Abstract
Werner syndrome (WRN) is an uncommon autosomal recessive disease whose phenotype includes features of premature aging, genetic instability, and an elevated risk of cancer. We used three different experimental strategies to show that WRN cellular phenotypes of limited cell division potential, DNA damage hypersensitivity, and defective homologous recombination (HR) are interrelated. WRN cell survival and the generation of viable mitotic recombinant progeny could be rescued by expressing wild-type WRN protein or by expressing the bacterial resolvase protein RusA. The dependence of WRN cellular phenotypes on RAD51-dependent HR pathways was demonstrated by using a dominant-negative RAD51 protein to suppress mitotic recombination in WRN and control cells: the suppression of RAD51-dependent recombination led to significantly improved survival of WRN cells following DNA damage. These results define a physiological role for the WRN RecQ helicase protein in RAD51-dependent HR and identify a mechanistic link between defective recombination resolution and limited cell division potential, DNA damage hypersensitivity, and genetic instability in human somatic cells.
Werner syndrome (WRN) was originally identified among adult siblings in a single family, all of whom displayed cataract formation, premature greying and loss of hair, and scleroderma-like skin changes (48). Further characterization of the clinical, pathological, and genetic aspects of this syndrome following Otto Werner's initial description has led to the recognition of Werner syndrome as an uncommon autosomal recessive disease whose phenotype includes features of premature aging, genetic instability, and an elevated risk of cancer (13, 18, 39).
The WRN gene (also referred to as RECQ3 or RECQL2) was identified by positional cloning in 1996 (51) and was found to encode a 162-kDa member of the human RecQ helicase family with 3′→5′ helicase and 3′→5′ exonuclease activities. Werner patient mutations truncate the WRN open reading frame and promote loss of the altered protein and both of its associated biochemical activities (4, 28, 42). Mutations in other human RecQ helicase genes have also been identified in patients with two other genetic instability and tumor predisposition syndromes, Bloom syndrome (12) and Rothmund-Thomson syndrome (24, 26).
Recently, a homologous recombination (HR) defect in WRN cell lines was identified that included a 25-fold reduction in the rate of generation of viable recombinant daughter cells together with a shift in molecular recombination products from conversion-type to crossover or “popout”-type recombinants that are normally less frequent (35). These analyses focused on spontaneous mitotic recombintion and did not further define the WRN recombination defect or indicate how HR, cell survival, and the response to DNA damage were interrelated in WRN cells. In the work reported here, three different experimental approaches were used to define the WRN recombination defect and the interrelationship of HR and cell survival following DNA damage in WRN cells. Expression of wild-type WRN protein or the bacterial resolvase protein RusA were both shown to rescue the WRN recombination defect and to improve cell survival following DNA damage. The dependence of WRN cellular phenotypes on RAD51-dependent HR function was demonstrated with a dominant-negative mammalian RAD51 protein (SMRAD51) (22). Expression of SMRAD51 suppressed HR in control and WRN cells as predicted while leading to markedly improved WRN cell survival after DNA damage.
These results confirm the presence of an HR defect in WRN cells, more clearly identify the HR stage and likely molecular intermediates or products involved, and demonstrate the interdependence of defective HR, reduced cell division potential, and DNA damage hypersensitivity following a loss of WRN function. These results define a physiological role for WRN. The results also suggest a model for WRN function that explains how WRN loss leads to reduced cell division, DNA damage hypersensitivity, and genetic instability and thus may act to promote disease pathogenesis.
MATERIALS AND METHODS
Plasmid DNAs.
Plasmid DNAs encoding wild-type or K577 M missense mutant forms of human WRN protein have been previously described (28). Oligonucleotide-mediated, site-directed mutagenesis (Transformer; Clontech) was used to introduce an E84A substitution in a wild-type WRN open reading frame. An EcoRI-BsrGI fragment containing the E84A substitution was then subcloned into a K577 M WRN expression vector to give a WRN open reading frame and protein that lacked helicase and exonuclease activities. The resulting plasmids were sequence verified and tested for expression by Western blot analysis after transient transfection (see below). The plasmids pPURO and pSMRad51 were kindly provided by Bernard Lopez (CNRS-CEA, Fontenay aux Roses, France) (22). Plasmids pMW400 and pMW436, expressing wild-type and D70N forms of RusA, respectively, were kindly provided by Matthew Whitby (Oxford University) (11). For expression in human cells, RusA open reading frames were transferred to pPURO, a pcDNA3.1 (Invitrogen) derivative.
Cell lines and transfection.
The control simian virus 40 (SV40)-transformed fibroblast cell lines GM639 and GM847 were obtained from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository (Camden, N.J.). The WRN SV40-transformed fibroblast cell lines WV1 and AG11395 (WS780) have been previously described (36, 38). Both lines contain truncating mutations in the WRN open reading frame (1) and lack WRN protein detectable by Western blot analysis. The generation of sublines containing chromosomally integrated copies of the recombination reporter plasmids pNeoA and pLrec (25) has been described previously (35). Cells were grown in Dulbecco modified Eagle's medium containing 4,500 mg of glucose/liter and supplemented with 10% fetal bovine serum (HyClone), penicillin G sodium (100 U/ml), and streptomycin sulfate (100 μg/ml) in a humidified 37°C, 7% CO2 incubator. The cultures were screened periodically and verified to be free of Mycoplasma infection by DAPI (4′,6′-diamidino-2-phenylindole) staining and fluorescence microscopy.
