Abstract
Interferon A (IFN-A) genes are differentially expressed after virus induction. The differential expression of individual IFN-A genes is modulated by the specific transcription activators IFN regulatory factor 3 (IRF3) and IRF-7 and the homeoprotein transcription repressor Pitx1. We now show that repression by Pitx1 does not appear to be due to the recruitment of histone deacetylases. On the other hand, Pitx1 inhibits the IRF3 and IRF7 transcriptional activity of the IFN-A11 and IFN-A5 promoters and interacts physically with IRF3 and IRF7. Pitx1 trans-repression activity maps to specific C-terminal domains, and the Pitx1 homeodomain is involved in physical interaction with IRF3 or IRF7. IRF3 is able to bind to the antisilencer region of the IFN-A4 promoter, which overrides the repressive activity of Pitx1. These results indicate that interaction between the Pitx1 homeodomain and IRF3 or IRF7 and the ability of the Pitx1 C-terminal repressor domains to block IFN-A11 and IFN-A5 but not IFN-A4 promoter activities may contribute to our understanding of the complex differential transcriptional activation, repression, and antirepression of the IFN-A genes.
Type I interferon (IFN) genes are expressed after virus induction in leukocytes, epithelial cells, and fibroblast cells. The individual subtypes of the IFN-A and IFN-B genes are transcribed at various levels depending on the cell type and on the inducing virus, reflecting differences in the transcriptional activity of the corresponding gene promoter (5, 15, 16). For instance, in L929 cells, the murine IFN-A11 and IFN-A5 genes are poorly expressed upon induction by Newcastle disease virus (NDV) whereas the IFN-A4 gene is strongly inducible.
Differential expression of the IFN genes is due to the presence of various combinations of enhancer, silencer, and antisilencer elements in their promoters. The weak inducibility of the IFN-A11 gene is due in part to substitutions that inactivate two of the four enhancer elements which are active within the proximal virus responsive element of the IFN-A4 promoter (VRE-A4) (1, 3) and to the presence of an active distal negative regulatory element (DNRE) located upstream of the VRE-A11 (14, 24). A DNRE is also present in the IFN-A4 promoter, but a central antisilencer region located between the distal silencer and the proximal VRE-A4 overrides the silencer activity.
We recently searched for factors that bound to the DNRE. One such factor was the homeoprotein Pitx1 (13). Pitx1 (pituitary homeobox 1) was initially described as a transcriptional activator of the pituitary pro-opiomelanocortin (POMC) gene and other pituitary genes (9, 35). Upon virus induction, we recently showed that Pitx1 negatively regulates the transcription of DNRE-containing IFN-A11 promoter but not IFN-A4 promoter because of the presence of the central antisilencer region. After virus induction, the expression of the Pitx1 antisense RNA leads to a significant increase of endogenous IFN-A gene transcription, is able to modify the pattern of differential expression of individual IFN-A genes, and derepresses IFN-A11 and IFN-A5 genes (13). These studies show that Pitx1, previously described as an activator, could also act as a repressor as it prevented the activation by viral infection of the IFN-A11 and IFN-A5 genes.
Induction of transcription of the IFN-A genes is mainly due to the transcription factors IRF3 and IRF7, which are activated and translocated to the nucleus after viral induction (17, 25, 26). In this report we deal with the mechanism by which Pitx1 represses the IFN-A11 and IFN-A5 genes. We show that this repression does not appear to be due to the recruitment of histone deacetylases but rather to a direct interaction with the activators IRF3 and IRF7. We also demonstrate that the domain of Pitx1 responsible for interaction with both IRF3 and IRF7 is different from that responsible for the repressing effect. IRF3 binds to the antisilencer region of the IFN-A4 gene, which is located in the vicinity of the Pitx1 binding site. We suggest that Pitx1 cannot repress the IFN-A4 gene because its interaction with IRF3 bound to the antisilencer may prevent its repressing interaction with IRF3 and IRF7 binding the proximal activator region. IRF3 bound to the antisilencer of IFN-A4 promoter may thus be considered as a trap for Pitx1. These results may contribute to our understanding of the complex differential transcriptional activation, silencing, and antisilencing of the IFN-A genes.
MATERIALS AND METHODS
DNA transfection, viral induction, and transfection assays.
L929 and HeLa S3 cells were transfected as previously described (14) and by the standard calcium phosphate precipitation method. NDV induction was carried out 48 h later. The mock-induced cells were set up as explained above except that no NDV was added. Cells were harvested 24 h postinduction, and cytoplasmic extracts were prepared. Luciferase activities were measured in cell lysates using commercial reagents (Promega). Transfection efficiency was determined by the β-galactosidase activity assay with a chemiluminescence kit (Tropix). In each experiment, a given construction was transfected in duplicate and two different clones of each construction were tested. For trichostatin A (TSA) (Sigma) experiments, TSA was diluted in culture medium and added to the cells at various final concentrations. The TSA was removed from the medium before virus infection (30).
EMSA.
Nuclear extracts were prepared as described previously (14). Maltose-binding protein (MBP)-Pitx1, nuclear extracts, and electrophoretic mobility shift assays (EMSA) were handled as described previously (13) with a 32P-end-labeled probe corresponding to the CE3 (5′-ACCAGGATGCTAAGCCTCTGTC-3′), PROX (5′-CCGAGTGCTGGGATTAAAGTGGTGCA-3′), PROX-M (5′-CCGAGTGCTGTTCTTAAAGTGGTGCA), DIST (5′-CATACATTGAGGATTAAAATAAATTG), and DIST-M (5′-CATACATTGATTCTTAAAATAAATTG) sequences. Competitor experiments were performed as described previously (13). IRF3 EMSA were performed with a sequence corresponding to the dimer of the PRDI sequence (5′-GAAAGTGAAAAGGAAAGTGAAAAG-3′), 4D, (5′-AGCAGTGAAACTGAAAGCAATGATTGAACC-3′), 11D (5′-AGCAATGAAAGAGAAAGCAATGATGGAACC-3′), INT (5′-AGCAGTGAAAGAGAAAGCAATGATTGAACC-3′), EXT (5′-AGCAATGAAACTGAAAGCAATGATGGAACC-3′), and VAU (5′-AGCAGTGAAACTTGAAGCAATGATTGAACC-3′) sequences. Five micrograms of L929 nuclear extracts was incubated with 0.1 mg of poly(dG-dC) · poly(dG-dC)/ml and labeled oligonucleotides in binding buffer (10 mM Hepes [pH 7.9], 45 mM KCl, 0.2 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 2% glycerol, with 3 mM N-ethylmaleimide [NEM]). NEM in many cases eliminates protein-DNA interactions (19). Electrophoresis was performed by 6% nondenaturating Tris-borate-EDTA polyacrylamide gel electrophoresis, and the gels were dried and subjected to autoradiography. For supershift experiments, nuclear extracts were incubated on ice with the specified antibodies anti-IRF3, -IRF7 (Santa-Cruz Biotechnology), -IRF1, and IRF2 (a gift from T. Taniguchi) for 1 h at 4°C prior to the addition of the labeled oligonucleotide.
Plasmid constructions.
Native IFN-A11 promoter, already described (13), was cloned into pBL-Luc vector. This vector was derived from pBLCAT3 reporter by replacing the chloramphenicol acetyltransferase gene with the luciferase fragment. IFN-A5 promoter (−723 to +1) was cloned with two primers by PCR and subcloned into pBL-Luc vector (−723A5wt-Luc). Two copies of the upstream activator sequence (UAS) were integrated upstream of the −119A4wt or −119A11wt by PCR, and the amplified products were digested with the appropriate enzymes and cloned into pBL-Luc vector. All constructions were checked by nucleotide sequencing on a double-stranded DNA template. N- and C-terminal deletions of Pitx1 were generated by PCR and subsequently subcloned in a Rous sarcoma virus-driven expression vector described elsewhere (34, 36). The Pitx1 fragments used in the Gal4DBD-Pitx1 fusions were generated by PCR with primers containing restriction sites and subsequently subcloned in-frame in the corresponding sites of a Gal4DBD vector. Site-directed mutagenesis was used (36) to convert the lysine at position 139 of Pitx1 (residue 50 of the homeodomain [HD]) to an alanine using the pALTER (Promega) system according to the manufacturer's recommendations. IRF3, a gift from J. Hiscott, was subcloned into the pcDNA plasmid (Invitrogen), and pcDNA-IRF7A expression vector was a gift from J. S. Pagano. HDAC1 transcription transduction vector was a gift from A. Harel-Bellan. Maltose binding protein (MBP) fusion constructs were made by PCR with primers containing restriction sites. Amplified products were digested with appropriate enzymes and cloned in-frame with MBP into the pMal-c vector (New England Biolabs).
