Abstract
Gene silencing associated with repeated DNA sequences has been reported for many eukaryotes, including plants. However, its biological significance remains to be determined. One important function that has been proposed is the suppression of transposons. Here, we address transposon suppression by examining the behavior of the tobacco retrotransposon Tto1 and endogenous retrotransposons in Arabidopsis. After an initial increase in copy number because of active transposition in the Arabidopsis genome, Tto1 became silent. The amount of transcript was reduced, and the inactivated Tto1 became methylated. This silencing correlated with an increase in copy number. These phenomena mimic repeat-induced gene silencing. The homozygous ddm1 (for decrease in DNA methylation) mutation of Arabidopsis results in genomic DNA hypomethylation and the release of silencing in repeated genes. To investigate the role of DNA methylation and the gene-silencing machinery in the suppression of Tto1, we introduced the ddm1 mutation into an Arabidopsis line carrying inactivated Tto1 copies. In the homozygous ddm1 background, Tto1 became hypomethylated and transcriptionally and transpositionally active. In addition, one of the newly isolated endogenous Arabidopsis retrotransposon families, named Tar17, also became hypomethylated and transcriptionally active in the ddm1 mutant background. Our results suggest that the inactivation of retrotransposons and the silencing of repeated genes have mechanisms in common.
INTRODUCTION
Suppression of repeated genes is found in several eukaryotes and is referred to as homology-dependent gene silencing. The silencing of repeated transgenes has been investigated extensively in plants (reviewed in Flavell, 1994; Matzke et al., 1996; Meyer and Saedler, 1996; Vaucheret et al., 1998; Grant, 1999; Selker, 1999). Silencing results from a decrease in the amount of mRNA at steady state and is often, but not always, accompanied by cytosine methylation of the affected genes. The DNA methylation associated with silencing is not restricted to repeated transgenes but is also found in repeated endogenous genes, such as the tryptophan biosynthetic gene PAI of Arabidopsis (Bender and Fink, 1995) and R of maize (Ronchi et al., 1995).
The biological significance of the silencing of repeated genes, however, remains to be determined. Silencing repeated genes should confer a selective advantage, given that it is a well-conserved biological phenomenon seen in plants, mammals, fungi, and other eukaryotes (Pal-Bhadra et al., 1997; Garrick et al., 1998). One possible function of DNA methylation and inactivation of repeated sequences is suppression of transposons within the genome (Yoder et al., 1997; Martienssen, 1998). Transposons induce detrimental changes in the host genes in several ways, one of which is the disruption of genes by insertion mutations. For Drosophila, in which DNA methylation does not occur, 80% of spontaneous mutations are caused by the insertion of retrotransposons (Green, 1988). In plants, a large part of the genome is occupied by retrotransposons. For example, 50 to 80% of the 2400-Mb maize genome and at least 40% of the 12,500-Mb fava bean genome are occupied by retrotransposons (Pearce et al., 1996; SanMiguel et al., 1996; SanMiguel and Bennetzen, 1998). However, only a small portion of spontaneous mutations in plants has been shown to be caused by retrotransposons (Wessler et al., 1995). In contrast to the retrotransposons of Drosophila, most plant retrotransposons are highly methylated, and they are almost always not transcribed (Bennetzen et al., 1994; Hirochika, 1997). Together, these results suggest that methylation is an effective strategy in retrotransposon suppression.
Because retrotransposons transpose by way of an RNA, inhibiting transcription by DNA methylation should be the most effective way to suppress the transposition of retrotransposons. In addition, because retrotransposons carry relatively strong promoter–enhancer elements (Pouteau et al., 1991; Hirochika et al., 1996a), the presence of a large number of retrotransposons in the genome could have a marked influence on the transcription of flanking genes. Thus, suppression of the promoter–enhancer activity by methylation should also be important for ensuring proper transcription of the nearby genes. Suppression of transcription may be more important than suppression of transposition, because most plant retrotransposons have already lost the ability to transpose, given an accumulation of mutations (Flavell et al., 1992; Voytas et al., 1992; Hirochika and Hirochika, 1993).
For genetic dissection of gene silencing and DNA methylation, Arabidopsis provides a good model system. Two types of Arabidopsis mutants with decreases in DNA methylation have been isolated: ddm1 mutants isolated from a population mutagenized by exposure to ethyl methanesulfonate (Vongs et al., 1993; Kakutani et al., 1995), and transgenic plants that express the DNA methyltransferase gene (MET1) in the antisense orientation (Finnegan and Dennis, 1993; Finnegan et al., 1996; Ronemus et al., 1996). Recently, several mutants modifying the silencing of repeated transgenes were also isolated, including modifiers of gene silencing such as the egs1 and egs2 mutations, which enhance silencing of rolB transgene (Dehio and Schell, 1994); suppressors of transgene silencing such as the sgs1 and sgs2 mutations, which release cosuppression (Elmayan et al., 1998); somniferous (som) mutations, which release the silencing of repeated hygromycin phosphotransferase (HPT) genes driven by the cauliflower mosaic virus 35S promoter (Mittelsten Scheid et al., 1998); and the altered transgene-silencing hog1, sil1, and sil2 mutants, which reduce the silencing of repeated chalcone synthase (CHS) transgenes (Furner et al., 1998).
For both repeated HPT and repeated CHS systems, the ddm1 mutation also releases transgene silencing (Furner et al., 1998; Mittelsten Scheid et al., 1998). Moreover, this mutation impairs silencing of the repeated endogenous PAI2 gene (Jeddeloh et al., 1998). Some of the som mutations are allelic to ddm1, and all the som mutants and the hog1 mutant cause hypomethylation of the genomic DNA as well as of the transgenes, although methylation is not affected by the sil1 or sil2 mutations (Furner et al., 1998; Mittelsten Scheid et al., 1998). Interestingly, sgs1 and sgs2 distinctly modify methylation of the transgene but not methylation of the host centromere (Elmayan et al., 1998).
In this study, we tested the hypothesis that both methylation of genomic DNA and the gene-silencing machinery are necessary for suppression of retrotransposons. An impaired gene-silencing system could be induced by the ddm1 mutation of Arabidopsis. Because no active endogenous retrotransposon has thus far been isolated from Arabidopsis, we used the tobacco retrotransposon Tto1 (Hirochika, 1993). After an initial increase in copy number, Tto1 became silent in the wild-type Arabidopsis genome. In the ddm1 mutant background, however, the Tto1 transcript accumulated, and the Tto1 copy number increased. In addition, a family of endogenous retrotransposons was transcriptionally reactivated in a ddm1 mutant background. These results strongly suggest that the gene-silencing machinery is effective in defending the genome against retrotransposons.