WRN or control cell lines that stably expressed SMRAD51 protein were generated by transfecting pSMRad51 or the control vector pPURO into cells by using SuperFect (Qiagen), followed by the selection of colonies able to grow in the presence of 0.4 to 1 μg of puromycin/ml. SMRAD51 protein expression was determined by Western blot analysis using an anti-Rad51 antibody (Oncogene Research) as previously described (22). WRN and RusA expression vectors were transiently transfected using SuperFect, and the transfection efficiency was monitored by green fluorescent protein fluorescence. WRN protein expression was monitored by Western blot analysis as previously described (28).
Cell survival assays.
Colony-forming efficiency (CFE) was determined by plating 100 or 1,000 cells in six-well plate wells (Falcon). Higher cell numbers were used in some cases when survival after DNA damage was studied. The cells were grown for 10 to 18 days and then fixed and stained with crystal violet prior to the determination of the fraction of cells plated that were able to form colonies of ≥6 or ≥50 cells. Colony size distributions (CSDs) were determined by allowing single cells to form colonies over the course of 10 to 18 days, followed by crystal violet staining and counting the number of cells in individual colonies. CSD data are reported as a cumulative probability distribution of the percentage of cells able to form colonies of greater than or equal to n cells after a defined growth interval (43).
Aqueous stocks of cis-platinum (cis-Pt) and hydroxyurea (HU) were prepared (1 mM cis-Pt or 1 M HU), filter sterilized through a 0.22-μm-pore-size filter, and stored at 4°C (cis-Pt) or −20°C (HU) until they were diluted just prior to use. Ionizing (γ) radiation sensitivity experiments were performed using a 137Cs source at a dose rate of 1 Gy/min. Cell survival and growth following DNA damage were determined by CFE or CSD assays as described above, in which mock-treated cells were used as controls to determine survival in the absence of DNA damage. The statistical significance of differences in growth or survival were determined as previously described (36) except that comparisons were made between treatments, not cell lines, and no time variable was involved.
Recombination assays.
The ability of cis-Pt or HU treatment to induce viable G418-resistant (neo+) mitotic recombinant colonies was determined by exposing cells containing pNeoA to 2 μM cis-Pt or 1 mM HU for 24 h, followed by 10 to 18 days of growth in the absence of treatment or selection to determine survival or in 400 to 600 μg of G418/ml to determine recombinant frequency. Recombination frequencies were corrected for both the background (untreated) neo+ colony frequency and for survival to give the number of induced neo+ colonies per 106 viable treated cells. Beta-galactosidase-positive (lac+) recombinant-cell frequencies were determined as previously described (35) and reported as the number of lac+ cells per 106 viable cells 24 h after cis-Pt or HU treatment as described above. The statistical significance of differences in the frequencies of recombinant neo+ colonies or lac+ cells was determined by applying one-way analysis of variance to the calculated frequencies for each cell line and testing all pairwise contrasts (35).
RESULTS
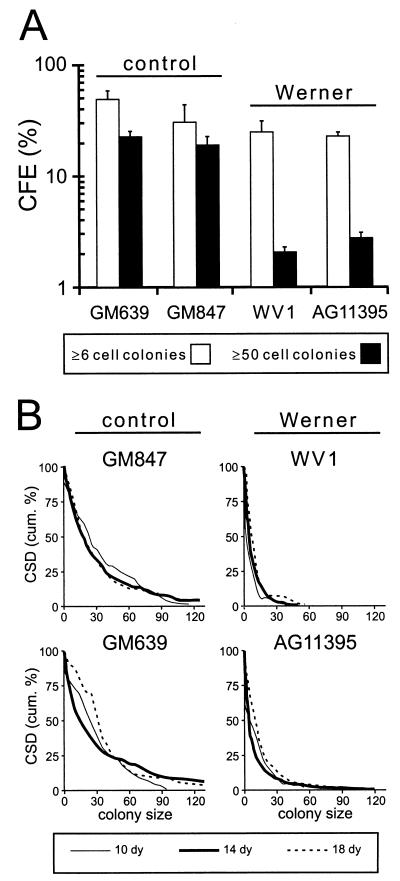
We first determined the ability of WRN and control SV40-transformed fibroblast cell lines to grow and form colonies in the absence of exogenous DNA damage. The WRN mutation status and WRN protein content of all four lines have been defined previously (1, 35, 36). Two assays were used to quantify cell division or growth potential: CFE and CSD assays. Both assays revealed an intrinsic growth deficit in WRN SV40-transformed fibroblast cell lines in the absence of exogenous DNA damage. WRN cell line CFEs were substantially lower than those of controls when colony formation was defined as ≥50 cells after 10 days of growth. This difference was less marked when the criterion for colony formation was ≥6 cells after 10 days of growth (Fig. 1A). In CSD assays, ∼6.5% of WRN colonies consisted of ≥25 cells after 10 days of growth in contrast to 41% of control colonies. Only one of the WRN cell lines, AG11395, was able to generate any large colonies after 18 days of growth (∼2% of colonies had >75 cells versus 10% in control lines) (Fig. 1B).
