Abstract
Human RNA-binding protein HuR, a nucleocytoplasmic shuttling protein, is a ubiquitously expressed member of the family of Hu proteins, which consist of two N-terminal RNA recognition motifs (RRM1 and RRM2), a hinge region, and a C-terminal RRM (RRM3). Although in vitro experiments showed indiscriminate binding of Hu proteins synthesized in bacterial systems to many different AU-rich elements (AREs), in vivo studies have pointed to a cytoplasmic role for HuR protein in antagonizing the rapid decay of some specific ARE-containing mRNAs, depending on physiological situations. By ectopically overexpressing HuR and its mutant derivatives in NIH 3T3 cells to mimic HuR upregulation of specific ARE-containing mRNAs in other systems, we have examined the in vivo ARE-binding specificity of HuR and dissected its functionally critical domains. We show that in NIH 3T3 cells, HuR stabilizes reporter messages containing only the c-fos ARE and not other AREs. Two distinct binding sites were identified within the c-fos ARE, the 5′ AUUUA-containing domain and the 3′ U-stretch-containing domain. These actions of HuR are markedly different from those of another ARE-binding protein, hnRNP D (also termed AUF1), which in vivo recognizes AUUUA repeats found in cytokine AREs and can exert both stabilizing and destabilizing effects. Further experiments showed that any combination of two of the three RRM domains of HuR is sufficient for strong binding to the c-fos ARE in vitro and to exert an RNA stabilization effect in vivo comparable to that of intact HuR and that the hinge region containing nucleocytoplasmic shuttling signals is dispensable for the stabilization effect of HuR. Our data suggest that the ARE-binding specificity of HuR in vivo is modulated to interact only with and thus regulate specific AREs in a cell type- and physiological state-dependent manner.
Regulation of the fate of mRNA in the cytoplasm, including RNA localization, translation, and turnover, has recently been recognized as an important control point for gene expression (43, 48). The adenylate and uridylate-rich elements (AREs) constitute one class of regulatory cis elements that influence the fate of cytoplasmic mRNA (50). AREs are the most commonly found and best-studied signals in the 3′ untranslated regions (UTRs) that target a variety of labile mRNAs for rapid degradation and in certain cases for translation blockade (8). The widespread occurrence of AREs in mRNAs coding for proteins with diverse functions points to a critical role of AREs in the regulation of many biological processes.
This view is further underscored by demonstrations that ARE-mediated mRNA decay is itself subject to regulation. For instance, upregulation of protein synthesis from ARE-containing mRNAs can be achieved by stabilization of the mRNAs under conditions of heat shock (20, 29), hypoxia (13, 31), UV irradiation (47), lipopolysaccharide or proinflammatory cytokine stimulation (32, 40, 44), oncogenic transformation (4), and cell proliferation (45). Moreover, observations (33, 39, 51) that the RNA-destabilizing functions of different AREs are differentially regulated add another layer of complexity to the mechanism(s) controlling ARE-mediated mRNA turnover.
Previously, AREs were assigned to three classes based on distinct sequence features and functional properties (reviewed in reference 8). Class I AREs, found in early-response gene mRNAs like c-fos and c-myc, contain one to three scattered copies of the pentanucleotide AUUUA embedded within U-rich regions; class II AREs, found only in cytokine mRNAs such as those for interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), and tumor necrosis factor alpha, contain multiple overlapping copies (five to eight copies) of the AUUUA motifs; and class III AREs, such as the one in c-jun mRNA, lack the hallmark AUUUA pentanucleotide but require U stretches and possibly other unknown features for their destabilizing function.
The cellular factors that bind to AREs and control their activities have been the subject of intense investigation recently. At least 14 such proteins, collectively termed ARE-binding proteins (ARE-BPs), have been identified (reviewed in references 7, 15, 33, and 49). Among them, hnRNP D (also termed AUF1), HuR (alternatively named HuA), and tristetraproline are the best-studied examples and have been demonstrated to alter the stability of ARE-containing mRNAs in vivo. Whereas tristetraproline has been found to play a role in ARE-mediated RNA destabilization (28), HuR has been identified as a trans factor that stabilizes ARE-containing mRNAs (19, 39, 45, 47). HnRNP D has dual roles in ARE-mediated mRNA decay, functioning either as a destabilizing or as a stabilizing factor depending on the cell type (33, 51). Recent in vitro studies support the idea that ARE-BPs may interact with the exosome complex to recruit it for degrading the body of the deadenylated mRNAs from the 3′ end to the 5′ end (10, 37).
HuR is a ubiquitously expressed member of the human ELAV family of RNA-binding proteins (36). The expression of three other ELAV members, including HuB, HuC, and HuD, is neuron specific (2). All four ELAV family proteins exhibit high sequence and structural similarity, and all contain three RNA recognition motifs (RRMs) (7). Between RRM2 and RRM3 is a less-conserved hinge region of ≈50 to 80 amino acids that was shown in the case of HuR to contain signals for nucleocytoplasmic shuttling (18). Many studies have indicated that ELAV family proteins upregulate gene expression by stabilizing ARE-containing mRNAs in vivo (reviewed in references 2, 7, and 26). The finding that expression of antisense RNA to HuR increases the decay of a specific ARE-containing mRNA is consistent with HuR's acting as an RNA-stabilizing factor (45, 47). The physiological significance of RNA stabilization by HuR has also been demonstrated by several studies that link HuR upregulation to cell growth and differentiation (reviewed in references 2 and 7).
In vitro binding studies with bacterially made ARE-BPs, e.g., HuR and AUF1 (hnRNP D), indicated high binding affinity for various AREs (5, 16, 17, 35, 36). In addition, recombinant ELAV proteins Hel-N1 and Hel-N2 were also shown to bind to a subset of polyadenylated mRNAs that contain an ARE (21, 22, 30). The in vivo implications of these studies are that ARE-BPs exert their function through rather indiscriminate direct binding to various AREs in vivo, which raises an issue concerning how various AREs can be regulated by these ARE-BPs in vivo.
However, several recent reports suggest that ARE-BPs may be modulated to recognize only a subset of AREs in given physiological conditions or a given cell type. First, we have shown that the in vivo actions on AREs of ectopically expressed hnRNP D are selective (51), in contrast to the indiscriminate binding of recombinant hnRNP D observed in vitro. We found that hnRNP D discriminates among the three classes of AREs, most effectively blocking rapid decay directed by class II AREs. Further experiments identified the overlapping AUUUA motifs, one critical characteristic of class II AREs, as the key feature recognized in vivo by hnRNP D for its stabilization effect. Second, upregulation of a specific Hu protein correlates well with stabilization of only a subset of ARE-containing mRNAs under various physiological conditions (2, 3, 25, 26). Third, Brennan et al. (6) showed that only c-fos mRNA is retained in the nucleus after leptomycin B treatment of cells, which results in increased HuR retention in the nucleus.