Recombinant protein production.
Escherichia coli strain BL21 was transformed with MBP fusion vectors (MBP-Pitx1, MBP-HD, MBP-N+HD, MBP-IRF3, and MBP-IRF7) derived from pMal-c (New England Biolabs). Colonies were grown in 1,000 ml to an optical density at 600 nm of 0.4 to 0.6. Induction of the expression of recombinant proteins and their purification were performed as recommended by the manufacturer. 35S-labeled translated Pitx1 (wild type and mutant), IRF3, IRF7, histone deacetylase 1 [HDAC1], and luciferase were obtained using the TNT-coupled transcription-translation rabbit reticulocyte lysate system (Promega).
Protein-protein interaction assay.
Protein-protein interaction assays were performed using MBP fusion proteins coupled to amylose-Sepharose beads (New England Biolabs) and 5 to 10 μl of in vitro-translated 35S-labeled protein incubated in the presence of 1× binding buffer (200 mM NaCl, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1-μg/ml leupeptin, 1-μg/ml pepstatin A, 0.25% bovine serum albumin) 2 h at 4°C with agitation and then centrifuged at 3,000 rpm in an Eppendorf microcentrifuge at room temperature. Beads were washed five times in binding buffer at room temperature; the protein complexes were released after boiling in Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Labeled proteins were visualized by autoradiography. For binding assays with nuclear extracts, 250 μg of L929 nuclear extracts induced by NDV for IRF3 or 250 μg of HeLa S3 expressing IRF7 and induced by NDV was incubated with MBP fusion proteins bound on beads for 4 h at 4°C with agitation in 250 μl of 1× binding buffer 20 mM HEPES KOH [pH 7.9], 50 mM KCl, 1 μM ZnSO 4, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.01% igepal, 20% glycerol, 1 μg of leupeptine/ml, 1 μg of pepstatine A/ml. The resulting binding complexes were washed in the same binding buffer for five times, and the bound proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred on a Hybond-polyvinylidene difluoride membrane and subjected to immunoblotting. Anti-IRF3 and anti-IRF7 antibodies (Santa-Cruz Biotechnology) were used. Western blot analysis was done using chemiluminescence as described by the manufacturer (Amersham). For coimmunoprecipitation experiments, each assay was carried out in 460 μl of buffer containing 20 mM HEPES KOH [pH 7.9], 50 mM KCl, 0.1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 5% glycerol containing 200 μg of L929 nuclear extracts induced by NDV for IRF3 or 100 μg of HeLa S3 expressing IRF7 and induced by NDV. Anti-IRF3 and anti-IRF7 antibodies (Santa-Cruz Biotechnology) were used for coimmunoprecipitation. An unrelated polyclonal immunoglobulin G (INC Technologies) was used as a negative control. After overnight incubation on a wheel at 4°C, 40 μl of protein A-Sepharose (Amersham) was added for 1 h at 4°C. The mixture was then centrifuged, and the pellets were washed four times in the same buffer at 4°C. Pitx1 was revealed by Western blotting using anti-Pitx1 antibody. Western blot analyses were performed as described previously.
RESULTS
Repression by Pitx1 of IFN-A promoter-mediated transcription does not require histone deacetylases.
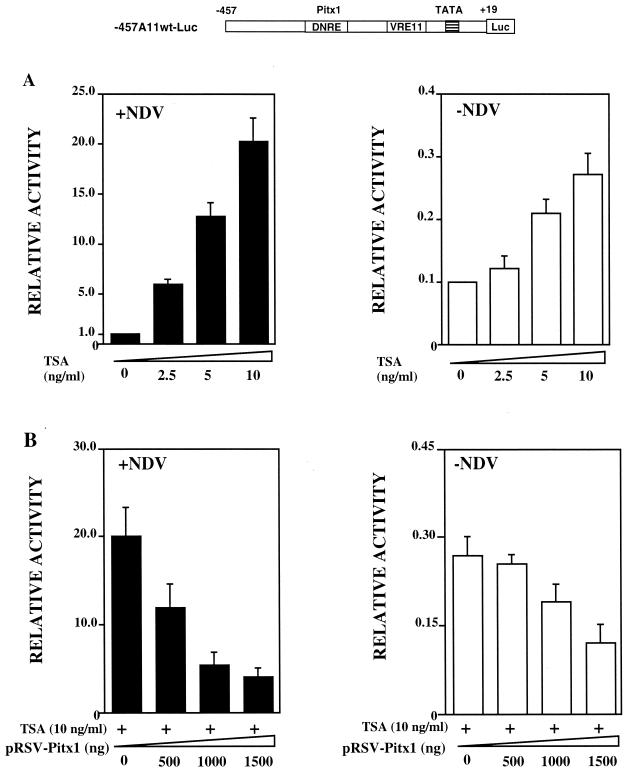
Since IFN-B promoter is known to be repressed by recruitment of corepressors, such as HDACs (30), we decided to determine whether Pitx1 repressed transcription by recruiting HDACs. It was first necessary to determine whether HDACs played any role in repression of the IFN-A promoters. This was accomplished by testing the effect of TSA, an inhibitor of HDACs, on the transcriptional activity of the IFN-A11 promoter. L929 cells were transiently transfected with a plasmid bearing a reporter luciferase (Luc) gene placed under the control of the IFN-A11 promoter (−457A11wt-Luc) and were incubated in the presence of various concentrations of TSA, with or without NDV. As expected, luciferase activity was poorly increased by the virus. Here we show that TSA was able to further increase the activity in a dose-dependent manner, particularly in the presence of virus (Fig. 1A). Complementary experiments using mutations that affected nucleotides which are involved in the repressive effect of the DNRE have been performed in the presence of TSA. After virus induction, the transcriptional activity of the promoters was released by these mutations, thus suggesting that the DNRE is still able to repress the native promoter even in the presence of TSA (data not shown). If the repressing effect of Pitx1 was mediated by HDACs, their inhibition by TSA should abolish this repressing effect. L929 cells were cotransfected with the −457A11wt-Luc reporter plasmid and increasing amounts of an expression vector containing Pitx1 cDNA. Cells were cultivated in the presence of 10 ng of TSA/ml, with or without NDV. The presence of TSA did not affect the ability of Pitx1 to repress transcription driven by the IFN-A11 promoter (Fig. 1B). We may conclude that (i) HDACs have an important function in the regulation of the IFN-A promoter activity, and (ii) the mechanism by which Pitx1 represses transcription appears to be independent of HDACs.
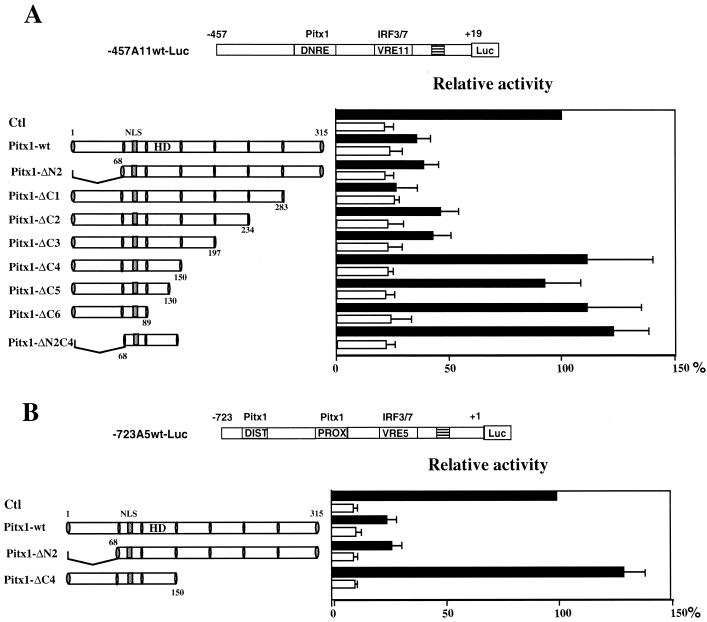
FIG. 1.
