Abstract
The peptidyl-prolyl isomerase (PPIase) cyclophilin A (Cpr1p) is conserved from eubacteria to mammals, yet its biological function has resisted elucidation. Unable to identify a phenotype that is suggestive of Cpr1p's function in a cpr1Δ Saccharomyces cerevisiae strain, we screened for CPR1-dependent strains. In all cases, dependence was conferred by mutations in ZPR1, a gene encoding an essential zinc finger protein. CPR1 dependence was suppressed by overexpression of EF1α (a translation factor that binds Zpr1p), Cpr6p (another cyclophilin), or Fpr1p (a structurally unrelated PPIase). Suppression by a panel of cyclophilin A mutants correlated with PPIase activity, confirming the relevance of this activity in CPR1-dependent strains. In CPR1+ cells, wild-type Zpr1p was distributed equally between the nucleus and cytoplasm. In contrast, proteins encoded by CPR1-dependent alleles of ZPR1 accumulated in the nucleus, as did wild-type Zpr1p in cpr1Δ cells. Transport kinetic studies indicated that nuclear export of Zpr1p was defective in cpr1Δ cells, and rescue of this defect correlated with PPIase activity. Our results demonstrate a functional interaction between Cpr1p, Zpr1p, and EF1α, a role for Cpr1p in Zpr1p nuclear export, and a biological function for Cpr1p PPIase activity.
The cyclophilins comprise a large family of proteins found in most organisms. They were discovered by virtue of their high-affinity interaction with cyclosporine (CsA), an immunosuppressive drug used widely to inhibit allograft rejection (20). The drug's immunosuppressive effect results from the formation of a CsA-cyclophilin complex which inhibits calcineurin, a phosphatase responsible for the activation of multiple cytokine genes in T cells (28). Cyclophilin A (CyPA; Cpr1p) binds to the human immunodeficiency virus type 1 (HIV-1) Gag protein (23) and is required for wild-type HIV-1 replication kinetics (7).
Cyclophilins are defined by a common eight-stranded beta-barrel structure. A solvent-exposed hydrophobic pocket is the binding site for proline-containing substrates, CsA, and HIV-1 Gag (6, 21, 35). CyPA consists of only this core domain, whereas other family members have additional functional domains. Paralogs are found in virtually every cellular compartment, with mammalian genomes encoding 15 cyclophilins (7).
High-level evolutionary conservation, together with a broad cellular and tissue distribution, suggests that cyclophilins perform an essential function in the cell. However, the biological function of the core cyclophilin domain is unknown. In vitro, CyPA accelerates the cis-to-trans isomerization of oligopeptide substrates containing proline, a rate-limiting step in the refolding of denatured proteins (27). In addition, transcription of some cyclophilin genes is increased in response to heat shock (12, 32), and some cyclophilins associate with known chaperones (1). These findings suggest that cyclophilins regulate protein folding in vivo. CyPA is required for normal growth of Cryptococcus neoformans (33), but deletion in a single Saccharomyces cerevisiae strain of all eight cyclophilins and all four members of another family of peptidyl-prolyl isomerases (PPIases), the FK506-binding proteins (FKBPs), yields a viable cell (12). Similarly, CyPA is not required for growth of human T cells (7) or murine embryonic stem cells (10).
We have endeavored to determine the biological function of CyPA. We were unable to find a phenotype associated with deletion of CPR1 in yeast cells that was suggestive of Cpr1p's natural function. We hypothesized that CPR1 carries out a critical function that is masked by the function of other genes, that mutation of these other genes would confer CPR1 dependence on the cell (synthetic lethality), and that identification of these genes would help elucidate the function of CPR1. Our screen identified ZPR1 as a synthetically lethal partner with CPR1. Zpr1p is an essential zinc-finger-containing protein that translocates to the nucleus in response to growth stimuli (15-17). Our results demonstrate that the PPIase activity of CyPA promotes proper subcellular localization of Zpr1p.
MATERIALS AND METHODS
Strains, plasmids, and media.
The yeast strains used in this study are listed in Table 1. Standard techniques were used in the construction, transformation, gene targeting, and growth of the yeast strains (25, 29). To construct strains HC22-1B, HC24-8A, HC70-8B, and HC72-7C, the carboxy terminus of Zpr1p, zpr1-1p, or zpr1-3p was tagged with green fluorescent protein (GFP) by using PCR-based insertion at the ZPR1 genomic locus as described previously (22).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade2 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| W303-1B | MATα ade2 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| W1536-5B | MATaade2 ade3 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| W1536-8B | MATα ade2 ade3 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| HC1-2B | MATaade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::TRP1 | This study |
| HT1 | MATα ade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::LEU2 | This study |
| GT1 | MATaade2 can1 his3 leu2 trp1 ura3 cpr1Δ | This study |
| GT2 | MATα ade2 can1 his3 leu2 trp1 ura3 cpr1Δ | This study |
| CCFY100 | MATaade2 ade3 can1 his3 leu2 trp1 ura3 HMRΔE::TRP1 rDNA::ADE2-CAN1 VRTEL::URA3 | 28 |
| GC1-1A | MATα ade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ HMRΔE::TRP1 rDNA::ADE2-CAN1 VRTEL::URA3 | This study |
| HC12-1A | MATaade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::TRP1 zpr1-1 (pCH1122-CPR1 [CPR1/ADE3/URA3/CEN]) | This study |
| HC17-5C | MATaade2 can1 his3 leu2 trp1 ura3 cpr1Δ::TRP1 zpr1-1 (pCH1122-CPR1 [CPR1/ADE3/URA3/CEN]) | This study |
| HM2 | MATα ade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::LEU2 zpr1-2 (pCH1122-CPR1 [CPR1/ADE3/URA3/CEN]) | This study |
| HM3 | MATα ade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::LEU2 zpr1-3 (pCH1122-CPR1 [CPR1/ADE3/URA3/CEN]) | This study |
| HT23 | MATaade2 ade3 can1 his3 leu2 trp1 ura3 cpr1Δ::TRP1 zpr1-1 (pBM272-CPR1 [GALp-CPR1/URA3/CEN]) | This study |
| HC21-11D | MATaade2 can1 his3 leu2 trp1 ura3 zpr1-1 | This study |
| HC22-1B | MATaade2 can1 his3 leu2 trp1 ura3 ZPR1-GFP::kanMX | This study |
| HC24-8A | MATaade2 can1 his3 leu2 trp1 ura3 cpr1Δ::LEU2 ZPR1-GFP::kanMX | This study |
| HC70-8B | MATα ade2 can1 his3 leu2 trp1 ura3 zpr1-1-GFP::kanMX | This study |
| HC72-7C | MATα ade2 can1 his3 leu2 trp1 ura3 zpr1-3-GFP::kanMX | This study |
The plasmids used in this study are listed in Table 2. The yeast CPR1 promoter (CPR1p) and terminator (CPR1t) were cloned as XbaI-NcoI and BamHI/XhoI fragments, respectively, into the vectors pRS414 and pRS415, resulting in plasmids pRS414-PT and pRS415-PT, respectively. Human CyPA (hCyPA) and the six hCyPA point mutants (6, 35) were cloned as NcoI/BamHI fragments into pRS414-PT and pRS415-PT between CPR1p and CPR1t.