Given the wide spectrum of processes in which ELAV proteins have a regulatory role, it is critical to investigate the protein domains of HuR that are necessary for its in vivo specificity. Some initial studies reached conflicting results concerning the contribution of structurally distinct domains of HuR to its function and binding specificity. Ma et al. (35) showed that RRM3 is capable of binding poly(A) in a filter-binding assay. However, in NIH 3T3 cells, HuR protects the body of mRNA from degradation instead of affecting poly(A) shortening (39). In addition, Fan and Steitz (19) suggested that RRM3 is necessary for HuR's stabilization effect, because its deletion slightly impedes HuR's ability to stabilize ARE-containing mRNA. In vitro RNA-binding results with recombinant HuD, an ELAV family protein closely related to HuR, indicated that while individual RRMs bind poorly to the ARE, a combination of RRM1 and either RRM2 or RRM3 allows binding with high affinity (38). On the other hand, in the case of HuC, in vitro results indicated that RRM1 is the major RNA-binding determinant (1).
To further our understanding of the in vivo binding specificity of HuR and the molecular mechanism by which HuR regulates ARE-mediated mRNA turnover, we examined the ability of HuR to discriminate between class I and class II AREs in NIH 3T3 cells, the identity of the target ARE(s), and how individual RMMs contribute to HuR's function in vivo. The results reveal several unexpected new facets of HuR's cytoplasmic role in antagonizing the rapid decay of ARE-containing mRNAs.
MATERIALS AND METHODS
Plasmid construction.
The construction of plasmids pSVα1/GAPDH, pBBB+AREc-fos, pBBB+AREI, pBBB+AREII, pBBB+ARE(II/I), pBBB+ARE(U→G), pBBB+AREc-myc, pBBB+AREGM-CSF, pBBB+AREGM3, pBBB+AREIL-3, pBFB, pTet-Myc-Ovep, pTet-Myc-HuR, pTet-Myc p37 (for hnRNP D), pT3AREfos (for transcribing c-fos ARE), pT3ARE5′ (for transcribing c-fos ARE domain I), and pT3ARE3′ (for transcribing c-fos ARE domain II) has been described previously (9, 11, 12, 39, 42, 51, 54). A series of four HuR deletion mutant cDNA fragments, including deletions of RRM1 (ΔI; amino acids 20 to 104 were deleted), RRM2 (ΔII; amino acids 105 to 185 were deleted), basic hinge region (ΔB; amino acids 186 to 242 were deleted), or RRM3 (ΔIII; amino acids 243 to 326 were deleted) were created by PCR and subcloned into the unique EcoRV site in the pTet-Myc-Ovep plasmid to create plasmids pTet-Myc-HuRΔI, pTet-Myc-HuRΔII, pTet-Myc-HuRΔB, and pTet-Myc-HuRΔIII, respectively.
To create pTet-Myc-HuRIII-tr, a partial truncation of RMM3 which left 10 amino acids of RRM3 fused to the hinge region, an XbaI linker (New England Biolab) containing stop codons in all three reading frames was inserted into the unique EcoNI (blunt ended with Klenow fragment) site located within the region encoding RRM3. The proper in-frame insertion of cDNAs for the above constructs was confirmed by DNA sequencing.
RNA blot analysis and preparation of NIH 3T3 cytoplasmic and nuclear extracts.
Cell culture, DNA transfection, isolation of total cytoplasmic RNA, Northern blot analysis, and lysate preparation were conducted as described previously (34, 41). Briefly, NIH 3T3 B2A2 cells stably harboring tetracycline-controlled transactivator were transfected for ≈16 h with a total of 20 μg of DNA which included 3 μg of pBBB+ARE and/or 3.5 μg of pBFB (42), 6 μg of pTet-Myc-HuR (39), pTet-Myc hnRNP D isoform p37 (51), or vector (pTet-Myc-Ovep) (39), 2 μg of pSVα1/GAPDH (11), and enough carrier plasmid pT3/T7α-18 (Gibco-BRL) to make a final amount of 20 μg of DNA. The cells were then serum starved for 25 h, followed by stimulation with 20% bovine serum (Gibco-BRL). Total cytoplasmic RNA was extracted at intervals for time course experiments. Gene-specific DNA probes were prepared by the method of random oligonucleotide priming for Northern blot analysis. The 32P-labeled probes were produced by inclusion of [α-32P]dCTP (>6,000 Ci/mmol; DuPont).
For lysate preparations, one plate of transfected cells (10-mm culture dish) from a parallel time course experiment was harvested for preparing cytoplasmic and nuclear extracts as described previously (39). Protein concentration was analyzed with the bicinchoninic acid protein assay reagent (Pierce). These protein samples were used in Western blot analysis and gel mobility shift and supershift assays.
Western blot analysis.
Cytoplasmic and nuclear lysates were resolved on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and analyzed with an ECL Western blotting kit (Amersham, Arlington Heights, Ill.). The blots were probed with specific antibodies as described in the legend to Fig. 4. The anti-HuR monoclonal antibody 3A2 (mouse immunoglobulin G [IgG]) was kindly provided by J. A. Steitz (20). The antibody for the Myc tag was obtained by collecting culture medium from hybridoma cells (9E10; American Type Culture Collection) (14) and used at a 1:100 dilution. The purified monoclonal antibody against α-tubulin (DM1A) was purchased from Sigma and was used at a 1:20,000 dilution as a positive control for cytoplasmic protein preparations. The monoclonal antibody against U1 70K (mouse IgG) was kindly provided by Sue Berget and used at a 1:100 dilution as a positive control for nuclear protein preparations.
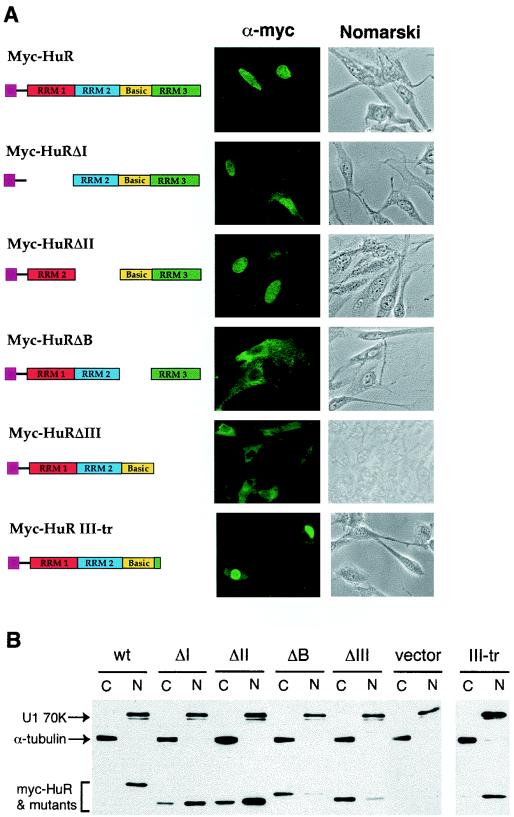
FIG. 4.