Effects of Pitx1 and TSA on the IFN-A11 promoter. The map of −457A11 wt-Luc is shown at the top. Distances from the transcription start site of IFN-A11 are indicated in base pairs. The DNRE and VRE-A11 are shown as open boxes. The TATA box is symbolized as a shaded box. The Luc gene is symbolized as a slightly larger open box. (A) L929 cells were transiently transfected with −457A11wt-Luc and treated with TSA for 48 h either in the presence (solid bars) or in the absence (open bars) of NDV. Luc activity was measured 72 h after transfection as described under Materials and Methods. All values are expressed relative to that of cells induced by the virus in the absence of TSA (left bar of left panel). The means and standard errors for Luc activity determined in at least five separate experiments are shown. (B) Cells were treated as for panel A except that they were cotransfected with increasing amounts of the expression vector pRSV-Pitx1. TSA promotes activation of the IFN-A11 promoter, particularly in the presence of virus. The repressing effect of Pitx1 is little affected by the presence of TSA.
If Pitx1 does not recruit a corepressor such as HDACs, it may directly inhibit an activator. Since IRF3 and IRF7 are the main activators of IFN-A genes, it appeared therefore to be of interest to determine whether the repressing effect of Pitx1 on the IFN-A11 and IFN-A5 promoters is mediated by interference with the transcriptional activities of IRF3 and IRF7.
Pitx1 inhibits the IRF3 and IRF7 transcriptional activity of the IFN-A11 and IFN-A5 promoters.
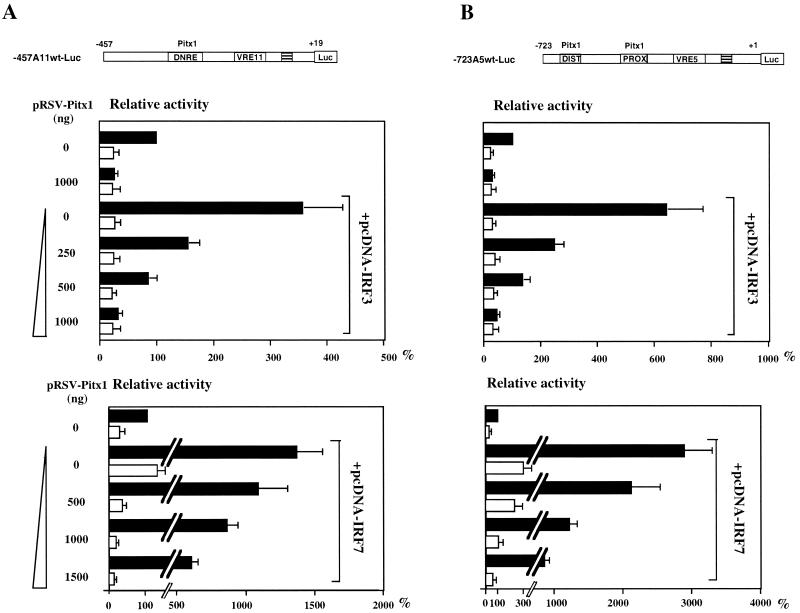
The effect of Pitx1 on the activation of the IFN-A11 promoter induced by overexpression of either IRF3 or IRF7 was tested by transfecting L929 cells bearing a −457A11wt-Luc reporter plasmid with a combination of Pitx1 and either IRF3 or IRF7 expression vectors. IRF3 was able to activate the IFN-A11-mediated transcription induced by NDV. Overexpression of Pitx1 repressed this activation by IRF3 in a dose-dependent manner (Fig. 2A, upper panel). IRF7 also stimulated IFN-A11 promoter-mediated transcription with and without virus induction (Fig. 2A, lower panel) but with a much stronger effect than IRF3. Pitx1 repressed the activation by IRF7, but its repressing effect was greater on activation by IRF3 than on that by IRF7 (Fig. 2A). Moreover, Pitx1 repressed in the absence of virus induction when IRF7 was used (Fig. 2A, lower panel), so Pitx1 may repress without virus-induced posttranslational modifications, such as phosphorylations.
FIG. 2.
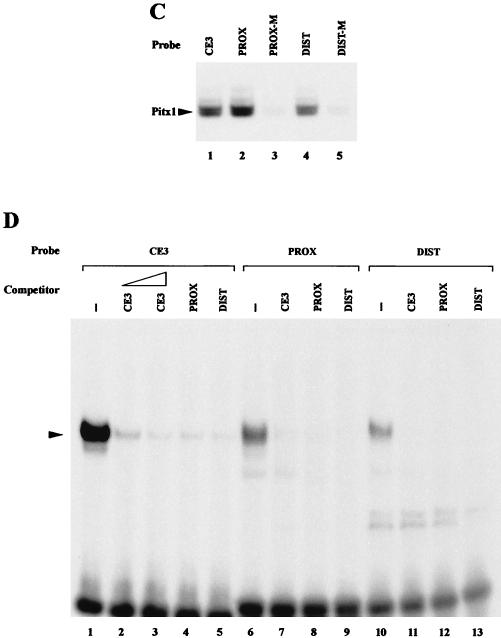
Effects of Pitx1 and IRF3 or IRF7 on IFN-A11 and IFN-A5 transcriptional activities. (A) Cells were transfected with −457A11wt-Luc, pRSV-Pitx1, and either pcDNA-IRF3 or pcDNA-IRF7. (B) Cells were transfected with the same plasmids, except that −723A5wt-Luc was used instead of −457A11wt-Luc. The distal (DIST) and proximal (PROX) Pitx1 binding sites as well as the VRE-A5 promoter are shown as open boxes. Pitx1 represses both IFN-A11 and IFN-A5 transcriptional activities even when IRF3 or IRF7 is overexpressed. (C) MBP-Pitx1 was used for EMSA with five oligonucleotides: wild-type CE3 (9), PROX and DIST probes (lanes 1, 2, and 4), and mutants for the Pitx1 binding sites PROX-M and DIST-M (lanes 3 and 5). (D) Nuclear extracts from the L929 cell line were incubated with the CE3, PROX, and DIST probes (lanes 1, 6, and 10, respectively) and a 50-fold molar excess of unlabeled CE3 (lane 2) or a 100-fold molar excess of unlabeled CE3, PROX, and DIST (lanes 3 to 5, 7 to 9, and 11 to 13, respectively).
Since we had previously shown that endogenous IFN-A5 gene expression was particularly sensitive to repression by Pitx1 (13), it was of interest to test the effect of Pitx1 on IRF3- and IRF7-induced IFN-A5 promoter (−723A5wt-Luc) activity. Results were very similar to those obtained with the IFN-A11 promoter (Fig. 2B). In view of the strong repressing effect of Pitx1 on the IFN-A5 promoter, it appeared of interest to demonstrate direct binding of Pitx1 to the IFN-A5 promoter. Two putative Pitx1 binding sites (TAATCC [distal, −684 to −679; proximal, −485 to −480, in the noncoding strand]) have been identified on the IFN-A5 promoter, but actual binding of Pitx1 to these sequences has never been shown. EMSA was performed by incubation of 32P-labeled probes containing either the proximal (PROX) or distal (DIST) putative Pitx1 binding site of IFN-A5 in the presence of recombinant Pitx1 fused to the MBP (MBP-Pitx1). Binding of Pitx1 to both the PROX and DIST probes was clearly observed. Point mutations within the consensus sequences of either of the two elements abolished Pitx1 binding (Fig. 2C). EMSA was used to test the DNA-binding properties of nuclear extracts from the L929 cell line. The CE3 element, which is known to bind Pitx1, was used as a positive control under conditions previously described (13). As shown in Fig. 2D (lane 1), a single band was observed with the CE3 probe. Similar electrophoretic mobility binding activity was observed with the PROX and DIST probes (lanes 6 and 10). The specificity of the DNA-binding activities of nuclear extracts was also demonstrated. Using CE3 as a probe (Fig. 2D, lanes 1 to 5), the binding activity was competed by CE3, PROX, and DIST, which were used as unlabeled competitors. Essentially identical results were obtained when the PROX and DIST sequences were used as probes (Fig. 2D, lanes 6 to 13). Indeed, the same specific binding of the major band was competed by CE3, PROX, and DIST, used as unlabeled competitors. Taken together, these results suggest that a protein binding to CE3, PROX, and DIST is related or identical to the Pitx1 protein. The strong and dose-dependent repression by Pitx1 of the activation by IRF3 and IRF7 suggested a direct interaction between Pitx1 and the two IRFs.