FIG. 1.
Intrinsic growth defect of SV40-transformed WRN cell lines in the absence of DNA damage. (A) CFE of SV40-transformed WRN and control fibroblast cell lines as determined by the ability of single cells to form colonies of ≥6 cells or ≥50 cells after 10 days of growth. The error bars indicate standard deviations for two independent determinations. (B) CSDs for the same four cell lines as determined by the ability of single cells to form colonies of greater than or equal to n cells after 10 to 18 days of growth. The CSD results are plotted as a cumulative distribution (43).
These results indicate that SV40 transformation in part suppresses the severe cellular growth deficit displayed by primary WRN fibroblast strains while leaving other aspects of the WRN cellular phenotype, such as genetic instability and selective DNA damage hypersensitivity, largely intact (see below). The ability of SV40 transformation to compensate in part for the loss of WRN RecQ helicase activity may reflect the strong helicase activity of SV40 large-T-antigen protein (14, 27). An alternative and more plausible explanation is that the presence of T antigen suppresses cellular responses to DNA damage in the absence of WRN function.
CFE and CSD assays were next used to determine the survival and growth of WRN and control cells following DNA damage. Work in several laboratories, including our own, had previously shown that WRN patient peripheral blood lymphocytes, B-lymphoblastoid cell lines, primary fibroblasts, and fibroblast cell lines display selective sensitivity to a limited spectrum of DNA-damaging agents, including DNA cross-linking agents, the DNA topoisomerase I inhibitor camptothecin, and 4-nitroquinoline 1-oxide (16, 17, 29, 30, 33, 34, 36). In light of the intrinsic growth defect of WRN SV40 fibroblast cell lines in the absence of DNA damage, we reexamined the sensitivity of WRN and control SV40 fibroblast cell lines to cis-Pt-induced DNA cross-links, to HU-mediated replication arrest, and to ionizing radiation damage (137Cs γ radiation) using CFE and CSD assays designed to minimize the intrinsic differences in WRN cell growth and division potential identified above.
WRN cell lines treated with cis-Pt had a statistically significant, dose-dependent reduction in survival compared with controls at all exposure times tested (6 to 30 h) in both CFE and CSD assays (Fig. 2; also see Fig. 4 and 5). In both assays, survival of WRN cells as a function of the cis-Pt dose displayed a steep slope that, in contrast to control cells, did not plateau at high cis-Pt doses or long exposure times (results not shown). The HU sensitivity of WRN cells was examined in light of reports of poor recovery of growth by fission yeast lacking the Rqh1 RecQ helicase following HU-mediated S-phase arrest (44) and a report of HU sensitivity in WRN lymphoblastoid cell lines and primary fibroblasts (31). Two independent SV40-transformed WRN fibroblast cell lines displayed dose- and time-dependent HU sensitivity in CSD, though not CFE, assays. The most pronounced effect was observed after treatment with 1 mM HU for 24 h (results not shown). Finally, a modest though reproducible and statistically significant sensitivity of SV40-transformed WRN fibroblast cell lines to ionizing radiation was observed. In CFE assays, there was significantly suppressed survival at radiation doses of ≥6 Gy (results not shown). The magnitude of the effect was similar to that recently reported for hTERT-immortalized WRN fibroblasts (50). In CSD assays, WRN cell lines displayed a reduction in the ability to form large colonies (≥25 cells) after 6 Gy of radiation.
FIG. 2.
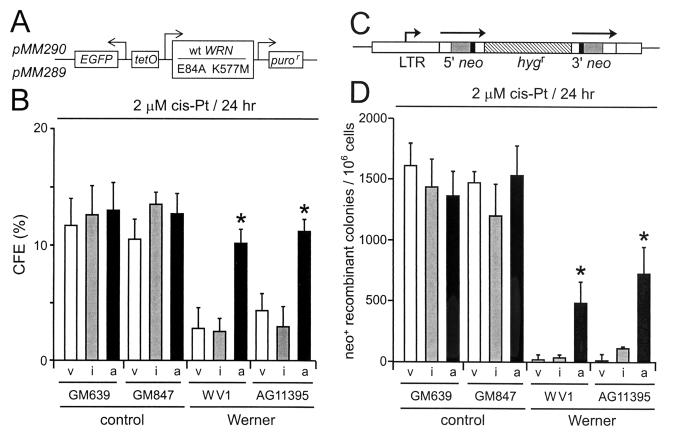
WRN protein rescues WRN cell survival and recombination after DNA damage. (A) Expression vectors encoding active (pMM290; wild-type [wt] WRN) or missense mutant (pMM289; E84A K577M) WRN protein lacking exonuclease and helicase activities. puror, puromycin resistance gene; EGFP, enhanced green fluorescence protein gene. (B) CFEs of WRN and control cell lines after transfection with control (pPURO) (v), WRN missense mutant (pMM289) (i), or active (pMM290) (a) expression vectors and cis-Pt treatment (2 μM; 24 h). (C) Structure of the direct-repeat recombination reporter plasmid pNeoA (25). The arrows indicate the extent of the neomycin phosphotransferase genes (neo); the solid boxes indicate the positions of inactivating linker insertions, the shaded boxes indicate the overlap region of 592 bp between linker insertion sites, and the hatched box indicates a hygromycin resistance gene (hygr). LTR, long terminal repeat. (D) Frequency of cis-Pt-induced neo+-recombinant colonies per 106 viable cells after transfection as described for panel B. ∗, statistically significant difference in survival or recombinant frequency. The error bars indicate standard deviations for two to five replicate experiments.