RESULTS
Transposition of Tto1 in Arabidopsis
The tobacco retrotransposon Tto1 is one of a few active plant retrotransposons (Hirochika, 1993). Tto1 is almost inactive in unstressed tissues but can be activated by tissue culture. Because transposition of Tto1 occurs concomitantly with a marked increase in Tto1 RNA, it appears to be primarily regulated at the transcriptional level. Being an autonomous element known to be active in the distantly related heterologous host rice (Hirochika et al., 1996a), the Tto1-1 clone was introduced into Arabidopsis by way of Agrobacterium-mediated transformation. Because the silencing of Tto1 was observed clearly in transgenic lines that carried a high copy number of transposed Tto1 copies, the main focus of this study was on transgenic lines with high copy numbers. Characterization of lines with low copy numbers will be described elsewhere (H. Okamoto and H. Hirochika, unpublished data).
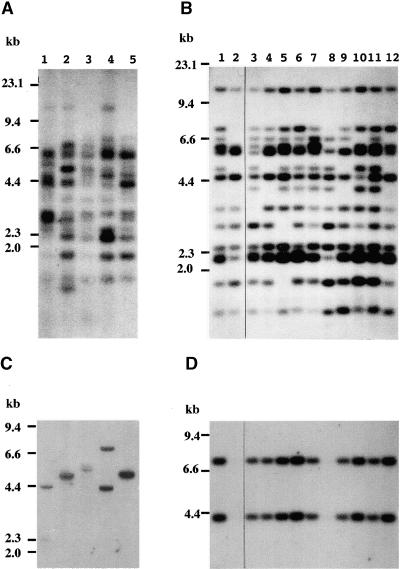
Transposition of Tto1 was assayed by using DNA gel blot analysis. As shown in Figure 1A, the copy number of Tto1 ranged from ∼10 to 15 in five independent T0 transgenic plants characterized in this study. The original copy number of Tto1 introduced into Arabidopsis was estimated by counting the copy number of the HPT gene placed downstream of Tto1 in the Ti plasmid. Only one or two copies of the HPT gene were detected in the five transgenic lines (Figure 1C), supporting the transposition of Tto1.
Figure 1.
DNA Gel Blot Analysis of Transposition of Tto1 in Transgenic Arabidopsis Lines.
(A) and (C) Analyses of five independent T0 transgenic lines.
(B) and (D) Analyses of the T1 progeny of one T0 line (lane 4 in [A]).
Genomic DNA was digested with EcoRV and hybridized with a Tto1-gag probe ([A] and [B]) or an HPT probe ([C] and [D]). DNA length markers are shown at left in kilobases.
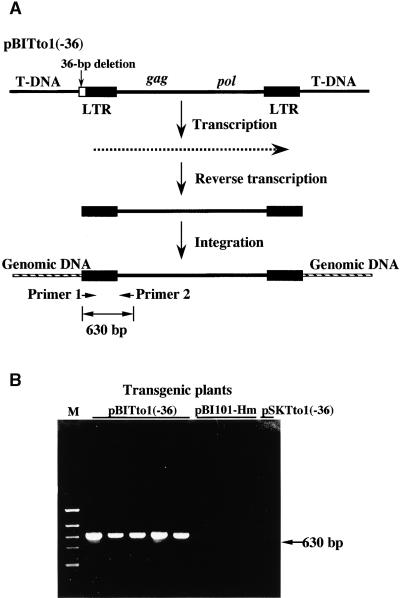
To further confirm transposition, the following experiments were performed. As shown in Figure 2A, the original Tto1 retrotransposon introduced into Arabidopsis carries a 36-bp deletion at the 5′ end. If transposition occurs, the 5′ end should be recovered (Boeke et al., 1985) and integrated into the genome. This experimental strategy has been used to demonstrate the transposition of Tnt1 (Lucas et al., 1995) and Tto1 (Hirochika et al., 1996a) in Arabidopsis and rice, respectively. The recovery of the deleted sequence was examined by polymerase chain reaction (PCR), using a primer pair in which one primer (primer 1) was complementary to the deleted sequence. A fragment of the expected length was amplified from all of the Tto1 transgenic plants (Figure 2B). From control plants transformed with the β-glucuronidase (GUS) gene or with the plasmid carrying Tto1 with the 36-bp deletion (pSKTto1[−36]), no fragment was amplified. These data, along with the data from DNA gel blot analysis, indicate that Tto1 had transposed in Arabidopsis. Presumably, the transposition of Tto1 occurred mainly in callus tissues during the transformation process, because the transcription of Tto1 is induced by tissue culture in Arabidopsis, as is the case with tobacco (H. Okamoto and H. Hirochika, unpublished data). The Tto1 copies detected in the T0 plant are stably inherited in the next generation, as shown in Figure 1B, and no new transposed copy was detected in the progeny. The latter result is consistent with the tissue culture–specific expression of Tto1 in tobacco.
Figure 2.
PCR Analysis of Transposition of Tto1 in T0 Transgenic Lines.
(A) Process of transposition and recovery of the 5′ deleted sequence expected from transposition. A derivative of Tto1-1 carrying a 36-bp deletion at the 5′ end (Tto1[−36]) was inserted into the T-DNA region of the vector plasmid, resulting in pBITto1(−36). In transgenic lines, the 5′ deleted sequence is expected to be recovered during transposition by means of an RNA. A 630-bp fragment was amplified only from transposed Tto1 copies by using primers 1 and 2, because primer 1 is homologous to the deleted sequence. LTR, long terminal repeat.
(B) Results of PCR analysis. Five T0 transgenic lines used in Figure 1A and control transgenic lines transformed with the vector plasmid (pBI101-Hm) were subjected to PCR analysis. The plasmid carrying Tto1(−36), pSKTto1(−36), was also used as a control. M, HincII-digested φX174 as a length marker.