TABLE 2.
Plasmids used in this study
| Plasmid(s) | Insert or marker | Copy no. | Source or reference |
|---|---|---|---|
| pRS plasmids | Stratagene | ||
| pCH1122 | ADE3/URA3 | CEN | 22 |
| pCH1122-CPR1 | CPR1/ADE3/URA3 | CEN | This study |
| pBM272 | GALp/URA3 | CEN | 20 |
| pBM272-CPR1 | GALp-CPR1/URA3 | CEN | This study |
| pRS414-PT | CPR1p-CPR1t/TRP1 | CEN | This study |
| pRS414-CPR1 | CPR1/TRP1 | CEN | This study |
| pRS414-hCyPA | hCyPA/TRP1 | CEN | This study |
| pRS414-H54Q | hCyPA-H54Q/TRP1 | CEN | This study |
| pRS414-H126Q | hCyPA-H126Q/TRP1 | CEN | This study |
| pRS414-ZPR1 | ZPR1/TRP1 | CEN | This study |
| pRS415-PT | CPR1p-CPR1t/LEU2 | CEN | This study |
| pRS415-CPR1 | CPR1/LEU2 | CEN | This study |
| pRS415-hCyPA | hCyPA/LEU2 | CEN | This study |
| pRS415-H54Q | hCyPA-H54Q/LEU2 | CEN | This study |
| pRS415-F55A | hCyPA-R55A/LEU2 | CEN | This study |
| pRS415-F60A | hCyPA-F60A/LEU2 | CEN | This study |
| pRS415-F113A | hCyPA-F113A/LEU2 | CEN | This study |
| pRS415-W121A | hCyPA-W121A/LEU2 | CEN | This study |
| pRS415-H126Q | hCyPA-H126Q/LEU2 | CEN | This study |
| pRS425-TEF1 | TEF1/LEU2 | 2μm | This study |
| YEplac181-CPR6 | CPR6/LEU2 | 2μm | This study |
| YEplac181-FPR1 | FPR1/LEU2 | 2μm | This study |
| pFPR4 | FPR4/LEU2 | 2μm | J. Heitman |
| pESS1 | ESS1/HIS3 | 2μm | S. Hanes |
Where indicated, synthetic complete medium (SC) plates were supplemented with 1 mg of 5-fluoroorotic acid (5-FOA) per liter, SC plates lacking arginine were supplemented with 60 mg of canavanine sulfate per liter, and yeast extract-peptone-dextrose plates were supplemented with 200 mM LiCl and 100 μg of CsA per ml. Strains were grown on plates containing up to the following maximum doses: 1 M sorbitol, 2 mM hydrogen peroxide, 0.2 mM t-butyl hydroperoxide, 50 μg of bleomycin per ml, 10 μg of actinomycin D per ml, 5 ng of rapamycin per ml, 100 ng of cycloheximide per ml, 16 mg of paromomycin per ml, 100 μg of hygromycin per ml, 10 μg of puromycin per ml, and 50 μg of brefeldin A per ml.
Survival experiments at 48°C and silencing experiments.
Log- and stationary-phase survival experiments at 48°C were done as described previously (14, 32) with strains W1536-5B and HC1-2B. Silencing experiments were carried out as described previously (26) with CCFY100 and GC1-1A cells.
Screen for synthetically lethal mutations.
A red/white colony-sectoring assay was performed twice to identify mutations that were synthetically lethal with the cpr1Δ strain (4). Strains HC1-2B and HT1 carrying pCH1122-CPR1 were mutagenized with ethyl methanesulfonate (EMS), and solid red colonies were checked for lethality on 5-FOA to identify strains that retained pCH1122-CPR1. Candidate strains were transformed with either pRS414-CPR1 or pRS415-CPR1 and retested for lethality on 5-FOA. Those that displayed 5-FOA resistance were considered CPR1-dependent strains.
Cloning of ZPR1 and high-copy-number suppressor screen.
Strain HC12-1A was transformed with a yeast genomic library (p366-based LEU2/CEN; ATCC 77162). Library plasmids that complemented the 5-FOA-sensitive, CsA- and LiCl-sensitive, and temperature-sensitive growth of HC12-1A were recovered and sequenced. Genetic linkage confirmed that a mutation in ZPR1 caused the synthetic lethality phenotype in the cpr1Δ strain.
To clone high-copy-number suppressors of slc1, strain HC12-1A was transformed with a high-copy-number yeast genomic library (YEplac181-based LEU2/2μm; provided by Joe Heitman). Library plasmids that complemented the 5-FOA-sensitive and CsA- and Li-sensitive phenotypes were recovered and sequenced.
Growth curves.
For Cpr1p depletion, strains HC1-2B (cpr1Δ ZPR1) and HT23 (cpr1Δzpr1-1), each carrying plasmid pBM272-CPR1, were grown to log phase (optical density at 600 nm [OD600], 0.3) in SC plus galactose and then diluted and shifted to SC plus glucose. For temperature shift, strains W303-1A (CPR1 ZPR1) and HC21-11D (CPR1zpr1-1) were grown to log phase (OD600, 0.3) at 25°C and then diluted and shifted to 37°C.
Fluorescence microscopy, glucose starvation experiments, and nuclear import assay.
Strains HC22-1B (CPR1 ZPR1-GFP), HC24-8A (cpr1Δ ZPR1-GFP), HC70-8B (CPR1zpr1-1-GFP), and HC72-7C (CPR1 zpr1-3-GFP) were grown in SC plus glucose. When plasmids were present, the appropriate dropout medium was used. At mid-log phase (OD600, 0.3), cells were examined by fluorescence microscopy. Only cells with nonfluorescing vacuoles were counted, and in the case of budded cells, only mother cells were counted. Cells were scored as nuclear if the nucleus was brighter than the surrounding cytoplasm and a clear nuclear-cytoplasmic boundary was visible. For each strain, approximately 80 to 200 cells in total were examined.
Starvation experiments were carried out as described previously (15). Strains HC22-1B and HC24-8A were grown in 20 ml of SC containing 2% glucose to mid-log phase (OD600, 0.3). Cells were pelleted, washed three times with water, resuspended in 20 ml of SC lacking glucose, and incubated at 30°C on a shaker for 18 h. After starvation, cells were refed with glucose to a final concentration of 2%. At the times indicated, 1-ml aliquots were pelleted and cells were prepared, examined, and scored as described above.
A nuclear import assay was carried out as described previously (30). Strains HC22-1B and HC24-8A were grown in 20 ml of SC plus glucose to mid-log phase (OD600, 0.3). Cells were pelleted and washed once with 1 ml of double-distilled water and once with 0.1% bovine serum albumin. The washed cell pellets were resuspended in 1 ml of SC lacking glucose but containing 10 mM sodium azide and 10 mM 2-deoxy-d-glucose and incubated for 45 min at 30°C to allow nucleocytoplasmic equilibration of the Zpr1p-GFP fluorescence. Cells were washed once with 1 ml of ice-cold double-distilled water, and transport of Zpr1p-GFP was initiated by resuspending cells in 100 μl of SC plus glucose. Every 3 min, cells were harvested, examined, and scored as described above.