Pitx1 interacts directly and specifically with both IRF3 and IRF7.
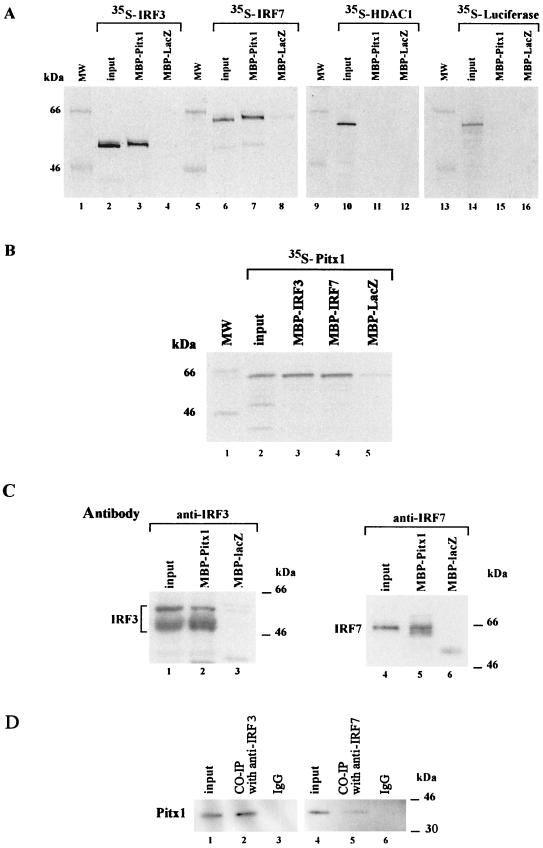
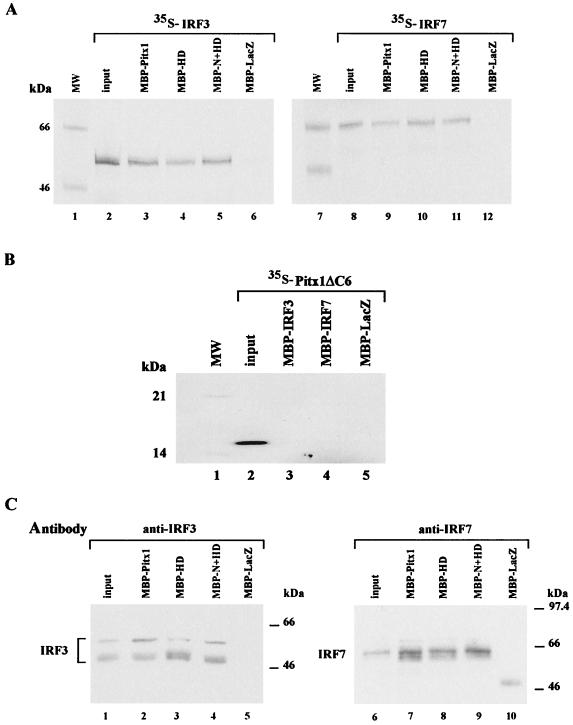
Whereas it is known that Pitx1, IRF3, and IRF7 have to bind DNA in order to exert their regulatory activities, a direct interaction between Pitx1 and either IRF3 or IRF7 has never been demonstrated. In order to test this hypothesis, we assessed whether Pitx1, IRF3, and IRF7 interact by use of the pull-down assay. Different recombinant MBP fusion proteins bound to agarose beads coated with maltose were incubated in the presence of 35S-labeled proteins. Results shown in Fig. 3A clearly demonstrate that both 35S-labeled IRF3 and 35S-labeled IRF7 interact with MBP-Pitx1. No IRF3 or IRF7 was detected when MBP-LacZ was used. In other experiments, 35S-labeled HDAC1 or luciferase did not interact with MBP-Pitx1. The interaction between Pitx1 and the two IRFs, IRF3 and IRF7, was confirmed by the reciprocal experiment. 35S-labeled Pitx1 interacted with either MBP-IRF3 or MBP-IRF7 (Fig. 3B). Since these experiments were performed with recombinant IRF3 and IRF7, it was of interest to determine whether the endogenous IRF3 or IRF7 of cells induced by NDV was able to interact with Pitx1. Nuclear extracts were prepared and incubated in the presence of MBP-Pitx1. The bound material was tested by Western blotting using either an anti-IRF3 antibody or an anti-IRF7 antibody. Both IRF3 and IRF7 were readily detectable. Neither IRF3 nor IRF7 was detected when MBP-LacZ was used (Fig. 3C). The different forms of IRF3 detected probably correspond to phosphorylated forms of this protein, as previously described (29, 37). IRF7 is spliced in several forms (39). Here we show that the spliced form of IRF7 detected corresponds to IRF7A. To confirm that Pitx1 and IRF3 and IRF7 can interact in vivo, nuclear extracts (Fig. 3D) were subjected to coimmunoprecitation. Western blot analysis of the immunoprecipitates revealed Pitx1 (Fig. 3D, lanes 2 and 5) in extracts immunoprecipitated with the antibodies anti-IRF3 and anti-IRF7 but not in control immunoglobulin G-treated extracts (Fig. 3D, lanes 3 and 6). We may conclude that Pitx1 specifically interacts with IRF3 and IRF7 but not with HDAC1. These results are in keeping with the fact that Pitx1 repressing activity does not depend on HDACs (Fig. 1B) but affects IRF3 and IRF7 activities (Fig. 2A and B).
FIG. 3.
Physical interaction of Pitx1 with IRF3 and IRF7. (A) Direct interactions were shown in pull-down assays performed using MBP fusion proteins (MBP-Pitx1 and MBP-LacZ, as the control) and in vitro-translated 35S-labeled IRF3, IRF7, HDAC1, and Luc proteins. An aliquot of input protein corresponding to 10% of labeled protein used in the assay is shown for comparison. (B) Pitx1 was labeled by in vitro translation and tested for binding to MBP-IRF3, MBP-IRF7, or MBP-LacZ. (C) MBP-Pitx1 and MBP-LacZ were incubated with nuclear extracts from NDV-induced cells. Western blot analysis of the proteins shows that both IRF3 and IRF7 were specifically bound to MBP-Pitx1. (D) Nuclear extracts from NDV-induced cells was used for coimmunoprecipitation (CO-IP), input, and control samples. Western blot analysis of the proteins shows that Pitx1 was specifically bound to both IRF3 and IRF7. MW, molecular mass markers.
Pitx1 trans-repression activity maps to the C-terminal domains, beyond the HD.
The trans-repression properties of Pitx1 on IFN-A promoters after virus induction have not yet been dissected. In order to identify the domain of Pitx1 involved in the repression, a series of Pitx1 mutants were tested in transfection assays. First, the transcriptional properties of the Pitx1 mutants were tested using simple IFN-A11 and IFN-A5 promoters. As shown in Fig. 4A and B, deletion of the N-terminal domain of Pitx1 (ΔN2) did not affect its ability to repress the two reporters after virus induction. However, with deletion of a 47-amino-acid region in the C-terminal domain between amino acids 150 and 197 of Pitx1 (ΔC4), transcription was no longer repressed. As expected, deletion of the N- and C-terminal domains of Pitx1 (ΔN2C4, which retains the HD) did not affect the repressive activity. Also, deletion within the HD (ΔC5 and ΔC6) abolished negative activity (Fig. 4A and B).