Tto1 Copies in the Transgenic Plants Are Silenced and Highly Methylated
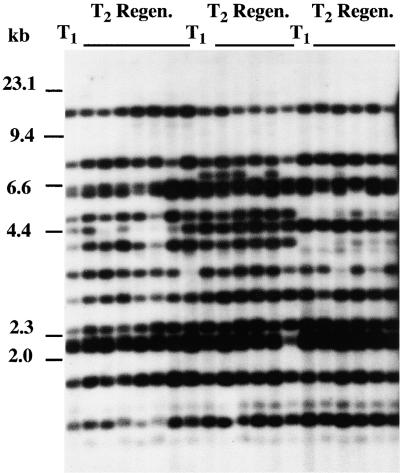
The above results suggest that the transposition of Tto1 can be reactivated in transgenic Arabidopsis plants by tissue culture, as is the case with the original host, tobacco. To test this hypothesis, the progeny of T1 plants were cultured to activate Tto1 and regenerated to measure the increase in copy number. The DNA of T1 plants derived from one transgenic line and of regenerated plants from cultured T2 plants was analyzed by gel blot analysis. If transposition is induced during tissue culture, new bands not found in the parental T1 plants should be detected. Contrary to our initial expectation, no new band was observed in the regenerated plants (Figure 3). Minor variations in the banding pattern can be explained by the segregation of Tto1 copies, because the regenerated plants were derived from pooled progeny. These results indicate that Tto1 in the T2 generation has become inactive.
Figure 3.
Effect of Tissue Culture on Transposition in the Progeny of Transgenic Lines.
Hypocotyl sections of pooled progeny (10 T2 plants from each T1 plant) of three T1 plants derived from one T0 line (Figure 1B, lanes 2, 4, and 5) were cultured on callus-inducing medium. Genomic DNA prepared from T1 plants (T1) and from regenerated plants (T2 Regen.) from induced calli was digested with EcoRV and subjected to DNA gel blot analysis with the Tto1 gag probe. DNA length markers are shown at left in kilobases.
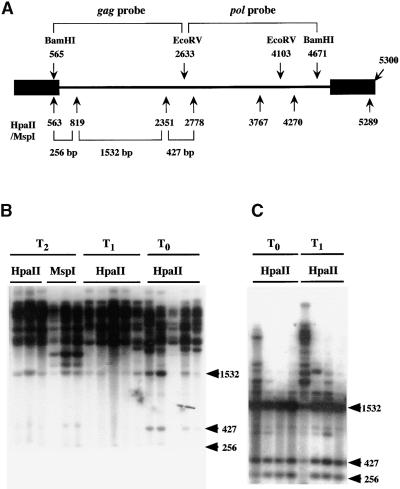
DNA methylation of inactivated Tto1 was then examined by DNA gel blot analysis with the methylation-sensitive restriction endonucleases HpaII and MspI. HpaII cleaves CCGG sites only when the C residue is not methylated, whereas MspI, an isoschizomer of HpaII, cleaves C5mCGG but not 5mCCGG. DNAs from three regenerated T2 plants, in which the Tto1 copies were silenced, were treated with either HpaII or MspI and subjected to DNA gel blot analysis with a Tto1 gag probe. If Tto1 is not methylated, three bands of 1532, 427, and 256 bp should be visible (Figure 4A). As shown in Figure 4B, only a weak band of 1532 bp was detected in the HpaII-digested samples. Other major bands were longer than expected, indicating that all of the Tto1 copies present in the genome of T2 generations are highly methylated. Even in the MspI-digested samples, major bands were longer than expected, indicating that the first C residue in the CCGG sequence is also methylated. Using a pol probe gave the same results (data not shown), indicating that the DNA methylation is not confined to a specific region of Tto1. Accordingly, the inactivation of Tto1 is associated with DNA methylation.
Figure 4.
Analysis of DNA Methylation of Tto1 in T0, T1, and T2 Transgenic Lines.
(A) Restriction maps of Tto1-1 and probes used for the analysis. Black rectangles denote the long terminal repeat.
(B) DNA gel blot analysis of methylation of Tto1 in transgenic lines having high copy numbers. Genomic DNA was digested with HpaII or MspI and analyzed by DNA gel blotting with the Tto1 gag probe. T2 DNA was prepared from the pooled progeny (four regenerated T2 plants from each T1 plant used in Figure 3). T1 DNA was prepared from the pooled progeny (five T1 plants) derived from each T0 line.
(C) DNA gel blot analysis of methylation of Tto1 in transgenic lines having low or medium copy numbers. Genomic DNA was digested with HpaII and analyzed by DNA gel blotting with the Tto1 gag probe. T1 DNA was prepared from the pooled progeny (10 T1 plants) derived from each T0 line. The copy numbers of Tto1 in transgenic lines (from left to right) are six, two, one, and one (data not shown). Only the left-most transgenic line carries transposed copies (four copies).
All of the DNA samples analyzed in (B) and (C) were shown to be equally well digested with the restriction enzyme by reprobing the blot with a single-copy sequence (m105; Pruitt and Meyerowitz, 1986) (data not shown). Lengths and positions of fragments expected from digestion of unmethylated Tto1 are shown at right in (B) and (C) in kilobases.
To determine when the DNA methylation had been induced, DNA from T0 and T1 plants was analyzed. As shown in Figure 4B, Tto1 copies in T0 and T1 plants were already highly methylated, although the extent of methylation in T0 plants was less than that in T1 plants. These results indicate that DNA methylation increased progressively through the generations. On the other hand, in the transgenic plants carrying a low copy number of Tto1 (one to two copies), Tto1 was almost unmethylated (Figure 4C), suggesting that increased copy number is a major factor in DNA methylation. In the transgenic line carrying six copies of Tto1, methylation of Tto1 was extensive (Figure 4C). The increase in copy number correlates with both the DNA methylation and the inactivation of Tto1.