Subcellular distributions and expression levels of human HuR in mouse NIH 3T3 cells. (A) Indirect immunofluorescence microscopy study with a monoclonal antibody against the Myc epitope tag (9E10) showing the subcellular distribution of HuR and its truncated mutants (depicted by the diagram). Both phase contrast (depicted as Nomarski) and immunofluorescence (depicted as α-Myc) views are shown. (B) Western blot analysis. Cytoplasmic (C) and nuclear (N) lysates prepared from NIH 3T3 cells transfected with individual plasmids containing Myc-tagged HuR or its truncated mutant derivatives were resolved on an SDS-12% polyacrylamide gel. The blots were probed with the 9E10 monoclonal antibody for HuR and its mutant proteins, a control monoclonal antibody against α-tubulin for the cytoplasmic lysate, and a control monoclonal antibody against U1 70K for the nuclear lysate.
Immunofluorescence microscopy study.
Indirect immunofluorescence was conducted as described previously (39). Briefly, NIH 3T3 B2A2 cells were grown on coverslips and transfected with plasmids expressing individual Myc-tagged HuR, a truncated mutant HuR, or vector only. After 48 h, cells were fixed with 100% methanol and permeabilized with 0.5% Triton X-100. Cells were stained with monoclonal antibody against the Myc epitope tag (9E10) as the primary antibody. The secondary antibody for anti-mouse IgG (purchased from Sigma) was coupled to fluorescein isothiocyanate. All images were viewed and captured with a Spot-Digital camera (Diagnostics) and processed for publication at 300 dots/inch with Adobe PhotoShop (version 4.0) software.
Analysis of RNA-protein interactions.
RNA probe synthesis, gel mobility shift assays, and antibody supershift assay were carried out as described previously (39, 54). In vitro transcription with pT3AREfos (for transcribing c-fos ARE), pT3ARE5′ (for transcribing c-fos ARE domain I), and pT3ARE3′ (for transcribing c-fos ARE domain II) was carried out as described previously (54). Briefly, cytoplasmic lysate (8 μg of protein) and 32P-labeled RNA (1 ng) were incubated at room temperature for 15 min in a buffer containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 40 mM KCl, 2 mM dithiothreitol, 10% glycerol, and 0.5% IGAPEL CA-630. Heparin (5 μg/ml, final concentration) and yeast total RNA (200 ng/ml, final concentration) were added to reduce nonspecific binding. The volume of each reaction was 10 μl. Subsequently, unbound RNA was digested by including 0.6 U of RNase T1 (Calbiochem, San Diego, Calif.) for 20 min at room temperature. RNA-protein complexes were resolved in nondenaturing polyacrylamide gels.
To perform gel mobility supershift analysis, following RNA-protein binding and RNase T1 digestion, 3 μl of a monoclonal antibody against the Myc tag (9E10) (Calbiochem, San Diego, Calif.) or 2 μl of a monoclonal antibody against human HuR (a gift from J. A. Steitz) was added to the binding reaction. After a 15-min incubation at room temperature, RNA-protein-antibody complexes were resolved on 6% nondenaturing polyacrylamide gels.
Nucleotide sequence accession number.
The accession number for the nucleotide sequence and predicted open reading frame of human HuR is U38175.
RESULTS
HuR specifically targets mRNA containing the c-fos ARE for stabilization in NIH 3T3 cells.
Recent studies have indicated that an increase in ELAV-like or Hu proteins in the cytoplasm leads to stabilization of specific ARE-containing mRNAs during cell growth, differentiation, and various stress responses (reviewed in references 2, 7, and 26). Ectopic expression of HuR in NIH 3T3 cells mimics upregulation of ELAV-like proteins in other systems in which the transcription-pulsing approach for monitoring mRNA decay kinetics is not readily feasible. Thus, our transcriptional pulsing systems (34, 53) provide a way to characterize the HuR binding targets in vivo and to dissect the functionally critical domains of HuR.
Previously, we demonstrated that ectopic expression of HuR in NIH 3T3 cells impedes mRNA decay directed by the c-fos ARE, an AUUUA-containing ARE, but does not affect mRNA decay directed by the c-jun ARE, which does not contain the AUUUA motif (39). A class I ARE, the c-myc ARE, and another class II ARE, the GM-CSF ARE, were chosen to explore the possibility that HuR's stabilization role in ARE-mediated decay is AUUUA specific.
The BBB+ARE plasmid, encoding the β-globin mRNA bearing an ARE in its 3′ UTR, was transiently cotransfected with a plasmid expressing an N-terminally Myc epitope-tagged HuR. BBB+ARE mRNA was transiently transcribed from the c-fos promoter after serum induction of the growth-arrested NIH 3T3 cells. The transcription pulse from the c-fos promoter allowed determination of the mRNA decay kinetics without the complications of adding transcription inhibitors (34, 41). A control plasmid encoding a stable message, termed α-globin/glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was included in the cotransfection to serve as an internal standard for normalization of the test messages. Our results showed that decay of either BBB+AREc-myc or BBB+AREGM-CSF mRNA was hardly affected by ectopic expression of HuR, whereas decay of BBB+AREc-fos mRNA (positive control) in the cytoplasm was dramatically impeded (Fig. 1). These results demonstrated that HuR does not stabilize all ARE-containing mRNAs.
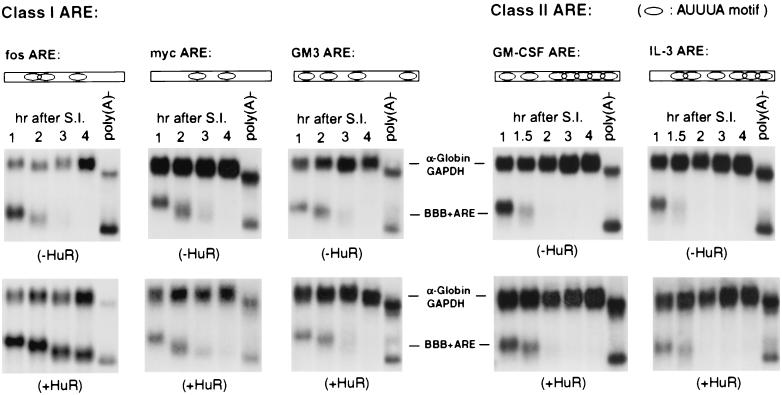
FIG. 1.