FIG. 4.
Mapping of the Pitx1 trans-repression domains. Cells were cotransfected with either −457A11wt-Luc (A) or −723A5wt-Luc (B) and with empty expression vector (Ctl) or vector encoding wild-type Pitx1 or various deletion mutants of Pitx1 (delimited by shaded vertical bars). The nuclear localization signal (NLS) and the HD of Pitx1 are shown. Luc activities are expressed relative to that of the induced activity of the −457A11wt-Luc or −723A5wt-Luc construct alone, both arbitrarily set at 100%. Assay conditions were as described in the legend to Fig. 1A.
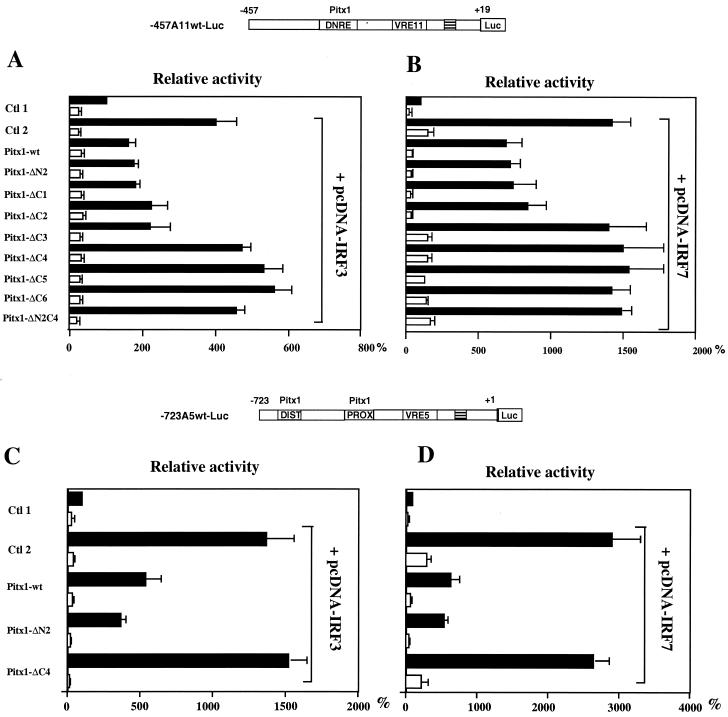
The abilities of these mutants to repress the IFN-A promoters in the presence of IRF3 and IRF7 were then evaluated. As well as for its activity on simple reporters (Fig. 4A and B), the amino acids 150 to 197 appeared to be necessary for trans repression on the IFN-A11 promoter after virus induction when IRF3 was overexpressed (Fig. 5A). Nevertheless, the amino acids 197 to 234 did appear to be necessary for trans repression when IRF7 was overexpressed with or without virus infection (Fig. 5B). The C-terminal domain of Pitx1 also seems to be important for trans repression of the IRF3- and IRF7-induced IFN-A5 promoter (−723A5wt-Luc) activity (Fig. 5C and D).
FIG. 5.
Mapping of the Pitx1 trans-repression domains involved in repression of IFN-A11 transcription activated by IRF3 or IRF7. (A) Cells were cotransfected with a combination of −457A11wt-Luc, pcDNA-IRF3, and progressively deleted forms of the wild-type Pitx1. Ctl1 and Ctl2 correspond to cells that were transfected with −457A11wt-Luc in combination with either empty overexpression vectors (Ctl 1) or pcDNA-IRF3 (Ctl 2). (B) The same experiment as for panel A except that pcDNA-IRF7 was used instead of pcDNA-IRF3. (C and D) The same experiments as for panels A and B except that −723A5wt-Luc was used instead of −457A11wt-Luc. The trans-repression domains of Pitx1 on the IFN-A11 promoter map to residues 150 to 197 (ΔC4) for IRF3 overexpression and to residues 197 to 234 (ΔC3) for IRF7 overexpression. The trans-repression domain of Pitx1 on the IFN-A5 promoter was more broadly defined but appeared to be the same.
These results suggest that Pitx1 trans-repression activity maps to the C-terminal domains, beyond the HD, and these domains have not yet been described as trans-regulator domains (see also Fig. 9B). We then decided to map the residues of Pitx1 that are critical for its interaction with IRF3 or IRF7. Such residues need not necessarily be the same as those which are critical for the repressing effect.
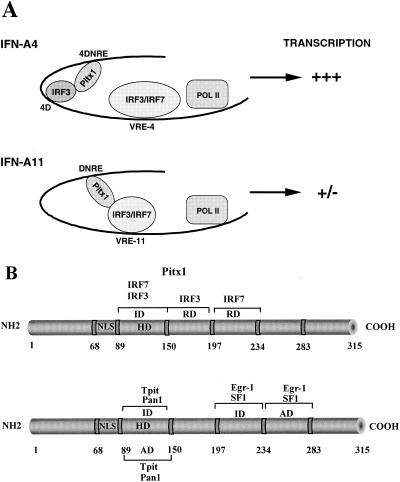
FIG. 9.
(A) Model for Pitx1 repression. Pitx1 represses transcriptional activity of the IFN-A11 promoter by its interaction with IRF3 or IRF7. Pitx1 does not repress IFN-A4 because IRF3 bound to its antisilencer interacts with Pitx1, thereby preventing it from interacting with IRF3 and IRF7 bound to the VRE-A4. (B) Schematic representations showing the trans-repression domains and the interaction domain of Pitx1 with IRF3 and IRF7 (upper diagram); the trans-activation domains and interaction domains of Pitx1 with SF1, Egr-1, Pan1, and Tpit are shown in the lower diagram. ID, interaction domain; RD, trans-repression domain; AD, activation domain. The various factors interacting with Pitx1 also are shown.
Pitx1 interacts with IRF3 and IRF7 through its HD.
The region of physical interaction between Pitx1 and IRF3 or IRF7 was identified using various truncated forms of MBP-Pitx1, containing the HD alone (MBP-HD, residues 84 to 153) or the HD and the N-terminal domain (MBP-N+HD, residues 1 to 153) in pull-down assay experiments with 35S-labeled IRF3 or IRF7. MBP-HD of Pitx1 is sufficient for interaction with either IRF3 or IRF7 with the same efficiency as was intact MBP-Pitx1. No interaction was detected in the presence of MBP-LacZ (Fig. 6A). The absence of interaction of either MBP-IRF3 or MBP-IRF7 with 35S-labeled Pitx1 lacking the HD confirmed that the HD was the part of Pitx1 responsible for interaction with IRFs (Fig. 6B). In order to determine whether endogenous IRF3 or IRF7 could interact with the HD of Pitx1, a nuclear extract induced with NDV was incubated in the presence of various MBP-truncated forms. The bound material was analyzed by Western blotting using an anti-IRF3 antibody or an anti-IRF7 antibody. The Pitx1 HD interacted with IRF3 and IRF7 with an affinity close to that of intact Pitx1. No interaction of either IRF3 or IRF7 with MBP-LacZ was observed (Fig. 6C). These results show that Pitx1 interacts with both IRF3 and IRF7 through its HD but exerts its repressing effect on IRF3 and IRF7 through a region adjacent to the HD, on its C-terminal side. It remained to be determined whether interaction of Pitx1 HD with IRF3 and IRF7 was necessary for its repressing activity.
FIG. 6.
Mapping of the Pitx1 domains, which interact with IRF3 or IRF7. (A) Interactions of IRF3 or IRF7 with Pitx1 were examined in pull-down assays, using MBP fusion proteins: MBP-Pitx1, MBP-HD (residues 84 to 153), MBP−N+HD (residues 1 to 153) and MBP-LacZ as a control. Proteins were incubated in the presence of in vitro-translated 35S-labeled IRF3 and IRF7 proteins as described for Fig. 3. (B) Pitx1 lacking the HD (Pitx1-ΔC6) protein was labeled by in vitro translation and tested for binding to MBP-IRF3, MBP-IRF7, or MBP-LacZ. (C) The interaction of endogenous IRF3 or IRF7 protein with Pitx1 was examined by incubating MBP fusion proteins (MBP-Pitx1, MBP-HD, MBP-N+HD, and MBP-LacZ) in the presence of nuclear extracts from NDV-induced cells. Binding of IRF3 or IRF7 to Pitx1 was analyzed by Western blotting using an anti-IRF3 or anti-IRF7 antibody. The HD is the region of Pitx1 that interacts with IRF3 and IRF7.