FIG. 4.
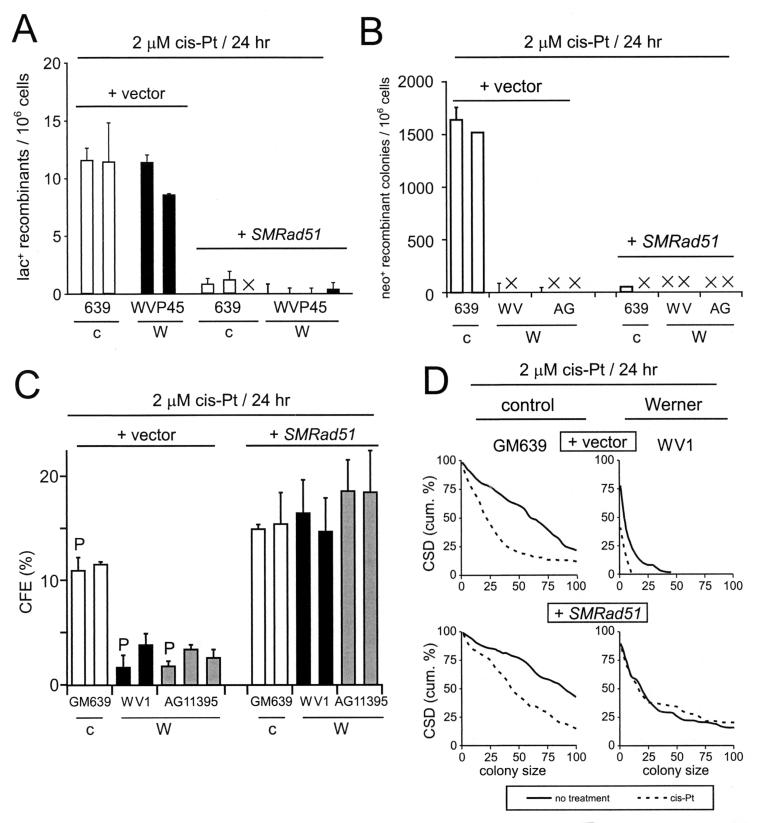
Recombination and cell survival after SMRAD51 protein expression. (A) Frequency of cis-Pt-induced lac+ recombinant cells per 106 viable cells generated by control (+ vector) or SMRAD51-expressing (+ SMRAD51) control (c) and WRN (W) sublines. ×, no cells observed. (B) Frequency of cis-Pt-induced neo+-recombinant colonies per 106 viable cells generated by control or SMRAD51-expressing sublines. ×, no or too few colonies to display using the scale shown (range, 0 to 40 colonies). (C) CFE of WRN and control sublines expressing control plasmid alone or SMRAD51 protein after cis-Pt treatment. The bars represent parental cell lines (P) or clonally derived sublines. The recovery of survival in WRN cells expressing SMRAD51 protein after cis-Pt treatment was highly statistically significant (P < 2 × 10−5 for all four comparisons; SMRAD51 protein-expressing cells versus the parental cell line or SMRAD51 protein-expressing cells versus the clonally derived subline within both WV1 and AG11395). (D) CSDs of WRN and control sublines expressing vector or SMRAD51 protein after cis-Pt treatment. The recovery of survival of WRN cells expressing SMRAD51 protein after cis-Pt treatment was again highly statistically significant (P < 5 × 10−7). cum., cumulative. The error bars in all cases indicate standard deviations for two to five replicate experiments.
FIG. 5.
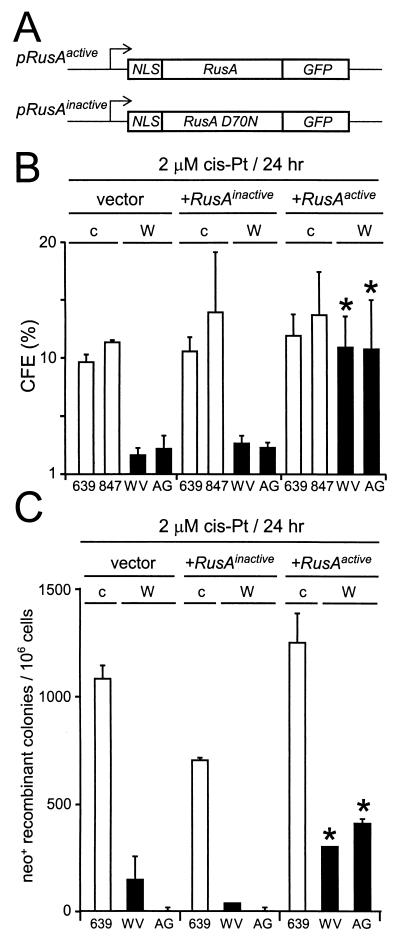
Expression of a bacterial resolvase protein improves WRN cell survival and recombination. (A) Vectors for expression of active or catalytically inactive (D70N substitution) forms of the bacterial resolvase protein RusA (NLS, nuclear localization signal; GFP, green fluorescent protein) (11). (B) Survival of control (c) or WRN (W) SV40-transformed fibroblast cell lines transfected with control (vector) plasmid or plasmid expressing inactive (+RusAinactive) or active (+RusAactive) RusA protein and treated with cis-Pt. (C) cis-Pt-induced neo+-recombinant colonies per 106 viable cells after transfection of control (c) or WRN (W) cells as described for panel B. ∗, statistically significant difference compared with control or RusAinactive-transfected cells (see the text). The error bars indicate standard deviations for two to five replicate experiments.