HuR selectively inhibits c-fos ARE-mediated and not other ARE-mediated mRNA decay. RNA blots showing decay of BBB+ARE mRNAs representing class I AREs (fos ARE, myc ARE, and GM3 ARE) and class II AREs (GM-CSF ARE and IL-3 ARE) in NIH 3T3 B2A2 cells expressing the control vector (−HuR, upper blots) or wild-type HuR (+HuR, lower blots). NIH 3T3 B2A2 cells were transiently cotransfected with one of the test BBB+ARE plasmids, as indicated above each blot, a control plasmid (pSVα-1/GAPDH), and either pTet-Myc-HuR or a control vector, as indicated under each blot. Total cytoplasmic mRNA was isolated at various times after serum stimulation of quiescent cells and analyzed by Northern blot analysis. Transcription of various BBB+ARE mRNAs was driven by the serum-inducible c-fos promoter. The control mRNA (α-globin/GAPDH) was expressed constitutively and served as an internal standard. The times given at the top of each blot correspond to hours after serum induction (S.I.). Nonpolyadenylated [poly(A)−] RNA was prepared in vitro by treating RNA samples from the 1-h time point with oligo(dT) and RNase H. Open rectangles and oval symbols depict AREs and AUUUA motifs, respectively.
To further test this notion, we chose two other AUUUA-containing AREs for further examination: the IL-3 ARE (representing another class II ARE) and the GM3 ARE (representing a class I ARE). The GM3 ARE is a mutant derivative of the GM-CSF ARE whose (AUUU)5, a key feature of class II AREs, has been changed to two dispersed AUUUA motifs, AUUUAUUAAUAUAAUUAUUUA, thus mimicking a class I ARE. Previously, we showed that the GM3 ARE displayed class I ARE decay kinetics, whereas the IL-3 ARE exhibited class II kinetics (52). As can be seen in Fig. 1, ectopic expression of HuR had little effect on the rapid decay of either BBB+AREGM3 or BBB+AREIL-3 mRNA. Western blot analysis showed that levels of HuR expression in the cytoplasm of the above transfections were similar (data not shown). Together, these results indicated that some specific cis-acting features in the c-fos ARE make it a target for HuR action in NIH 3T3 cells.
c-fos ARE contains two distinct HuR binding sites.
As an initial step to identifying key sequence features of the c-fos ARE that are functionally important for the in vivo stabilization effects of HuR, we performed a gel mobility shift assay. The c-fos ARE consists of two functionally distinct subregions, a 5′ AUUUA-containing core domain, designated domain I, and a 3′ U-rich domain, designated domain II (Fig. 2A). Domain I by itself functions as a potent destabilizing element, whereas domain II alone has no destabilization effect but does enhance the RNA-destabilizing function of domain I (9).
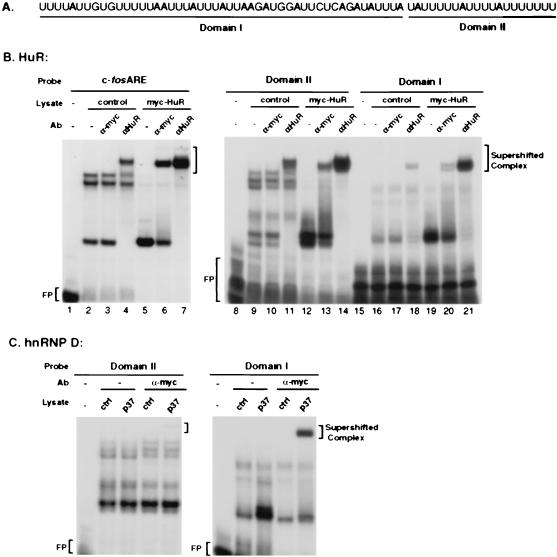
FIG. 2.
In vitro analysis of interactions between ectopically expressed human HuR or the hnRNP D p37 isoform with the c-fos ARE or its domain I or domain II. (A) RNA sequence of human c-fos ARE, with domain I and domain II underlined. (B) Gel mobility shift analysis of interactions between HuR and c-fos ARE, domain I, or domain II. (C) Gel mobility shift analysis of interactions between hnRNP D p37 isoform and c-fos ARE domain I or domain II. 32P-labeled RNA transcribed in vitro from the T3 promoter-driven c-fos ARE, domain I, or domain II DNA templates was incubated with cytoplasmic lysates from NIH 3T3 B2A2 cells expressing Myc-tagged HuR (B) or Myc-tagged hnRNP D p37 isoform (C). The RNA-protein complexes were then digested with RNase T1. The digestion mixtures were left alone (−) or further incubated in the presence of the 9E10 monoclonal antibody (Ab) against the Myc epitope tag (α-Myc) or a monoclonal antibody against human HuR (α-HuR) as indicated on the top of each lane. The final reaction mixtures were resolved by electrophoresis in a 6% native polyacrylamide gel. Antibody-supershifted complexes, Myc-HuR- and HuR-containing complexes, and free probe (FP) are indicated.
In vitro gel mobility shift assays were carried out with crude cytoplasmic extracts prepared from nontransfected NIH 3T3 cells or cells overexpressing Myc epitope-tagged HuR and three 32P-labeled RNA substrates: intact c-fos ARE, domain I, and domain II. Figure 2B shows that RNA-protein complexes were readily detected with these three RNA substrates with cytoplasmic lysate from nontransfected cells (Fig. 2B, lanes 2, 9, and 16). In contrast, when cytoplasmic lysate prepared from cells overexpressing Myc-HuR was used, a dramatic change in complex formation occurred in all three cases (Fig. 2B, compare lanes 2 and 5, 9 and 12, and 16 and 19). Ectopic overexpression of HuR led to the loss of several slow-migrating ribonucleoprotein (RNP) complexes detected in nontransfected lysates with c-fos ARE or its domain II as the probe (Fig. 2B, compare lanes 2 and 5 and 9 and 12). In all three cases, Myc-tagged HuR expression enhanced the formation of a faster-migrating complex which could be supershifted by monoclonal antibodies against either the Myc epitope tag or endogenous HuR (Fig. 2B, lanes 5, 6, and 7 for c-fos ARE; lanes 12, 13, and 14 for domain II; and lanes 19, 20, and 21 for domain I).
It appears that both Myc-HuR and endogenous HuR support the formation of RNP complexes that comigrate in these native gels with all three probes. Antibody supershift assays showed that Myc-HuR accounted for approximately 10 to 40% of the HuR-ARE RNP complexes (Fig. 2B, lanes 6, 13, and 20). In contrast, the c-jun ARE, whose destabilizing function is not affected by HuR overexpression, did not form any RNP complex with HuR that could be supershifted by anti-HuR antibody (39) (data not shown).