Western blot analysis.
Yeast lysates (OD600, 0.3) were probed with rabbit polyclonal antibodies to either hCyPA (Affinity Bioreagents), yeast Cpr1p (provided by Joe Heitman), GFP (provided by Pamela Silver), or yeast Zpr1p (amino acids 100 to 118; Research Genetics, Inc.).
RESULTS
Characterization of the cpr1Δ strain.
To learn about Cpr1p function, we deleted CPR1 from the yeast genome. Consistent with previous findings (12), our cpr1Δ strain grew on medium containing CsA and lithium chloride, whereas the wild-type strain did not (data not shown). The cpr1Δ strain behaved like the wild type when tested for growth at 17, 30, or 37°C or in the presence of sorbitol, hydrogen peroxide, t-butyl hydroperoxide, bleomycin, actinomycin D, rapamycin, cycloheximide, paromomycin, hygromycin, puromycin, or brefeldin A (data not shown). It displayed no growth disadvantage when cocultured with the wild type (data not shown), and unlike what has been reported previously (14, 32), no difference in the abilities of wild-type and cpr1Δ strains to survive at 48°C in log or stationary phase was observed (data not shown). It was reported that Cpr1p overexpression decreased silencing of a marker in ribosomal DNA (2), but we detected no differences in silencing between CPR1+ and cpr1Δ strains bearing reporters in ribosomal DNA, the HMRΔE locus, and a telomere (data not shown).
Isolation of CPR1 synthetic-lethality mutants.
To help elucidate CyPA's native function, we performed a screen of synthetic lethality of cpr1Δ strains to identify mutants that require CPR1 for viability. Strains HC1-2B and HT1, which are ade2 ade3 strains harboring genomic cpr1Δ mutations, were transformed with the plasmid pCH1122-CPR1 (CPR1/ADE3/URA3). A colony-sectoring assay was used to monitor plasmid loss: the starting strain was red (ade2), and loss of pCH1122-CPR1 generated white (ade2 ade3) sectors. Strains which required the pCH1122-CPR1 plasmid, on the other hand, retained ADE3 and formed solid red colonies.
Two separate screens were performed (Table 3). Approximately 5 × 105 cells were treated with EMS to achieve 20% survival (first screen) or to achieve 7.5% survival (second screen). Solid red colonies were checked for sensitivity to 5-FOA to identify strains that retained pCH1122-CPR1. Those that were 5-FOA sensitive were considered candidate CPR1-dependent strains. Candidate strains were transformed with either pRS414-CPR1 or pRS415-CPR1 and retested for sensitivity to 5-FOA. Those that displayed 5-FOA resistance were considered CPR1-dependent strains. Of the approximately 1.4 × 105 colonies surviving EMS treatment, 1,180 were solid red, 121 were 5-FOA sensitive, and three were CPR1-dependent strains (Table 3 and Fig. 1A).
TABLE 3.
Screens for CPR1-dependent mutants
| Screen no. | Results of screen
|
||||
|---|---|---|---|---|---|
| No. of cells treated with EMS | No. of cells surviving EMS treatment | No. of solid red colonies | No. of S-FOA-sensitive colonies | No. of CPR1-dependent colonies | |
| 1 | 500,000 | 100,000 | 620 | 82 | 1a |
| 2 | 500,000 | 37,500 | 560 | 39 | 2b |
Strain HC12-1A (zpr1-1).
Strains HM2 (zpr1-2) and HM3 (zpr1-3).
FIG. 1.
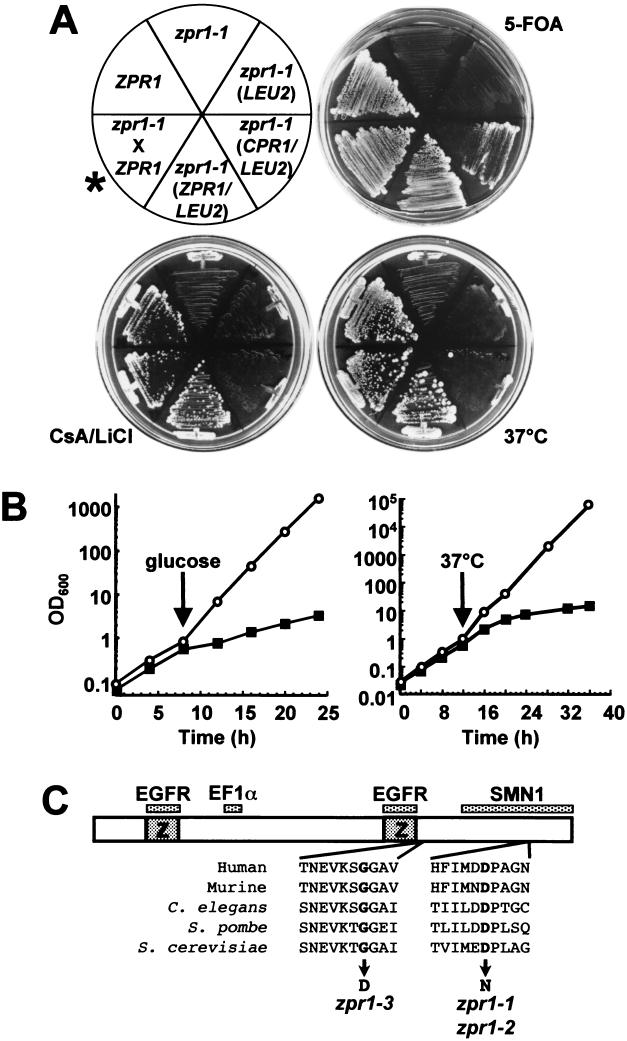
The slc1-1 strain bears a mutant allele of ZPR1 (zpr1-1) that confers CPR1 dependence and temperature sensitivity. (A) ZPR1 cpr1Δ (HC1-2B) and zpr1-1 cpr1Δ (HC12-1A) strains containing the pCH1122-CPR1 plasmid (CPR1/ADE3/URA3) and the indicated plasmid-borne genes (given in parentheses) were grown on 5-FOA, on CsA and LiCl, or at 37°C. The zpr1-1 strain (HC12-1A) was crossed with a ZPR1 cpr1Δ strain (HT1), and the resulting diploid was 5-FOAR CsA/LiClR and was able to grow at 37°C, indicating that the zpr1-1 mutation is recessive (asterisk). (B) (Left) ZPR1 cpr1Δ (HC1-2B; open circles) and zpr1-1 cpr1Δ (HT23; closed squares) strains containing the plasmid pBM272-CPR1 (GALp-CPR1) were grown to log phase in SC plus galactose and shifted to SC plus glucose at the indicated time. (Right) Wild-type (W303-1A; open circles) and zpr1-1 (HC21-11D; closed squares) cells were grown to log phase at 25°C in yeast extract-peptone-dextrose and shifted to 37°C at the indicated time. Culture densities were monitored by measuring the OD600 at the times indicated. (C) Schematic diagram of Zpr1p showing the locations of the zinc fingers (Z) and the interaction sites of the epidermal growth factor receptor (EGFR), EF1α, and SMN1, the mutations encoded by zpr1-1, zpr1-2, and zpr1-3;the sequences of the same regions from Zpr1p homologues are also indicated. C. elegans, Caenorhabditis elegans.