In order to determine whether the reduced cell division potential and DNA damage hypersensitivity of WRN cell lines resulted from a loss of WRN function, we expressed active or missense mutant forms of WRN protein in WRN and control cells and then quantified cell survival after cis-Pt treatment. Only wild-type WRN protein possessing helicase and exonuclease activities significantly improved the survival and growth of WRN cells prior to or after DNA damage by cis-Pt treatment (P ≤ 0.0001; Fig. 2A and B and results not shown). The generation of viable neo+ recombinant colonies after cis-Pt damage was also significantly improved when wild-type WRN protein was expressed in WRN cells prior to cis-Pt damage (P = 0.003 to 0.0001) (Fig. 2C and D). In contrast, expression of a missense mutant form of WRN lacking helicase and exonuclease activities (mutant WRN) did not significantly improve survival or recombination in WRN cells and did not confer a dominant-negative or toxic phenotype in control cells (Fig. 2B and D) (P = 0.15 to 0.45). In these experiments, the transfection efficiencies and levels of expression of wild-type and mutant WRN proteins, respectively, were comparable (results not shown). The coordinate recovery of cell survival and the ability to generate viable recombinant colonies by WRN cells after expressing wild-type WRN protein indicates that defective HR, limited cell division potential, and DNA damage hypersensitivity are interrelated and are likely to result directly from the loss of WRN function.
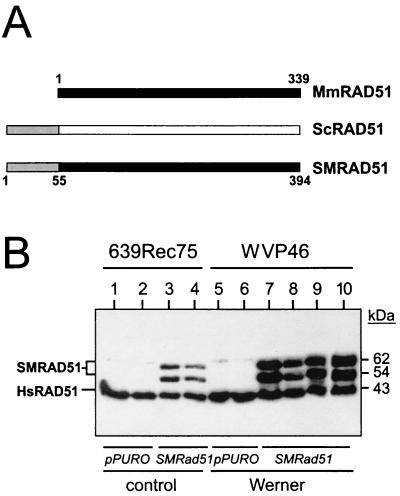
Two additional experimental strategies were used to demonstrate a role for WRN in recombination. First, the dependence of WRN cellular phenotypes on HR function was determined by using a dominant-negative form of mammalian RAD51 protein to suppress HR in WRN and control cells. Second, we determined whether expression of a bacterial resolvase protein, RusA, in WRN cells led to increased cell survival and the generation of viable recombinant daughter cells. The dominant-negative form of RAD51 used in these experiments consisted of the first 55 amino acid residues of Saccharomyces cerevisiae RAD51 fused to the N terminus of the 339-residue murine Rad51 open reading frame (SMRAD51) (Fig. 3A). SMRAD51 separates the viability and intrachromosomal recombination functions of mammalian RAD51 protein and had been shown previously to suppress the generation of RAD51-dependent recombinants in rodent cells (22).
FIG. 3.
Dominant-negative mammalian RAD51 protein and expression in stable cell lines. (A) Mus musculus (Mm), S. cerevisiae (Sc), and chimeric Mm-Sc (SM) RAD51 proteins (the numbers indicate amino acid residues) (22). Filled and open boxes represent the M. musculus and S. cerevisiae RAD51 open reading frames, respectively. Shaded segments represent the 55 N-terminal amino acid residues of yeast RAD51 that have been fused in frame to mouse RAD51 to generate the chimeric SMRAD51 protein. (B) Western blot analysis of expression of endogenous (HsRAD51) or stably expressed SMRAD51 proteins in representative control (639Rec75) or WRN (WVP46) SV40-transformed fibroblast-derived sublines (35). Lanes 1, 2, 5, and 6 were clonally derived from control plasmid (pPURO) transfections. The two SMRAD51 protein bands arise from alternative initiation or stable degradation (22). Multiple independent sublines were generated for use in subsequent experiments.