Together, these results showed that Myc-HuR recognizes and binds to c-fos ARE domain II and, to a lesser extent, to domain I and that endogenous HuR and ectopic Myc-HuR are able to outcompete nearly all other ARE-binding activities in the lysate for RNP complex formation (Fig. 2, compare lanes 2, 5, and 7; lanes 9, 12, and 14; and lanes 16, 19, and 21). We conclude that HuR has two distinct binding sites in the c-fos ARE, a strong binding site in domain II and a weaker binding site in domain I. This ARE-binding property of HuR is very different from that of hnRNP D, which specifically binds AREs with the AUUUA repeats found in class II AREs (51).
HuR binding to c-fos ARE domain II contributes to RNA stabilization in vivo.
What might be the significance of HuR binding to domain II of c-fos ARE, a region without AUUUA motifs? Our previous mutagenesis studies showed that the destabilizing function of the c-fos ARE results from the interplay between domains I and II, with domain I being the essential core region whose RNA-destabilizing function is enhanced by domain II (9).
The phenotypes of two of the critical mutant c-fos AREs illustrate this point. First, when c-fos domain II was placed immediately upstream of domain I, the resulting mutant ARE [fos ARE(II/I)] retained the full destabilizing ability of the wild-type c-fos ARE (Fig. 3A, upper right panel) (9). Second, changing eight uridylate residues surrounding the three AUUUA motifs in domain I to guanylates completely abolished the RNA-destabilizing function of domain I (9). Intriguingly, these U-to-G changes in domain I had no effect on the destabilizing function when domain II was present [fos ARE(U→G), Fig. 3A, upper left panel].
FIG. 3.
Stabilizing effects of HuR and hnRNP D on mRNA decay directed by the c-fos ARE mutant derivatives. RNA blots showing (A) the effect of ectopically expressed HuR on the destabilizing function of two mutant derivatives of c-fos ARE, ARE(U→G) and ARE(II/I), and (B) the effect of ectopically expressed hnRNP D on the destabilizing function of the c-fos ARE domain I. RNA isolation, nonpolyadenylated [poly(A)−] RNA preparation and time course experiments were carried out as described in the legend to Fig. 1. The control mRNA (α-globin/GAPDH) was expressed constitutively and served as an internal standard. The times given at the top of each blot correspond to hours after serum induction (S.I.).
It appears that domain II can compensate for the negative effect of U-to-G changes in domain I. We hypothesized that domain II in these two mutant AREs, ARE(II/I) and ARE(U→G), should bind HuR in vivo. If this were the case, ectopic expression of Myc-HuR should stabilize both BBB+ARE(II/I) and BBB+ARE(U→G) mRNAs. As shown in Fig. 3A, ectopic expression of HuR dramatically impeded the decay of β-globin mRNA bearing either of the two AREs. These in vivo functional data indicate that the interaction between HuR and domain II is sufficient for stabilization of mRNA containing the c-fos ARE.
HuR displays an ARE-binding preference distinct from that of hnRNP D/AUF1.
The binding preference of HuR for domain II of the c-fos ARE over domain I is different from that of hnRNP D, which exhibits a binding preference for AUUUA pentanucleotides in class II AREs. Previously, we showed that the hnRNP D p37 isoform binds the c-fos ARE (51). Since the c-fos ARE domain I contains three AUUUA motifs, it is possible that p37 binds preferentially to domain I. Gel mobility shift assays were carried out with the lysate prepared from NIH 3T3 cells overexpressing the p37 isoform, with c-fos ARE domain I or II as the probe. Figure 2C shows that hnRNP D p37 interacted strongly only with the AUUUA-containing domain I but barely bound to domain II. These results clearly demonstrated that these two different ARE-binding proteins, when synthesized in mammalian cells and studied in the context of crude cytoplasmic extracts, exhibit distinct binding preferences.
The significance of the p37 binding to domain I detected above was studied further by time course experiments in NIH 3T3 cells. Previously, we showed that the c-fos ARE domain I alone can function as a potent RNA-destabilizing element (9). The in vitro interaction detected between domain I and p37 suggested that ectopic expression of p37 should result in a significant stabilization effect on the decay of BBB mRNA bearing c-fos ARE domain I. As shown in Fig. 3B, p37 overexpression indeed impeded the decay of β-globin mRNA bearing c-fos ARE domain I, whereas transfection of the cloning vector had no effect on mRNA decay. Taken together, our results showed that HuR and hnRNP D attenuate the RNA destabilization function of c-fos ARE in distinct manners, which were not obvious from in vitro studies with recombinant proteins made in bacteria.
Basic hinge region is necessary but not sufficient for nuclear localization of HuR and is not required for message stabilization by HuR.
We wanted to identify the HuR domains that are functionally important for the stabilization effects. A series of four HuR deletion mutants, including deletions of RRM1 (ΔI), RRM2 (ΔII), the basic hinge region (ΔB), and RRM3 (ΔIII), were created (Fig. 4A). cDNA clones encoding the mutant HuR proteins were subcloned into the pTet-Myc-Ovep plasmid, whose constitutive expression in the absence of tetracycline is driven by the tetracycline-off promoter. The Myc epitope tag was fused in frame to the N terminus of all the deletion mutants to facilitate detection. NIH 3T3 B2A2 cells were grown on coverslips and transiently transfected with Myc-HuR or one of the mutant derivative constructs. Indirect immunofluorescence was first performed with monoclonal antibody against the Myc epitope tag (9E10) to evaluate the subcellular distributions of HuR and its deletion mutants. The results showed that the ΔI and ΔII mutants were found mainly in the nucleus, with a modest level also detected in the cytoplasm, whereas the ΔB and ΔIII mutants were found nearly exclusively in the cytoplasm.
Western blot analysis was performed to provide a complementary evaluation of the relative distributions of HuR and its mutants between the nucleus and the cytoplasm. Both cytoplasmic and nuclear extracts were prepared from transfected NIH 3T3 B2A2 cells. Western blot analysis with a monoclonal antibody against the Myc tag (Fig. 4B) showed that whereas wild-type HuR was present mainly in the nucleus, all four mutants were detected in both the cytoplasm and nucleus. ΔI and ΔII retained the shuttling signals, showing much higher levels of nuclear localization than ΔB and ΔIII, which lacked the shuttling signal. Conversely, ΔB and ΔIII had relatively higher levels of distribution in the cytoplasm than ΔI and ΔII. No cross-contamination between the nuclear extract and cytoplasmic extract was detected, as demonstrated by detection of α-tubulin only in the cytoplasmic extracts and U1 70K splicing factor only in the nuclear preparations. Together, these results showed that both the hinge region and RRM3 are necessary for nuclear localization
These results are in large part consistent with the study by Fan and Steitz (18), showing that the hinge region connecting RRM2 and RRM3 contains a signal for nucleocytoplasmic shuttling. However, unlike the RRM3-truncated mutant in their study, our ΔIII mutant, even though retaining the basic hinge region containing the shuttling signal, was no longer localized in the nucleus. One difference is that their ΔIII construct had a Flag epitope tag fused immediately downstream of the hinge region. It is possible that this extra tag sequence somehow influenced recognition of the shuttling signal in the hinge region. To test this possibility, we created a partial truncation of RMM3 which left 10 amino acids of RRM3 fused to the hinge region. As shown in Fig. 4, this minor modification dramatically restored the localization of HuR in the nucleus. We conclude that nuclear localization of HuR requires the presence of both the hinge region and an immediate downstream region in RRM3.