Demethylation of Silenced Tto1 by the ddm1 Mutation
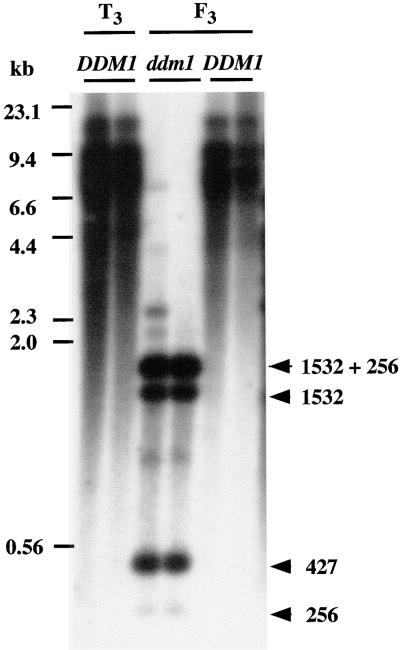
The results mentioned above suggest that the multiple-copy Tto1 was suppressed by a mechanism similar to repeat-induced gene silencing associated with DNA methylation. To test this possibility, the ddm1 mutation of Arabidopsis, which is known to cause genomic DNA hypomethylation (Vongs et al., 1993) and release of gene silencing (Furner et al., 1998; Mittelsten Scheid et al., 1998), was introduced into the transgenic line carrying silenced Tto1 copies by crossing one T2 plant with the ddm1 homozygous mutant. Because the ddm1 mutation is recessive (Vongs et al., 1993; Kakutani et al., 1999), homozygous mutant lines (119 and 120) were screened among F2 plants as described in the Methods, and F3 plants of these two lines were used in the subsequent experiments. Controls were T3 plants (plants 121 and 122) originating from the transgenic plant by selfing, and F3 sibling lines (123 and 124) in which the ddm1 mutation was not introduced. DNA methylation of Tto1 in these lines was examined by using HpaII. As shown in Figure 5, three clear bands of 1.8, 1.5, and 0.4 kb were detected in the two ddm1 homozygous lines but not in control plants. These results indicate that the ddm1 mutation causes the loss of methylation in most HpaII sites in Tto1.
Figure 5.
Analysis of DNA Methylation of Tto1 in the Wild Type and ddm1 Mutant.
Genomic DNAs prepared from two T3 plants (T3; lines 121 and 122), and from F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families (from a cross T2 plant [Tto1 DDM1] × [ddm1]) were digested with EcoRV and analyzed by DNA gel blotting with the Tto1 gag probe. All of the DNA samples were found to be equally well digested by the restriction enzyme by reprobing the blot with the single-copy sequence discussed in the legend to Figure 4 (data not shown). DNA length markers are shown at left in kilobases. Expected lengths of fragments are shown at right in kilobases.
Reactivation of Transcription and Transposition of Silenced Tto1 by the ddm1 Mutation
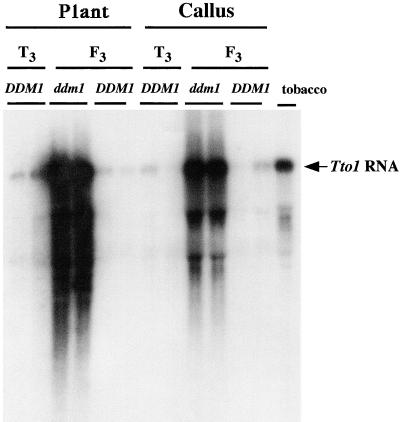
Transcription is the first step in the transposition of retrotransposons and has been shown to be the major regulatory step for the well-characterized plant retrotransposons, such as Tto1 (Hirochika, 1993), Tnt1 (Pouteau et al., 1991), and Tos17 (Hirochika et al., 1996b). Because DNA methylation within promoters represses the transcription of cellular genes (Kass et al., 1997), transcripts of the inactivated and the demethylated Tto1 were compared (Figure 6) by using F3 plants derived from mutant lines (119 and 120), sibling lines (123 and 124), and wild-type lines (121 and 122). Because the transcription of Tto1 is activated in tissue culture in tobacco (Hirochika, 1993; Takeda et al., 1999) and also in Arabidopsis (H. Okamoto and H. Hirochika, unpublished data), normal plant tissues and cultured cells were analyzed for its presence. Cultured cells of the ddm1 mutants showed clear induction of Tto1 RNA having the expected length, but in quantities two- to threefold more than that induced in tobacco. However, the methylated Tto1 in the wild-type background was transcribed very little in both cultured cells and normal plant tissues. The ddm1-induced strong RNA signal was observed in both callus and normal plant tissues, correlating with the Tto1 hypomethylation in both tissues.
Figure 6.
Analysis of Tto1 RNA in the Wild Type and ddm1 Mutant.
Total RNA was prepared from whole plants or calli of T3 plants (T3; lines 121 and 122) and F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families. As a control, total RNA from tobacco BY2 cells (Nagata et al., 1981) was analyzed. Twenty micrograms of total RNA was loaded on the gel.
To examine the transpositional activity in tissue culture, calli were induced from F3 plants derived from mutant lines (119 and 120) and wild-type lines (121 and 122) and then were cultured for 3 months. Weak, smeared bands detected by DNA gel blot analysis were induced in the mutant lines but not in the wild type (data not shown). Smeared bands could be explained by transposition occurring independently in each cell of the mutant calli. To show newly transposed Tto1 copies individually, calli were divided into small pieces, and each piece was cultured for one more month separately from the others—a process that reduced heterogeneity.
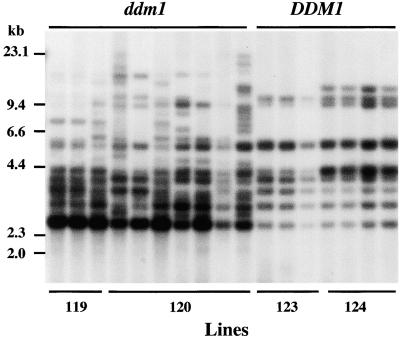
Analysis for the copy of Tto1 in these partially cloned calli (Figure 7) showed no new bands in calli derived from the wild type. Some minor differences observed among calli were also observed in the original plants. In ddm1 calli derived from lines 119 and 120, new bands that had not been detected in the original plants were visible. In addition, a 2.6-kb band that was present in wild-type calli was much stronger in the ddm1 calli. This length of band, which is comparable to that of the left end fragment of the linear Tto1 molecule generated by digestion with EcoRV (Figure 4A), suggests the presence of a Tto1 linear molecule.
Figure 7.
Reactivation of Tto1 Transposition by the ddm1 Mutation in Calli.
Calli were induced from F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families and cultured for 3 months. Induced calli were smashed into pieces, and each piece was cultured separately for one more month. DNA from each callus was digested with EcoRV and analyzed by DNA gel blotting with the Tto1 gag probe. DNA length markers are shown at left in kilobases.
