Abstract
The SSU processome is required for production of the small ribosomal subunit RNA, the 18S rRNA. Specifically, the U3 small nucleolar RNA (snoRNA) component of the SSU processome is essential for the formation of the conserved central pseudoknot and for cleavages of the pre-rRNA, both of which are required for 18S maturation. To further elucidate how these events are mediated, we examined the regulatory and mechanistic roles of the U3 specific proteins: Imp3p, Imp4p, and Mpp10p. We found that these proteins demonstrated an interdependence with respect to their stability and to their association with the U3 snoRNA. Because mutations in the U3 snoRNA that disrupt pre-rRNA processing confer similar defects on growth and pre-rRNA processing as do carboxy-terminal truncations of Mpp10p, we hypothesized that Mpp10p may be involved in maintaining U3 snoRNA-pre-rRNA base pairing. However, combining the two mutations resulted in a more pronounced cleavage defect at site A2, suggesting that Mpp10p is also required at an additional mechanistic step. Furthermore, heterologous complementation experiments demonstrate that the last 95 amino acids of yeast Mpp10p are specifically required for growth and pre-rRNA processing at low temperatures.
Eukaryotic ribosomes contain four RNAs: 25S, 18S, 5.8S, and 5S. The 5S rRNA is transcribed by RNA polymerase III, whereas the three remaining rRNAs are transcribed as a 35S polycistronic precursor by RNA polymerase I. Contained within the 35S precursor are the 18S rRNA, the RNA component of the small subunit, and the 5.8S and 25S rRNAs, which are components of the large subunit. The 35S pre-rRNA undergoes extensive cleavage, processing, and modification so that the rRNAs contained within the precursor are liberated and incorporated into the correct ribosomal subunit (Fig. 1A) (reviewed in references 16 and 30). Many of the processes leading to the maturation of the rRNAs are carried out in the nucleolus by a myriad of small nucleolar RNPs (snoRNPs). Specifically, maturation of the 18S, 5.8S, and 25S rRNAs requires the box C/D snoRNPs, the box H/ACA snoRNPs, and RNase mitochondrial RNA processing (MRP). The RNA molecules found in all box C/D snoRNPs contain conserved sequence elements, boxes C and D. Most of the box C/D snoRNPs are required to guide the 2′-O-ribose methylation of specific sites on the nascent 35S rRNA precursor.
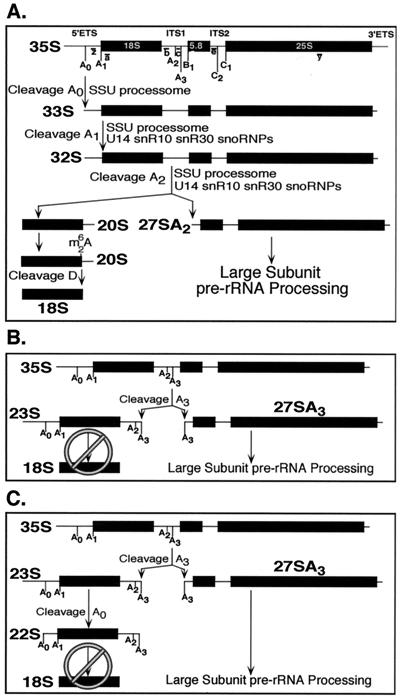
FIG. 1.
Normal and defective pre-rRNA processing in Saccharomyces cerevisiae. (A) The normal pathway for pre-rRNA processing. The 35S pre-rRNA is transcribed by RNA polymerase I and undergoes extensive modifications and processing, which results in the production of the 18S, 5.8S, and 25S rRNAs. The processing sites of the initial transcript are designated (reviewed in references 16 and 30). This figure has been adapted from reference 16. The relative positions of the oligonucleotide probes (a, b, c, e, and y) used in Northern blot analysis are indicated in lowercase letters. (B) Defective pre-rRNA processing as a result of depletion of SSU processome components. Decreased or absent cleavage at the A0, A1, and A2 sites in the pre-rRNA results in the accumulation of the 23S precursor and reduced accumulation of the 18S rRNA. (C) Defective pre-rRNA processing in yeast strains rendered cold sensitive by mutations in either the U3 snoRNA or Mpp10p. Defective processing at sites A1 and A2 results in the accumulation of the 22S precursor.
There are, however, two box C/D snoRNPs, U3 and U14, that are evolutionarily conserved and are required for cleavage events leading to the production of mature 18S rRNA. Until a short time ago, the U3 snoRNP was thought to consist of the U3 snoRNA (333 nucleotides) and 11 proteins. The proteins common to U3 and all other box C/D snoRNPs include Nop1p, Nop5/58p, Nop56p, and Snu13p (10, 23, 29, 32), and the proteins specific to U3 include Sof1p, Mpp10p, Lcp5p, Imp3p, Imp4p, Dhr1p, and Rrp9p (7, 9, 15, 18, 31, 35). More recently, however, it has become evident that the label “snoRNP” may not be entirely accurate. Because the U3 processing complex runs at 80S on sucrose gradients and contains at least 28 proteins (8), it has been renamed the SSU processome.
All specific, essential protein components of the SSU processome, including the U3 snoRNA itself, are required for cleavage at sites A0 and A1 of the 5′ external transcribed spacer (ETS) and at site A2 in internal transcribed spacer 1 (ITS1) (Fig. 1A) (14). The U3-mediated cleavage at site A2 results in the separation of the RNAs destined for the small ribosomal subunit from the RNAs destined for the large ribosomal subunit. Mutations in both U3 and Mpp10p cause cold sensitivity; a study of these mutations demonstrated that the role of the SSU processome in cleavage at A0 could be separated from its role in cleavage at A1 and A2 (13, 17). Since cold sensitivity is often a hallmark of defects in the assembly of large complexes (12, 28), these results suggest that the mutations in the U3 snoRNA and the Mpp10 protein disrupt assembly steps that are vital to the function of the SSU processome. It has been proposed that the mutations in the U3 snoRNA disrupt the U3 snoRNA-pre-rRNA base pairing that is necessary for formation of the central pseudoknot (13). The cold-sensitive nature of truncations of Mpp10p, however, may be linked to its interactions with the other U3 specific proteins, Imp3p and Imp4p. U3 is essential for two base-pairing interactions that result in formation of the evolutionarily conserved central pseudoknot in the SSU rRNA and are required for cleavage of the pre-rRNA (4, 5, 13, 19, 22, 24).