We next addressed whether the nucleocytoplasmic shuttling signal is required for the RNA stabilization effect of HuR. The decay of β-globin mRNA bearing the c-fos ARE (BBB+AREc-fos) was monitored in NIH 3T3 B2A2 cells constitutively expressing Myc-HuR or the ΔB mutant lacking the hinge region containing the shuttling signal. The results (Fig. 5, upper panel, ΔB) showed that rapid decay of BBB+AREc-fos mRNA in the cytoplasm was significantly impeded by overexpressing the ΔB mutant. The stabilization effect seen with ΔB was indistinguishable from that of wild-type HuR, demonstrating that the shuttling signal is not necessary for RNA stabilization.
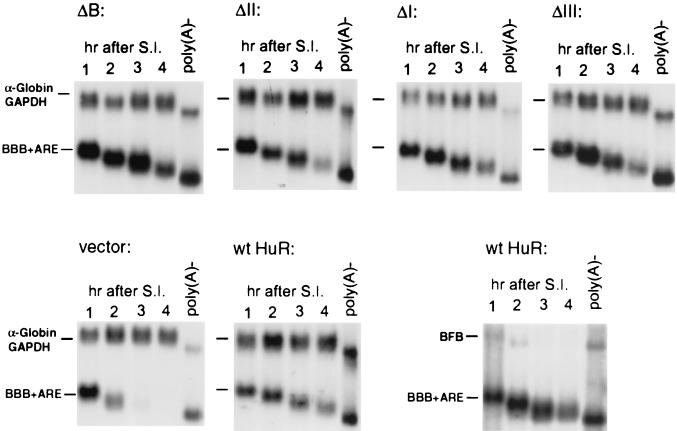
FIG. 5.
Evaluation of the contribution of individual RRMs and hinge region of HuR to its stabilization effect. RNA blots showing stabilization effects of various truncated HuR mutants on decay of β-globin mRNA containing the c-fos ARE (BBB+ARE) (upper panels) and stabilization effect of intact HuR on decay of BBB+ARE mRNA or BFB mRNA, a hybrid message containing the c-fos protein-coding region determinant of instability (lower panels). RNA isolation, nonpolyadenylated [poly(A)−] RNA preparation, and time course experiments were carried out as described in the legend to Fig. 1. The control mRNA (α-globin/GAPDH) was expressed constitutively and served as an internal standard. The times given at the top of each blot correspond to hours after serum induction (S.I.).
Combinations of any two of the three HuR RRMs are sufficient for significant RNA stabilization in NIH 3T3 cells.
We then systematically evaluated the contribution of individual RRMs for HuR's stabilization function. The stability of BBB+AREc-fos mRNA was determined in NIH 3T3 B2A2 cells ectopically expressing ΔI, ΔII, or ΔIII. As shown in Fig. 5, the stability of BBB+AREc-fos mRNA increased dramatically in all cases tested. As a control, we also determined the decay of BBB+AREc-fos mRNA along with a second mRNA, termed BFB mRNA, in B2A2 cells that overexpressed wild-type HuR. BFB mRNA is a hybrid message made between β-globin and c-fos mRNAs previously shown to be targeted for rapid decay by the c-fos protein-coding determinant (42). The results illustrated in Fig. 5 showed that decay of BFB mRNA was not affected by HuR, whereas BBB+AREc-fos mRNA was stabilized, demonstrating that the RNA stabilization effect by HuR is specific for the ARE. Together, these results indicate that any combination of two of the three RRMs in HuR is sufficient for RNA stabilization in NIH 3T3 cells.
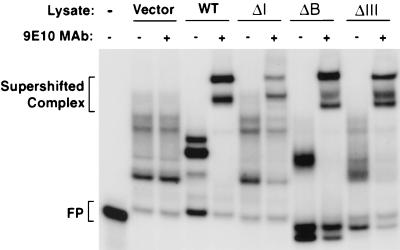
To ask whether the above stabilization effects displayed by HuR mutants in NIH 3T3 cells correlate with their ability to interact with the c-fos ARE, we performed gel mobility shift assays as well as antibody supershift assays with the c-fos ARE as the probe and control cytoplasmic extract or extracts containing ectopically expressed wild-type HuR or ΔI, ΔB, or ΔIII mutant protein. The results in Fig. 6 showed that all mutant proteins supported significant RNP complex formation with the c-fos ARE probe and that the complex was supershifted by antibody against the Myc tag on HuR or its mutant derivatives. These data provide further evidence for a correlation between mRNA stabilization by HuR in NIH 3T3 cells and formation of a specific ARE-protein complex with cytoplasmic extracts in vitro.
FIG. 6.
In vitro analysis of interactions between ectopically expressed human HuR and its mutant derivatives with the c-fos ARE. 32P-labeled ARE RNA transcribed in vitro from a T3 promoter-driven template was incubated with cytoplasmic lysates from NIH 3T3 B2A2 cells expressing no ectopic protein (vector), Myc-tagged HuR (wild type [WT]), or a mutant protein (ΔI, ΔB, or ΔIII). The RNA-protein complexes were then digested with RNase T1. The digestion mixtures were left alone (−) or further incubated in the presence of the 9E10 monoclonal antibody (MAb) against the Myc epitope tag. The final reaction mixtures were resolved by electrophoresis in a native polyacrylamide gel. Antibody-supershifted complexes and free probe (FP) are indicated.
DISCUSSION
Previously, both gel shift and UV cross-linking experiments showed that recombinant HuR made in bacteria binds strongly and rather nonselectively to a wide variety of AREs and mRNAs containing AREs (35, 36). These observations did not explain how up- or downregulation of mammalian ELAV-like proteins, including HuR, correlates well in vivo with the specific regulation of only particular ARE-containing mRNAs under given physiological conditions (reviewed in references 2, 7, and 26). This is a particularly relevant issue because other ARE-containing mRNAs are also present. Does HuR have any effect on the stability of these other ARE-containing mRNAs? If not, why is its effect so selective in a given physiological state?
To address this critical issue, we examined the effect of HuR ectopic expression on mRNA decay directed by various AREs in NIH 3T3 cells undergoing G0 to G1 transition following serum stimulation. Ectopic expression of HuR in NIH 3T3 cells mimics upregulation of HuR under other physiological situations, where transcription pulsing approaches for monitoring decay kinetics are not readily available. Our results demonstrate a highly selective action of HuR in antagonizing the destabilization function of the c-fos ARE in NIH 3T3 cells.
