Abstract
Metazoan replication-dependent histone mRNAs end in a conserved stem-loop rather than in the poly(A) tail found on all other mRNAs. The 3′ end of histone mRNA binds a single class of proteins, the stem-loop binding proteins (SLBP). In Xenopus, there are two SLBPs: xSLBP1, the homologue of the mammalian SLBP, which is required for processing of histone pre-mRNA, and xSLBP2, which is expressed only during oogenesis and is bound to the stored histone mRNA in Xenopus oocytes. The stem-loop is required for efficient translation of histone mRNAs and substitutes for the poly(A) tail, which is required for efficient translation of other eucaryotic mRNAs. When a rabbit reticulocyte lysate is programmed with uncapped luciferase mRNA ending in the histone stem-loop, there is a three- to sixfold increase in translation in the presence of xSLBP1 while xSLBP2 has no effect on translation. Neither SLBP affected the translation of a luciferase mRNA ending in a mutant stem-loop that does not bind SLBP. Capped luciferase mRNAs ending in the stem-loop were injected into Xenopus oocytes after overexpression of either xSLBP1 or xSLBP2. Overexpression of xSLBP1 in the oocytes stimulated translation, while overexpression of xSLBP2 reduced translation of the luciferase mRNA ending in the histone stem-loop. A small region in the N-terminal portion of xSLBP1 is required to stimulate translation both in vivo and in vitro. An MS2-human SLBP1 fusion protein can activate translation of a reporter mRNA ending in an MS2 binding site, indicating that xSLBP1 only needs to be recruited to the 3′ end of the mRNA but does not need to be directly bound to the histone stem-loop to activate translation.
Replication-dependent histone mRNAs are unique among metazoan mRNAs because they are not polyadenylated but end, instead, in a highly conserved stem-loop (35). In contrast, histone mRNAs from fungi and plants are polyadenylated. Histone pre-mRNA processing requires only a single endonucleolytic cleavage to form the 3′ end of the mature mRNA immediately after a highly conserved stem-loop structure (16). Most of the regulation of histone mRNA levels is mediated by the stem-loop and occurs at the posttranscriptional level, via regulation of mRNA processing and stability (22, 44). It is likely that the stem-loop also fulfills the essential functions that are performed by the poly(A) tail on other mRNAs. One of these functions is to enhance the efficiency of translation (27, 52). For example, the stem-loop has been shown to promote localization of histone mRNAs to polyribosomes (56). mRNAs ending in the histone stem-loop at the 3′ end are translationally more active than messages ending in other stem-loops when transfected into Chinese hamster ovary cells (15).
By using the yeast three-hybrid system, proteins that bind to the stem-loop have been isolated from several species (34, 55, 64, 66). Although only a single SLBP is present in mammals (34, 66), Drosophila (55), and Caenorhabditis elegans, frog oocytes (46) express two SLBPs, called xSLBP1 and xSLBP2 (64). The centrally located RNA binding domain (RBD) is highly conserved between xSLBP1 and xSLBP2 (64). The two frog SLBPs otherwise are not similar in structure, suggesting that they may have distinct activities in histone mRNA metabolism. xSLBP2 is expressed only during oogenesis and is confined to the cytoplasm (64). xSLBP1 is the orthologue of the human and mouse SLBPs that regulate replication-dependent histone mRNA metabolism (69). xSLBP1 is expressed throughout development and is located both in the nucleus and in the cytoplasm (64).
Translation in eucaryotes is accomplished by a number of trans-acting factors that interact with the 5′ end of polyadenylated mRNAs (reviewed in references 17, 23, and 52). Initiation factor eIF-4E binds the m7G(5′)ppp(5′)N cap structure. Initiation factor eIF-4G is associated with the 5′ end of the message via its interaction with eIF-4E and also serves as a scaffold protein for binding of eIF3. eIF-4A, an RNA helicase, also binds to eIF-4G and, together with eIF-4E, makes up the eIF-4F complex. The 40S ribosomal subunit is bound to the mRNA 5′ end by virtue of its interaction with eIF3. This binding promotes proper localization of the preinitiation complex, allowing recruitment of the initiator tRNA and also is necessary for joining of the 60S and 40S subunits.
Recently, it has been shown that the 3′ untranslated region (UTR) of many mRNAs contains sequence elements required for regulation of translation (8, 9, 43, 50, 75). The 3′ end of mRNA participates synergistically with the 5′ end in translation of polyadenylated mRNAs (14). The poly(A) tail, which is usually 50 to 200 residues long, binds to the poly(A) binding protein (PABP). PABP mediates bridging of the 5′ and 3′ ends of the message by interacting directly with eIF-4G (25, 29, 59, 60, 68). It is believed that the resulting circular mRNP structure promotes translation reinitiation by keeping the ribosomes that are coming off the 3′ end of the message in close proximity to the 5′ initiation site (27). There is also evidence that PABP enhances binding of eIF-4E to the cap structure, promoting the initial rounds of translation (6, 67). By binding to the poly(A) tail and the cap/eIF-4F complex, PABP may also help protect both ends of the mRNA from degradation (5, 68). Degradation of the polyadenylated mRNAs may require breaking of the interaction between the 3′ and 5′ ends of the message (4, 61), allowing decapping of the mRNA, which is often followed by 5′-to-3′ degradation.
Histone mRNA and protein synthesis occurs very early during oogenesis in Xenopus laevis and is completed by the end of stage II (1, 72). The histone mRNA is then stored in an inactive form for the remainder of oogenesis, while the histone protein is stored in the germinal vesicle complexed with acidic proteins, nucleoplasmin and N1/N2 (11). During late oogenesis (stages IV to VI), when no histone protein synthesis is occurring, the bulk of the histone mRNA is bound to xSLBP2 and only 10% can be immunoprecipitated with anti-xSLBP1 antibodies (64). Translation of histone mRNA is reactivated at oocyte maturation (1), which can be induced by treatment of stage VI oocytes with progesterone. During oocyte maturation, xSLBP2 is degraded and xSLBP1 then binds to the histone mRNA (64). These observations suggest that regulation of histone message translation may depend on which xSLBP is bound to the stem-loop and that xSLBP1 may actively promote histone mRNA translation.
Here we report that xSLBP1, but not xSLBP2, can stimulate translation of a luciferase mRNA that ends in the histone stem-loop in vitro and in vivo. A 10- to 15-amino-acid region in the N-terminal portion of xSLBP1 is responsible for translation activation. These data provide direct evidence that the translation of histone mRNA in Xenopus oocytes is controlled by the two xSLBPs: xSLBP2, a histone mRNA-specific masking factor, and xSLBP1, a histone mRNA-specific translation activator.
MATERIALS AND METHODS
Construction of SLBP variants.
All of the SLBPs were expressed from the modified p64T vector pXFRM, previously described, which provides 5′ and 3′ β-globin UTRs and a poly(A) tail for efficient translation of in vitro-transcribed RNA in oocytes (26, 64). The chimeric SLBPs were constructed as described previously (26). We have recently determined that xSLBP2 contains an additional 23 amino acids at the amino terminus compared to the xSLBP2 sequence previously reported (64) (accession no. AF106799); these additional amino acids were introduced by PCR into the xSLBP2 expression vector. The original xSLBP2 cDNA from the two-hybrid vector pGAD10 was used as the template; the forward primer was used to introduce a BspHI site that contains the new initiation codon at the beginning of xSLBP2. The resulting PCR product was digested with BspHI and XbaI and subcloned into pXFRM digested with NcoI and XbaI.
Mutant forms xSLBP65-69/5A, xSLBP70-74/5A, xSLBP75-79/5A, xSLBP80-84/5A, and xSLBP85-89/5A were generated by PCR with xSLBP1 as the template. The 5′ end of the forward and reverse primers started with the sequences CAGCAGCA and CTGCTGC, respectively, which together resulted in a substitution of five alanines. The xSLBP1 deletion mutant forms Δ68-1-1, Δ68-1-0, Δ81-1-1, Δ81-1-0, Δ1-1-0, and Δ13 were generated by PCR with xSLBP1 as the template. Human SLBP (hSLBP) and the mutant forms hSLBPRR/KK and hSLBPYY/SS were constructed as described previously (12).
The MS2 and MS2-hSLBP vectors were generated by subcloning of the MS2 coding sequence from a vector provided by Joseph Viradell (Albert Einstein School of Medicine) into the pFastBac-HTa vector (HTa-MS2). Human SLBP was subcloned into HTa-MS2 from the previously described pXFRM-hSLBP vector (12). The MS2 and MS2-hSLBP coding sequences were subcloned into pXFRM from vectors HTa-MS2 and HTa-MS2-hSLBP.
Construction of reporter mRNAs.