Few analyses have addressed the regulatory or mechanistic roles of the SSU processome proteins. We focused on the U3 specific proteins Imp3p, Mpp10p, and Imp4p and found that the Imp3 protein regulates SSU processome activity by mediating the interactions of the Imp4 and Mpp10 proteins with the U3 snoRNA. In addition, Mpp10p regulates SSU processome activity by controlling the stability of the Imp3 and Imp4 proteins. Genetic analyses of strains carrying mutations in both the U3 snoRNA and Mpp10p, which cause cold sensitivity, indicated that together they are required to promote cleavage at site A2. Furthermore, heterologous complementation experiments revealed that species-specific sequences at the carboxy terminus of Mpp10p are required for its function in yeast. Together, these results suggest that Imp3p, Mpp10p, and Imp4p form a closely linked functional unit that is essential for the action of the SSU processome in ribosome biogenesis.
MATERIALS AND METHODS
Yeast protocols and media.
Yeast transformations were performed using a standard lithium acetate protocol (11). In general, yeast were grown under standard media conditions with YPD (1% yeast extract, 2% peptone, 2% dextrose) and YPGR (1% yeast extract, 2% peptone, 2% galactose, 2% raffinose). Yeast selective media SC −leucine (Clontech), SC −tryptophan (Clontech), SC −methionine −tryptophan (−met −trp), SC −leucine −uracil (Clontech), and SC −leucine −uracil −histidine (Clontech) were supplemented with 2% dextrose or with 2% galactose and 2% raffinose. 5-Fluoroorotic acid was used to select against yeast harboring a functional URA3 gene (6, 25). All solid media were prepared with 2% Bacto agar.
Construction of U3/Mpp10p cold-sensitive strains.
Strain YKW100 (a ura3-52 his3-Δleu2 lys2-801amber trp1-Δ63 u3aΔ UASGAL:U3A::URA3 u3bΔ::LEU2) is a haploid segregant resulting from the mating of JH84 (14, 22) and YPH499 (26). Homologous recombination of the HIS3 gene following nucleotide 1494 of the MPP10 genomic locus, in strain YKW100, resulted in the creation of strain YKW200 (a ura3-52 his3-Δ leu2 lys2-801amber trp1-Δ63 u3aΔ UASGAL:U3A::URA3 u3bΔ::LEU2 mppcs1::HIS3). Expression of mpp10cs1 results in the production of an mpp10p that is truncated by 95 amino acids (aa) at the carboxy terminus.
The XhoI-EcoRI U3 fragments were excised from pRU3*[wt], pRU3*[JH125], and pRU3*[JH137] (36) and cloned into the XhoI-EcoRI sites of pRS314 (26) to create pTRP-U3tag[wt], pTRP-U3tag[cs1], and pTRP-U3tag[cs2], respectively. The pTRP-U3tag series of plasmids were transformed into YKW100 and YKW200 and maintained on SC −trp minimal medium.
Construction of strains for analysis of Mpp10p-Imp3p-Imp4p interactions with the U3 snoRNA.
pGAL1::IMP3 was transformed with p415GPD-IMP4-HA, and pGAL1::IMP4 was transformed with p415GPD-IMP3-HA (18). The strains were maintained on SC −leu GAL/RAF.
Construction of Mpp10p complementation strains.
Using the BLAST algorithm, the yeast Mpp10 protein sequence was compared to all Drosophila melanogaster cDNAs available at the Berkeley Drosophila genome project. The database search revealed a cDNA (LD02752) with significant similarity to yeast Mpp10p. This cDNA clone was obtained from Research Genetics Inc. and was then fully sequenced in both directions by primer walking (W. M. Keck DNA Sequencing Facility, Yale University).
The LD02752 XbaI-XhoI fragment, containing the entire dMPP10 cDNA, was cloned into XbaI and XhoI sites of the p415GPD vector (Ampr LEU2 ARS/CEN) (21) to form p415GPD-dMPP10. To create p415GPD-d-yMPP10, the XbaI-SacII amino-terminal dMPP10 fragment and the SacII-XhoI carboxy-terminal yMPP10 fragment were cloned into XbaI-XhoI-cut p415GPD. The oligonucleotide primers YMPP10.30 (5′ GAG CCG CGG AGG TAA AGC AGA AAA GGA CGG A) and YMPP10.31 (5′GTC GCT CGA GTC AAA GTT TTA TAT TTG TGC T) were used with the pGAD3-MPP10 (9) template to amplify the SacII-XhoI carboxy-terminal yMPP10 fragment by PCR.
To create a yeast strain expressing the dMpp10 protein or the d-yMpp10 fusion protein, the vectors created above were shuffled (6, 25) into pGAL1::MPP10. These strains and a strain containing p415GPD-yMPP10 (17) were examined for conditional growth by plating 10-fold serial dilutions on YPD.
Northern blot analysis.
Total RNA was extracted by the hot-acid-phenol method as previously described (3, 9). For high-molecular-weight RNA species, 5 μg of total RNA was separated on formaldehyde-1.25% agarose gels. For low-molecular-weight RNAs, 2 μg of total RNA was separated on an 8% denaturing polyacrylamide gel. RNA was transferred to Zeta probe-GT nylon membranes (Bio-Rad) and UV cross-linked and hybridized as previously described (9).
The following large rRNA oligonucleotide probes were used: a (5′CAT GGC TTA ATC TTT GAG AC), b (5′GCT CTT TGC TCT TGC C), c (5′ATG AAA ACT CCA CAG TG), d (5′CCA GTT ACG AAA ATT CTT G), e (5′GGC CAG CAA TTT CAA GT), and y (5′GCC CGT TCC CTT GGC TGT G); the small RNA oligonucleotide probes included 5.8S rRNA (5′TTT CGC TGC GTT CTT CAT C) and U6 (5′AAA ACG AAA TAA ATC TCT TTG). The untagged U3 snoRNA was detected using probe SD14, and the tagged U3 snoRNA was detected using probe SD13 (22).
Immunoprecipitations.