The physical interaction between the Pitx1 HD and IRF3 or IRF7 is necessary for trans repression by the Pitx1 C-terminal domains.
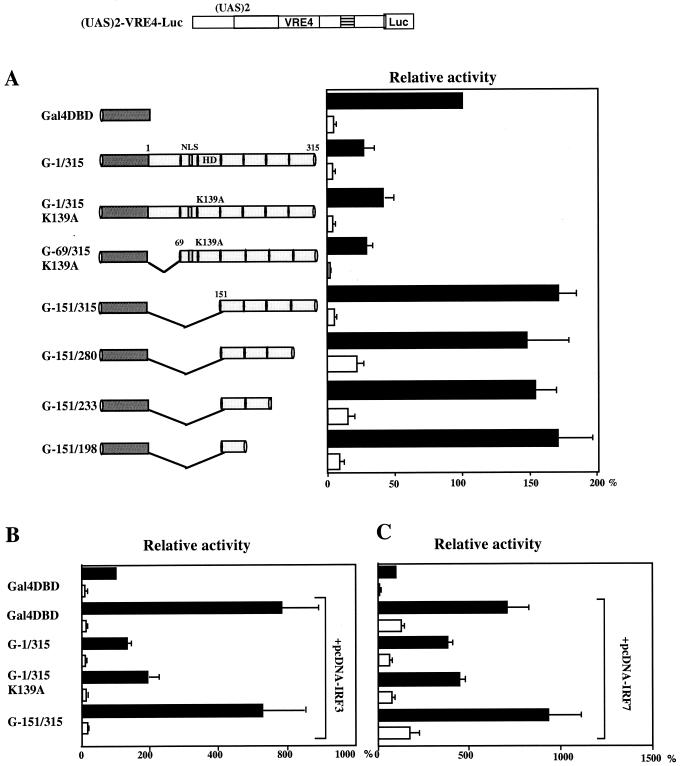
Since the HD of Pitx1 was responsible for binding to DNA and for interaction with IRF3 and IRF7, it was not possible to delete it and thereby determine whether suppression of interaction with IRFs led to concomitant suppression of the repressing activity. To circumvent this problem, we fused Pitx1 cDNA to the DNA binding domain (DBD) of Gal4 and tested the repressing activity of the fusion protein on a Luc reporter gene containing Gal4 binding sites (upstream activating sequence [UAS]) placed upstream of a proximal IFN-A4 promoter [(UAS)2-VRE4-Luc], containing the VRE-A4 but no Pitx1 binding site. The Gal4-Pitx1 (G-1/315) fusion protein was able to repress efficiently the (UAS)2-VRE4-Luc gene. Deletion mapping showed that the repressing effect of Pitx1 require residues 69 to 150, which include the entire HD (residues 90 to 150). Replacement of the lysine residue normally present at position 139 of Pitx1 with an alanine residue has been shown to abolish DNA binding (36). This replacement had no effect on the repressing activity of the Gal4-Pitx1 fusion protein (Fig. 7A). With experiments similar to those described above, we showed that the HD was also necessary for the repressing activity of Pitx1 in cells overexpressing IRF3 or IRF7 (Fig. 7B and C). In summary, we have shown that the interaction of the HD of Pitx1 with IRF3 or IRF7 is critical for the repressing activity of Pitx1. However, the repressing activity of Pitx1 is not confined to the HD itself but extends towards the adjacent C-terminal region.
FIG. 7.
The Pitx1 HD which interacts with IRF3 or IRF7 is critical for trans repression by the C-terminal domains. (A) Cells were cotransfected with (UAS)2-VRE4-Luc and one of various expression vectors encoding progressively deleted forms of the Pitx1 cDNA fused to the Gal4 DBD. In some constructs, the lysine at position 139 of Pitx1 was converted to an alanine (K139A) by site-directed mutagenesis. UAS is the Ga14 binding site. VRE4 is the virus-responsive element of IFN-A4. (B and C) Experiments similar to those for panel A except that cells were also cotransfected with pcDNA-IRF3 and pcDNA-IRF7, respectively. Deletion of the HD of Pitx1 abolishes trans repression.
IRF3 binds the central antisilencer region 4D of the IFN-A4 gene promoter.
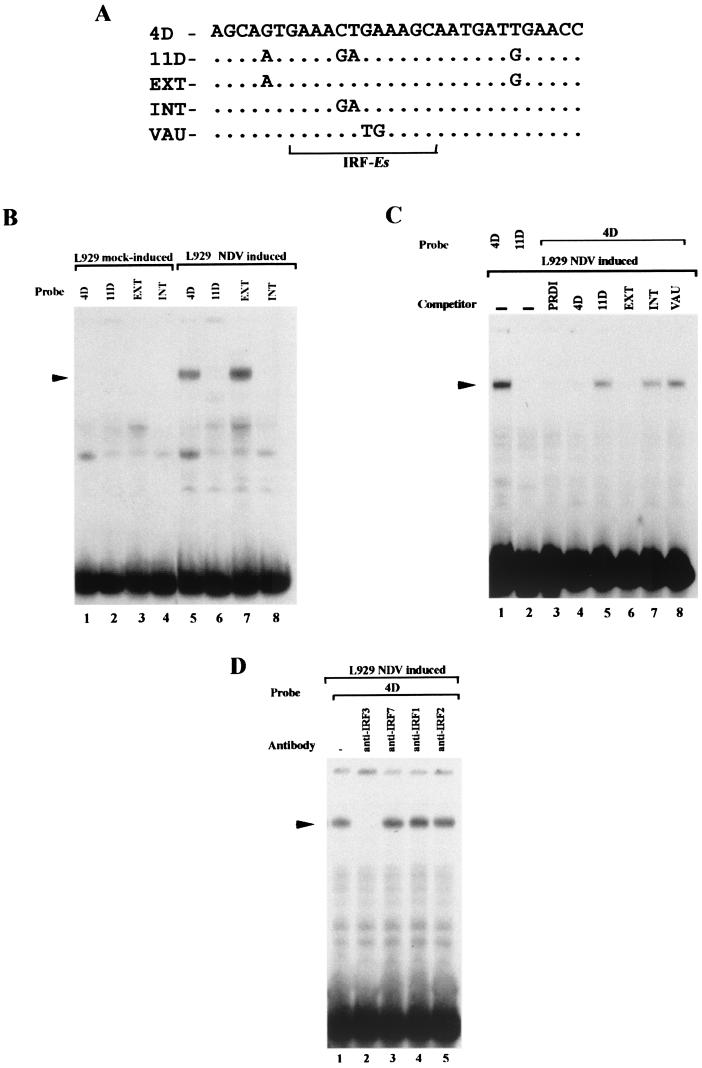
We have previously shown that the distal DNRE found in the IFN-A4 promoter (4DNRE) is able to reduce the transcriptional activity of minimal proximal VRE-A4 promoters and has been considered a silencer (14). The fact that the intact IFN-A4 gene promoter remains highly inducible upon virus induction whereas the intact IFN-A11 gene promoter is poorly expressed has also been explained by the presence of a central antisilencer region (4D; between −260 and −212). The related element present in the IFN-A11 promoter (11D; between −146 and −199) has no antisilencer activity, and there are only four substitutions between the 4D and 11D elements. DNase 1 footprinting analysis of the IFN-A4 promoter exhibited a protection (−193/−164) with nuclear extracts from uninduced and NDV-induced L929 cells on both the coding and noncoding strands (data not shown). We have designated the protected region in the IFN-A4 promoter as 4D. The IFN-A4 promoter differs from that from IFN-A11 in that it possesses within its antisilencer a putative IRF binding site (IRF-Es) (Fig. 8A) (32) in close proximity to its Pitx1 binding site and in that it is not repressed by Pitx1 (13). The IRF-Es within the 4D antisilencer element share two nucleotide substitutions in the corresponding nonfunctional element 11D (Fig. 8A). Factors binding to 4D and not to 11D elements were identified by EMSAs with nuclear extracts from uninduced and NDV-induced L929 cells (Fig. 8B). The EXT element, which contains the two nucleotide substitutions outside of the IRF-Es, and INT, which contains the two substitutions within the IRF-Es, were also used as probes. Alkylation by NEM, which in many cases eliminates some protein-DNA interactions (19), was used. Nonrelevant complexes were observed before virus induction when 4D, 11D, EXT, and INT were used as probes (Fig. 8B, lanes 1 to 4). After virus induction, a lower-mobility complex was detected by the use of 4D and EXT probes (Fig. 8B, lanes 5 and 7), whereas the formation of this complex is absent with use of 11D and INT probes (Fig. 8B, lanes 6 and 8). So in NDV-induced nuclear extract, a protein complex binds the 4D element but not the 11D element, and since it could not bind the INT element, this complex seems to bind the IRF-Es. The specificity of the formation of this complex was demonstrated using unlabeled positive regulatory domain I (PRDI) (which contains IRF-Es), 4D, 11D, INT, EXT, and VAU (a mutant of the IRF-Es that is known to not bind IRFs) elements (Fig. 8C). In the presence of unlabeled 11D, INT, and VAU elements, no competition was observed (Fig. 8C, lanes 5, 7, and 8), whereas there is a competition with PRDI, 4D, and EXT (Fig. 8C, lanes 3, 4, and 6). These results are in keeping with the fact that this protein complex binds the IRF-Es. In EMSA with nuclear extracts from induced L929 cells using 4D as a probe, several antibodies against IRF factors (anti-IRF3, -IRF7, -IRF1, and -IRF2) were used (Fig. 8D). The antibody anti-IRF3 (Fig. 8D, lane 2) but not antibodies anti-IRF7, anti-IRF1, and anti-IRF2 was able to inhibit the formation of the complex of interest.