The mutations in these three strains were shown to be recessive by backcross to HC1-2B or HT1 and the subsequent demonstration of 5-FOA resistance of the resulting diploids (Fig. 1A). Each backcrossed diploid was sporulated and microdissected. The ratio of sectoring to nonsectoring red spores and of 5-FOAS to 5-FOAR spores was 2:2, indicating that in each case, synthetic lethality was caused by a mutation in a single gene (data not shown). The three slc (synthetically lethal with cyclophilin) mutant strains fell into a single complementation group and were designated slc1-1, slc1-2, and slc1-3. As expected, the slc1 mutant strains failed to grow when CPR1 expression was turned off (Fig. 1B). The slc1-1 and slc1-2 strains, but not the slc1-3 strain, were found to be severely growth impaired at 37°C (Fig. 1 and data not shown).
The slc1-1 mutation was identified by transformation with a yeast genomic plasmid library and by selection for growth on 5-FOA, on CsA and LiCl (i.e., pCH1122-CPR1 is no longer required), and at 37°C. Library plasmids were rescued from transformants that met these criteria, sequenced, and found to contain the ZPR1 gene (Fig. 1A). Subcloning and linkage analysis confirmed that a mutation in ZPR1 caused the synthetic-lethality phenotype with the cpr1Δ mutation. The slc1-1 and slc1-2 alleles (hereafter referred to as zpr1-1 and zpr1-2), which were isolated independently in separate mutagenesis experiments, had identical guanine-to-adenine mutations at position 1330 encoding an aspartic acid-to-asparagine change. The slc1-3 allele (hereafter referred to as zpr1-3) had a guanine-to-adenine mutation at position 1010 encoding a glycine-to-aspartic acid change (Fig. 2C). Finally, we confirmed the previous report that ZPR1 is an essential gene in S. cerevisiae (17).
FIG. 2.
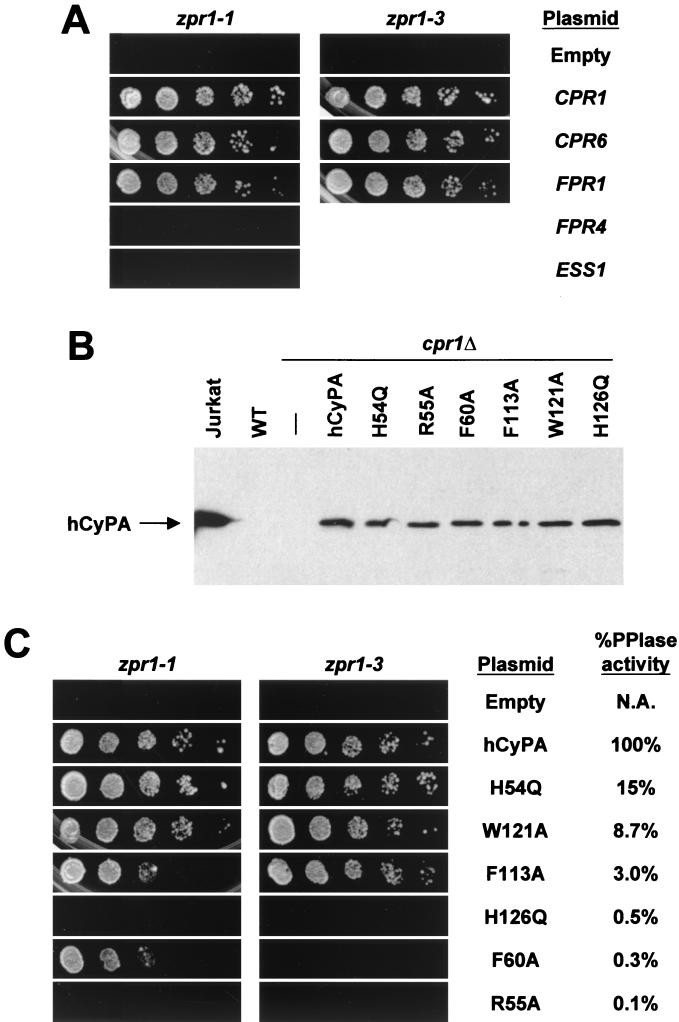
PPIase activity is required in zpr1 mutant strains. (A) zpr1-1 cpr1Δ (HC12-1A) and zpr1-3 cpr1Δ (HM3) cells, each containing the plasmid pCH1122-CPR1 (CPR1/URA3) and empty vector, pRS415-CPR1, or 2μm vectors containing the indicated PPIase, were plated onto 5-FOA in fivefold serial dilutions. (B) Lysates of wild-type (W1536-5B; WT), cpr1Δ (HC1-2B), and cpr1Δ strains expressing wild-type and mutant hCyPA proteins from the plasmid pRS415 were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with anti-hCyPA antibody. Jurkat T-cell lysate was loaded as a control for hCyPA expression. (C) zpr1-1 cpr1Δ (HC12-1A) and zpr1-3 cpr1Δ (HM3) cells, each containing the plasmid pCH1122-CPR1 (CPR1/URA3) and empty vector (pRS415) or pRS415 containing the indicated PPIase, were plated onto 5-FOA in fivefold serial dilutions. Percentages of in vitro PPIase activity relative to that of wild-type hCyPA (35) are indicated. N.A., not applicable.
High-copy-number plasmids bearing CPR6 or FPR1 are substitutes for CPR1 in zpr1 mutant strains.
To find high-copy-number suppressors of CPR1 dependence, the zpr1-1 strain HC12-1A was transformed with a 2μm yeast genomic library. Two clones rescued 5-FOA sensitivity and CsA and Li sensitivity but not temperature sensitivity, indicating that they were suppressors of the zpr1-1 strain's CPR1 dependence but likely did not carry the wild-type CPR1 or ZPR1 genes. Sequencing of the plasmids isolated from these clones revealed that one contained the CPR6 gene while the other contained FPR1. These plasmids carrying CPR6 and FPR1 were also introduced into zpr1-3, and both conferred 5-FOAR growth (Fig. 2A). Cpr6p, another member of the cyclophilin family in S. cerevisiae, is localized to the cytoplasm (12). Fpr1p is a member of theFKBPs, another family of proteins with in vitro PPIase activity, and is also localized to the cytoplasm (12). Unlike Cpr6p and Fpr1p, however, overexpression of Fpr4p (a nuclear FKBP) or Ess1p (a nuclear PPIase of the parvulin family) did not suppress the CPR1 dependence of the zpr1-1 strain (Fig. 2A).
zpr1 mutant strains require cyclophilin's PPIase activity.