Whole-cell yeast extracts were prepared and immunoprecipitations were carried out as previously described (18). For RNA coimmunoprecipitations, 50 μl of the anti-yMpp10 polyclonal antibody (9) or 200 μl of the anti-HA monoclonal 12CA5 antibody (culture supernatant) was used. For protein coimmunoprecipitations, 10 μl of the anti-yMpp10 polyclonal antibody (9) or 10 μl of the anti-yImp4 polyclonal antibody (J. Gallagher and S. Baserga, unpublished data) was used. All antibodies were bound to 2.5 mg of protein A-Sepharose CL-4B (Amersham Pharmacia Biotech).
Western blot analysis.
Western blot analyses were performed as previously described (17). Briefly, proteins were separated on sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The anti-Mpp10, anti-Imp3 (Gallagher and Baserga, unpublished), and anti-Imp4 polyclonal antibodies were used at a concentration of 1:10,000 following immunoprecipitation. The anti-Imp3 and anti-Imp4 antibodies were used at a dilution of 1:2,000 on analysis of total protein. The anti-Nop1 monoclonal B15 (2) was used at a dilution of 1:5,000 following immunoprecipitation but at 1:10,000 otherwise.
RESULTS
Mpp10p is required for the stability of SSU processome components Imp3p and Imp4p.
To examine the relationship between the Imp4, Imp3, and Mpp10 proteins, each protein was depleted and Western blot analyses were performed on cell extracts (Fig. 2A, lanes 2 to 4). Surprisingly, Mpp10p depletion resulted in the reduction of Imp3p and Imp4p to undetectable levels (lane 4). However, Imp3p and Imp4p depletion did not affect Mpp10p levels (lanes 2 and 3). Likewise, Imp4 depletion did not affect Imp3 levels and Imp3p depletion did not affect Imp4p levels (lanes 2 and 3, respectively). Levels of the Nop1 protein (fibrillarin) were not affected by depletion of either Imp4p, Imp3p, or Mpp10p (lanes 2 to 4). These results suggest that there is an interdependence among the Imp4, Imp3, and Mpp10 proteins for the maintenance of protein levels.
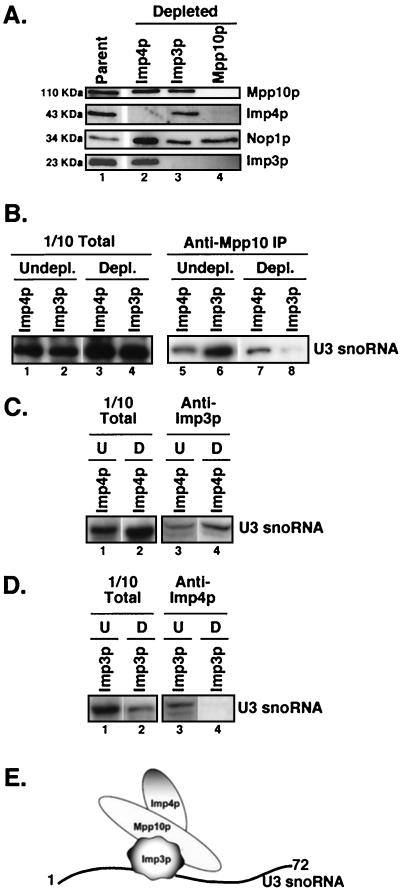
FIG. 2.
Imp3p, Mpp10p, and Imp4p stability and U3 snoRNA association are interdependent. (A) Western blots for Mpp10p, Imp4p, Nop1p, and Imp3p on extracts prepared from cells depleted of Imp4p, Imp3p, or Mpp10p. The approximate mobility on sodium dodecyl sulfate-polyacrylamide gels of each protein is indicated. (B) Anti-Mpp10p immunoprecipitation (IP) on yeast depleted of Imp3p and Imp4p. Mpp10p-associated U3 snoRNA was analyzed by Northern blotting. Totals represent the RNA from 1/10 the amount of extract used for immunoprecipitations. (C) Anti-HA-Imp3p immunoprecipitation on yeast depleted of Imp4p. Imp3p-associated U3 snoRNA was analyzed by Northern blotting. U indicates that Imp4p was not depleted, and D indicates that Imp4p was depleted. Totals represent the RNA from 1/10 the amount of extract used for immunoprecipitations. (D) Anti-HA-Imp4p immunoprecipitation on yeast depleted of Imp3p. Imp4p-associated U3 snoRNA was analyzed by Northern blotting. U indicates that Imp3p was not depleted, and D indicates that Imp3p was depleted. Totals represent the RNA from 1/10 the amount of extract used for immunoprecipitations. (E) A model for the relative arrangement of the Imp3, Imp4, and Mpp10 proteins with respect to the U3 snoRNA. This diagram is based on the results in panels A to D and on data in reference 36.
Imp3p is required for association of Mpp10p and Imp4p with the U3 snoRNA.
Through depletion and RNA coimmunoprecipitation experiments, we determined which of the three proteins were required to mediate interaction with the U3 snoRNA (Fig. 2B to D). Depletion of Imp3p but not Imp4p inhibited the ability of Mpp10p to coimmunoprecipitate the U3 snoRNA (Fig. 2B, lanes 7 and 8). As might be expected from these results, depletion of Imp4p did not impair the ability of Imp3p to coimmunoprecipitate the U3 snoRNA (Fig. 2C, lane 4) but depletion of Imp3p affected the ability of Imp4p to coimmunoprecipitate the U3 snoRNA (Fig. 2D, lane 4). Since depletion of Mpp10p results in decreased levels of both Imp3p and Imp4p (see above), the ability of Imp3p and Imp4p to associate with the U3 snoRNA in the absence of Mpp10p could not be determined. These results predict a model in which the putative RNA binding protein Imp3p (18) binds to the U3 snoRNA, Mpp10p binds to Imp3p, and Imp4p binds to Mpp10p (Fig. 2E). Because it has been demonstrated that nucleotides 1 to 72 of the U3 snoRNA are required for Mpp10p association (36) and because sequences within these nucleotides are required for base-pairing interactions, it is tempting to speculate that the Imp3 protein associates with these nucleotides and thus mediates the interaction between the U3 snoRNA and Mpp10p.
Creation of a system to study the role of Mpp10p in the SSU processome.