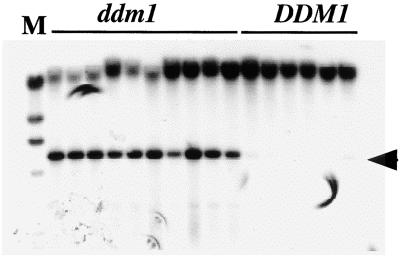
To test for the possibility of linearity, undigested genomic DNA was analyzed (Figure 8). A strong DNA band of the size of full-length linear Tto1 (5.3 kb) was detected for the ddm1 calli, but only trace amounts of it were evident for wild-type calli, indicating that the linear molecule accumulated in the ddm1 calli. Comparing the intensity of the band with that of the copy number control yielded an estimate of four to eight copies of the linear molecule per cell (data not shown). The linear molecule is believed to be a precursor for the transposition of the Ty1 retrotransposon of yeast (Eichinger and Boeke, 1988), of retroviruses (Fujiwara and Mizuuchi, 1988), and of the tobacco retrotransposon Tnt1 (Feuerbach et al., 1997). The structural analysis of the linear Tto1 DNA molecule is under way.
Figure 8.
Analysis of the Linear Molecule of Tto1 in the ddm1 Mutant.
Total DNAs prepared from calli of F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families were analyzed without restriction digestion by DNA blotting with the Tto1-gag probe. The linear Tto1 molecule is indicated by an arrow. M, HindIII-digested λ DNA as a length marker.
Although the finding was not directly comparable with the above data in tissue culture, we could not detect retrotransposition in normal plant tissue. The copy number of Tto1 in F3 plants of lines 119 and 120 was assayed by DNA gel blotting. Despite assaying >80 plants, no evidence of transposition was obtained (data not shown), even though Tto1 RNA accumulated in normal plant tissues (Figure 6). A tissue-specific control mechanism in steps after transcription (e.g., translation and integration) may also be involved.
Reactivation of Transcription of an Endogenous Retrotransposon by the ddm1 Mutation
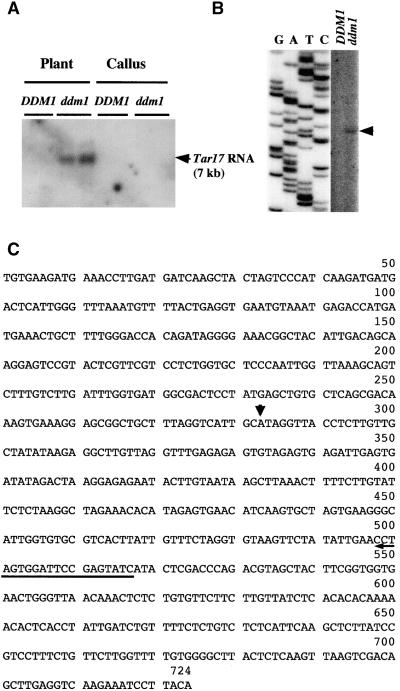
The above results demonstrate that DNA methylation and gene-silencing machinery are effective against exogenous retrotransposons. This raises the possibility that endogenous retrotransposons may have been silenced by similar mechanisms. In fact, transcripts of endogenous retrotransposons were generally not detected (Konieczny et al., 1991; H. Hirochika, unpublished data). To test whether the gene-silencing mechanism also functions for endogenous transposons, the expression of retrotransposons was surveyed by reverse transcription–PCR (Hirochika, 1993) with the ddm1 mutant. cDNA prepared from cultured cells of the ddm1 mutant was used as a template for PCR amplification with degenerate primers corresponding to the conserved pol region. Analysis of 18 sequences determined 10 families of retrotransposons: Ta3 and Ta4 (Konieczny et al., 1991), Tar1 (Hirochika and Hirochika, 1993), Tar8, Tar13, Tar17, and Tar26 to Tar29. Tar1 to Tar3 (Hirochika and Hirochika, 1993) and Tar4 to Tar25 (H. Hirochika, unpublished data) have been isolated by PCR with use of genomic DNA as a template. Using RNA gel blot analysis to examine the transcriptional activation of these retrotransposons in the ddm1 mutant, we detected the RNA band only with the Tar17 probe. The Tar17-specific RNA band of ∼7 kb was detected in RNA from leaf tissues of the ddm1 mutant but not callus tissue (Figure 9A). In the DNA databases, the complete Tar17 sequence was found (GenBank accession number AC006841, the bacterial artificial chromosome sequence of chromosome 2). The total length of Tar17 is 7788 bases. The 5′ and 3′ long terminal repeat (LTR) sequences are 724 bases long and differ at only five base positions. The internal region encodes one uninterrupted open reading frame of 1333 amino acids. These features indicate that Tar17 was transposed relatively recently. To show that the RNA detected by gel blot analysis is not due to read-through from the flanking sequence, we used primer extension analysis (Figure 9B). The signal was detected only for the ddm1 mutant, and transcription was shown to start within the LTR (Figure 9C).
Figure 9.
Activation of Transcription of the Endogenous Retrotransposon Tar17 by the ddm1 Mutation.
(A) RNA gel blot analysis. Total RNAs prepared from whole plants or calli of ddm1/ddm1 (ddm1) and DDM1/DDM1 (DDM1) were analyzed by using the Tto1 gag probe.
(B) Primer extension analysis. Ten micrograms of total RNA extracted from whole plants was hybridized with the 5′ end-labeled oligonucleotide. The hybrids were extended with reverse transcriptase, and the cDNA products were electrophoresed on a sequencing gel alongside a sequencing reaction using the same primer, as described previously (Hirochika, 1993). One band (indicated by an arrowhead) was detected in the ddm1 mutant only.
(C) Nucleotide sequence of the LTR. The 5′ end of Tar17 RNA, determined by primer extension, is indicated by an arrowhead. The position of the oligonucleotide used for primer extension analysis is indicated by an arrow.
DNA gel blot analysis showed that Tar17 is demethylated in the ddm1 mutant (data not shown). DNA gel blot analysis using a methylation-insensitive enzyme showed two other weakly hybridizing signals. By using recombinant inbred lines from Lister and Dean (1993), their position was mapped to chromosomes 2 (Tar17c) and 5 (Tar17b) (see Methods). The mapping information is available in the database at Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/new_ri_map.html). From the map position and sequence similarity, Tar17c is likely to be T16I21.11 in a sequenced bacterial artificial chromosome (accession number AC006570). Expression of T16I21.11 was also ddm1-specific (data not shown). These results indicate that the DDM1 gene product is necessary for suppressing Tar17 members in the wild type. Despite the transcriptional activation, no evidence for transpositional activation was obtained (data not shown), perhaps because of the accumulation of mutations, which led to loss of function of the encoded proteins.