FIG. 8.
EMSA showing binding of IRF3 to the antisilencer of the IFN-A4 promoter. (A) Nucleotide sequences of the oligonucleotides used. 4D is the sequence found in the antisilencer of IFN-A4; 11D is the corresponding sequence in IFN-A11; EXT, INT, and VAU are forms of antisilencer mutated either inside or outside the IRF binding site (IRF-Es, underlined). (B) L929 nuclear extracts were subjected to alkylation and incubated in the presence of oligonucleotides labeled with [γ-32P]ATP. The nucleoprotein complexes were resolved by EMSA. Specific protein binding was observed using the antisilencer 4D after virus induction (lanes 5 and 7) but not the IFN-A11 corresponding region after virus infection. (C) A nuclear extract was incubated in the presence of 32P-labeled 4D oligonucleotide and an excess of one of the various unlabeled oligonucleotides. Nucleoprotein complexes were examined as for panel B. Binding to the 4D region is abolished by an excess of the 4D oligonucleotide. (D) Nucleoprotein complex formation was inhibited by the addition of an antibody against IRF3 but not by antibody against IRF7, IRF1, or IRF2.
In conclusion, a complex which specifically binds the 4D antisilencer element, which prevents silencing in the intact IFN-A4 promoter, is related or identical to IRF3.
DISCUSSION
Pitx1 is a silencing and quenching repressor of IFN-A genes.
Most of the research on eukaryotic genes expression has been focused on the transcriptional activation mechanisms (33). Although activation of gene expression is essential, it becomes clear now that transcriptional repression is at least as important. Indeed, as an inherent feature, those gene systems which are positively regulated may require an inhibition mechanism to account for the following: temporal or spatial expression during development, cell or tissue specificity, modulation of gene expression depending on environmental changes, and rapid expression shutdown after induction (2, 4, 7, 11, 23). Different mechanisms of transcriptional repression have been proposed. The first mechanism, called squelching, involves direct interaction between repressors and activators without binding to DNA. The second mechanism is competition for overlapping DNA-binding sites between repressors and activators or basal transcription factors. The third mechanism is direct repression, also sometimes termed silencing by analogy with enhancing (position- and orientation-independent regulatory element), by which the repressor blocks transcription by interaction with basal transcription factors. The fourth mechanism is quenching, by which the repressor blocks transcription by specific interaction with activators and coactivators. For silencing and quenching, the repressor acts by nonoverlapping DNA-binding activity. The DNRE of IFN-A genes exerts an inhibitory effect on proximal VRE-A promoters after virus induction, whatever its orientation or position, and is therefore considered a silencer (14, 24). One property of the DNRE is that its silencing activity is strictly dependent upon the presence of a functional VRE-A and does not affect heterologous promoters. It was therefore not surprising that Pitx1, which binds the DNRE, could not repress heterologous promoters. We had concluded that Pitx1 interfered with specific activators bound to the VRE-A and did not affect the basal transcriptional machinery. In the present study we identify these activators as IRF3 and IRF7. According to the definitions stated above, Pitx1 should be considered as a repressor including both silencing and quenching activities.
The repressing effect of Pitx1 on the IFN-A11 gene is likely to be independent of the chromatin structure. We show that Pitx1 is not able to recruit HDACs to the IFN-A promoter (Fig. 1B). However, HDACs must be important regulators of IFN-A11 gene transcription, since TSA, an inhibitor of HDACs, leads to the activation of the gene. Since HDACs do not bind DNA directly, they must be recruited to the IFN-A11 promoter by an unknown factor, distinct from Pitx1.
IRF3 bound to the antisilencer of the IFN-A4 gene as a molecular trap for Pitx1.
Whereas Pitx1 represses the expression of the IFN-A11 gene, it has no effect on that of the IFN-A4 gene. This is because the IFN-A4 promoter contains an antisilencer which overrides the activity of the DNRE of the IFN-A4 gene. What is the mechanism by which the antisilencer overrides the DNRE and the effect of Pitx1 bound to it? We have identified an IRF binding site on the antisilencer close to the Pitx1 element. We show that IRF3 binds this IRF binding site. We hypothesize that the close proximity of the antisilencer with the Pitx1 binding site would favor the interaction of Pitx1 with IRF3 bound to the antisilencer rather than its interaction with IRF3 and IRF7 bound to the more distant VRE-A4. In the absence of antisilencer, as in the IFN-A11 gene, Pitx1 is free to interact with IRF3 and IRF7 bound to VRE-A11 and to repress transcription. IRF3 bound to the antisilencer may be considered a molecular trap maintaining the IFN-A4 gene in a highly inducible state (Fig. 9A).
Pitx1 is both a repressor and an activator of gene expression.
Pitx1 which is present in numerous tissues, was initially described as an activator of genes expressed in the pituitary gland, such as those encoding POMC, luteinizing hormone (LHβ), and growth hormone (35). We have recently found that Pitx1 was a repressor of IFN-A gene expression (13). Targeted inactivation of the Pitx1 gene in mice leads to skeletal abnormalities (10). Whether these developmental skeletal abnormalities result from the loss of activation or from the loss of repression of target genes by Pitx1 remains unknown.
We have mapped functional negative domains, C-terminal domains consisting of amino acids 150 to 197 for virus induction or IRF3 activation and amino acids 197 to 234 for IRF7 activation (Fig. 9B). The Pitx1 HD is involved in physical interaction with IRF3 or IRF7 (Fig. 9B), and this interaction is critical for trans repression by the Pitx1 C-terminal domains (Fig. 7). Pitx1 can modulate differently IFN-A gene and POMC or other pituitary gene expression. The activity of Pitx1 as positive regulator of the transcription is synergized by cell-restricted transcription factors to confer pituitary-, lineage-, and promoter-specific expression. The opposite functions of Pitx1 factor may be due to the multiple positive and negative trans-regulatory domains and interaction regions. Indeed, several known transcriptional interaction factors act in synergy with Pitx1: Pan1 (21, 22) and Tpit (8) for the POMC promoter, SF-1 and Egr-1 (34-36) for the LHβ promoter. The Pitx1 C-terminal region is involved in transcriptional activation (amino acids 234 and 283) with SF1 or Egr-1, and the Pitx1 HD is involved in transcriptional synergism with Tpit and Pan1 (Fig. 9B). The Pitx1 HD is involved in physical interaction with Tpit and Pan1, and amino acids 197 to 234 interact with SF1 and Egr-1. Therefore, the opposite functions of the Pitx1 factor are due to the different positive and negative trans-regulatory domains.