To further elucidate the mechanism by which the early steps of pre-rRNA processing occur, we created a series of strains to study the genetic interactions between the U3 snoRNA and Mpp10p (Fig. 3A). Initially, two haploid parent strains were created in which one copy of the U3 gene was disrupted and the other was under the control of a galactose- inducible/glucose-repressible promoter (Fig. 3A). The first parent strain contains the wild-type genomic copy of MPP10 (YKW100), and the second contains mpp10cs1 (YKW200) (Fig. 3A). Both parent strains were transformed with plasmids containing U3 under its endogenous promoter (pTRP-U3tag) (Fig. 3A). The exogenously expressed U3 contains a sequence-specific tag and either is wild type or is one of two cold-sensitive box A mutants: U3wt, U3cs1, or U3cs2 (Fig. 3B) (13, 22). Because strains expressing either U3 cold-sensitive allele behaved identically in subsequent analyses, we have included the results from only one in most cases. The resulting strains, U3wt/MPP10, U3wt/mpp10cs1, U3cs1/MPP10, U3cs1/mpp10cs1, U3cs2/MPP10, and U3cs2/mpp10cs1, were further analyzed for defects in growth and pre-rRNA processing.
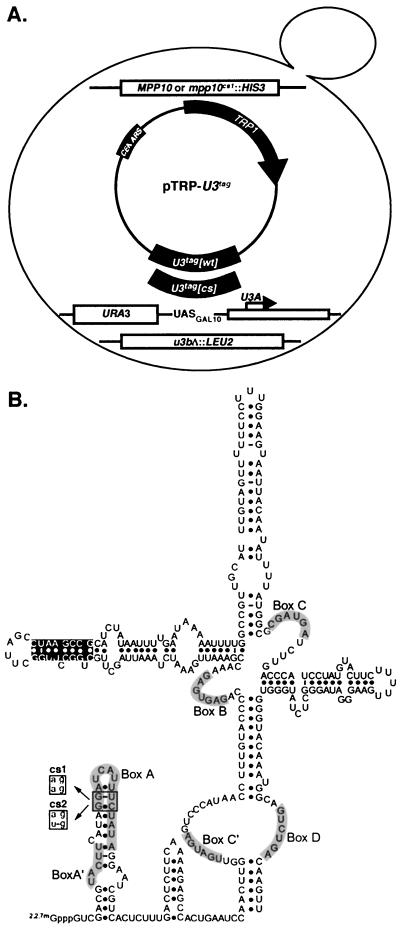
FIG. 3.
Features of the strains constructed to test the genetic interactions between mutations in two components of the SSU processome which confer cold sensitivity. (A) Strain construction and design. The haploid yeast strain contains a disruption of one U3 gene, while the other has been placed under the control of a galactose-inducible/glucose-repressible promoter (GAL10). Strains bear either a wild-type MPP10 gene or a genomic integration of mpp10cs1. mpp10cs1 represents, at the protein level, a carboxy-terminal truncation of 95 aa and confers cold sensitivity. Plasmids expressing either wild-type or mutated tagged U3 snoRNAs were also transformed into the MPP10 or mpp10cs1 strains. U3tag[cs] represents either U3cs1 or U3cs2; they each confer cold sensitivity. (B) Schematic of the in vitro secondary structure of the U3 snoRNA. The location of the sequence-specific tag (black shading) and the U3 snoRNA mutations which confer cold sensitivity (cs1 and cs2) are indicated. Adapted from references 19 and 22.
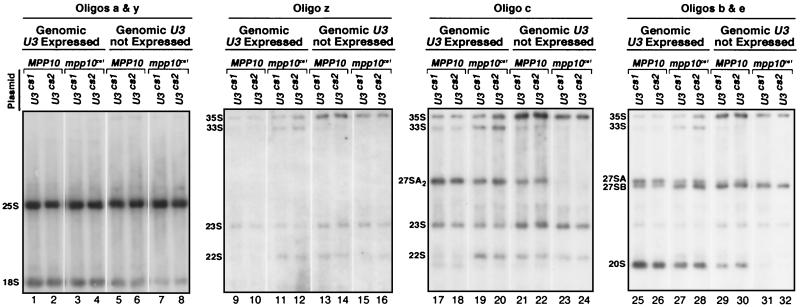
Due to the presence of a specific sequence tag in the exogenously expressed U3 snoRNAs (Fig. 3B), their levels and stability can be monitored independently of the endogenous U3 (Fig. 4A). As expected, Northern blot analysis with probes specific to genomically encoded (untagged) U3 indicated that growth of the strains in galactose promotes expression of the genomically encoded U3 while growth in glucose represses such expression (Fig. 4A, compare lanes 1 to 6 to lanes 7 to 12). Use of an oligonucleotide probe specific to the plasmid-borne U3 revealed that its levels increase in the absence of genomic U3 expression, in contrast to the U6 snRNA, whose levels remain constant (Fig. 4A). These results indicate that during glucose repression, only plasmid-borne U3 is expressed and that the U3cs molecules accumulate to the same levels as the endogenous U3 (Fig. 4A, lanes 7 to 12).
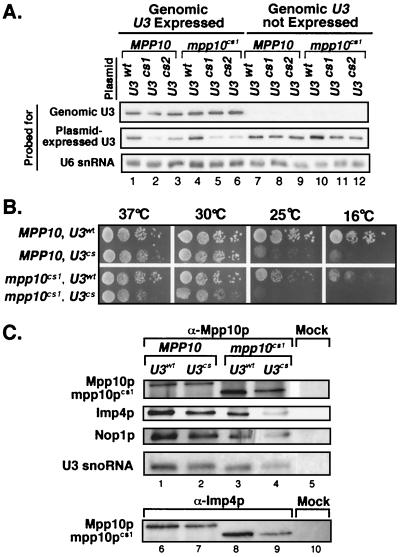
FIG. 4.