DISCUSSION
Regulation of Transposons and Gene Silencing
In most plant species, a large part of the genome consists of retrotransposons (Pearce et al., 1996; SanMiguel et al., 1996; SanMiguel and Bennetzen, 1998). The results presented in this report indicate that DDM1 gene function is necessary to suppress retrotransposons, both the endogenous Tar17 family and Tto1 introduced from tobacco.
Two types of gene-silencing mechanisms have been observed in plants: transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS). TGS affects transcriptional initiation, whereas PTGS affects the stability of the transcript. TGS is meiotically heritable, whereas PTGS is reset after meiosis and recurs every generation at some stage of plant development (reviewed in Vaucheret et al., 1998). The DDM1 gene is necessary for TGS, whereas the EGS1, EGS2, SGS1, and SGS2 genes affect PTGS.
We have shown here that the increase in copy number correlates with DNA methylation and inactivation of Tto1. This suggests that the increased copy number is a primary cause for silencing and methylation of Tto1. A similar correlation was previously observed in the case of Mutator elements of maize (Bennetzen, 1987), although the regulatory mechanism involved there is still unclear. Several lines of evidence suggest that the inactivation of multiple-copy Tto1 is similar to TGS. The inactivation of Tto1 was observed in both callus and plant tissues. During propagation, the methylation spread progressively, and the silencing strengthened. Finally, the silencing was released by the ddm1 mutation. The increasing methylation progressing through generations and being associated with silencing has been reported for TGS in Arabidopsis (Luff et al., 1999). It should be noted that increased methylation during a progression of generations and even during the development of an individual plant was also observed in the case of Mutator (reviewed in Bennetzen et al., 1993).
Much evidence suggests that PTGS is effective in defending plants against RNA viruses (Covey et al., 1997; Ratcliff et al., 1997; Al-Kaff et al., 1998; reviewed in Vaucheret et al., 1998). Similarly, the results presented here suggest that TGS is effective in protecting the plant genome against transposons. Several properties of TGS are useful for defense against retrotransposons. If TGS were a defense against parasites in the genome, for example, its meiotically heritable property would be important; indeed, the silenced state would be maintained at every stage of development to maintain genome structure and proper gene expression.
To date, release of TGS by the ddm1 mutation has been reported in three systems: clustered p35S::HPT (Mittelsten Scheid et al., 1998), clustered CHS genes (Furner et al., 1998), and PAI genes (Jeddeloh et al., 1998). In all of these, silencing was induced by a tandem repeat or inverted repeat at one locus. In the Tto1 system reported here, all or most copies are scattered throughout the genome (data not shown). If the primary function of TGS is to defend against retrotransposons, silencing the induction of multiple unlinked copies would be important because retrotransposons generally jump to an unlinked position, resulting in dispersed repeats. In this context, one should note the propagation of the silenced state and DNA methylation in trans from the PAI1-PAI4 inverted repeat to single-copy PAI genes (Luff et al., 1999) and from the CHS transgene repeat to the endogenous CHS gene (Furner et al., 1998).
Control of Retrotransposons
We previously showed that the tobacco retrotransposon Tto1 can transpose in rice (Hirochika et al., 1996a). Here, we show that Tto1 can transpose in another heterologous plant, Arabidopsis. After the initial active transposition in the Arabidopsis genome, however, Tto1 became hypermethylated and silent. We provide here direct evidence of repeat-induced silencing of a plant retrotransposon; Lucas et al. (1995) have suggested that the tobacco retrotransposon Tnt1 in Arabidopsis is also silenced.
Although we have shown here that the Tar17 family became transcriptionally active in a ddm1 mutant background, the other nine families of endogenous retrotransposons did not show transcriptional activation, or at least not to a detectable amount when RNA gel blot analysis was used. One possibility is that in most retrotransposons, the LTR function may have become inefficient because of the accumulation of mutations. Alternatively, even if the endogenous retrotransposons are released from silencing, the conditions for induction of transcription may not have been appropriate. In fact, transcription of the tobacco retrotransposons Tnt1 and Tto1 is activated only under stress conditions or in specific tissues (Pouteau et al., 1991; Grandbastien, 1998; Takeda et al., 1998, 1999). Interestingly, the amount of expression of Tar17 was greater in normal plant tissue than in callus tissue.
Unlike most other plant species, only a small part of the genome in Arabidopsis consists of retrotransposons, and most of the endogenous retrotransposon families are present in only a few copies in the genome. How then is a retrotransposon of low copy number silenced? Because retrotransposons have two identical promoter–enhancer elements, we can speculate that a single-copy retrotransposon is also a target for gene silencing, even if an increase in copy number makes silencing more efficient. In fact, tandemly arrayed two transgene copies were shown to be silenced and methylated in Arabidopsis (Assaad et al., 1993). In the original tobacco hosts, the copy number of Tto1 is ∼30, and Tto1 copies are not silenced (Hirochika, 1993). Even in the cell line BY2, in which the copy number was increased up to 300, Tto1 copies are actively transcribed. We do not know how Tto1 copies escape gene silencing in tobacco. One possible explanation is that silencing is not induced due to the heterogeneity of sequences in each member of Tto1, which has been reported for Tnt1 (Casacuberta et al., 1995). Alternatively, the suppression of retrotransposons may not be as efficient in tobacco as in Arabidopsis, reflecting its large genome.
Plant Gene Expression, DNA Methylation, and Suppression of Repeat Elements
In addition to the release of gene silencing, the ddm1 mutation induces several kinds of developmental abnormalities by causing heritable change at other loci (Kakutani et al., 1996; Kakutani, 1997). Similar developmental abnormalities were induced in MET1 antisense lines (Finnegan et al., 1996; Ronemus et al., 1996). At least some of the ddm1-induced phenotypes seem to result from epigenetic change, because all of four independently established late-flowering traits were mapped to the same chromosomal location near the FWA locus (Kakutani, 1997). These results could be interpreted to mean that DNA methylation is important for tissue-specific expression of plant genes during development. Alternatively, the change in genomic DNA methylation might cause activation of endogenous repeat elements, which disturbs the normal expression of nearby plant genes.