Pitx1 is not the first example of a transcription factor that can be either a repressor or an activator, depending on the promoter to which it is bound. Engrailed, another homeoprotein, represses Hsp70 gene transcription (20) and activates directly a Polycomb group gene, polyhomeotic, during embryogenesis (28). IRF2 represses the IFN-B gene (27) and activates the VCAM-1 gene (6). IRF7, which contains both repressor and activation domains (12, 18), represses the Epstein-Barr Virus EBNA-11 Q promoter (39) and activates the IFN-A and -B genes (26) as well as the Tap-2 gene (38), presumably by distinct mechanisms.
Pitx1 represses the IFN-A11 and IFN-A5 promoters by interacting with IRF3 and IRF7. It would be of interest now to determine the domains of IRF3 and IRF7 which interact with Pitx1 and whether Pitx1 can also repress the activation by IRFs of others genes.
Acknowledgments
We thank J. Hiscott, J. S. Pagano, A. Harel-Bellan and T. Taniguchi for kindly providing IRF3, IRF7, HDAC1 cDNA, and IRF1/IRF2 antibodies respectively. We are grateful to E. Bonnefoy for discussion and encouragements, and E. Prieto for photographs. This work at UPR-2228 was supported by the Centre National de la Recherche Scientifique (CNRS), the Université René Descartes Paris V and grants from the Association de Recherche contre le Cancer (ARC, contrat 9994), Ligue Régionale contre le Cancer and CNRS PCV program (PCV098-33). M-L Island is a recipient of the Association de Recherche contre le Cancer and T Mesplede is a recipient of the Ligue Nationale contre le Cancer.
REFERENCES
- 1.Braganca, J., P. Genin, M. T. Bandu, N. Darracq, M. Vignal, C. Casse, J. Doly, and A. Civas. 1997. Synergism between multiple virus-induced factor-binding elements involved in the differential expression of interferon A genes. J. Biol. Chem. 272:22154-22162. [DOI] [PubMed] [Google Scholar]
- 2.Clark, A. R., and K. Docherty. 1993. Negative regulation of transcription in eukaryotes. Biochem. J. 296:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genin, P., J. Braganca, N. Darracq, J. Doly, and A. Civas. 1995. A novel PRD I and TG binding activity involved in virus-induced transcription of IFN-A genes. Nucleic Acids Res. 23:5055-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodbourn, S. 1990. Negative regulation of transcriptional initiation in eukaryotes. Biochim. Biophys. Acta 1032:53-77. [DOI] [PubMed] [Google Scholar]
- 5.Hiscott, J., K. Cantell, and C. Weissmann. 1984. Differential expression of human interferon genes. Nucleic Acids Res. 12:3727-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jesse, T. L., R. LaChance, M. F. Iademarco, and D. C. Dean. 1998. Interferon regulatory factor-2 is a transcriptional activator in muscle where it regulates expression of vascular cell adhesion molecule-1. J. Cell. Biol. 140:1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson, A. D. 1995. The price of repression. Cell 81:655-658. [DOI] [PubMed] [Google Scholar]
- 8.Lamolet, B., A. M. Pulichino, T. Lamonerie, Y. Gauthier, T. Brue, A. Enjalbert, and J. Drouin. 2001. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell 104:849-859. [DOI] [PubMed] [Google Scholar]
- 9.Lamonerie, T., J. J. Tremblay, C. Lanctot, M. Therrien, Y. Gauthier, and J. Drouin. 1996. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10:1284-1295. [DOI] [PubMed] [Google Scholar]
- 10.Lanctot, C., A. Moreau, M. Chamberland, M. L. Tremblay, and J. Drouin. 1999. Hindlimb patterning and mandible development require the Ptx1 gene. Development 126:1805-1810. [DOI] [PubMed] [Google Scholar]
- 11.Levine, M., and J. L. Manley. 1989. Transcriptional repression of eukaryotic promoters. Cell 59:405-408. [DOI] [PubMed] [Google Scholar]
- 12.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320-34327. [DOI] [PubMed] [Google Scholar]
- 13.Lopez, S., M. L. Island, J. Drouin, M. T. Bandu, N. Christeff, N. Darracq, R. Barbey, J. Doly, D. Thomas, and S. Navarro. 2000. Repression of virus-induced interferon A promoters by homeodomain transcription factor Ptx1. Mol. Cell. Biol. 20:7527-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez, S., R. Reeves, M. L. Island, M. T. Bandu, N. Christeff, J. Doly, and S. Navarro. 1997. Silencer activity in the interferon-A gene promoters. J. Biol. Chem. 272:22788-22799. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald, N. J., D. Kuhl, D. Maguire, D. Naf, P. Gallant, A. Goswamy, H. Hug, H. Bueler, M. Chaturvedi, J. de la Fuente, H. Ruffner, and C. Weissmann. 1990. Different pathways mediate virus inducibility of the human IFN-alpha 1 and IFN-beta genes. Cell 60:767-779. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis, T., L. A. Whittemore, W. Du, C. M. Fan, A. D. Keller, V. J. Palombella, and D. Thanos. 1992. Positive and negative control of human interferon-beta gene expression, p. 1193-1220. In S. L. McKnight and K. R. Yamamoto, (ed.), Transcriptional regulation, vol. II. C. Press, New York, N.Y.
- 17.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marie, I., E. Smith, A. Prakash, and D. E. Levy. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkuma, Y., M. Horikoshi, R. G. Roeder, and C. Desplan. 1990. Engrailed, a homeodomain protein, can repress in vitro transcription by competition with the TATA box-binding protein transcription factor IID. Proc. Natl. Acad. Sci. USA 87:2289-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulin, G., M. Lebel, M. Chamberland, F. W. Paradis, and J. Drouin. 2000. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 20:4826-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulin, G., B. Turgeon, and J. Drouin. 1997. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17:6673-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renkawitz, R. 1990. Transcriptional repression in eukaryotes. Trends Genet. 6:192-197. [DOI] [PubMed] [Google Scholar]
- 24.Roffet, P., S. Lopez, S. Navarro, M. T. Bandu, C. Coulombel, M. Vignal, J. Doly, and G. Vodjdani. 1996. Identification of distal silencing elements in the murine interferon-A11 gene promoter. Biochem. J. 317:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 26.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 27.Senger, K., M. Merika, T. Agalioti, J. Yie, C. R. Escalante, G. Chen, A. K. Aggarwal, and D. Thanos. 2000. Gene repression by coactivator repulsion. Mol. Cell 6:931-937. [DOI] [PubMed] [Google Scholar]
- 28.Serrano, N., and F. Maschat. 1998. Molecular mechanism of polyhomeotic activation by Engrailed. EMBO J. 17:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 30.Shestakova, E., M. T. Bandu, J. Doly, and E. Bonnefoy. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 75:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simeone, A., D. Acampora, A. Mallamaci, A. Stornaiuolo, M. R. D'Apice, V. Nigro, and E. Boncinelli. 1993. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 12:2735-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka, N., T. Kawakami, and T. Taniguchi. 1993. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tjian, R., and T. Maniatis. 1994. Transcriptional activation: a complex puzzle with few easy pieces. Cell 77:5-8. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay, J. J., and J. Drouin. 1999. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol. Cell. Biol. 19:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay, J. J., C. Lanctot, and J. Drouin. 1998. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol. Endocrinol. 12:428-441. [DOI] [PubMed] [Google Scholar]
- 36.Tremblay, J. J., A. Marcil, Y. Gauthier, and J. Drouin. 1999. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 18:3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J. Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]