Several lines of experiments support the idea that the stabilization by HuR that we observed is not a nonspecific effect resulting simply from overexpression of an RNA-binding protein. First, we showed in the same experiment (Fig. 5, lower panel, right) that rapid decay of a control transcript containing the c-fos coding determinant (BFB) was not affected by HuR overexpression. Second, we showed in Fig. 6 that all HuR mutants supported significant formation of RNP complexes with the c-fos ARE, which could be supershifted by monoclonal antibody against Myc-tagged HuR, to an extent similar to that of the wild-type HuR. In contrast, no such supershifted complexes could be detected in the control lysate. Third, we showed in Fig. 1 that only the c-fos ARE was affected by overexpression of HuR and not the other AREs tested. It is plausible that ectopically expressed HuR displaces other destabilizing ARE-BPs, e.g., tristetraproline and hnRNP D, leading to stabilization. This interpretation is consistent with the results in Fig. 2, showing that ectopically expressed HuR outcompetes other endogenous ARE-BPs in the formation of gel-retarded RNP complexes with the c-fos ARE RNA (Fig. 2B). It will be interesting to pinpoint the potential destabilizing ARE-BP that is displaced by HuR in a future study.
In contrast to the relaxed binding specificity for AREs observed in vitro with recombinant proteins synthesized in bacteria, previously we showed that hnRNP D discriminates among three different classes of ARE in vivo (51). It most effectively downregulates the destabilizing function of class II AREs via recognition of multiple overlapping AUUUA motifs. Consistent with the results with hnRNP D, in this study we show that HuR, an ARE-BP structurally and functionally distinct from hnRNP D, also displays a highly selective action in antagonizing ARE-mediated decay in vivo. Recombinant HuR and hnRNP D made in bacteria show a significant overlap in terms of ARE binding in vitro, implying that these two structurally distinct ARE-BPs not only cannot discriminate among different classes of ARE but may in fact compete with each other. However, our results showed that in NIH 3T3 cells, HuR only blocks the function of c-fos ARE, a class I ARE, whereas hnRNP D is most effective in antagonizing class II ARE-mediated mRNA decay, e.g., GM-CSF ARE (51). Moreover, when RNA-binding specificity was studied with crude mammalian cytoplasmic extracts, both mammalian HuR (this study) and mammalian hnRNP D (51) showed a discriminating ARE-binding specificity that was not displayed by bacterial ARE-BPs. The present results provide further evidence that the broad ARE-binding specificity exhibited in vitro by individual ARE-BPs made in bacteria does not reflect their binding specificity in vivo. This suggests that caution should be applied to the interpretation of in vitro binding experiments with bacterial ARE-BPs.
The issue becomes identification of the molecular basis for the discrepancy between the in vivo functional data and the in vitro results. The studies by Steitz's group may offer some clues. They identified and characterized several protein ligands of HuR (6). When nuclear export mediated by CRM1 was blocked by leptomycin B, the association of HuR with two of the four ligands, pp32 and APRIL, in the nucleus was increased. This led to specific retention of an ARE-containing c-fos mRNA in the nucleus. It is striking to note that in our study, the destabilizing function of c-fos ARE was specifically impeded by HuR in NIH 3T3 cells. These data provide evidence for modulation of HuR's affinity for its target mRNAs by HuR ligands in vivo. The highly selective action of HuR on a specific ARE observed in this study may be a manifestation of modulation of HuR activity by its interacting protein ligands. On the other hand, the discrepancy between in vivo and in vitro observations could also be due to the lack of posttranslational modification, such as methylation or phosphorylation, of recombinant ARE-BPs expressed in bacterial systems. This possibility gains further support from a recent study showing that AMP-activated kinase, known as a cellular sensor of metabolic stress, regulates the HuR binding activity in the cytoplasm (46).
Although predominantly nuclear, HuR shuttles between the nucleus and the cytoplasm via a novel shuttling sequence, HNS, located in the hinge region between its second and third RRMs (18). HuR's ability to shuttle has led to the suggestion that HuR may initially become associated with target ARE-containing mRNAs in the nucleus and accompany them into the cytoplasm, where HuR is then able to exert its stabilizing actions (7, 26). However, our experiments showed that when nuclear localization of HuR is impeded by deleting the HNS or the third RRM, the mutants remain capable of blocking rapid decay directed by the c-fos ARE. Accordingly, gel shift experiments showed that such mutant HuRs still bind the c-fos ARE in a way that is indistinguishable from that of wild-type HuR (Fig. 6).
These results are in contrast to the case of hnRNP D. When we mutated hnRNP D to knock down its shuttling ability without affecting its ARE-binding ability, the nonshuttling mutant hnRNP D no longer had a stabilizing effect on the ARE-containing mRNA (unpublished data). Intriguingly, the ability of this mutant hnRNP D to block ARE-mediated RNA decay was regained by fusing a heterologous shuttling domain to restore the nucleocytoplasmic shuttling. These observations indicate that nuclear association with target mRNAs is necessary for the stabilizing effects of hnRNP D but not HuR; HuR has the potential to find and stabilize its target mRNAs in the cytoplasm. One possibility is that HuR is required for efficient nuclear export of its target ARE-containing mRNA as well as for protecting mRNA from nuclear degradation prior to its export to the cytoplasm (7, 26).
It is worth noting that when RRM3 was deleted from HuR, the truncated HuR lost its ability to enter the nucleus even though it retained the HNS shuttling signal. Fusing either a Flag epitope tag (18) or part of the HuR RRM3 (this study) immediately C-terminal to HNS was sufficient to restore shuttling of the resultant HuR mutants. This suggests that HNS requires some downstream peptide sequence in order to function in nucleocytoplasmic shuttling. Although deleting any one of the four HuR domains did not profoundly affect the ability of the corresponding mutant proteins to impede c-fos ARE-mediated decay, it should be noted that the cytoplasmic levels of these truncated HuR proteins were significantly higher than the level of wild-type HuR, which was barely detected under the same conditions (Fig. 4). It is possible that all four domains are required for optimal ARE-binding affinity and/or specificity. It will be interesting to determine if these truncated HuRs are able to discriminate among different AREs in NIH 3T3 cells.
It is also worth pointing out that there are several discernible differences between HuR and hnRNP D. First, although widely expressed, the HuR and hnRNP D proteins do vary in concentration among different tissues (23, 27). Second, sucrose gradient fractionation studies show that HuR and hnRNP D have very different subcellular profiles (20, 24). Whereas hnRNP D cosediments with two distinct complexes with molecular masses of ≈300 kDa and >700 kDa, HuR apparently comigrates with protein complexes of over 700 kDa. Third, cellular stresses that induce HuR redistribution between the cytoplasm and the nucleus do not cause similar redistribution of hnRNP D (45).