We examined a panel of CyPA mutants, all of which have in vitro PPIase activity reduced to different degrees. The hCyPA coding sequence and the coding sequences of six previously characterized hCyPA mutants (6, 35) were each cloned into plasmid pRS415 under control of the yeast CPR1 promoter for expression in yeast (Fig. 2B). These plasmids were introduced into zpr1-1 and zpr1-3 strains, and the transformants were assayed on 5-FOA. In both strains, hCyPA conferred a degree of 5-FOA resistance similar to that seen with yeast Cpr1p. This was expected, because hCyPA and yeast Cpr1p are 65% identical at the amino acid level. The point mutants with greater in vitro PPIase activity conferred a higher degree of 5-FOA resistance on the zpr1-1 and zpr1-3 strains than did mutants with lower PPIase activity (Fig. 2C). This finding, together with the isolation of two PPIases, Cpr6p and Fpr1p, from a library screen to identify suppressors of CPR1 dependence, suggests that zpr1 mutant strains require PPIase activity.
Overexpression of EF1α suppresses zpr1-associated phenotypes.
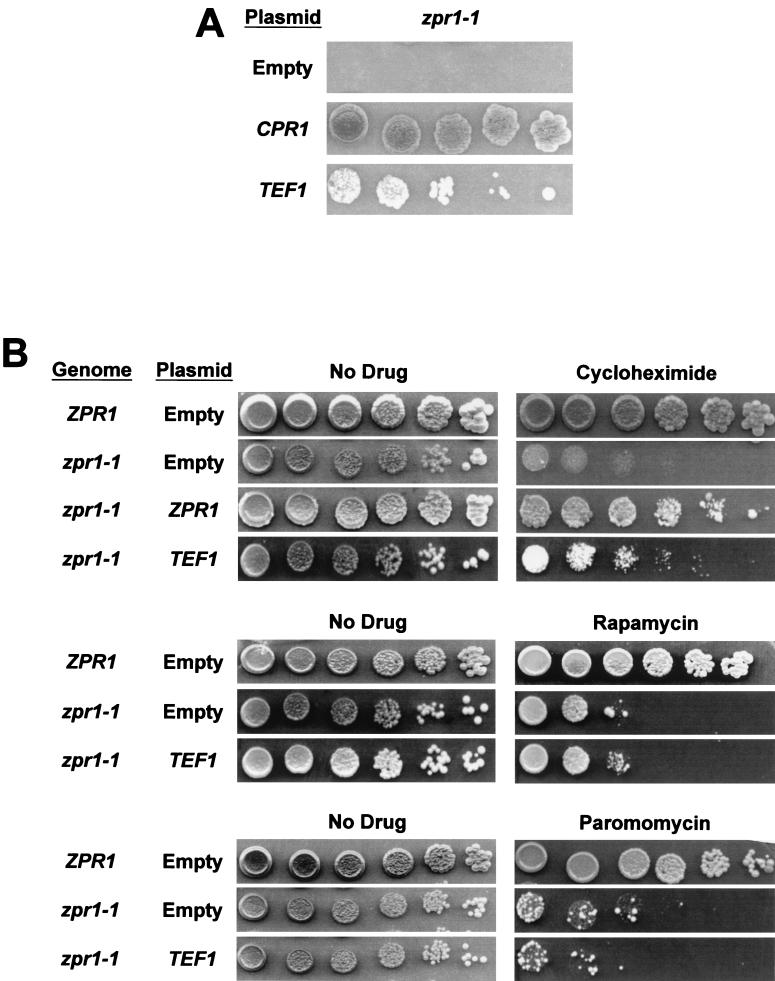
It has been shown that Zpr1p and the translation elongation factor EF1α, encoded in S. cerevisiae by the identical genes TEF1 and TEF2, physically interact with one another and that cells expressing a mutant form of Zpr1p that does not interact with EF1α are growth impaired (17). In yeast two-hybrid experiments, we confirmed this interaction but were unable to detect an interaction between EF1α and Cpr1p (data not shown). We observed, however, that overexpression of EF1α suppressed the CPR1 dependence of the zpr1-1 strain (Fig. 3A). Overexpression of EF1α did not rescue temperature sensitivity of the zpr1-1 strain or the lethality of a zpr1Δ strain (data not shown).
FIG. 3.
Overexpression of TEF1 suppresses CPR1 dependence and cycloheximide sensitivity of the zpr1-1 strain. (A) Strain HC17-5C zpr1-1 cpr1Δ cells with pCH1122-CPR1 (CPR1/ADE3/URA3) were transformed with empty vector, pRS415-CPR1 (CEN), or pRS425-TEF1 (2μm) and then serially diluted 1:5, plated onto 5-FOA plates, and grown for 5 days at 30°C. (B) ZPR1 (W303-1A) and zpr1-1 (HC21-11D) cells containing empty vector, pRS414-ZPR1 (CEN), or pRS425-TEF1 (2μm) were plated onto SC containing 40 ng of cycloheximide per ml, 1 ng of rapamycin per ml, or 4 mg of paromomycin per ml in fivefold serial dilutions.
Because EF1α is an essential translation factor, we looked at the sensitivity of the zpr1-1 mutant strain to three translation inhibitors, each of which functions via a distinct mechanism. The zpr1-1 strain was hypersensitive to cycloheximide, rapamycin, and paromomycin (Fig. 3B). As with CPR1 dependence, the cycloheximide hypersensitivity of the zpr1-1 strain was suppressed by overexpression of TEF1. This effect was specific, as overexpression of TEF1 did not suppress the sensitivity of the zpr1-1 strain to rapamycin or paromomycin. Although TEF1 overexpression suppressed CPR1 dependence and cycloheximide sensitivity of the zpr1-1 strain, it failed to suppress temperature sensitivity of the zpr1-1 strain or to rescue the viability of a zpr1Δ strain (data not shown). These results indicate that Cpr1p, Zpr1p, and EF1α are functionally related molecules and that Cpr1p and EF1α contribute to upstream events that promote the essential function of Zpr1p.
Absence of Cpr1p alters nuclear localization of Zpr1p.
The requirement for PPIase activity in zpr1 mutants suggests that Cpr1p regulates the folding of Zpr1p and that the two proteins directly interact. However, we were unable to detect an interaction in yeast two-hybrid, glutathione-S-transferase pull-down, and coimmunoprecipitation experiments with Zpr1p-protein A fusions (data not shown). Another possibility is that Cpr1p contributes to the stability of Zpr1p, but we observed no difference in steady-state Zpr1p levels in cpr1Δ cells compared to levels in wild-type cells (data not shown).