Although some of the ddm1-induced developmental abnormalities are inherited as dominant traits, others are inherited as recessive traits (Kakutani et al., 1996; Kakutani, 1997). Activation of repeat elements could cause not only overexpression but also suppression of nearby plant genes. In fact, in the hcf106-mum1 allele of maize, in which a nonautonomous Mutator1 (Mu1) transposable element is inserted in the promoter region, the gene becomes epigenetically silent only when Mu1 becomes active and hypomethylated in the presence of the autonomous MuDR elsewhere in the genome (i.e., in “Mu-on” plants) (Barkan and Martienssen, 1991; Martienssen, 1996). Examining the activity of DNA-type transposons in the Arabidopsis genome (Surzycki and Belknap, 1999) under the ddm1 mutant background should also be interesting.
The transcript level of retrotransposon-related endogenous retroviruses, which are normally transcriptionally silent, has recently been shown to be increased in mouse embryos that are deficient in DNA methyltransferase-1 (Walsh et al., 1998). Because the DDM1 gene encodes a protein similar to the chromatin-remodeling factor SWI2/SNF2 (Jeddeloh et al., 1999), the primary effect of the ddm1 mutation possibly involves the chromosomal epigenetic state rather than DNA methylation. Interestingly, retrotransposon I of Drosophila is also suppressed by homology-dependent gene silencing, despite the lack of detectable genomic DNA methylation (Chaboissier et al., 1998; Jensen et al., 1999). The results of Walsh et al. (1998) mentioned above, however, suggest that DNA methylation is directly involved in suppression of retroelements, at least in the mouse system.
In summary, TGS, and possibly DNA methylation, may have evolved to support symbioses of the plant genes with the transposons. Comparative studies of repeat-induced gene silencing and control of transposons would lead to a better understanding of the plant genome.
METHODS
Construction of Plasmids
The plasmid used in transformation of the plants (Arabidopsis thaliana) was constructed as follows. The deleted form of Tto1-1, Tto1(−36), was derived from the plasmid pSKTto1(−36) (Hirochika et al., 1996a). The HindIII-PstI fragment carrying the 5′ part of Tto1-1 (−36) and the PstI-XhoI fragment carrying the 3′ part of Tto1-1(−36) were mixed and ligated with the largest HindIII-SalI fragment of pBI101-Hm (Akama et al., 1992) to construct pBITto1(−36). pBI101-Hm is a derivative of pBI101 (Jefferson et al., 1987) and carries the hygromycin phosphotransferase (HPT) gene in addition to the neomycin phosphotransferase (NPTII) gene as a selective marker gene. The construct was introduced into Agrobacterium tumefaciens EHA101 (Hood et al., 1986).
Transformation and Tissue Culture
Agrobacterium-mediated transformation of Arabidopsis (ecotype Wassilewskija) was performed as described (Akama et al., 1992). Transformed calli (T0 generation) were selected in the presence of kanamycin (50 μg/mL) and hygromycin (20 μg/mL). Calli were induced from transgenic lines by culturing hypocotyl sections on callus-inducing medium (Valvekens et al., 1988) at 22°C and transferred onto fresh media every month. To ensure cell activity, calli were transferred twice (1 week apart) onto fresh media before extraction of RNA and DNA. For production of regenerated plants, hypocotyl sections of pooled T2 transgenic plants were cultured on callus-inducing medium for 12 days and transferred onto shoot-inducing medium to regenerate plants.
Extraction of Nucleic Acids and RNA and DNA Gel Blot Hybridization
Extraction of total RNA and DNA, blotting, preparation of probes, and hybridization were performed as described previously (Hirochika et al., 1996a).
Introduction of ddm1 Mutation into a Transgenic Arabidopsis Line with Silenced Tto1
A transgenic line with silenced Tto1 was crossed with a decreased DNA methylation ddm1 plant that had been backcrossed six times (Kakutani et al., 1996). From the F2 progeny, homozygous ddm1 mutants were selected by examining the methylation status of the genomic DNA, using gel blot analysis with the methylation-sensitive restriction enzyme HpaII. Of the 21 F2 progeny, two plants showed hypomethylation in all copies of Tto1 sequences and endogenous rDNA sequences (Vongs et al., 1993), indicating that they were ddm1 homozygotes. DDM1 plants from this segregating family were used as controls.
Polymerase Chain Reaction Analysis of Transposition
The oligonucleotide primers LTR-11 (5′-TGTTAGTTTTTCCAACAATTATGGT-3′, corresponding to nucleotides 1 to 25 of Tto1-1) and Tnt2-gag (5′-GGATGAATAGTACTCGTACGTATG-3′, corresponding to nucleotides 630 to 606; Hirochika et al., 1996a) were used to amplify the 5′ end recovered after transposition of Tto1(−36) carried on the plasmid pBITto1(−36).
Amplification and Cloning of Reverse Transcriptase Domain of Retrotransposons
The genomic DNA or the first-strand cDNA was amplified by using degenerate primers corresponding to QMDVKT and YVDDM, as described previously (Hirochika, 1993). The cDNA was generated from poly(A)+ RNA isolated from callus tissues of the ddm1 mutant by using random primers and a cDNA cycle kit (Invitrogen, Chatsworth, CA). Polymerase chain reaction (PCR) products were cloned into the HincII site of M13mp18.
Restriction Fragment Length Polymorphism Mapping of Tar17 Family Members by Using Recombinant Inbred Lines
During DNA gel blot filter analysis using EcoRV, the Tar17 probe detected one strong signal (Tar17a) and two weak signals (Tar17b and Tar17c). All of them showed polymorphism between Columbia and Landsberg erecta ecotypes (the Tar17a signal was not detected in the Landsberg erecta ecotype). Map positions were determined by using 96 recombinant inbred lines constructed by Lister and Dean (1993).
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre (Colney, UK) and the Arabidopsis Biological Stock Center (Ohio State University, Columbus) for recombinant inbred mapping and the bacterial artificial chromosome clones. This study was supported by a project grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan and a grant for the enhancement of Center-of-Excellence, the special coordination funds for promoting science and technology in Japan.