The Luc-polyA, Luc-SL, and Luc-TL constructs, in which the firefly luciferase coding region is under the control of the T7 promoter, have been described previously (15). The pRL-CMV vector (Promega) was used as a template to amplify the Renilla luciferase coding region by PCR. An NcoI site was introduced at the initiation codon. After digestion with NcoI and XbaI, the Renilla PCR product was subcloned into the homologous sites in the Luc-SL vector to generate the R-Luc-SL vector. The polyadenylated chloramphenicol acetyltransferase (CAT-A50) coding region, previously subcloned into pBluescript II (Stratagene), was digested with HindIII and EcoRI and subcloned into pGEM-3Zf (Promega). To generate the Luc-MS2 vector, two complementary oligonucleotides encoding the MS2 binding site were annealed and ligated into the Luc-SL vector digested with BamHI and AflII.
In vitro transcription.
SLBP plasmids were linearized with EcoRI and transcribed with SP6 RNA polymerase in the presence of a cap analogue (New England Biolabs). The luciferase plasmids with different 3′ ends were linearized with AflII (R-Luc-SL, Luc-SL, Luc-TL, and Luc-MS2) prior to transcription with T7 RNA polymerase (New England Biolabs) in the presence or absence of the cap analogue (15). The CAT-A50 and Luc-polyA plasmids were linearized with NdeI prior to transcription with T7 RNA polymerase. Transcription reaction mixtures were treated with RQ1 DNase (Promega), and the RNA was purified on G-50 microcolumns (Pharmacia).
In vitro translation.
Plasmids encoding SLBP, MS2, and MS2-hSLBP under control of the SP6 promoter in the pXFRM vector (64) (final concentration, 20 ng/μl) were incubated for 1 h in a micrococcal nuclease-treated transcription-translation coupled rabbit reticulocyte lysate (RRL) system (Promega) in the presence of SP6 RNA polymerase and [35S]methionine. A fraction of this reaction mixture (5 μl) was added to a mixture (10 μl) containing fresh micrococcal nuclease-treated RRL, [35S]methionine, uncapped CAT-A50 mRNA (20 ng/μl), and one of the firefly luciferase uncapped transcripts (20 ng/μl). After incubation at 30°C for 1 h, the in vitro translation reaction mixture was resolved by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and protein synthesis was quantified on a Storm 840 PhosphorImager with the ImageQuant program (Molecular Dynamics).
Injection of oocytes.
Ovaries were removed from adult female frogs (NASCO) and treated for 2 h in 0.2% collagenase in OR-2 solution at 27°C. After removal of the collagenase, the oocytes were allowed to recover at 18°C in OR-2 solution for at least 24 h. Stage VI oocytes were injected in the cytoplasm with 30 nl of capped SLBP mRNAs (50 ng/μl) and incubated in OR-2 solution for 8 to 16 h at 18°C. Control oocytes were injected with 30 nl of water. Capped firefly and Renilla luciferase mRNAs were injected in the cytoplasm (30 nl, 50 ng/μl, equimolar concentrations) and harvested 12 h later. Twelve to 15 oocytes were pooled for each experiment. The oocytes were homogenized (2.5 μl per oocyte) in 20 mM Tris (pH 7.5)-1 mM phenylmethylsulfonyl fluoride (Invitrogen)-1× protease inhibitor cocktail (Sigma). These lysates were then used for RNA preparation, mobility shift assays, luciferase assays, and Western blotting.
Luciferase assay.
Luciferase was measured with a Monolight 2010 luminometer (Analytical Luminescence Laboratory) after dilution of the in vitro translation reaction mixture in 1× luciferase lysis buffer 1/25 (Analytical Luminescence Laboratory). Lysates equivalent to two oocytes were diluted in 1× passive luciferase buffer 1/25 (Promega). Renilla and firefly luciferase activities were measured with a dual luminometer (Lmax; Molecular Devices).
RNA extraction and analysis.
Total RNA was extracted from oocyte lysates with Trizol reagent (Invitrogen) and 1 oocyte equivalent of total RNA was hybridized to 5 ng of probe. The S1 probe used to determine luciferase RNA levels was generated by linearization of the Luc-SL vector with DraII, which cuts in the luciferase coding region. The 3′ end was labeled with the Klenow fragment of DNA polymerase I (New England Biolabs) and [α-32P]dCTP. Hybridization was done at 56°C, and S1 digestion was performed as previously described (12, 18). The protected fragments were resolved on a 10% polyacrylamide-7 M urea gel and detected by autoradiography.
Antibodies.
The RBD was deleted by PCR from the glutathione S-transferase (GST)-xSLBP2 fusion protein (64) to give the GST-xSLBP2-RBD fusion protein. xSLBP2 antibodies were purified as previously described, with the GST-xSLBP2-RBD fusion protein expressed in bacteria (64). Antibodies generated against xSLBP1 expressed in baculovirus were affinity purified with immobilized full-length xSLBP1.
Western blot assays.
The lysate from four oocytes was diluted in 0.5 ml of 20 mM Tris (pH 7.5)-1 mM phenylmethylsulfonyl fluoride (Invitrogen)-1× protease inhibitor cocktail (Sigma) and centrifuged for 5 min at 14,000 rpm in an Eppendorf microcentrifuge. The clear supernatant was precipitated with 1 ml of acetone and centrifuged for 15 min at 14,000 rpm. The pellet was resuspended in 1% SDS-5% glycerol-0.06 M Tris (pH 6.8)-5% β-mercaptoethanol and boiled; proteins were resolved by SDS-10% PAGE and transferred to nitrocellulose. The filter was incubated with the appropriate affinity-purified antibody, and the bound antibodies were detected by chemiluminescence with SuperSignal (Pierce).
Mobility shift assay.
Oocyte lysate equivalent to one oocyte was mixed on ice with buffer (final concentrations: 70 mM KCl, 14 mM HEPES [pH 7.9], 150 mM EDTA [pH 8.0], 7% glycerol, 5 mM dithiothreitol, and 1 μg of tRNA per μl) and 5 ng of a 30-nucleotide (nt) synthetic radiolabeled stem-loop RNA that was prepared as previously described (70). The complexes were resolved on a 10% native polyacrylamide gel and were visualized by autoradiography.
RESULTS
Since SLBP is the only known protein that specifically binds the 3′ end of histone mRNA, it is an obvious candidate to mediate the effect of the stem-loop on translation of histone mRNA. There are two SLBPs in frog oocytes, each of which binds the stem-loop with similar affinity (26, 64). Each of these proteins consists of about 280 amino acids, with the RBD located in the center of the protein.
Only xSLBP1 activates translation of Luc-SL mRNA in vitro.
To investigate the possible role of SLBP in translation, we used both an in vitro system, the RRL, and an in vivo system, injection of reporter mRNAs into Xenopus oocytes. We used three reporter luciferase mRNAs, which are shown in Fig. 1A. The first (Luc-SL) ends in the histone stem-loop, the second (Luc-TL) ends in a GNRA tetraloop that contains the same stem and flanking sequences as the histone stem-loop but that does not bind SLBP, and the third (Luc-polyA) ends in a 50-nt poly(A) tail. We also used an mRNA encoding the Renilla luciferase that ended in a histone stem-loop (R-Luc-SL) as an internal control for the in vivo experiments. We determined the effect of expression SLBPs on the translation of the various reporter luciferase mRNAs in vitro and in vivo.
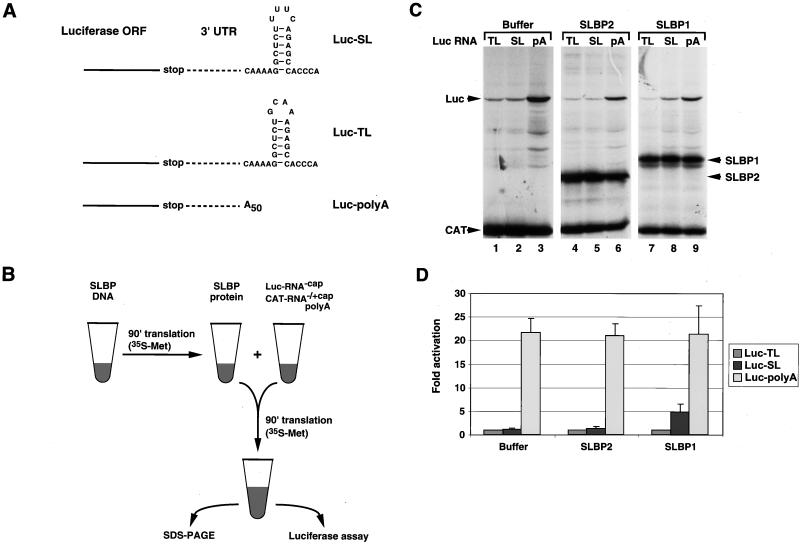
FIG. 1.