Expression of mpp10cs1 and U3cs together exacerbates growth defects in the cold without disrupting the SSU processome. (A) Repression of the genomic U3 gene by growth in glucose reveals normal levels of expression of the plasmid-expressed U3 snoRNA. The U3wt/MPP10, U3wt/mpp10cs1, U3cs1/MPP10, U3cs1/mpp10cs1, U3cs2/MPP10, and U3cs2/mpp10cs1 strains were either grown in galactose (Genomic U3 Expressed) or in glucose to repress the expression of the genomic U3 gene (Genomic U3 not Expressed). Total RNA was analyzed by Northern blotting with oligonucleotide probes specific to either the U3 snoRNA expressed from the genome, the tag in the plasmid-expressed U3 snoRNA, or the endogenous U6 snRNA. (B) Combining mutations in Mpp10p and the U3 snoRNA which cause cold sensitivity exacerbates growth defects. Serial dilutions of the U3wt/MPP10, U3cs1/MPP10, U3wt/mpp10cs1, and U3cs1/mpp10cs1 strains were grown at 37, 30, 25, and 16°C on glucose to repress the expression of the genome-encoded U3 snoRNA. (C) The SSU processome is not disrupted by coexpression of mpp10cs1 and U3cs. Anti-Mpp10p and anti-Imp4p immunoprecipitations were performed on extracts prepared from U3wt/MPP10, U3cs2/MPP10, U3wt/mpp10cs1, and U3cs2/mpp10cs1 strains. Coimmunoprecipitation of the Imp4, Nop1, Mpp10, and Mpp10cs1 proteins was examined by Western blotting. Coimmunoprecipitation of the U3 snoRNA was examined by Northern blotting. For the Western blot analysis, an extract from the U3cs2/mpp10cs strain was used for mock immunoprecipitation (beads alone). For the Northern blot analysis, an extract from the U3wt/MPP10 strain was used for mock immunoprecipitation (beads alone).
Combining the mpp10cs1 and U3cs mutations exacerbates growth and pre-rRNA processing defects.
Previous work on the U3cs mutations and mutations similar to mpp10cs1 indicated that both conferred slow growth at low temperatures (13, 17). Interestingly, quantitative analysis of the growth defects exhibited by these strains revealed that the U3cs allele affects growth more at low temperatures than does the mpp10cs1 allele (Fig. 4B). Strains expressing both the U3cs and mpp10cs1 alleles demonstrated a synergistic effect; growth was worse than for either single mutation alone (Fig. 4B). Because the enhanced growth defect could be explained by either of two alternative hypotheses, i.e., that either SSU processome integrity is destabilized in the double mutant or SSU processome function is further impaired in the double mutant, we examined both possibilities.
To verify that the increased severity of growth defect in the double mutant was not merely due to instability of the SSU processome, we examined the interactions between U3 proteins and between the Mpp10 protein and the U3 snoRNA (Fig. 4C). While it appears that the amount of coimmunoprecipitation between the SSU processome components (Mpp10p, Nop1p, Imp4p, and the U3 snoRNA) decreases in the double-mutant strain (Fig. 4C, lanes 4 and 9) compared to the singly mutated strains (lanes 2 and 7 or 3 and 8) or the unmutated strain (lanes 1 and 6), it should be taken into consideration that the double mutants are severely defective in pre-rRNA processing (see below) and therefore ribosome biogenesis. These defects would eventually result in reduced ribosome levels and reduced protein levels. For example, the amount of mpp10pcs1 that is immunoprecipitated by anti-Mpp10 antibodies (lane 4) is reduced in the double mutant because overall protein levels are lower. Because coimmunoprecipitation is dependent on levels of Mpp10p, reduced levels of Mpp10p result in the appearance of equivalently reduced levels of coimmunoprecipitating SSU processome components. As a result, the coimmunoprecipitation experiments indicate that the interactions between Mpp10p or mpp10pcs1 and Imp4p, Nop1p, U3wt, and U3cs remained intact under our conditions.
Northern blot analysis of pre-rRNA processing was used to examine the effects of the U3 snoRNA and Mpp10p cold-sensitive mutants on SSU processome function (Fig. 5). Single and double mutants (U3cs1/MPP10, U3cs2/MPP10, U3cs1/mpp10cs1 and U3cs2/mpp10cs1) were grown at the semipermissive temperature of 21°C for 42 h under conditions where genomic U3 is either expressed or repressed. Consistent with the defects in growth of the U3cs and mpp10cs1 strains (Fig. 4B), pre-rRNA processing was more severely impaired in U3cs mutants than in the mpp10cs1 mutant (Fig. 5; refer to Fig. 1A to C for processing intermediates). The U3cs mutants showed a stronger accumulation of the 35S and 23S pre-rRNA precursors but a decrease in accumulation of the 33S, 27SA2, 22S, and 20S pre-rRNA precursors compared to the mpp10cs1 mutant (Fig. 5, lanes 13, 14, 21, 22, 29, and 30 compared to lanes 11, 12, 19, 20, 27, and 28). These results indicate that the cold-sensitive mutations in the U3 snoRNA result in slight defects in cleavage at A0 and reduced cleavage of the pre-rRNA at sites A1 and A2 whereas the Mpp10p mutation results in reduced cleavage only at sites A1 and A2. In either case, pre-rRNA processing is only partially affected by each of the single mutations (Fig. 5, controls are shown in the first two lanes of each panel).
FIG. 5.
Expression of mpp10cs1 and U3cs together exacerbates pre-rRNA processing defects. The U3cs1/MPP10, U3cs1/mpp10cs1, U3cs2/MPP10, and U3cs2/mpp10cs1 strains were grown at 21°C either in galactose (Genomic U3 Expressed) or in glucose to repress the expression of the genomic U3 gene (Genomic U3 not Expressed). Total RNA was collected and analyzed by Northern blotting. The same membrane was sequentially hybridized with oligonucleotide probes (shown in Fig. 1A) designed to detect both mature and precursor rRNAs.
Like the enhanced growth defects (Fig. 4B), pre-rRNA processing was also more affected in the double mutants (U3cs/mpp10cs1). While mutations in the U3 snoRNA or Mpp10p alone conferred moderate defects in processing at A0, A1, or A2, when combined they conferred more severe defects at A2. Specifically, the 27SA2 and 20S precursors, both of which require cleavage at A2, were not present at the semipermissive temperature (Fig. 5, lanes 15, 16, 23, 24, 31, and 32).
Species-specific sequences in the carboxy-terminal domain of Mpp10 are functionally required for pre-rRNA processing.