Stable WRN and control sublines that expressed SMRAD51 protein and contained the chromosomally integrated recombination reporter plasmid pNeoA or pLrec were generated (25) (Fig. 3B). These reporter plasmids consist of genetically inactive direct repeats of the neomycin phosphotransferase (pNeoA) or β-galactosidase (pLrec) gene that can give rise to neo+ colonies or lac+ cells following recombination. An important distinction between these two reporters, despite their similar structures, is the requirement for cell growth to reveal neo+ recombinants as colonies of neo+ cells (25, 35). The expression of SMRAD51 protein strongly suppressed spontaneous and cis-Pt-induced lac+-recombinant generation in WRN and control cells (Fig. 4A) and neo+-colony formation in control cells (Fig. 4B) while significantly improving the survival of WRN cells after cis-Pt treatment as measured by CFE (Fig. 4C) (P < 10−4 for all comparisons between Rad51-expressing cells and non-Rad51-expressing cells within each Werner line) or CSD (Fig. 4D) (P < 5 × 10−7) assays. The improved survival of WRN cells following the suppression of RAD51-dependent HR pathways indicates that HR function is important for the generation of WRN cellular phenotypes such as reduced cell division potential and reduced survival after DNA damage.
In order to better delineate the molecular nature of RAD51-dependent in vivo HR products that might be responsible for WRN cellular phenotypes, we determined whether expression of the bacterial resolvase protein RusA could rescue WRN cell survival and recombination (Fig. 5A). RusA is a 120-residue protein that was originally identified as a suppressor of deficiencies in the Escherichia coli bacterial resolvase protein RuvC (23, 40, 41, 49). RusA can bind a variety of different DNA junction structures, though it efficiently cleaves only four-way Holliday junctions in a metal ion- and sequence-dependent reaction (23, 40, 41). The expression of active RusA protein in WRN cells significantly improved both cell survival and the generation of neo+ recombinants following DNA damage (Fig. 5B and C) (P = 0.007 and 3.7 × 10−5, respectively).
These results are reminiscent of work with Schizosaccharomyces pombe, in which RusA expression was shown to partially suppress defects in cell viability and recombination in cells lacking the sole fission yeast RecQ helicase, Rqh1 (11). RusA protein was also recently shown to suppress a late-stage meiotic recombination defect in S. pombe cells lacking the Mus81 protein (2), a putative recombination resolution activity in both yeast and mammalian cells (19). The ability of RusA expression to restore WRN cell survival and the generation of viable neo+-recombinant colony formation following DNA damage suggests that at least a portion of the postulated unresolved recombination products in WRN cells contain Holliday junctions.
DISCUSSION
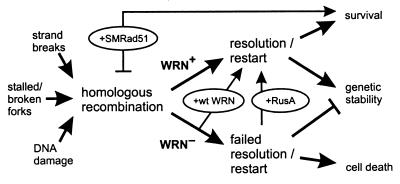
The experiments described above reveal the dependence of WRN cellular phenotypes on RAD51-mediated recombination function and the importance of successful recombination resolution to ensure cell survival after DNA damage. These experimental results establish a physiological role for WRN protein in recombination resolution in human somatic cells and indicate that WRN cellular phenotypes arise from a recombination defect rather than hyperrecombination, as has been widely assumed. The identification of a role for WRN in recombination resolution also provides insight into the molecular nature of in vivo substrates for WRN function and suggests a mechanistically coherent model that indicates how loss of WRN function suppresses cell division, confers DNA damage hypersensitivity, and promotes genetic instability in human somatic cells (Fig. 6).
FIG. 6.
Model of WRN function and origins of WRN cellular phenotypes. DNA damage, replication, or repair can initiate HR (left) (8, 21, 46). WRN promotes HR resolution or replication restart to insure cell viability and genetic stability (WRN+ arrow). In the absence of WRN (WRN−), HR resolution and/or replication restart fails, leading to mitotic arrest, cell death, and genetic instability. Experimental tests of this model are shown in ovals: reexpressing WRN protein (+wt WRN) (Fig. 2) improves both cell survival and the recovery of viable mitotic recombinants, as does expression of the bacterial resolvase protein RusA (+RusA) (Fig. 5). The dependence of WRN phenotypes on RAD51 pathway function and products can be revealed by expressing a dominant-negative form of mammalian RAD51 protein (+SMRAD51) (Fig. 3 and 4) that suppresses mitotic recombination in WRN and controls cells while improving WRN cell survival after cis-Pt damage. Anticipated consequences of survival in the absence of HR function are mutagenesis and genetic instability (45, 46).
The identification of a role for WRN in the resolution of RAD51 pathway HR products is one of the most intriguing aspects of the results presented here. While a detailed picture of recombination resolution has been developed for bacteria and phage (41, 49) and eukaryotic recombination resolution is likely to use a similar functional “core” (9), the identities of eukaryotic recombination resolution proteins remain in large part obscure (6, 19, 23). The idea that WRN could play a role in recombination resolution is biochemically plausible. WRN possesses or could recruit additional biochemical activities to promote recombination, replication restart, or other aspects of cellular DNA metabolism, such as telomere maintenance, that may require resolution. WRN can bind and unwind model Holliday junctions in vitro (7) and has been shown to interact physically or functionally with replication protein A, proliferating cell nuclear antigen, DNA polymerase δ, the p53 protein, topoisomerases I and III, the FEN-1/flap endonuclease, RAD51, and the nonhomologous-end-joining (NHEJ) components Ku and DNA-PK (4, 42). While WRN alone could unwind and resolve many conversion-type intermediates, it would need to act with other proteins to cleave Holliday junctions (2, 5, 6, 19, 23, 40). It is not clear how many Holliday junction endonucleases are present in mammalian cells or how important a role they play in recombination resolution, as these cells normally generate comparatively few crossover-type recombinants (20).