SLBP stimulates histone mRNA translation in vitro. (A) Structure of the 3′ end of the luciferase reporter mRNAs. The Luc-SL mRNA ends in the histone stem-loop that binds SLBP, the Luc-TL mRNA ends in a stem-loop that does not bind SLBP, and the Luc-polyA mRNA ends in a poly(A) tail 50 nt long. ORF, open reading frame. (B) Schematic of the in vitro translation assay. An aliquot of RRL was incubated either with buffer or with SLBP DNA in the presence of T7 RNA polymerase and [35S]methionine. After incubation for 90 min, an aliquot of the lysate was mixed with a fresh aliquot of reticulocyte lysate together with a luciferase uncapped reporter mRNA and a polyadenylated uncapped CAT mRNA as an internal standard. After incubation for 90 min at 30°C, the reaction products were analyzed by SDS-PAGE and the in vitro-synthesized proteins were detected by autoradiography. An aliquot of the assay was analyzed for luciferase activity by luminometry. (C) Lysates containing no SLBP (lanes 1 to 3), xSLBP2 (lanes 4 to 6), or xSLBP1 (lanes 7 to 9) were incubated with the standard polyadenylated CAT mRNA and the Luc-TL (lanes 1, 4, and 7), Luc-SL (lanes 2, 5, and 8), or Luc-polyA (lanes 3, 6, and 9) mRNA. The reaction products were analyzed by gel electrophoresis, and the proteins were detected by autoradiography. (D) The autoradiogram (panel C) was quantified, and relative luciferase activity was calculated. In addition, an aliquot of each reaction mixture was analyzed for luciferase activity with a luminometer. Quantification of the results by PhosphorImager and by luciferase assay yielded identical results. Panel D shows the results of an average of five experiments with five different batches of RRL, and the error bars represent the standard deviation. The fold activation is the result of luciferase activity from the Luc-SL and Luc-polyA mRNAs divided by the luciferase activity from the Luc-TL mRNA.
A schematic of the in vitro assay is shown in Fig. 1B. Briefly, xSLBP1, xSLBP2, or any one of various chimeric or mutated proteins was first expressed in an RRL. This served as the source of SLBP for the in vitro assays. After incubation for 90 min, equal amounts of these lysates were added to a translation reaction mixture containing uncapped, polyadenylated CAT mRNA and a luciferase reporter mRNA, also uncapped. Thus, within each experiment, the different reporter mRNAs were tested with the same amount and preparation of SLBP.
The reticulocyte lysate contains very small amounts of SLBP (66). Expression of SLBP in the lysate from synthetic mRNA results in the synthesis of sufficient SLBP to bind exogenous stem-loop RNA in a mobility shift assay (12, 66). Enough SLBP is produced by this approach to bind all of the reporter mRNAs added to the lysate. Since the reticulocyte lysate largely responds to the cap at the 5′ end of the mRNA, we used uncapped mRNAs in these experiments. There was no effect of any of the proteins (or different 3′ ends) when we used capped mRNAs expressed from the same constructs (data not shown). We included an uncapped polyadenylated CAT mRNA both to provide an internal control for translation efficiency and as a competitor for translation initiation factors to maximize the effect of the 3′ end of the luciferase mRNA (49). We analyzed the expression of the xSLBPs, CAT, and luciferase proteins by gel electrophoresis, followed by autoradiography (Fig. 1C). We also assayed luciferase activity with a luminometer (Fig. 1D).
We first tested the effects of xSLBP1 and xSLBP2 on the translation of the Luc-SL, Luc-TL, and Luc-polyA mRNAs in vitro. When either buffer or in vitro-synthesized xSLBP2 was added to the reticulocyte lysate containing different reporter mRNAs, the Luc-SL and Luc-TL mRNAs were translated with similar efficiencies (Fig. 1C, lanes 1 and 2 and 4 and 5, respectively). The Luc-polyA mRNA was translated about 20-fold more efficiently than the mRNAs ending in a stem-loop (Fig. 1C, lane 3). In the presence of xSLBP2, the absolute amounts of translation of all of the mRNAs (including the CAT competitor) were reduced (Fig. 1C, lanes 4 to 6) because of the presence of the additional competitor SLBP mRNA. The relative amounts of luciferase expression compared to CAT expression determined by PhosphorImager analysis was essentially the same for all three luciferase mRNAs (Fig. 1C, lanes 1 to 3 versus lanes 4 to 6). In contrast, when xSLBP1 was included in the final reaction mixture, there was a four- to fivefold increase in the translation of the Luc-SL mRNA (Fig. 1C, lane 8 versus lane 5), relative to the Luc-TL mRNA. There was no change in the translation of the Luc-polyA mRNA relative to that of the Luc-TL mRNA in the presence of xSLBP2 (Fig. 1C, lanes 4 and 6) or xSLBP1 (Fig. 1C, lanes 7 and 9).
To determine whether the effect of xSLBP1 was dependent on the interaction of xSLBP1 with the stem-loop, we replaced xSLBP1 with in vitro-synthesized hSLBP (the orthologue of xSLBP1) or two hSLBP mutant forms (hSLBPRR/KK and hSLBPYY/SS, Fig. 2A). Addition of hSLBP stimulated translation of the Luc-SL mRNA four- to fivefold, the same level as xSLBP1 (Fig. 1D). In the mutants, two conserved arginines at positions 10 and 11 in the RBD were replaced with lysines (hSLBPRR/KK) or the tyrosines at positions 24 and 27 were replaced with two serines (hSLBPYY/SS). Neither of these proteins binds the stem-loop with high affinity, as determined by mobility shift assay (12). Both hSLBPRR/KK and hSLBPYY/SS failed to stimulate translation of the Luc-SL mRNA (Fig. 2A).
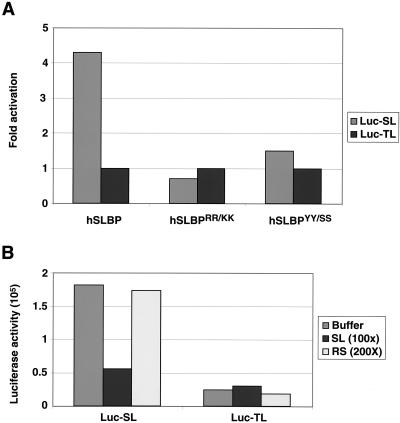
FIG. 2.
SLBP must bind mRNA to stimulate translation. (A) Lysates containing hSLBP or two mutant hSLBPs that do not bind the stem-loop were incubated with the standard polyadenylated CAT mRNA and the Luc-TL or Luc-SL mRNA and assayed for luciferase activity. hSLBPRR/KK has arginines 10 and 11 in the RBD changed to lysines, and hSLBPYY/SS has tyrosines 24 and 27, also in the RBD, changed to serines (12). (B) Lysates containing xSLBP1 were incubated with the standard polyadenylated CAT mRNA and the Luc-TL or Luc-SL mRNA in the presence of either excess 30-nt RNA containing the histone stem-loop (SL) or with the reverse stem-loop (RS) and then assayed for luciferase activity.
Competition experiments were also carried out to demonstrate that SLBP must bind the Luc-SL mRNA to stimulate translation. Short, uncapped RNA oligoribonucleotides containing either the stem-loop (SL) or the reverse stem (RS), which does not bind SLBP (45, 70), were added together with Luc-SL or Luc-TL mRNA to the translation reaction mixtures supplemented with xSLBP1. A 100-fold molar excess of the 30-mer SL RNA effectively inhibited the stimulation of translation of Luc-SL mRNA in the reaction mixtures containing xSLBP1 (Fig. 2B). Addition of a 200-fold molar excess of the RS oligoribonucleotide had no affect on the stimulation of translation of Luc-SL mRNA (Fig. 2B). Neither competitor oligoribonucleotide had any effect on the translation of the Luc-TL mRNA (Fig. 2B). We conclude that the stimulation of the translation of Luc-SL mRNA requires that it form a complex with xSLBP1.
xSLBP1 activates translation of Luc-SL mRNA in vivo.
We also tested whether xSLBP1 could stimulate translation in vivo by injecting similar reporter mRNAs into Xenopus oocytes after overexpression of different SLBPs in vivo. In this case, the reporter mRNAs were capped (since uncapped mRNAs are very unstable in the oocytes) but they were otherwise identical to the three mRNAs used in the in vitro studies (Fig. 1A). To allow us to readily measure the relative translation of two different mRNAs in the same oocytes, we constructed a reporter mRNA that encoded the Renilla luciferase (R-Luc-SL) in addition to the reporter mRNAs encoding firefly luciferase. We then independently assayed the Renilla luciferase and firefly luciferase activities from the same samples, providing an internal control for the activity of the reporter mRNAs (39, 74).
Stage VI oocytes were injected (T0, Fig. 3A ) in the cytoplasm with capped mRNA encoding an SLBP or with buffer. Synthesis of SLBP was allowed to proceed for 12 to 16 h prior to cytoplasmic injection of equimolar amounts of the different capped firefly luciferase reporter mRNAs and the R-Luc-SL mRNA at time T1. At time T2, 12 to 16 h after injection of the reporter mRNAs, the oocytes were harvested. The activities of the two luciferases were measured by luminometry, and the SLBP levels were measured by Western blotting and, in some cases, by mobility shift assay. For each experiment shown, the same batch of oocytes was used. There was variability in the magnitude of the activation observed among different batches of oocytes, but the qualitative effects of the SLBPs were identical.