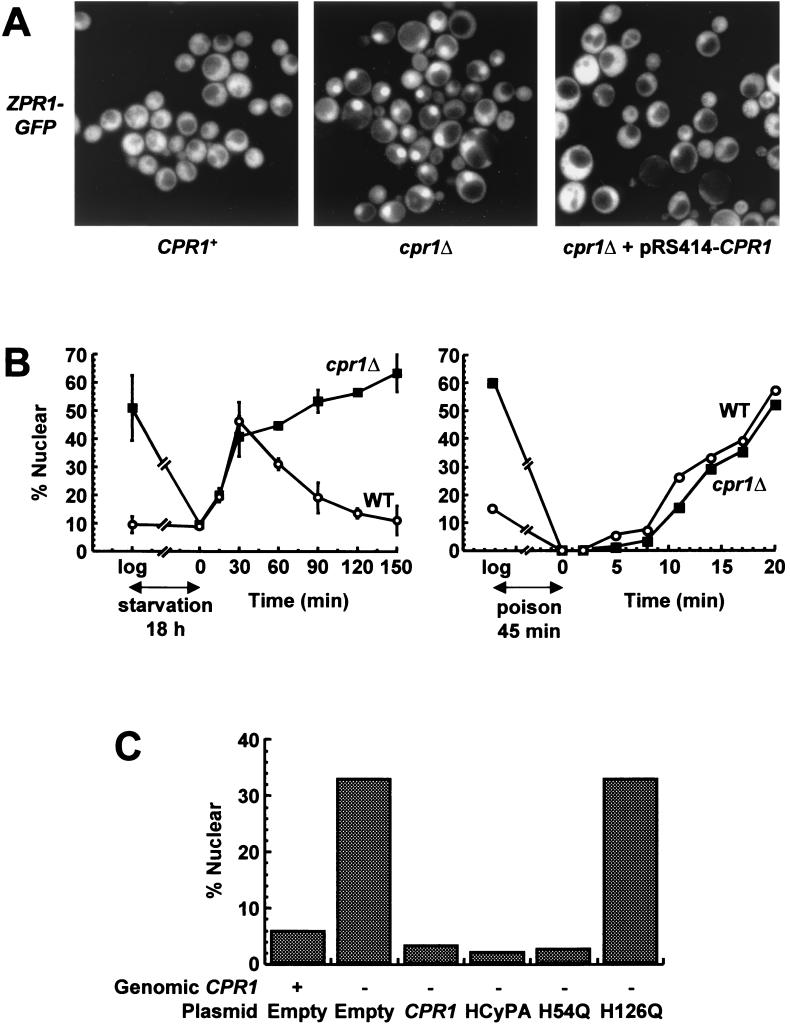
Previous studies have shown that stimulation of mammalian cells with serum triggers Zpr1 translocation to the nucleus (15). It has also been observed that Zpr1p localizes to the nuclei of yeast cells growing in log phase, redistributes to the cytoplasm when cells are deprived of glucose, and translocates to the nucleus when glucose is restored (15). To determine if the absence of Cpr1p had an effect on the localization of Zpr1p, we constructed CPR1+ and cpr1Δ strains of S. cerevisiae carrying a ZPR1-GFP fusion gene integrated at the genomic locus. These cells were grown to log phase and examined by fluorescence microscopy. In wild-type cells, Zpr1p-GFP did not accumulate in the nucleus and was instead evenly distributed between the nucleus and the cytoplasm, with less than 1% of cells displaying predominantly nuclear fluorescence (Fig. 4A). In contrast, nuclear localization of Zpr1p-GFP was prominent in cpr1Δ cells. When Cpr1p was expressed from a plasmid in the cpr1Δ ZPR1-GFP strain, Zpr1p-GFP no longer accumulated in the nucleus and was evenly distributed between the nucleus and the cytoplasm. Western blot analysis using an anti-GFP antibody demonstrated that the diffuse fluorescence observed in wild-type cells was not due to the degradation of the Zpr1p-GFP fusion protein with concomitant diffusion of the GFP moiety (data not shown). Thus, the nuclear accumulation of Zpr1p-GFP can be attributed to the absence of Cpr1p.
FIG. 4.
The absence of Cpr1p and PPIase activity results in nuclear accumulation of Zpr1p. (A) CPR1 ZPR1-GFP (HC22-1B) cells containing pRS414 and cpr1Δ ZPR1-GFP (HC24-8A) cells containing either pRS414 or pRS414-CPR1 were grown to log phase in SC lacking tryptophan and examined by fluorescence microscopy. (B) (Left) Wild-type (HC22-1B) and cpr1Δ (HC24-8A) cells expressing Zpr1p-GFP were grown to log phase in SC plus glucose, starved in glucose-free SC for 18 h, and then refed glucose. (Right) Wild-type (HC22-1B) and cpr1Δ (HC24-8A) cells expressing Zpr1p-GFP were grown to log phase, poisoned with sodium azide-deoxyglucose for 45 min, washed, and resuspended in SC plus glucose. The percentages of cells displaying predominantly nuclear fluorescence at the indicated times are shown. (C) CPR1 ZPR1-GFP (HC22-1B) cells containing empty vector (pRS414) and cpr1Δ ZPR1-GFP (HC24-8A) cells containing either empty vector (pRS414) or pRS414 bearing the indicated PPIases were grown to log phase in SC lacking tryptophan and examined by fluorescence microscopy. The percentages of cells displaying predominantly nuclear fluorescence are shown.
Glucose starvation significantly decreased the nuclear localization of Zpr1p-GFP in the cpr1Δ strain (Fig. 4B). Under these conditions, cells stopped dividing, but there was no loss in viability (data not shown). When the cells of both wild-type and cpr1Δ strains were refed glucose after starvation, the percentage of cells with nuclear fluorescence increased immediately and with identical kinetics for both strains (40.9% in the cpr1Δ mutant and 46.3% in the wild type 30 min after refeeding). Between 30 and 120 min after refeeding, however, the percentages diverged, with the cpr1Δ strain maintaining a significantly higher level of Zpr1p-GFP nuclear localization than that of the wild-type strain (63.4% in the cpr1Δ mutant and 11.1% in the wild type).
This pattern suggested that Cpr1p was either inhibiting the nuclear import or promoting the nuclear export of Zpr1p. A nuclear import assay in which import is inhibited by poisoning cells with sodium azide and 2-deoxyglucose (30) showed identical rates of increase in Zpr1p nuclear fluorescence in both the wild-type and cpr1 strains after import was reinitiated by the removal of the poison and the addition of glucose (Fig. 4B). We conclude that the major effect of Cpr1p is to promote nuclear export of Zpr1p rather than to inhibit Zpr1p's nuclear import.
To determine if Cpr1p exports Zpr1p by working in concert with some of the known mediators of nuclear export, we looked for functional interactions between Cpr1p or Zpr1p and either Crm1p, Rat7p, Los1p, or Msn5p (18, 31). Neither temperature-sensitive alleles of CRM1 or RAT7 nor the deletion of LOS1 or MSN5 had any effect on the localization of Zpr1p, indicating that these molecules do not regulate the export of Zpr1p (data not shown). Yeast strains carrying the crm1-T539C allele are sensitive to leptomycin B, a potent inhibitor of Crm1p function (24). The cpr1Δ mutation did not confer hypersensitivity to leptomycin B in crm1-T539C cells, nor did it confer synthetic lethality in crm1, rat7, los1, or msn5 mutant strains (data not shown). Taken together, these data indicate that Cpr1p and Zpr1p1 do not functionally interact with these export molecules.