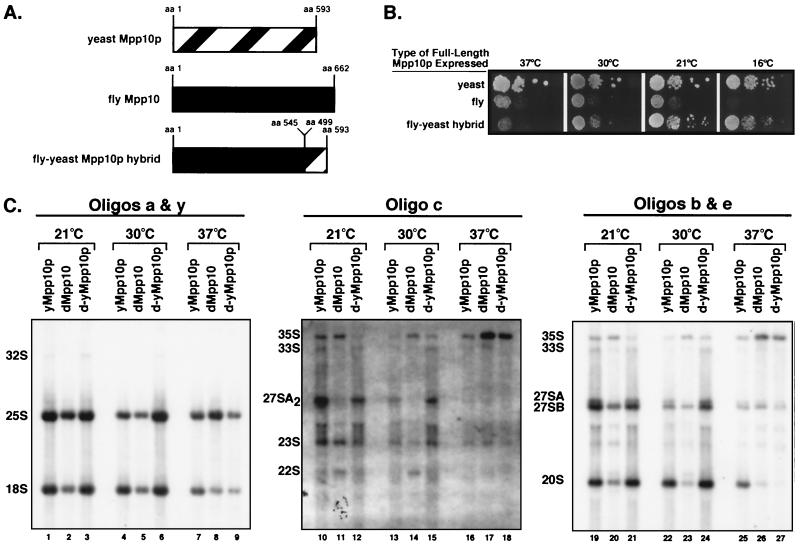
Mpp10p is conserved from yeast to humans (9). As a result of the Drosophila genome project, a putative Drosophila homolog of Mpp10p was revealed (dMpp10) (Fig. 6). While this homolog is 25% identical and 42% similar to the yeast protein throughout its length, there are regions of stronger conservation (Fig. 6). For example, the region of yMpp10p between aa 320 and 495 appears to have the strongest similarity to dMpp10. Interestingly, the similarity between the two proteins begins to diverge near the site of truncation (aa 498) in mpp10pcs1 (see above). For this reason, we were interested in determining if expression of dMpp10, in a strain where no other Mpp10p is being expressed, would result in cold-sensitive growth and defective pre-rRNA processing, as was observed for strains expressing mpp10pcs1 (see above). Additionally, we created a fly-yeast hybrid Mpp10p to determine if mutant phenotypes could be suppressed by restoration of a yeast-like carboxy-terminal domain (Fig. 7A).
FIG. 6.
Alignment of the yeast (yMpp10p) and Drosophila (dMpp10) Mpp10 proteins. Black shading indicates identity and open boxes indicate similarity between the two sequences.
FIG. 7.
Species-specific sequences in Mpp10p are required for pre-rRNA processing. (A) Schematic representation of the yeast, fly, and fly-yeast hybrid Mpp10 proteins. The yeast sequence (aa 499 to 593) swapped with the fly sequence at the carboxy terminus (aa 545) is indicated. (B) The carboxy terminus of yeast Mpp10p restores growth at low temperatures when swapped onto the fly Mpp10. Serial dilutions of yeast expressing either yeast Mpp10p, fly Mpp10, or the fly-yeast Mpp10p hybrid were grown at 37, 30, 21, and 16°C. (C) The carboxy terminus of yeast Mpp10p restores pre-rRNA processing at low temperatures when swapped onto the fly Mpp10. Strains expressing yeast Mpp10p, fly Mpp10, and the fly-yeast Mpp10p hybrid were grown at 37, 30, and 21°C. Total RNA was analyzed by Northern blotting. Mature and precursor rRNAs were detected by sequential hybridizations of the same membrane with specific oligonucleotide probes (shown in Fig. 1A).
Yeast expressing fly Mpp10 as their sole source of Mpp10 grew slowly at 37, 30, and 21°C but did not grow at 16°C (Fig. 7B), indicating that expression of fly Mpp10 results in both cold and heat sensitivity. Most striking were the results with the strains expressing the fly-yeast Mpp10p hybrid. While heat-sensitive growth was not alleviated, swapping the fly carboxy terminus with the yeast carboxy terminus restored growth at low temperatures (Fig. 7B). These results indicate that amino acids in the carboxy terminus of yeast Mpp10p are required to abrogate the cold-sensitive defect caused by the fly Mpp10.
In agreement with growth defects observed for yeast expressing dMpp10, Northern blot analysis indicated that pre-rRNA processing is also affected at all temperatures (Fig. 7C). When the last 95 aa of the yMpp10p carboxy terminus was swapped onto dMpp10 (d-yMpp10p), normal pre-rRNA processing was restored at lower temperatures (Fig. 7C). Like the mpp10cs1 strain, which is defective for processing at sites A1 and A2, yeast expressing the dMpp10 protein accumulated the 22S precursor but lost the 27SA2 and 20S precursors when grown at low temperatures (Fig. 7C, lanes 11, 14, 20, and 23; see Fig. 1A and C for processing intermediates). Expression of the Drosophila-yeast hybrid protein restored precursor levels to wild-type levels at 21 and 30°C (compare lanes 12, 15, 21, and 24 to lanes 10, 13, 19 and 22). As expected, dMpp10-related processing defects were quite severe at 37°C and resulted in complete loss of processing at sites A0, A1, and A2 (lanes 17 and 26). However, they could not be rescued by swapping the yeast carboxy terminus (lanes 18 and 27). Therefore the carboxy-terminal 95 aa of yeast Mpp10p is necessary for pre-rRNA processing at lower temperatures. Importantly, this 95-aa sequence can confer the ability to grow and process pre-rRNA in the cold to Mpp10 proteins from other species when they are expressed in yeast.
DISCUSSION
Our work addresses the complex roles of the Imp3, Mpp10, and Imp4 proteins in ribosome biogenesis. The three proteins are functionally dependent on one another and are involved in key steps of SSU processome activity. Imp3p is essential for the interaction of the Mpp10 and Imp4 proteins with the U3 snoRNA, while Mpp10p is essential for the stability of both Imp3p and Imp4p. To more fully understand the mechanism by which Mpp10p, an essential component of the SSU processome, functions within the cell, we also examined the genetic interaction between base-pairing-defective U3 snoRNAs and the carboxy-terminal truncated Mpp10p. These analyses revealed that while mutation of either processome component results in modest defects in cleavage at sites A0, A1, and A2, combination of these mutations results in a strikingly drastic reduction in cleavage at site A2 only. Additionally, it appears that sequences in the carboxy terminus of Mpp10p function in a species-specific manner to achieve maturation of the small subunit rRNA.