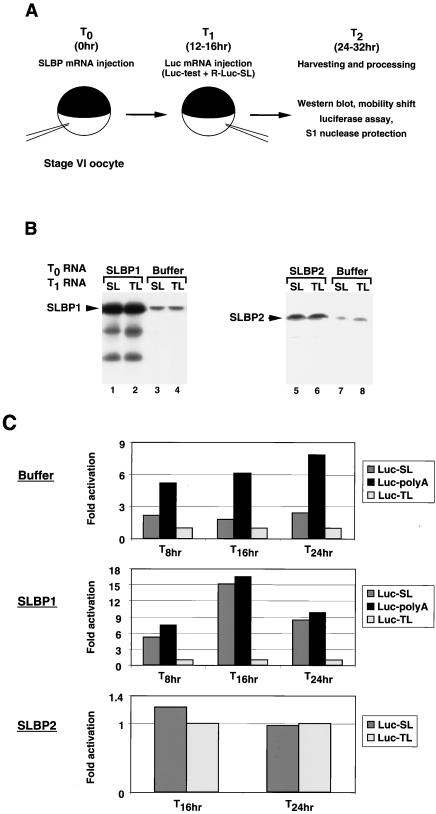
FIG. 3.
SLBP stimulates translation of Luc-SL mRNA in vivo. (A) Schematic of the in vivo assay. Xenopus oocytes were injected either with buffer or with a synthetic mRNA encoding an SLBP (T0). At 12 to 16 h later (T1), the oocytes were injected with the reporter firefly luciferase mRNA (Luc-test) mixed with a synthetic mRNA encoding Renilla luciferase ending in the histone stem-loop (R-Luc-SL). After incubation for an additional 12 to 16 h (T2), the oocytes were harvested and assayed for both luciferase activity and SLBP expression by Western blotting and by mobility shift assay with a radiolabeled stem-loop as the probe. The stability of reporter mRNAs was determined by S1 nuclease protection with a radiolabeled DNA fragment complementary to the 3′ end of the Luc-SL mRNA. (B) Oocytes were injected at T0 with buffer (lanes 3, 4, 7, and 8) or with xSLBP1 (lanes 1and 2) or xSLBP2 (lanes 5 and 6) and then at T1 with either the Luc-SL mRNA (lanes 1, 3, 5, and 7) or Luc-TL mRNA (lanes 2, 4, 6, and 8), together with R-Luc-SL mRNA. Oocytes were collected and lysed 8, 16, and 24 h later (T2 = 8, 16, and 24 h). Total oocyte protein was resolved by gel electrophoresis, transferred to nitrocellulose, and xSLBP1 (lanes 1 to 4) or xSLBP2 (lanes 5 to 8) was detected with appropriate antibodies. Panel B shows the Western blots of samples collected at T2 = 16 h. (C) Oocytes were injected with buffer (top), xSLBP1 mRNA (middle), or xSLBP2 mRNA (bottom). After 16 h, the oocytes were injected with the R-Luc-SL mRNA together with the Luc-SL, Luc-TL, or Luc-polyA mRNA. At 8, 16, or 24 h later, oocytes were harvested and assayed for Renilla and firefly luciferase activities. The ratio of Luc-TL luciferase activity to Renilla luciferase activity was determined, and this value was set at 1. The activity of the Luc-SL and Luc-polyA mRNAs was expressed relative to that of the Luc-TL mRNA.
To control for the stability of the mRNAs, we measured the level of luciferase mRNAs by S1 nuclease protection at the end of the incubation (see Fig. 7D). The ability of each of these reporter mRNAs (which were capped) to program luciferase activity in the reticulocyte lysate was also determined. All three reporter mRNAs had similar activities in the absence of competitor mRNAs or SLBPs, since the cap dominates the in vitro translation activity (data not shown) (14, 40).
FIG. 7.
Effect of xSLBP1 deletion mutant forms on translation activation in vivo. (A) Oocytes were injected with xSLBP1 deletion mutant mRNAs (Fig. 6A) or buffer at T0. At T1, they were injected with the R-Luc-SL mRNA together with the Luc-SL or Luc-TL mRNA. Oocytes were harvested at T2 and assayed for Renilla and firefly luciferase activities as described in Fig. 3C. The results shown are the averages of two experiments with two different batches of oocytes, with the error bars representing the standard deviations. (B) One oocyte equivalent of lysate from oocytes injected with mRNAs encoding the mutant SLBPs described in panel A was mixed with 5 ng of radiolabeled stem-loop RNA and analyzed by gel electrophoresis. The xSLBP/SL complex was detected by autoradiography. (A) Oocytes injected with buffer (lanes 9 to 11) or mRNAs encoding mutant SLBPs containing the intact C-terminal region of xSLBP1 (lanes 3 to 8) were analyzed by Western blotting as described in Fig. 3B. (B) Total RNA from oocytes injected as described in panel A and harvested at T2 = 32 h was hybridized to 3′-end-radiolabeled DNA complementary to the 3′ end of Luc-SL mRNA and subjected to S1 nuclease treatment. The protected DNA fragment was resolved by PAGE and detected by autoradiography. The Luc-SL mRNA protects a fragment 523 nt long that maps to the expected 3′ end, whereas Luc-TL mRNA protects a 508-nt fragment that maps to the start of the loop, where the sequences of Luc-SL and Luc-TL diverge.
We tested the effect of increased expression of xSLBP1 or xSLBP2 in vivo on the translation of the three luciferase reporter mRNAs. The xSLBP1 and xSLBP2 protein levels (measured by Western blotting) were each increased more than fivefold over endogenous levels after injection of the SLBP mRNA (Fig. 3B). The exogenous SLBPs persisted at the same levels at the end of the incubation, regardless of the nature of the luciferase transcripts injected at T1 (Fig. 3B, lanes 3 and 4 and 7 and 8). We measured the activities of both the Renilla and firefly luciferases and normalized the firefly luciferase activity from each reporter mRNA by adjusting to a constant amount of Renilla luciferase activity. We then set the firefly luciferase activity observed with the Luc-TL mRNA at 1 and expressed the activity of the Luc-SL and Luc-polyA mRNAs relative to the activity of the Luc-TL mRNA.
There is some free xSLBP1 and xSLBP2 present in the cytoplasm of Xenopus oocytes, as assayed by mobility shift assay, with a larger amount of free xSLBP1 (64). Thus, when the reporter mRNAs are injected into oocytes without expression of exogenous SLBPs, the activity measured is affected by the presence of the endogenous SLBPs. Figure 3C shows a time course of the relative expression of the reporter mRNAs following injection at T1 into the cytoplasm of oocytes that had previously been injected at T0 with buffer (top), xSLBP1 mRNA (middle), or xSLBP2 mRNA (bottom). In oocytes injected with buffer, translation of Luc-SL mRNA was two- to threefold greater than translation of Luc-TL mRNA (Fig. 3C, top). The Luc-polyA mRNA was translated two to three times better than the Luc-SL mRNA and six- to eightfold better than the Luc-TL mRNA. This stimulation of translation of the Luc-SL mRNA is likely due to the endogenous xSLBP1 in stage VI oocytes, and the translation of the Luc-polyA mRNA is probably activated by embryonic PABP (63). When xSLBP1 was overexpressed in the oocytes prior to injection of the reporter mRNAs, the translation of the Luc-SL mRNA was stimulated to a level similar to that observed with the Luc-polyA mRNA, increasing more than 15-fold at 16 h compared to the translation of the Luc-TL mRNA in this batch of oocytes (Fig. 3C, middle). Note that what is shown in Fig. 3C is relative luciferase activity. The absolute amount of activity of both luciferases increased at each time. In contrast, when xSLBP2 protein was overexpressed, the Luc-SL and Luc-TL mRNAs were translated to the same extent (Fig. 3C, bottom), while translation of the Luc-polyA mRNA was not affected (data not shown). The reduction in translation of the Luc-SL mRNA likely reflects the ability of overexpressed xSLBP2 to compete with the endogenous pool of xSLBP1 for the injected Luc-SL mRNA.
To rule out the possibility that these results were due to different stabilities of the reporter mRNAs, we measured the levels of the reporter mRNAs with an S1 nuclease protection assay (see Fig. 7D, lanes 1 and 2 and 13 and 14). These results demonstrate that xSLBP1 can activate translation of a capped reporter mRNA that ends in the histone stem-loop in frog oocytes. In the presence of overexpressed xSLBP1, the translational efficiency of Luc-SL is similar to the translational efficiency mediated by a poly(A) tail (Fig. 3C, middle), suggesting that, in frog oocytes, xSLBP1 is as effective in activating translation as is PABP.
The N-terminal domain of xSLBP1 is required for activation of translation.