There are clearly differences in the ways in which HuR and hnRNP D modulate the metabolism of ARE-containing mRNAs. For example, Wang et al. (45) showed that the fluctuation of cytoplasmic levels of HuR during the cell cycle in colorectal carcinoma RKO cells correlates well with changes in the stability of cyclin A and cyclin B1 mRNAs, both of which are ARE-containing transcripts. Intriguingly, they also noted that neither the level nor the subcellular localization of hnRNP D varied throughout the cell cycle. The authors also showed that cyclin D1 mRNA, another ARE-containing message, is bound to endogenous hnRNP D and not to HuR, although bacterial HuR binds the cyclin D1 transcript in vitro. That study and our results support the idea that the function of HuR and hnRNP D in mRNA turnover is differentially regulated to target different AREs in a given physiological state of mammalian cells.
In summary, our results sheds light on the mechanisms by which up- or downregulation of ARE-containing mRNAs induced by physiological stimuli is exerted through one particular ARE-BP. Understanding how cells achieve differential regulation of the stability of various ARE-containing mRNAs via various ARE-BPs remains an interesting challenge.
Acknowledgments
We thank R. Kulmacz for critical reading of the manuscript and valuable comments, S. Berget for anti-U1 70K monoclonal antibody, and J. A. Steitz for anti-HuR monoclonal antibody. Special thanks go to S. Peng for technical assistance.
This work was supported by a grant from the National Institutes of Health (GM 46454) to A.-B.S.
REFERENCES
- 1.Abe, R., E. Sakashita, K. Yamamoto, and H. Sakamoto. 1996. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 24:4895-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic, D., and J. D. Keene. 1997. Insights from model systems—embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 61:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranda-Abreu, G. E., L. Behar, S. Chung, H. Furneaux, and I. Ginzburg. 1999. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J. Neurosci. 19:6907-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwater, J. A., R. Wisdom, and I. M. Verma. 1990. Regulated mRNA stability. Annu. Rev. Genet. 24:519-541. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., T. Giordano, G. Brewer, and J. S. Malter. 1999. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 27:1464-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-Y., and A.-B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C.-Y. A., T.-M. Chen, and A.-B. Shyu. 1994. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol. Cell. Biol. 14:416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. Y., and A. B. Shyu. 1994. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 14:8471-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. Y., N. Xu, and A. B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claffey, K. P., S. C. Shih, A. Mullen, S. Dziennis, J. L. Cusick, K. R. Abrams, S. W. Lee, and M. Detmar. 1998. Identification of a human VPF/VEGF 3′ untranslated region mediating hypoxia-induced mRNA stability. Mol. Biol. Cell 9:469-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cravchik, A., and A. Matus. 1993. A novel strategy for the immunological tagging of cDNA constructs. Gene 137:139-143. [DOI] [PubMed] [Google Scholar]
- 15.Dean, J. L., R. Wait, K. R. Mahtani, G. Sully, A. R. Clark, and J. Saklatvala. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMaria, C. T., and G. Brewer. 1996. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271:12179-12184. [DOI] [PubMed] [Google Scholar]
- 17.DeMaria, C. T., Y. Sun, L. Long, B. J. Wagner, and G. Brewer. 1997. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J. Biol. Chem. 272:27635-27643. [DOI] [PubMed] [Google Scholar]
- 18.Fan, X. C., and J. A. Steitz. 1999. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95:15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, F. B., C. C. Carson, T. Levine, and J. D. Keene. 1994. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries with neuronal RNA-binding protein Hel-N1. Proc. Natl. Acad. Sci. USA 91:11207-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, F. B., and J. D. Keene. 1996. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J. Cell Sci. 109:579-589. [DOI] [PubMed] [Google Scholar]
- 23.Gouble, A., and D. Morello. 2000. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene 19:5377-5384. [DOI] [PubMed] [Google Scholar]
- 24.Grosset, C., C.-Y. A. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A.-B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 25.Jain, R. G., L. G. Andrews, K. M. McGowan, P. H. Pekala, and J. D. Keene. 1997. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol. 17:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 96:5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafon, I., F. Carballes, G. Brewer, M. Poiret, and D. Morello. 1998. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene 16:3413-3421. [DOI] [PubMed] [Google Scholar]
- 28.Lai, W. S., E. Carballo, S. J. R., E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larola, G., R. Guesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 30.Levine, T. D., F. Gao, P. H. King, L. G. Andrews, and J. D. Keene. 1993. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 13:3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy, N. S., S. Chung, H. Furneaux, and A. P. Levy. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273:6417-6423. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, T., C. Gueydan, G. Huez, J. J. Toulme, and V. Kruys. 1998. Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 273:13781-13786. [DOI] [PubMed] [Google Scholar]
- 33.Loflin, P. A., C.-Y. A. Chen, and A.-B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loflin, T. L., C.-Y. A. Chen, N. Xu, and A.-B. Shyu. 1999. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods 17:11-20. [DOI] [PubMed] [Google Scholar]
- 35.Ma, W.-J., S. Chung, and H. Furneaux. 1997. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 25:3564-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S., D. G. Myszka, M. Yu, S. J. Littler, and I. A. Laird-Offringa. 2000. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol. Cell. Biol. 20:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, S.-P., C.-Y. Chen, N. Xu, and A.-B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shyu, A.-B., J. A. Garcia-Sanz, and E. Mullner (ed.). 1996. Analysis of mRNA decay in mammalian cells. Academic Press, London, United Kingdom.
- 42.Shyu, A.-B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos mRNA is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 43.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 44.Suk, K., and K. L. Erickson. 1996. Differential regulation of tumour necrosis factor-alpha mRNA degradation in macrophages by interleukin-4 and interferon-gamma. Immunology 87:551-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, W., M. C. Caldwell, S. Lin, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, W., J. Fan, X. Yang, S. Furer-Galban, I. Lopez de Silanes, C. von Kobbe, J. Guo, S. N. Georas, F. Foufelle, D. G. Hardie, D. Carling, and M. Gorospe. 2002. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 22:3425-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson, M. F., and A. B. Shyu. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23:775-787. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, G. M., and G. Brewer. 1999. Identification and characerization of proteins binding A+U-rich elements. Methods 17:74-83. [DOI] [PubMed] [Google Scholar]
- 50.Wilusz, C. W., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 51.Xu, N., C. Chen, and A. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21:6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, N., C.-Y. A. Chen, and A.-B. Shyu. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, N., P. Loflin, C.-Y. A. Chen, and A.-B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You, Y., C. Y. Chen, and A.-B. Shyu. 1992. U-rich sequence-binding proteins interacting with a 20-nucleotide U-rich sequence in the 3′ untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol. Cell. Biol. 12:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]