Zpr1p localization is dependent on CyPA PPIase activity.
To determine whether the PPIase activity of Cpr1p is required for proper localization of Zpr1p, we expressed wild-type hCyPA and the H54Q and H126Q mutant proteins in the cpr1Δ ZPR1-GFP strain. hCyPA behaved exactly like yeast Cpr1p by preventing the nuclear accumulation of Zpr1p-GFP (2.2% of cells displayed nuclear fluorescence) (Fig. 4C). The H54Q mutant protein molecule, which retains 15% of the wild-type protein's in vitro PPIase activity, also behaved like wild-type yeast Cpr1p and wild-type hCyPA by preventing the nuclear accumulation of Zpr1p-GFP (2.7% of cells). However, the H126Q mutant protein molecule, which retains only 0.5% of the wild-type protein's in vitro PPIase activity, behaved like a CPR1 deletion mutant and allowed for the nuclear accumulation of Zpr1p-GFP (33% of cells). Thus, increased nuclear accumulation of Zpr1p correlates with a decrease in PPIase activity, suggesting that Cpr1p uses its PPIase activity to regulate the cellular localization of Zpr1p.
Zpr1 mutant proteins accumulate in the nucleus in the presence of Cpr1p.
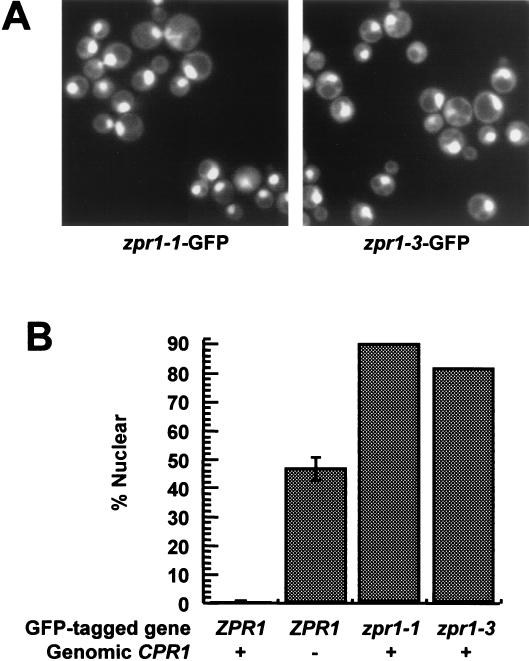
Because Cpr1p promotes nuclear export of Zpr1p, we hypothesized that Zpr1p carries out an essential function in the cytoplasm and that synthetic lethality might result if the proteins encoded by zpr1-1 and zpr1-3 were less easily exported to the cytoplasm. This model predicts that even in the presence of Cpr1p, the mutant Zpr1 proteins would accumulate in the nucleus to a greater extent than would wild-type Zpr1p. To test this, we constructed strains that express zpr1-1-GFP and zpr1-3-GFP fusion genes integrated at the ZPR1 locus. Both strains were CPR1 dependent, and the zpr1-1-GFP strain was temperature sensitive at 37°C. Consistent with our hypothesis, the zpr1-1-GFP and zpr1-3-GFP strains displayed high percentages of cells with intense nuclear fluorescence (90.0% in the zpr1-1-GFP strain and 83.0% in the zpr1-3-GFP strain) (Fig. 5A and B).
FIG. 5.
The zpr1-1 and zpr1-3 mutant proteins accumulate in the nucleus. (A) zpr1-1-GFP (HC70-8B) and zpr1-3-GFP (HC72-7C) cells were grown to log phase in SC plus glucose and examined by fluorescence microscopy. (B) The percentage of these cells displaying nuclear fluorescence is shown along with the percentages from CPR1 (HC22-1B) and cpr1Δ (HC24-8A) cells expressing ZPR1-GFP.
DISCUSSION
Cpr1p regulates the nuclear export of Zpr1p.
Our results suggesting a novel role for Cpr1p as a regulator of Zpr1p nuclear export add to the growing body of evidence that cyclophilins regulate the subcellular localization of proteins. Drosophila melanogaster NinaA binds Rh1 rhodopsin and mediates its trafficking within photoreceptor cells of the retina (3, 11). Yeast strains lacking Cpr1p fail to import fructose-1,6-bisphosphatase into specialized vesicles for targeting to the vacuole (9).
Nuclear accumulation of Zpr1p might result from abnormalities in Zpr1p conformation that result from lack of Cpr1p. We were unable to detect a direct interaction between Cpr1p and the proteins encoded by either wild-type or mutant alleles of ZPR1. Since Zpr1p may be an isomerase substrate, its interaction with Cpr1p may be too transient to be detected by the methods we attempted here. This is in contrast to recent reports with Itk and HIV-1 Gag, isomerase substrates that form stable complexes with CyPA (5, 8). Alternatively, the effects of Cpr1p on Zpr1p nuclear export might be indirect, via an unknown export factor, but mutations in several known mediators of nuclear export, CRM1, RAT7, LOS1, and MSN5, did not affect the localization of Zpr1p (data not shown).
The findings that wild-type Zpr1p concentrates in the nucleus in the absence of Cpr1p and that the alleles conferring synthetic lethality are concentrated in the nucleus to a greater extent suggest that the conditional alleles increase the requirement for an interaction that is already present with the wild type. If Zpr1p is required in the cytoplasm for viability, then perhaps quantities of wild-type Zpr1p sufficient for viability are exported in the absence of Cpr1p due to overlapping function from Cpr6p or Fpr1p. In contrast, sufficient quantities of the proteins encoded by zpr1-1 and zpr1-3 are exported to the cytoplasm only when Cpr1p is expressed. Attempts to verify this model by fusing a nuclear export signal to the mutant protein encoded by zpr1-1 were unsuccessful in that the cells still required Cpr1p (data not shown). However, interpretation of this experiment is complicated by the finding that zpr1-1p-GFP-nuclear export signal expression levels were significantly lower than those for either zpr1-1p-GFP or the wild type, Zpr1p-GFP.
CyPA PPIase activity is biologically relevant.
We demonstrated that CyPA point mutants suppress CPR1 dependence of zpr1 mutant strains in a PPIase-correlative manner. Furthermore, overexpression of Cpr6p or, more significantly, Fpr1p, also suppresses CPR1 dependence. In the absence of immunosuppressive drug ligands, the only known property that the cyclophilin and FKBP families of proteins share is PPIase activity. These findings parallel previous reports of functional overlap between structurally distinct families of PPIases, namely the suppression by CPR1 overexpression of an ess1 mutant strain's conditional lethality (2, 34). These data, along with recent work describing regulation of the tyrosine kinase Itk by CyPA (8), suggest that PPIase activity of CyPA is biologically relevant.