Our results suggest a mechanistically coherent model of WRN function and WRN disease pathogenesis that has four key features (Fig. 6). First, it is postulated that one important physiological role for WRN is to resolve recombination products generated during recombination, DNA repair, replication restart, or other aspects of cellular DNA metabolism. Second, RAD51 is postulated to generate an in vivo substrate(s) for WRN function from these nucleic acid metabolic processes, at least a portion of which contain Holliday junctions. Third, a failure to resolve RAD51-dependent recombination products is postulated to lead directly or indirectly to mitotic arrest, cell death, or genetic instability, where the details may be heavily dependent upon or conditioned by cell lineage properties. Fourth, unresolved recombination products that escape mitotic or programmed cell death are likely to promote mutations, gene rearrangements, or genetic instability when captured by error-prone or nonconservative repair pathways, such as NHEJ.
This model is strongly supported by the experimental results outlined above (Fig. 2 to 5). It is also consistent with what is known about human repair and recombination pathways and indicates how WRN is likely to interact with other DNA repair pathways or proteins, e.g., NHEJ, the BRCA1 and -2 proteins, and the MRE11/RAD50/NBS1 complex. Defects in these pathways and proteins also promote genetic instability with an elevated risk of cancer (10, 32, 45-47). For example, recent results indicate that WRN may play a role in NHEJ (4), and links between HR and NHEJ can be experimentally revealed (37). However, the close resemblance of WRN cellular phenotypes to those displayed by other mammalian HR mutants (46) indicates that if WRN does play a role in NHEJ, this role is likely to be subservient to its role(s) in HR.
A final point that argues strongly for the validity of the model depicted in Fig. 6 is that it provides a quantitative and mechanistically consistent explanation for WRN genetic instability data. This includes well-established results, e.g., the deletion mutator phenotype of WRN cell lines (15), as well as—perhaps most tellingly—previously paradoxical results, such as a surprisingly low loss of heterozygosity frequency in WRN cell lines that display both a deletion mutator phenotype and chromosomal instability (3, 15). It should be possible to build a more detailed predictive model from the one shown in Fig. 6 by adding information on the identity of in vivo substrates, the nucleic acid metabolic and DNA sequence contexts in which these substrates arise (8, 21), and how WRN acts with other proteins to resolve these substrates (6, 19). The resulting detailed picture will provide additional insight into WRN functional pathways and their roles to insure genetic stability and the survival of human somatic cells.
Acknowledgments
We thank our colleagues Bonny Brewer, Larry Loeb, Nancy Maizels, Martin Poot, Gerry Smith, and Peter Rabinovitch for helpful discussions, for communicating results prior to publication, and for reading manuscript drafts. Mike Moser generated several of the WRN expression constructs, and Alden Hackmann generated graphics.
This work was supported by grants from the NCI and from the Nippon Boehringer Ingelheim Virtual Research Institute of Aging to R.J.M., Jr., by an NCI R29 grant to M.J.E., and by grants from the Ligue Nationale Contre le Cancer and the Philippe Foundation to Y.S.
REFERENCES
- 1.Bennett, S. E., A. Umar, J. Oshima, R. J. Monnat, Jr., and T. A. Kunkel. 1997. Mismatch repair in extracts of Werner syndrome cell lines. Cancer Res. 57:2956-2960. [PubMed] [Google Scholar]
- 2.Boddy, M. N., P.-H. L. Gaillard, W. H. McDonald, P. Shanahan, J. R. I. Yates, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 3.Brooks-Wilson, A. R., M. J. Emond, and R. J. Monnat, Jr. 1997. Unexpectedly low loss of heterozygosity in genetically unstable Werner syndrome cell lines. Genes Chromosomes Cancer 18:133-142. [PubMed] [Google Scholar]
- 4.Brosh, R. M., Jr., and V. A. Bohr. 2002. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol. 37:491-506. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X.-B., R. Melchionna, C.-M. Denis, P.-H. L. Gaillard, A. Blasina, I. V. de Weyer, M. N. Boddy, P. Russell, J. Vialard, and C. H. McGowan. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8:1117-1127. [DOI] [PubMed] [Google Scholar]
- 6.Constantinou, A., A. A. Davies, and S. C. West. 2001. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104:259-268. [DOI] [PubMed] [Google Scholar]
- 7.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, Jr., V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 9.Cromie, G. A., J. C. Connelly, and D. R. F. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 10.D'Amours, D., and S. P. Jackson. 2002. The MRE11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 11.Doe, C. L., J. Dixon, F. Osman, and M. C. Whitby. 2000. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19:2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, N. A., J. Groden, T.-Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 13.Epstein, C. J., G. M. Martin, A. L. Schultz, and A. G. Motulsky. 1966. Werner's syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine 45:177-221. [DOI] [PubMed] [Google Scholar]
