. Mayfield. 2001. Interorganellar crosstalk: new perspectives on signaling from the chloroplast to the nucleus. Genome Biol. 2:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullerwell, C. E., and M. W. Gray. 2004. Evolution of the mitochondrial genome: protist connections to animals, fungi and plants. Curr. Opin. Microbiol. 7:528-534. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith, T. 2000. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5:174-182. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryotic family tree. J. Eukaryot. Microbiol. 46:347-366. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith, T. 1992. The number of symbiotic origins of organelles. Biosystems 28:91-108. [DOI] [PubMed] [Google Scholar]
- 9.Douce, R., and J. Joyard. 1981. Does the plastid envelope derive from the endoplasmic reticulum? Trends Biochem. Sci. 6:237-239. [Google Scholar]
- 10.Douzery, E. J., E. A. Snell, E. Bapteste, F. Delsuc, and H. Philippe. 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA 101:15386-15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fey, V., R. Wagner, K. Brautigam, and T. Pfannschmidt. 2005. Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56:1491-1498. [DOI] [PubMed] [Google Scholar]
- 12.Gray, M. W., B. F. Lang, and G. Burger. 2004. Mitochondria of protists. Annu. Rev. Genet. 38:477-524. [DOI] [PubMed] [Google Scholar]
- 13.Hackett, J. D., H. S. Yoon, M. B. Soares, M. F. Bonaldo, T. L. Casavant, T. E. Scheetz, T. Nosenko, and D. Bhattacharya. 2004. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr. Biol. 14:213-218. [DOI] [PubMed] [Google Scholar]
- 14.Harper, J. T., E. Waanders, and P. J. Keeling. 2005. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 55:487-496. [DOI] [PubMed] [Google Scholar]
- 15.Knappe, S., U. I. Flügge, and K. Fischer. 2003. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131:1178-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang, M., K. E. Apt, and P. G. Kroth. 1998. Protein transport into “complex” diatom plastids utilizes two different targeting signals. J. Biol. Chem. 273:30973-30978. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, J. D. 2003. The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 39:4-11. [Google Scholar]
- 18.Reumann, S., K. Inoue, and K. Keegstra. 2005. Evolution of the general protein import pathway of plastids (review). Mol. Membr. Biol. 22:73-86. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Ezpeleta, N., H. Brinkmann, S. C. Burey, B. Roure, G. Burger, W. Loffelhardt, H. J. Bohnert, H. Philippe, and B. F. Lang. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15:1325-1330. [DOI] [PubMed] [Google Scholar]
- 20.Saldarriaga, J. F., F. J. Taylor, P. J. Keeling, and T. Cavalier-Smith. 2001. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 53:204-213. [DOI] [PubMed] [Google Scholar]
- 21.Schlichting, R., and H. Bothe. 1993. The cyanelles (organelles of a low evolutionary scale) possess a phosphate-translocator and a glucose-carrier in Cyanophora paradoxa. Bot. Acta 106:428-434. [Google Scholar]
- 22.Schnepf, E., and M. Elbrächter. 1999. Dinophyte chloroplasts and phylogeny—a review. Grana 38:81-97. [Google Scholar]
- 23.Strand, A. 2004. Plastid-to-nucleus signalling. Curr. Opin. Plant Biol. 7:621-625. [DOI] [PubMed] [Google Scholar]
- 24.Yoon, H. S., J. D. Hackett, C. Ciniglia, G. Pinto, and D. Bhattacharya. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21:809-818. [DOI] [PubMed] [Google Scholar]