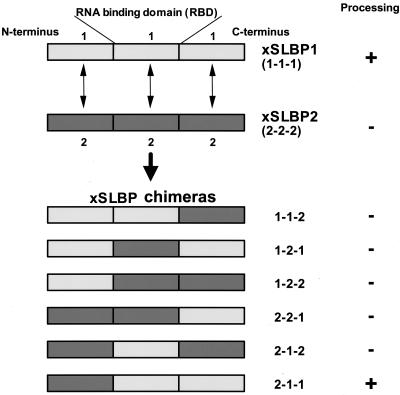
The two Xenopus SLBPs can be arbitrarily divided into three domains: the RBD and the flanking N- and C-terminal domains (Fig. 4). The Xenopus SLBPs are only similar in the RBD, which is located in the center of each protein (64). These domains were exchanged between the frog SLBPs to generate six chimeras that bind the histone stem-loop with similar affinities (26) (unpublished results). To localize the region of xSLBP1 required for translation, we tested the abilities of these chimeric proteins to stimulate translation of the Luc-SL mRNA in vitro. A similar approach previously allowed us to determine the regions of xSLBP1 required for pre-mRNA processing (26).
FIG. 4.
Xenopus xSLBPs and the chimeric proteins. xSLBP1 and xSLBP2 were arbitrarily divided into three domains, the central 73-amino-acid RBD and the N- and C-terminal domains. These domains were interchanged to give the six chimeric proteins shown at the bottom. The activity of the proteins in processing histone pre-mRNA in vivo and in vitro is indicated (26).
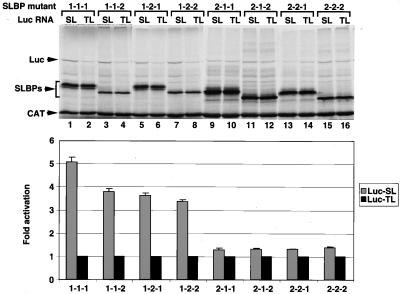
The various xSLBP chimeras were synthesized in the RRL, and the lysates containing these proteins was added to fresh lysate together with the CAT mRNA competitor and either Luc-TL or Luc-SL reporter mRNA as described in Fig. 1A. As previously shown (Fig. 1C), xSLBP1 stimulated translation of the Luc-SL mRNA (Fig. 5, lanes 1 and 2) while xSLBP2 did not (Fig. 5, lanes 15 and 16). The three chimeras that contained the amino-terminal portion of xSLBP1 (1-1-2, 1-2-1, and 1-2-2) stimulated translation of the Luc-SL mRNA to similar extents (Fig. 5, lanes 3 to 8). Those chimeras containing the amino-terminal portion of xSLBP2 (2-1-1, 2-1-2, and 2-2-1) had no effect on the translation of the Luc-SL mRNA (Fig. 5, lanes 9 to 14). Similar results were obtained both by quantifying luciferase protein by PhosphorImager or luciferase activity by luminometry (Fig. 5, bottom). We conclude that the N-terminal region of xSLBP1 contains the region required for translational activation in vitro. Similar results were obtained when these chimeras were analyzed in vivo (data not shown).
FIG. 5.
Assay of the effects of xSLBP chimeras on translation in vitro. Lysates containing the xSLBP chimeras shown in Fig. 4 were incubated with the standard polyadenylated CAT mRNA and the Luc-TL or Luc-SL mRNA and assayed for luciferase activity. The reaction mixtures were analyzed by gel electrophoresis, and the proteins were detected by autoradiography (top). Luciferase activity was assayed by luminometry, and the fold activation was expressed as the ratio of Luc-SL activity to Luc-TL activity (bottom). The bar graph shows the averages of two experiments with two different batches of RRL, and the error bars represent the standard deviations.
Localization of the translation activation domain in the amino-terminal region of xSLBP1.
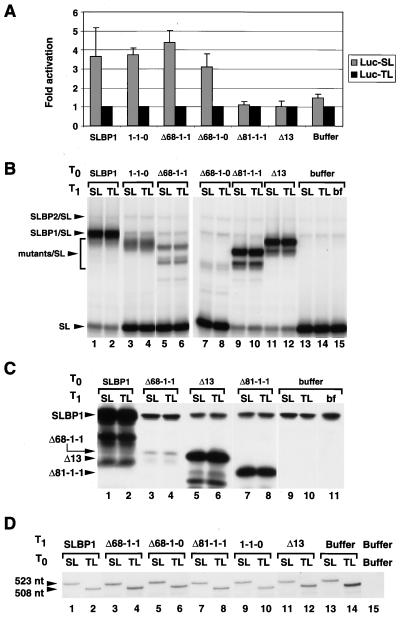
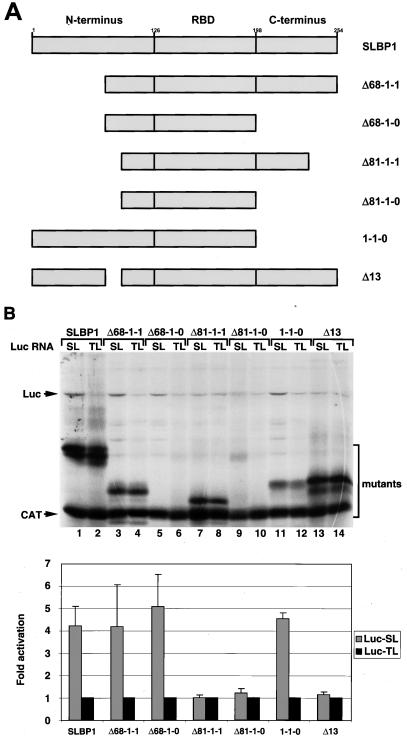
To demarcate the boundaries of the xSLBP1 translation activation domain and define a minimal protein required for activation of translation, we generated a number of deletions in the N- and C-terminal regions of xSLBP1 (Fig. 6A). We tested the ability of each deletion mutant to stimulate translation of the Luc-SL mRNA both in vitro and in vivo. A protein missing the first 68 amino acids (Δ68-1-1) was as effective as full-length xSLBP1 in stimulating translation in vitro (Fig. 6B, lanes 1 to 4). Deletion of 13 additional amino acids (Δ81-1-1) resulted in a loss of translation activation (Fig. 6B, lanes 7 and 8). Deletion of the entire C-terminal portion of xSLBP1 (1-1-0 and Δ68-1-0) had no effect on activation of translation (Fig. 6B, compare lanes 3 and 4 with lanes 5 and 6, lanes 7 and 8 with lanes 9 and 10, and lanes 1 and 2 with lanes 11 and 12).
FIG. 6.
Effect of xSLBP1 deletion mutant forms on translation activation in vitro. (A) Deletions in the amino- and carboxy-terminal regions of xSLBP1. (B) Lysates containing the xSLBP1 deletion mutant forms shown in panel A were incubated with the standard polyadenylated CAT mRNA and the Luc-TL or Luc-SL mRNA. Protein synthesis (top) and luciferase activity (bottom) were assayed as described in Fig. 1C and D, respectively. The bar graph shows the averages of two experiments with two different batches of RRL, and the error bars represent the standard deviations.
We tested the same deletion mutants in vivo with essentially similar results (Fig. 7A). Mutant forms 1-1-0, Δ68-1-1, and Δ68-1-0 stimulated translation in vivo to extents similar to that of xSLBP1, whereas the Δ81-1-1 deletion was inactive. We assayed for the presence of these proteins in Xenopus oocytes by measuring RNA binding activity (Fig. 7B) and, when possible, by Western blotting (Fig. 7C). Mutant forms Δ68-1-0 and 1-1-0 are not detectable by Western blotting because the antibody against xSLBP1 recognizes only the C-terminal portion of the protein. The Δ81-1-1 mutant form was clearly overexpressed (Fig. 7B, lanes 9 and 10, and C, lanes 7 and 8;) in the oocytes. The amounts of the Δ68-1-1 and Δ68-1-0 proteins active in stem-loop binding were greater than the store of endogenous xSLBPs (Fig. 7B, lanes 5 to 8 versus lane 15), although they were overexpressed to lesser extents than the other proteins. The 131-amino-acid mutant form Δ68-1-0 represents the smallest xSLBP1 protein we tested that is capable of activating translation of a reporter gene ending in the stem-loop.
The data provided by the deletion mutants suggested that the critical region for translation activation is located between amino acids 69 and 81. To confirm the importance of these residues, we deleted amino acids 69 to 81 from the full-length protein (Fig. 6A, Δ13). The Δ13 mutant did not stimulate translation of the Luc-SL mRNA in vitro (Fig. 6B, lanes 13 and 14) or in vivo (Fig. 7A). The Δ13 protein was clearly overexpressed in the oocytes, as shown by both mobility shift assay (Fig. 7B, lanes 11 and 12) and Western blotting (Fig. 7C, lanes 5 and 6). Note that the deletion of 13 amino acids caused a major change in the mobility of the protein on SDS-polyacrylamide gels both after synthesis in vitro (Fig. 6B, lanes 13 and 14) and after synthesis in vivo (Fig. 7C, lanes 5 and 6). xSLBP1 has aberrant mobility on SDS-PAGE (apparent molecular mass of 45 kDa for a 31-kDa protein) (64). Deletion of these 13 amino acids resulted in mobility similar to that expected for the 30-kDa Δ13 protein.