Overexpression of Fpr4p and Ess1p did not suppress the CPR1 dependence of the zpr1-1 strain. This could be due to less efficient isomerase activity, greater substrate specificity, or suboptimal cellular localization of these PPIases. Subcellular localization may be the most likely explanation, as both Fpr4p and Ess1p are nuclear PPIases whereas Cpr1p, Cpr6p, and Fpr1p are cytoplasmic (12).
ZPR1 was the only gene identified in our screen. Others have reported synthetic lethality between CPR1 or FPR1 and temperature-sensitive alleles of ess1 (2). Perhaps these ess1 alleles would be obtained with further screening, but it may be impossible to isolate these ess1 alleles by using the methods that we employed: another group similarly failed to isolate these alleles in a screen for mutants conferring synthetic lethality with FPR1 (13).
ZPR1 and rRNA.
It was previously reported that depletion of Zpr1p resulted in reduction of steady-state 35S pre-rRNA levels in Schizosaccharomyces pombe (15). We observed in S. cerevisiae that zpr1-1 cells at the nonpermissive temperature had steady-state levels of 35S pre-rRNA that are similar to those of the wild type, as determined by either Northern blotting or real-time PCR (data not shown). Similarly, processing rates of 35S pre-rRNA, as determined by metabolic labeling, were not altered (data not shown). A possible explanation for the differences between our results and those previously reported is that the authors of the previous report depleted Zpr1p in a 12-h promoter shutoff experiment whereas we used a mutant protein that is inactivated immediately by temperature shift.
Functional interactions between Cpr1p, Zpr1p, and EF1α.
Finally, we have established functional relationships between Cpr1p, Zpr1p, and EF1α. EF1α, the nuclear export factor Los1p, and the aminoacylation cofactor Arc1p functionally interact to facilitate nuclear export of tRNA (19). However, los1Δ mutations were not synthetically lethal with cpr1Δ or zpr1-1 mutations, Los1p overexpression did not suppress CPR1 dependence of the zpr1-1 strain, and overexpression of Cpr1p or Zpr1p did not suppress LOS1 dependence of an arc1Δ strain (data not shown). The hypersensitivity of the zpr1-1 strain to translation inhibitors such as cycloheximide and the rescue of this cycloheximide hypersensitivity by the overexpression of EF1α suggest that Zpr1p may also function in translation. It is interesting that these three molecules have highly conserved homologs in mammalian systems, suggesting that our findings for S. cerevisiae may be replicated in higher organisms.
Acknowledgments
We are grateful to Rodney Rothstein, Aaron Mitchell, Fred Chang, Hamish Young, Steve Goff, Marian Carlson, Liza Pon, Michael Rout, Joe Heitman, Steve Hanes, Pamela Silver, Charles Cole, Karsten Weis, Ed Hurt, Anita Hopper, Minoru Yoshida, Dev Mangroo, Michael Rosbash, Molly Fitzgerald-Hayes, Naz Erdeniz, Teresa Lamb, Tari Suprapto, Caterina Strambio-de-Castillia, and Robert Reid for strains, plasmids, reagents, and advice. We thank David Sayah and Rose Cohen for technical assistance.
This research was supported by the Sandler Program for Asthma Research and by the NIH (AI36199).
REFERENCES
- 1.Andreeva, L., R. Heads, and C. J. Green. 1999. Cyclophilins and their possible role in the stress response. Int. J. Exp. Pathol. 80:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arevalo-Rodriguez, M., M. E. Cardenas, X. Wu, S. D. Hanes, and J. Heitman. 2000. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 19:3739-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, E. K., N. J. Colley, and C. S. Zuker. 1994. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 13:4886-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosco, D. A., E. Z. Eisenmesser, S. Pochapsky, W. I. Sundquist, and D. Kern. 2002. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. USA 99:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazin, K. N., R. J. Mallis, D. B. Fulton, and A. H. Andreotti. 2002. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc. Natl. Acad. Sci. USA 99:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. R., D. Y. Cui, G. G. Hung, and H. L. Chiang. 2001. Cyclophilin A mediates Vid22p function in the import of fructose-1,6- bisphosphatase into Vid vesicles. J. Biol. Chem. 276:48017-48026. [DOI] [PubMed] [Google Scholar]
- 10.Colgan, J., M. Asmal, and J. Luban. 2000. Isolation, characterization and targeted disruption of mouse ppia: cyclophilin A is not essential for mammalian cell viability. Genomics 68:167-178. [DOI] [PubMed] [Google Scholar]
- 11.Colley, N. J., E. K. Baker, M. A. Stamnes, and C. S. Zuker. 1991. The cyclophilin homolog ninaA is required in the secretory pathway. Cell 67:255-263. [DOI] [PubMed] [Google Scholar]
- 12.Dolinski, K., S. Muir, M. Cardenas, and J. Heitman. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolinski, K. J., and J. Heitman. 1999. Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics 151:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, B., R. S. Haltiwanger, and B. Futcher. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galcheva-Gargova, Z., L. Gangwani, K. N. Konstantinov, M. Mikrut, S. J. Theroux, T. Enoch, and R. J. Davis. 1998. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell 9:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galcheva-Gargova, Z., K. N. Konstantinov, I. H. Wu, F. G. Klier, T. Barrett, and R. J. Davis. 1996. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 272:1797-1802. [DOI] [PubMed] [Google Scholar]
- 17.Gangwani, L., M. Mikrut, Z. Galcheva-Gargova, and R. J. Davis. 1998. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J. Cell Biol. 143:1471-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorsch, L. C., T. C. Dockendorff, and C. N. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosshans, H., E. Hurt, and G. Simos. 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 14:830-840. [PMC free article] [PubMed] [Google Scholar]
- 20.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 21.Kallen, J., C. Spitzfaden, M. G. Zurini, G. Wider, H. Widmer, K. Wuthrich, and M. D. Walkinshaw. 1991. Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature 353:276-279. [DOI] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 23.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 24.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid, R. J. D., M. Lisby, and R. Rothstein. 2001. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350:258-277. [DOI] [PubMed] [Google Scholar]
- 26.Roy, N., and K. W. Runge. 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10:111-114. [DOI] [PubMed] [Google Scholar]
- 27.Schiene, C., and G. Fischer. 2000. Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 10:40-45. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber, S. L., and G. R. Crabtree. 1992. The mechanism of action of cyclosporin A and FK506. Immunol. Today 13:136-142. [DOI] [PubMed] [Google Scholar]
- 29.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 30.Shulga, N., P. Roberts, Z. Gu, L. Spitz, M. M. Tabb, M. Nomura, and D. S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strom, A. C., and K. Weis. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:3008.1-3008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes, K., M. J. Gething, and J. Sambrook. 1993. Proline isomerases function during heat shock. Proc. Natl. Acad. Sci. USA 90:5853-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, P., M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, X., C. B. Wilcox, G. Devasahayam, R. L. Hackett, M. Arevalo-Rodriguez, M. E. Cardenas, J. Heitman, and S. D. Hanes. 2000. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 19:3727-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]