The carboxy-terminal truncation of Mpp10p, which confers cold sensitivity, results in defective processing at sites A1 and A2. Drosophila Mpp10, when expressed in yeast, also results in cold-sensitive growth and defects in processing at sites A1 and A2. Interestingly, the growth and processing defects attributed to dMpp10 expression were suppressed by swapping the fly and yeast carboxy termini. Since this portion of Mpp10p contains a coiled-coil domain, it is possible that a specific protein interacts with this region and is involved in processing at these sites. Because depletion of Utp16p or Dhr1p also results in the loss of cleavage at sites A1 and A2 (7; B. Mitchell and S. Baserga, unpublished data), their ability to interact with Mpp10p was examined in the two-hybrid system. Unfortunately, neither protein was found to interact with Mpp10p (data not shown). To date, Mpp10p has been shown to interact only with Imp3p and Imp4p (18).
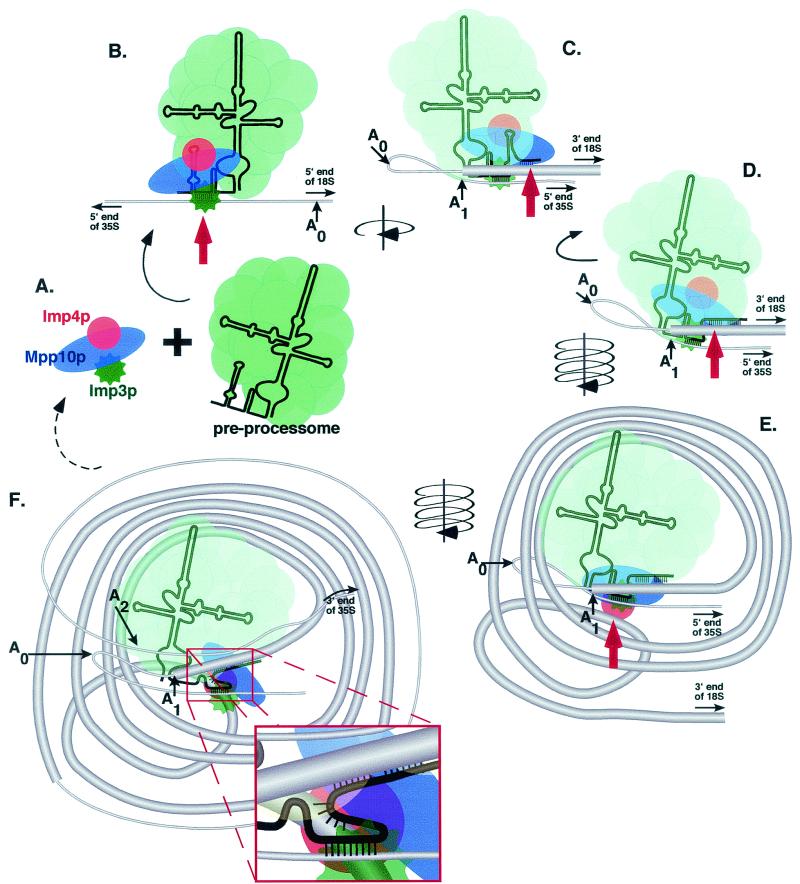
Considerable evidence suggests that the Imp3, Imp4, and Mpp10 proteins exist as an interdependent unit within the cell (Fig. 8A). The three proteins copurify in the absence of other SSU processome components (F. Dragon, S. Wormsley, and S. Baserga, unpublished data). By binding to (18) and stabilizing Imp3p and Imp4p, Mpp10p may function as the key regulatory subunit within this complex. In human cells, Mpp10 was first identified as a protein phosphorylated at specific sites during mitosis (34). As is the case with other types of cell cycle-dependent protein-protein interactions (reviewed in reference 1), it may be that phosphorylation of Mpp10 renders it defective for interaction with Imp3p and Imp4p. This, in turn, may result in their degradation. Since the Imp proteins are essential for SSU processome function (18), we propose that their degradation ultimately leads to the rapid inhibition of pre-rRNA processing that is observed at the onset of mitosis in vertebrate cells (27).
FIG. 8.
A model for the role of Mpp10p, Imp3p, and Imp4p in SSU processome activity. Thin gray lines, thick gray lines, and thin black lines indicate the transcribed spacers, the 18S rRNA, and the U3 snoRNA, respectively. The Imp4, Mpp10, and Imp3 proteins are represented by a pink circle, a blue oval, and an irregular green circle, respectively. The remaining SSU processome proteins are represented as yellow spheres. Black arrows indicate movement of the processome, and red arrows indicate the major processing step depicted in the panel. (A) The Imp3, Mpp10, and Imp4 proteins associate with one another prior to their association with the SSU processome. Imp3p then directs Mpp10p and Imp4p to the U3 snoRNA and the SSU processome to its binding site in the 5′ ETS. (B) The pre-rRNA is folded around the processome, and box A′ of the U3 snoRNA base pairs with the 18S rRNA. (C) This step probably facilitates the binding interaction between the 5′ end of box A and adjacent regions on the 18S rRNA. (D and E) The pre-rRNA is subsequently wound around the SSU processome, bringing the 3′ side of the central pseudoknot close to the RNA binding protein Imp4p. (F) Imp4p binding aligns the 18S rRNA with its binding site at the 3′ end of box A (illustrated in the inset). Additional winding of the pre-rRNA around the processome brings the A2 cleavage site close to the A0 and A1 cleavage sites.
The first 70 nucleotides of the U3 snoRNA are essential for SSU processome function (22) and contain sequences that base pair with the pre-rRNA (4, 5). Recent evidence has demonstrated that this region of the U3 snoRNA is also required for Mpp10p association (36). Our results indicate, however, that it is the putative RNA binding protein Imp3p (18) that is required for Mpp10p and Imp4p to associate with the U3 snoRNA. Therefore, we suggest that it is the Imp3 protein that directly associates with the first 70 nucleotides of the U3 snoRNA and thereby directs the preassembled Imp3p/Mpp10p/Imp4p complex to the U3 preprocessome. Since Imp3p bears the rRNA binding domain of the S4 family of ribosomal proteins (18) and probably associates with the region of the U3 snoRNA containing homology to the 5′ ETS, it is the best candidate to guide or facilitate binding of the U3 snoRNA to the pre-rRNA (Fig. 8B).