The amount of the Luc-SL and Luc-TL mRNAs present in the oocytes after the incubation was determined by an S1 nuclease protection assay (Fig. 7D). A probe labeled in the coding region of the Luc-SL gene is protected by the Luc-SL mRNA to the 3′ end of the mRNA and by the Luc-TL mRNA to the place in the 3′ UTR where the Luc-TL sequence differs from the Luc-SL sequence. Since there is a single labeled nucleotide in each probe, the intensity of the protected fragment is proportional to the amount of RNA present in the sample. The amounts of both mRNAs were similar in the oocytes injected with each of the mutant SLBPs, demonstrating that the differences in translation efficiency were not due to variations in reporter mRNA stability.
Amino acids 70 to 84 are essential for translation.
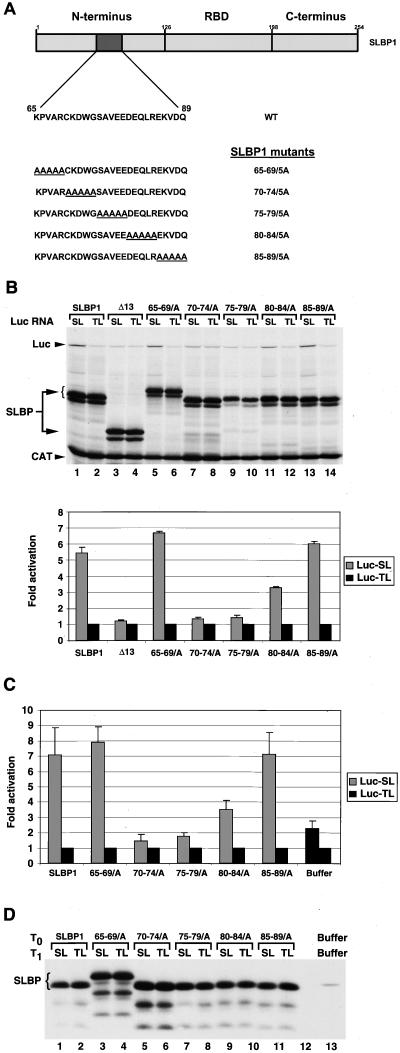
To precisely define the amino acids in xSLBP1 required for translation activation, we generated alanine scanning mutations in each of which a stretch of five amino acids was replaced with alanines spanning residues 65 to 89 (Fig. 8A), leaving the rest of xSLBP1 intact. The effects of these mutant forms on translation activation were analyzed with both in vitro (Fig. 8B) and in vivo (Fig. 8C) assays. All of the mutant forms were expressed to similar extents in vitro (Fig. 8B) and in vivo (Fig. 8D). Note that one of the mutant proteins, SLBP65-69/5A, when expressed both in vitro (Fig. 8B, lanes 5 and 6) and in vivo (Fig. 8D, lanes 3 and 4), has slower mobility than full-length SLBP. The xSLBP65-69/5A and xSLBP85-89/5A mutant proteins, which have mutations outside the region deleted in the Δ13 mutant protein, had translation activity similar to that of wild-type xSLBP1 in vitro (Fig. 8B, lanes 5 and 6 and 13 and 14) and in vivo (Fig. 8C). Luciferase synthesis was increased six- to sevenfold in these experiments from the Luc-SL mRNA compared to the Luc-TL mRNA. The xSLBP70-74/5A and xSLBP75-79/5A mutant proteins each were inactive in translation in vitro (Fig. 8B, lanes 7 to 10) and in vivo (Fig. 8C). Expression of xSLBP70-74/5A and xSLBP75-79/5A in oocytes reduced translation of the Luc-SL mRNA, consistent with these mutant proteins competing with the endogenous xSLBP1 protein for binding of the Luc-SL mRNA but being unable to activate translation. The xSLBP80-84/5A mutant protein has about 50% of the activity of wild-type xSLBP1 in activation of translation in vitro (Fig. 8B, lanes 11 and 12) and in vivo (Fig. 8C). Thus, the 10-amino-acid region of xSLBP1 between amino acids 70 and 80 is essential for the activation of translation of Luc-SL mRNA in both assays.
FIG. 8.
Alanine scanning of the xSLBP1 translation activation domain. (A) Amino acids 65 to 89 in the N-terminal region of xSLBP1 were replaced sequentially, five residues at a time, with five alanines (underlined). WT, wild type. (B) Lysates containing the five-alanine substitutions shown in panel A were incubated with the standard polyadenylated CAT mRNA and the Luc-TL or Luc-SL mRNA. Protein synthesis (top) and luciferase activity (bottom) were assayed as described in Fig. 1C and D, respectively. The bar graph shows the averages of two experiments with two different batches of RRL, and the error bars represent the standard deviations. (C) Oocytes injected at T0 with the xSLBP1 five-alanine substitution mRNAs and then again at T1 = 16 h with Luc-SL or Luc-TL mRNA in combination with R-Luc-SL mRNA were harvested at T2 = 32 h, processed, and analyzed for luciferase activity as described in Fig. 1C. The averages of two experiments with two different batches of oocytes are shown. The error bars represent the standard deviations. (D) A fraction corresponding to one oocyte worth of the same lysate used for the luciferase assay of panel C was used for detection of the xSLBP1 five-alanine substitution mutant proteins. The Western blot shown was performed as described previously. Oocytes injected at T0 and T1 with buffer show the level of endogenous xSLBP1 protein (lane 13).
The MS2-SLBP fusion protein activates translation of Luc-MS2 mRNA.
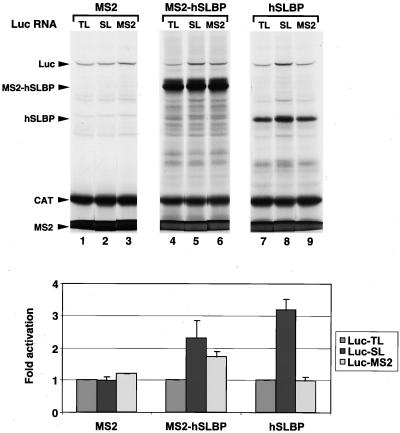
To determine whether SLBP has to be bound to the histone stem-loop or simply physically associated with the mRNA to activate translation, we constructed a fusion protein that had the entire hSLBP fused to the MS2 protein (MS2-hSLBP). We also constructed a luciferase reporter gene that had a binding site for the MS2 protein at the 3′ end of the mRNA. We expressed the MS2-hSLBP, the hSLBP, and the MS2 protein in the reticulocyte lysate and tested each for the ability to activate translation of the Luc-MS2, Luc-SL, or Luc-TL mRNA (Fig. 9). The MS2-hSLBP fusion protein activated translation of both the Luc-MS2 and Luc-SL mRNAs 1.5- to 2-fold (range of three independent experiments), whereas it had no effect on the translation efficiency of the Luc-TL mRNA. The fusion of the MS2 protein to the hSLBP reduced its activity on the Luc-SL mRNA, and the fusion protein was equally active on the Luc-SL and Luc-MS2 mRNAs. Not surprisingly, in vitro-synthesized hSLBP failed to affect translation of the Luc-TL and Luc-MS2 mRNAs, although it activated Luc-SL mRNA translation nearly threefold. Control protein MS2 failed to activate translation of any of the reporter mRNAs tested (Fig. 9, lanes 1 to 3). Thus, the MS2-hSLBP can activate translation even though the SLBP is not directly bound to the mRNA. The reduced activity of this hybrid protein compared with that of the hSLBP, observed both on the Luc-SL and Luc-MS2 mRNAs, is likely due to the reduced activity of the hybrid protein in activating translation. Most likely, this is a nonspecific effect of the fusion of the MS2 protein to the SLBP that may well cause steric problems in interactions with the translation initiation machinery. We have no evidence that the SLBP RBD-stem-loop complex plays a direct role in translation. When we expressed the SLBP-MS2 fusion in oocytes, we also saw a lower, but equivalent, activity of this protein in activation of translation of the capped Luc-MS2 and Luc-SL transcripts (data not shown). Thus, the translation activation region of SLBP must be localized to the mRNA to mediate activation of translation (Fig. 3A and 9, lanes 7 and 9 versus lane 8).
FIG. 9.
Translation activation by MS2-hSLBP fusion protein in vitro. Lysates containing MS2 (lanes 1 to 3), MS2-hSLBP fusion protein (lanes 4 to 6), or hSLBP (lanes 7 to 9) were incubated with the standard polyadenylated CAT mRNA and the Luc-TL, Luc-SL, or Luc-MS2 mRNA. Protein synthesis (top) and luciferase activity (bottom) were assayed as described in Fig. 3C and D, respectively. The bar graph shows the averages of three experiments with two different batches of RRL, and the error bars represent the standard deviations.