- 14.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 15.Fukuchi, K., G. M. Martin, and R. J. Monnat, Jr. 1989. Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl. Acad. Sci. USA 86:5893-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebhart, E., R. Bauer, U. Raub, M. Schinzel, K. W. Ruprecht, and J. B. Jonas. 1988. Spontaneous and induced chromosomal instability in Werner syndrome. Hum. Genet. 80:135-139. [DOI] [PubMed] [Google Scholar]
- 17.Gebhart, E., M. Schinzel, and K. W. Ruprecht. 1985. Cytogenetic studies using various clastogens in two patients with Werner syndrome and control individuals. Hum. Genet. 70:324-327. [DOI] [PubMed] [Google Scholar]
- 18.Goto, M. 1997. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech. Ageing Dev. 98:239-254. [DOI] [PubMed] [Google Scholar]
- 19.Haber, J. E., and W. D. Heyer. 2001. The fuss about Mus81. Cell 107:551-554. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. D., and M. Jasin. 2001. Double-strand break induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29:196-201. [DOI] [PubMed] [Google Scholar]
- 21.Klein, H. L., and K. N. Kreuzer. 2002. Replication, recombination and repair: going for the gold. Mol. Cell 9:471-480. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, S., and B. S. Lopez. 2000. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant negative forms. EMBO J. 19:3090-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilley, D. M. J., and M. F. White. 2001. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2:433-443. [DOI] [PubMed] [Google Scholar]
- 24.Lindor, N. M., Y. Furuichi, S. Kitao, A. Shimamoto, C. Arndt, and S. Jalal. 2000. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 90:223-228. [DOI] [PubMed] [Google Scholar]
- 25.Meyn, M. S. 1993. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science 260:1327-1330. [DOI] [PubMed] [Google Scholar]
- 26.Mohaghegh, P., and I. D. Hickson. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10:741-746. [DOI] [PubMed] [Google Scholar]
- 27.Monnat, R. J., Jr. 1992. Werner syndrome: molecular genetics and mechanistic hypotheses. Exp. Gerontol. 27:447-453. [DOI] [PubMed] [Google Scholar]
- 28.Moser, M. J., A. S. Kamath-Loeb, J. E. Jacob, S. E. Bennett, J. Oshima, and R. J. Monnat, Jr. 2000. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 28:648-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogburn, C. E., J. Oshima, M. Poot, R. Chen, K. E. Hunt, K. A. Gollahon, P. S. Rabinovitch, and G. M. Martin. 1997. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum. Genet. 101:121-125. [DOI] [PubMed] [Google Scholar]
- 30.Okada, M., M. Goto, Y. Furuichi, and M. Sugimoto. 1998. Differential effects of cytotoxic drugs on mortal and immortalized B-lymphoblastoid cell lines from normal and Werner's syndrome patients. Biol. Pharm. Bull. 21:235-239. [DOI] [PubMed] [Google Scholar]
- 31.Pichierri, P., A. Franchitto, P. Mosesso, and F. Palitti. 2001. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12:2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11:S52-S59. [DOI] [PubMed] [Google Scholar]
- 33.Poot, M., K. A. Gollahon, and P. S. Rabinovitch. 1999. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 104:10-14. [DOI] [PubMed] [Google Scholar]
- 34.Poot, M., J. S. Yom, S. H. Whang, J. T. Kato, K. A. Gollahon, and P. S. Rabinovitch. 2001. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 15:1224-1226. [DOI] [PubMed] [Google Scholar]
- 35.Prince, P. R., M. J. Emond, and R. J. Monnat, Jr. 2001. Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 15:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince, P. R., C. E. Ogburn, M. J. Moser, M. J. Emond, G. M. Martin, and R. J. Monnat, Jr. 1999. Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum. Genet. 105:132-138. [DOI] [PubMed] [Google Scholar]
- 37.Richardson, C., and M. Jasin. 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genome integrity in mammalian cells. Mol. Cell. Biol. 20:9068-9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito, H., and R. E. Moses. 1991. Immortalization of Werner syndrome and progeria fibroblasts. Exp. Cell Res. 192:373-379. [DOI] [PubMed] [Google Scholar]
- 39.Schellenberg, G. D., T. Miki, C.-E. Yu, and J. Nakura. 2001. Werner syndrome, p. 785-797. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular basis of inherited disease. McGraw-Hill, New York, N.Y.
- 40.Sharples, G. J. 2001. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 39:823-834. [DOI] [PubMed] [Google Scholar]
- 41.Sharples, G. J., S. M. Ingleston, and R. G. Lloyd. 1999. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 181:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, J.-C., and L. A. Loeb. 2000. The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 16:213-220. [DOI] [PubMed] [Google Scholar]
- 43.Smith, J. R., O. M. Pereira-Smith, and E. L. Schneider. 1978. Colony size distributions as a measure of in vivo and in vitro aging. Proc. Natl. Acad. Sci. USA 75:1353-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 46.van Gent, D. C., J. H. J. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 47.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 48.Werner, O. 1985. On cataract in conjunction with scleroderma. Adv. Exp. Med. Biol. 190:1-14. [PubMed] [Google Scholar]
- 49.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]
- 50.Yannone, S. M., S. Roy, D. W. Chan, M. B. Murphy, S. Huang, J. Campisi, and D. J. Chen. 2001. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 276:38242-38248. [DOI] [PubMed] [Google Scholar]
- 51.Yu, C.-E., J. Oshima, Y.-H. Fu, E. M. Wijsman, F. Hisama, S. Ouais, J. Nakura, T. Miki, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]