The cold-sensitive growth and defects in RNA processing conferred by the mutations in the U3 snoRNA and Mpp10p (13, 17) suggested that they may be involved in an identical step: for example, in pre-rRNA-U3 snoRNA base pairing. If this were indeed the case, combining the mutations should not have an additive effect on either growth or pre-rRNA processing defects. In contrast, our results suggest that the two mutations together do indeed cause increased growth defects at low temperatures. Surprisingly, increased defects in pre-rRNA processing were observed specifically at the A2 cleavage site in the pre-rRNA but not at A1 and A0. Complete loss of the 27SA2 precursor in the presence of both the U3 and Mpp10p mutations demonstrates a strong combinatorial defect in cleavage at site A2 and is consistent with a model in which U3 and Mpp10p are involved at different steps required for this cleavage event.
Imp4p is a member of a superfamily of RNA binding proteins and coimmunoprecipitates all of the pre-rRNA precursors that contain SSU processome cleavage sites (33). Since Mpp10p interacts with Imp4p (18) and mediates its interaction with the U3 snoRNA, it seems probable that Imp4p uses its RNA binding domain to contact the pre-rRNA directly. In light of these results, it is tempting to speculate that the 3′ portion of box A does in fact base pair with nucleotides 1139 to 1143 of the 18S rRNA, as previously proposed (13), and that Imp4p is responsible for bringing the two RNA molecules into contact (Fig. 8E and F). Previous reports of the inability to suppress the 3′ box A mutations by the creation of compensatory mutations in the rRNA (24) can thus be explained: mutation of the rRNA disrupts the Imp4p binding site. Therefore, we propose that in the double-mutant strains, the nature of the truncated Mpp10p alters the ability of the Imp4 protein to interact with the pre-rRNA and mutations in the U3 snoRNA prevent RNA-RNA base pairing.
While the results discussed above suggest a model in which cleavage at A2 is tightly linked to the formation of the central pseudoknot of the 18S rRNA, they do not address the process by which A2 is brought into the active site of the SSU processome. Recent evidence has revealed that components of the SSU processome are required for formation of the “terminal knobs” that were first described by Oscar Miller over 30 years ago (20). It may be that the pre-rRNA is wound around the SSU processome in a fashion similar to the way the DNA is wound around the histones (Fig. 8E and F). Such a process may explain how the 3′ portion of the central pseudoknot is first brought into position for Imp4p binding (Fig. 8E) and how subsequent cleavage at A2 is dependent on formation of the pseudoknot. It is possible that following pseudoknot formation, the pre-rRNA continues to wrap around the processome until the A2 cleavage site is precisely positioned at the active site (Fig. 8F). In this model, positioning of the cleavage sites is partially dependent on pseudoknot formation. Thus, it appears that the cleavages may act, in part, as a checkpoint, signaling through the release of the rRNA that the chaperone role of the SSU processome has been accomplished and that the pseudoknot has been appropriately formed.
Acknowledgments
We thank J. Aris for providing the Nop1p monoclonal antibody B15, S. Gibson for creating the dMpp10 complementation strain, and C. Potter for assisting in the identification of Drosophila MPP10 and critical reading of the manuscript.
K.A.W. was supported in part by NIH National Research Service Award GM07499-2. J.E.G.G. was supported in part by NIH National Research Service Awards GM07223-25 and F0001345. This work was supported by NIH GM-52581 to S.J.B. S.J.B. is a member of the Yale Cancer Center.
REFERENCES
- 1.Adams, P. D. 2001. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta 1471:M123-M133. [DOI] [PubMed] [Google Scholar]
- 2.Aris, J. P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 4.Beltrame, M., and D. Tollervey. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S synthesis. EMBO J. 14:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrame, M., and D. Tollervey. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 7.Colley, A., J. D. Beggs, D. Tollervey, and D. L. Lafontaine. 2000. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol. Cell. Biol. 20:7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragon, F., P. A. Compagnone-Post, J. E. G. Gallagher, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S rRNA biogenesis. Nature 417:967-970. [Advance online publication 9 June 2002 (doi:10.1038/nature 00769).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar, D. A., S. Wormsley, T. M. Agentis, and S. J. Baserga. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier, T., T. Bergès, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 12.Guthrie, C., H. Nashimoto, and M. Nomura. 1969. Structure and function of Escherichia coli ribosomes. VII. Cold-sensitive mutations defective in ribosome assembly. Proc. Natl. Acad. Sci. USA 63:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, J. M. X. 1996. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 259:645-654. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, J. M. X., and M. Ares, Jr. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen, R., D. Tollervey, and E. C. Hurt. 1993. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 12:2549-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S. J., and S. J. Baserga. 1997. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl. Acad. Sci. USA 94:13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Méreau, A., R. Fournier, A. Grégoire, A. Mougin, P. Fabrizio, R. Luhrmann, and C. Branlant. 1997. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP; protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 273:552-571. [DOI] [PubMed] [Google Scholar]
- 20.Miller, O. L., Jr., and B. R. Beatty. 1969. Visualization of nucleolar genes. Science 164:955-957. [DOI] [PubMed] [Google Scholar]
- 21.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 22.Samarsky, D. A., and M. J. Fournier. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 18:3431-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma, K., and D. Tollervey. 1999. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 19:6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirri, V., P. Roussel, and D. Hernandez-Verdun. 2000. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 148:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss, E. J., and C. Guthrie. 1991. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 5:629-641. [DOI] [PubMed] [Google Scholar]
- 29.Tollervey, D., H. Lehtonen, M. Carmo-Fonseca, and E. C. Hurt. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10:573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 31.Venema, J., H. R. Vos, A. W. Faber, W. J. Van Venrooij, and H. A. Raué. 2000. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA 6:1660-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins, N. J., V. Ségault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 33.Wehner, K. A., and S. J. Baserga. 2002. The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]
- 34.Westendorf, J. M., K. N. Konstantinov, S. Wormsley, M. D. Shu, N. Matsumoto-Taniura, F. Pirollet, F. G. Klier, L. Gerace, and S. J. Baserga. 1998. M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell 9:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiederkehr, T., R. F. Prétôt, and L. Minvielle-Sebastia. 1998. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4:1357-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wormsley, S., D. A. Samarsky, M. J. Fournier, and S. J. Baserga. 2001. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA 7:904-919. [DOI] [PMC free article] [PubMed] [Google Scholar]