DISCUSSION
There are two classes of cellular mRNAs in metazoans that are distinguished by their 3′ ends. The bulk of the mRNAs end in poly(A) tails, while the mRNAs from the replication-dependent histone genes, the products of about 70 genes in mice (65) and humans (2), lack poly(A) tails but end in a conserved stem-loop. The stem-loop sequence has been conserved in the histone mRNAs in all metazoans.
Translation of polyadenylated mRNAs in eukaryotes requires both the 5′ end, which contains the 7-methyl-G cap, and the poly(A) tail (29, 58, 60). In the competition for limiting translation initiation factors in the cell (31, 51), it is necessary for the cap to be recognized by eIF-4E, which in turn binds to eIF-4G, and for the poly(A) tail-bound PABP to interact with eIF-4G, effectively circularizing the mRNA (58, 68). In this paper, we provide evidence that efficient translation of histone mRNA may utilize a similar mechanism; the xSLBP1 bound to the stem-loop at the 3′ end of the histone mRNA, a functional homologue of the poly(A) tail, is essential for a high rate of translation.
Control of gene activity during early embryogenesis requires translation regulation of mRNAs that are synthesized and stored in the oocyte (10, 30, 32, 53, 54, 57). This is particularly true of organisms like Xenopus and Drosophila, where there is no transcription in the embryo until after the midblastula transition (41, 42, 73). Histone mRNAs accumulate early in oogenesis and are stored for the remainder of oogenesis (72); translation is activated during oocyte maturation (1, 71). Oocyte-specific xSLBP2 plays a major role in the storage of the histone mRNA in a silent form (64) and probably prevents xSLBP1 (which accumulates later in oogenesis) from activating translation of histone mRNA prior to oocyte maturation.
xSLBP1 bound to the 3′ end of histone mRNA stimulates translation.
To demonstrate a role for SLBP and the stem-loop in translation in vitro, it was necessary to establish a system in which the added mRNAs were competing for initiation factors (49) and the 3′ end of the mRNA could contribute to the translation activity. To do this, we used uncapped reporter mRNAs, since there is a large amount of eIF-4E activity in the reticulocyte lysates and capped mRNAs that are not polyadenylated are translated well because of the cap (6, 24, 38, 49). We also included an excess of competitor mRNA, a CAT mRNA that was uncapped but was polyadenylated. Since the reticulocyte lysate does not contain significant amounts of SLBP as assayed by mobility shift assays (66), we were able to develop a translation system that was dependent on exogenous SLBP. Since SLBP synthesized by in vitro translation is highly active in RNA binding and the recombinant baculovirus SLBPs vary in activity and solubility (unpublished results), we expressed the SLBP by translating it in the lysate prior to addition of the reporter mRNAs.
The 3′ end of the mRNA is important for translation under these conditions. The poly(A) tail had a significant effect on translation (Fig. 2D) as a result of the large amounts of PABP present in the lysate, as has been previously reported for the reticulocyte lysate under competitive conditions (6, 28, 40). The translation of the Luc-SL mRNA was specifically stimulated by the addition of either xSLBP1 (Fig. 1D) or hSLBP (Fig. 2A). Neither protein affected translation of an equally stable reporter mRNA that ended in a stem-loop structure that does not bind SLBP (Luc-TL).
SLBP enhances translation of histone mRNA via a novel domain.
In the heterologous RRL, xSLBP1 activated translation while xSLBP2 had no effect on translation of reporter mRNAs ending in the stem-loop. We used this assay to determine the precise region of xSLBP1 required for translation in the reticulocyte lysate. The 15 amino acids in the N-terminal region of xSLBP1 required for translation are identical in xSLBP1 and the mammalian SLBPs, consistent with this activation being physiologically important in mammalian cells. There is not a similar sequence in the Drosophila (55) or C. elegans SLBP (33, 37), and the Drosophila SLBP is not active in translation of the Luc-SL mRNA in the reticulocyte lysate (unpublished results).
One of the limitations of the in vitro assay is that it measures the effect of xSLBP1 on translation of uncapped mRNAs. We developed an in vivo assay to demonstrate that xSLBP1 can activate translation of capped reporter mRNAs in frog oocytes. This assay is complicated by the presence of some xSLBP1 in the frog oocyte that is capable of binding exogenous mRNA. In oocytes that do not express exogenous SLBP, the luciferase mRNA ending in the stem-loop (Luc-SL) was translated more efficiently than the Luc-TL mRNA that does not bind SLBP (Fig. 3C, top). Again, the Luc-polyA mRNA was translated better than the other reporter mRNAs, probably because of the large amount of free embryonic PABP in the oocyte (63). Overexpression of xSLBP1 in oocytes stimulated translation of the Luc-SL mRNA up to the same level as the Luc-polyA mRNA, suggesting that xSLBP1 may be a functional homologue of PABP. The same 15-amino-acid region was required for complete activation of translation in vitro and in vivo (Fig. 8B and C).
Role of xSLBP2 in histone mRNA translation activity.
The other SLBP present in frog oocytes, xSLBP2, was inactive in translation, as predicted from its proposed role in storing histone mRNA in an inactive mRNP (64). Is xSLBP2 a translational repressor, or does it simply serve to keep xSLBP1 from binding the reporter mRNA? It is not possible to definitively answer this question on the basis of the data presented here. However, it is clear that xSLBP2 had no effect on the basal translation of Luc-SL mRNA in the reticulocyte lysate (Fig. 2D). Since this is a heterologous artificial system, any inhibitor activity might not be observed, particularly if translational silencing requires other oocyte-specific factors. Expression of xSLBP2 does reduce the translation of the Luc-SL mRNA in oocytes (Fig. 3C). However, this effect could be explained by the exogenous xSLBP2 competing with endogenous xSLBP1 for binding to the Luc-SL mRNA. The reduction observed is to the same “basal” level of translation observed with the Luc-TL mRNA, which does not bind either xSLBP. In addition, expression of xSLBP1 mutant proteins inactive in translation (the Δ13 and Δ81-1-1 proteins or some of the alanine scanning mutants) also reduced the translation of the Luc-SL mRNA, consistent with xSLBP2 being a passive “masker” of translation in both of these assays.
It is certainly possible that storage of histone mRNA in a translationally inactive form might require factors other than xSLBP2. Repression of translation of other oocyte mRNAs requires the FRGY2 protein (36), and the stored inactive mRNP is assembled in a process that depends on the coupling of transcription and processing of the mRNA (7). There are many endogenous mRNAs that are not translated efficiently in frog oocytes but, instead, are stored in an inactive mRNP and contain short poly(A) tails (13). Translation of these mRNAs may be actively inhibited by a complex bound to the cytoplasmic polyadenylation element, consisting of cytoplasmic polyadenylation element B and maskin (21, 53), as well as other general mRNA binding proteins (20, 53). In this complex, maskin competes with eIF-4G for binding to eIF-4E, ultimately preventing the eIF3-associated 40S ribosomal subunit from binding to the mRNA.
Mechanisms for activation of histone mRNA translation.
It is possible that the stored histone mRNA is silenced by other factors in addition to xSLBP2. However, it is likely that translational activation of histone mRNA requires the recruitment of xSLBP1 to the mRNA. In addition to exchange of the xSLBPs, the histone mRNAs undergo other structural changes during oogenesis, the addition of a short poly(A) tail early in oogenesis and its subsequent removal at oocyte maturation (3). This modification may also play a role in translational control in vivo.
The role of the 3′ end in translation initiation of polyadenylated mRNAs and in rotavirus mRNAs is reasonably well understood. PABP binds to eIF-4G through a region comprising RRM1 and RRM2 in the N-terminal domain of PABP (25, 29), resulting in circularization of the mRNA. This event promotes formation of the preinitiation complex, as well as recycling of ribosomes from the termination site (6, 27, 67). Rotavirus mRNAs end in a consensus sequence that is bound by the NSP3 viral protein (48). The NSP3 protein interacts with the 40-amino-acid PABP binding domain in eIF-4G, with a motif that has no obvious similarity to the eIF-4G binding domain in PABP (47, 62), although it binds to the same site (19). There is no similarity between the sequence required for translation activation by SLBP and the eIF-4G binding sequences in either PABP or NSP3. Further data are required to determine whether SLBP promotes translation by interacting directly with the translation initiation complex, possibly by binding eIF-4G. Alternatively, SLBP may play a pivotal role in recruiting other factors required for activation of translation via mRNA circularization.
Acknowledgments
We thank Tom Ingledue for many of the chimeric SLBP constructs, Zbigniew Dominski and Bob Duronio for critical reading of the manuscript, and Handan Kaygun for providing the MS2-hSLBP fusion construct.
This work was supported by National Institutes of Health grant GM58921 to W.F.M.
REFERENCES
- 1.Adamson, E. D., and H. R. Woodland. 1977. Changes in the rate of histone synthesis during oocyte maturation and very early development of Xenopus laevis. Dev. Biol. 57:136-149. [DOI] [PubMed] [Google Scholar]
- 2.Albig, W., and D. Doenecke. 1997. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101:284-294. [DOI] [PubMed] [Google Scholar]
- 3.Ballantine, J. E., and H. R. Woodland. 1985. Polyadenylation of histone mRNA in Xenopus oocytes and embryos. FEBS Lett. 180:224-228. [DOI] [PubMed] [Google Scholar]
- 4.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81:179-183. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, P., S. W. Peltz, and J. Ross. 1989. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 9:659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., Y. M. Michel, and K. M. Kean. 2000. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 28:4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet, P., and A. P. Wolffe. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77:931-942. [DOI] [PubMed] [Google Scholar]
- 8.Castagnetti, S., M. W. Hentze, A. Ephrussi, and F. Gebauer. 2000. Control of oskar mRNA translation by Bruno in a novel cell-free system from Drosophila ovaries. Development 127:1063-1068. [DOI] [PubMed] [Google Scholar]
- 9.Dahanukar, A., J. A. Walker, and R. P. Wharton. 1999. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4:209-218. [DOI] [PubMed] [Google Scholar]
- 10.De Moor, C. H., and J. D. Richter. 1999. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 18:2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dilworth, S. M., S. J. Black, and R. A. Laskey. 1987. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell 51:1009-1018. [DOI] [PubMed] [Google Scholar]
- 12.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol. 21:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dworkin, M. B., and E. Dworkin-Rastl. 1985. Changes in RNA titers and polyadenylation during oogenesis and oocyte maturation in Xenopus laevis. Dev. Biol. 112:451-457. [DOI] [PubMed] [Google Scholar]
- 14.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 15.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24:1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gick, O., A. Krämer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 18.Graves, R. A., S. E. Wellman, I.-M. Chiu, and W. F. Marzluff. 1985. Differential expression of two clusters of mouse histone genes. J. Mol. Biol. 183:179-194. [DOI] [PubMed] [Google Scholar]
- 19.Groft, C. M., and S. K. Burley. 2002. Recognition of eIF-4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 9:1273-1283. [DOI] [PubMed] [Google Scholar]
- 20.Groisman, I., Y. S. Huang, R. Mendez, Q. P. Cao, W. Theurkauf, and J. D. Richter. 2000. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103:435-447. [DOI] [PubMed] [Google Scholar]
- 21.Hake, L. E., and J. D. Richter. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79:617-627. [DOI] [PubMed] [Google Scholar]
- 22.Harris, M. E., R. Böhni, M. H. Schneiderman, L. Ramamurthy, D. Schümperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentze, M. W. 1997. Translation—elF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 24.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingledue, T. C., Z. Dominski, R. Sanchez, J. A. Erkmann, and W. F. Marzluff. 2000. Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA 6:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson, A. 1996. PolyA metabolism and translation: the closed loop model, p. 451-480. In J. W. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Jacobson, A., and M. Favreau. 1983. Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 11:6353-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler, S. H., and A. B. Sachs. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khaleghpour, K., Y. V. Svitkin, A. W. Craig, C. T. DeMaria, R. C. Deo, S. K. Burley, and N. Sonenberg. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7:205-216. [DOI] [PubMed] [Google Scholar]
- 31.Laskey, R. A., A. D. Mills, J. B. Gurdon, and G. A. Partington. 1977. Protein synthesis in oocytes of Xenopus laevis is not regulated by the supply of messenger RNA. Cell 1:345-351. [DOI] [PubMed] [Google Scholar]
- 32.Marello, K., J. LaRovere, and J. Sommerville. 1992. Binding of Xenopus oocyte masking proteins to mRNA sequences. Nucleic Acids Res. 20:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, F., F. Michel, D. Zenklusen, B. Müller, and D. Schümperli. 2000. Positive and negative mutant selection in the human histone hairpin-binding protein using the yeast three-hybrid system. Nucleic Acids Res. 28:1594-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, F., A. Schaller, S. Eglite, D. Schümperli, and B. Müller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto, K., F. Meric, and A. P. Wolffe. 1996. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA—role of the cold shock domain, tail domain, and selective RNA sequence recognition. J. Biol. Chem. 271:22706-22712. [DOI] [PubMed] [Google Scholar]
- 37.Michel, F., D. Schümperli, and B. Muller. 2000. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA 6:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-poly(A) synergy in mammalian cell-free extracts: investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268-32276. [DOI] [PubMed] [Google Scholar]
- 39.Minshall, N., G. Thom, and N. Standart. 2001. A conserved role of a DEAD box helicase in mRNA masking. RNA 7:1728-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munroe, D., and A. Jacobson. 1990. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10:3441-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newport, J., and M. A. Kirschner. 1982. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 3:687-696. [DOI] [PubMed] [Google Scholar]
- 42.Newport, J., and M. W. Kirschner. 1982. A major developmental transition in early Xenopus embryos. I. Characterization and timing of cellular changes at the midblastula stage. Cell 30:675-686. [DOI] [PubMed] [Google Scholar]
- 43.Ostareck, D. H., A. Ostareck-Lederer, M. Wilm, B. J. Thiele, M. Mann, and M. W. Hentze. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89:597-606. [DOI] [PubMed] [Google Scholar]
- 44.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey, N. B., J.-H. Sun, and W. F. Marzluff. 1991. Different complexes are formed on the 3′ end of histone mRNA in nuclear and polysomal extracts. Nucleic Acids Res. 19:5653-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettitt, J., C. Crombie, D. Schümperli, and B. Müller. 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 115:857-866. [DOI] [PubMed] [Google Scholar]
- 47.Piron, M., T. Delaunay, J. Grosclaude, and D. Poncet. 1999. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 73:5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poncet, D., C. Aponte, and J. Cohen. 1993. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J. Virol. 67:3159-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 50.Puoti, A., M. Gallegos, B. Zhang, M. P. Wickens, and J. Kimble. 1997. Controls of cell fate and pattern by 3′ untranslated regions: the Caenorhabditis elegans sperm/oocyte decision. Cold Spring Harbor Symp. Quant. Biol. 62:19-24. [PubMed] [Google Scholar]
- 51.Richter, J. D., and L. D. Smith. 1981. Differential capacity for translation and lack of competition between mRNAs that segregate to free and membrane-bound polysomes. Cell 27:183-191. [DOI] [PubMed] [Google Scholar]
- 52.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 53.Stebbins-Boaz, B., Q. Cao, C. H. De Moor, R. Mendez, and J. D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 4:1017-1027. [DOI] [PubMed] [Google Scholar]
- 54.Stutz, A., B. Conne, J. Huarte, P. Gubler, V. Völkel, P. Flandin, and J. D. Vassalli. 1998. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 12:2535-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, J.-H., D. R. Pilch, and W. F. Marzluff. 1992. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 20:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tafuri, S. R., and A. P. Wolffe. 1993. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4). J. Biol. Chem. 268:24255-24261. [PubMed] [Google Scholar]
- 58.Tarun, S. Z., Jr., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 59.Tarun, S. Z., Jr., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 60.Tarun, S. Z., Jr., S. E. Wells, J. A. Deardorff, and A. B. Sachs. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94:9046-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 62.Vende, P., M. Piron, N. Castagné, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voeltz, G. K., J. Ongkasuwan, N. Standart, and J. A. Steitz. 2001. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 15:774-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Z.-F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, Z.-F., T. Krasikov, M. R. Frey, J. Wang, A. G. Matera, and W. F. Marzluff. 1996. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over one megabase. Genome Res. 6:688-701. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 67.Wei, C. C., M. L. Balasta, J. Ren, and D. J. Goss. 1998. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37:1910-1916. [DOI] [PubMed] [Google Scholar]
- 68.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 69.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams, A. S., and W. F. Marzluff. 1995. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 23:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodland, H. R. 1980. Histone synthesis during the development of Xenopus. FEBS Lett. 121:1-7. [DOI] [PubMed] [Google Scholar]
- 72.Woodland, H. R., and E. D. Adamson. 1977. The synthesis and storage of histones during oogenesis of Xenopus laevis. Dev. Biol. 57:118-135. [DOI] [PubMed] [Google Scholar]
- 73.Woodland, H. R., J. M. Flynn, and A. J. Wyllie. 1979. Utilization of stored mRNA in Xenopus embryos and its replacement by newly synthesized transcripts: histone H1 synthesis using interspecies hybrids. Cell 18:165-171. [DOI] [PubMed] [Google Scholar]
- 74.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, B. L., M. Gallegos, A. Puoti, E. Durkin, S. Fields, J. Kimble, and M. P. Wickens. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390:477-484. [DOI] [PubMed] [Google Scholar]