References
- Akama, K., Shiraishi, H., Ohta, S., Nakamura, K., Okada, K., and Shimura, Y. (1992). Efficient transformation of Arabidopsis thaliana: Comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 12 7–11. [DOI] [PubMed] [Google Scholar]
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279 2113–2115. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Tucker, K.L., and Signer, E.R. (1993). Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22 1067–1085. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Martienssen, R.A. (1991). Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J., and Fink, G.R. (1995). Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83 725–734. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L.. (1987). Covalent DNA modification and the regulation of Mutator element in maize. Mol. Gen. Genet. 208 45–51. [Google Scholar]
- Bennetzen, J.L., Springer, P.S., Cresse, A.D., and Hendricks, M. (1993). Specificity and regulation of the Mutator transposable element system in maize. Crit. Rev. Plant Sci. 12 57–95. [Google Scholar]
- Bennetzen, J.L., Schrick, K., Springer, P.S., Brown, W.E., and SanMiguel, P. (1994). Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37 565–576. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., Garfinkel, D.J., Styles, C.A., and Fink, G.R. (1985). Ty elements transpose through an RNA intermediate. Cell 40 491–500. [DOI] [PubMed] [Google Scholar]
- Casacuberta, J.M., Vernhettes, S., and Grandbastien, M.A. (1995). Sequence variability within the tobacco retrotransposon Tnt1 population. EMBO J. 14 2670–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier, M.C., Bucheton, A., and Finnegan, D.J. (1998). Copy number control of a transposable element, the I factor, a LINE-like element in Drosophila. Proc. Natl. Acad. Sci. USA 95 11781–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey, S.N., Al-Kaff, N.S., Langara, A., and Turner, D.S. (1997). Plants combat infection by gene silencing. Nature 387 781–782. [Google Scholar]
- Dehio, C., and Schell, J. (1994). Identification of plant genetic loci involved in post-transcriptional mechanisms for meiotically reversible transgene silencing. Proc. Natl. Acad. Sci. USA 91 5538–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, D., and Boeke, J.D. (1988). The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: Cell-free Ty1 transposition. Cell 54 955–966. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach, F., Drouaud, J., and Lucas, H. (1997). Retrovirus-like end processing of the tobacco Tnt1 retrotransposon linear intermediates of replication. J. Virol. 71 4005–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., and Dennis, E.S. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 98 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell, A.J., Dunbar, E., Anderson, R., Pearce, S.R., Hartley, R., and Kumar, A. (1992). Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 20 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell, R.B. (1994). Inactivation of gene expression in plants as a consequence of novel sequence duplications. Proc. Natl. Acad. Sci. USA 91 3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, T., and Mizuuchi, K. (1988). Retroviral DNA integration: Structure of an integration intermediate. Cell 54 497–504. [DOI] [PubMed] [Google Scholar]
- Furner, I.J., Sheikh, M.A., and Collett, C.E. (1998). Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 149 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick, D., Fiering, S., Martin, D.I., and Whitelaw, E. (1998). Repeat-induced gene silencing in mammals. Nature Genet. 18 56–59. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M.-A. (1998). Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3 181–187. [Google Scholar]
- Grant, S.R. (1999). Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell 96 303–306. [DOI] [PubMed] [Google Scholar]
- Green, M.M. (1988). Mobile elements and spontaneous gene mutation. In Eukaryotic Transposable Elements as Mutagenic Agents, M.E. Lambert, J.F. McDonald, and I.B. Weinstein, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 41–50.
- Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H. (1997). Retrotransposons of rice: Their regulation and use for genome analysis. Plant Mol. Biol. 35 231–240. [PubMed] [Google Scholar]
- Hirochika, H., and Hirochika, R. (1993). Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn. J. Genet. 68 35–46. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Otsuki, H., Yoshikawa, M., Otsuki, Y., Sugimoto, K., and Takeda, S. (1996. a). Autonomous transposition of the tobacco retrotransposon Tto1 in rice. Plant Cell 8 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996. b). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, E.E., Helmer, G.L., Fraley, R.T., and Chilton, M.-D. (1986). The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBio542 outside of T-DNA. J. Bacteriol. 168 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Bender, J., and Richards, E.J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet. 22 94–97. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., Gassama, M.P., and Heidmann, T. (1999). Taming of transposable elements by homology-dependent gene silencing. Nature Genet. 21 209–212. [DOI] [PubMed] [Google Scholar]
- Kakutani, T. (1997). Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J. 12 1447–1451. [DOI] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J., and Richards, E. (1995). Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K., and Richards, E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Munakata, K., Richards, E.J., and Hirochika, H. (1999). Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass, S.U., Pruss, D., and Wolffe, A.P. (1997). How does DNA methylation repress transcription? Trends Genet. 13 444–449. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., Voytas, D., Cummings, M., and Ausubel, F. (1991). A superfamily of retrotransposable elements in Arabidopsis thaliana. Genetics 127 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4 745–750. [DOI] [PubMed] [Google Scholar]
- Lucas, H., Feuerbach, F., Kunert, K., Grandbastien, M.-A., and Caboche, M. (1995). The tobacco retrotransposon Tnt1 transposes in Arabidopsis thaliana. EMBO J. 14 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff, B., Pawlowski, L., and Bender, J. (1999). An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3 505–511. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. (1996). Epigenetic silencing of Mu transposable elements in maize. In Epigenetic Mechanisms of Gene Regulation, A. Riggs, R. Martienssen, and V. Russo, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 593–608.
- Martienssen, R. (1998). Transposons, DNA methylation and gene control. Trends Genet. 14 263–264. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J.M., and Eggleston, W.B. (1996). Paramutation and transgene silencing: A common response to invasive DNA? Trends Plant Sci. 1 382–388. [Google Scholar]
- Meyer, P., and Saedler, H. (1996). Homology-dependent gene silencing in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 23–48. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., Afsar, K., and Paszkowski, J. (1998). Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Okada, K., Takebe, I., and Matsui, C. (1981). Delivery of tobacco mosaic virus RNA into plant protoplasts mediated by reverse-phase evaporation vesicles (liposomes). Mol. Gen. Genet. 184 161–165. [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. (1997). Cosuppression in Drosophila: Gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell 90 479–490. [DOI] [PubMed] [Google Scholar]
- Pearce, S.R., Harrison, G., Li, D., Heslop-Harrison, J.S., Kumar, A., and Flavell, A.J. (1996). The Ty1-copia group retrotransposons in Vicia species: Copy number, sequence heterogeneity and chromosomal localisation. Mol. Gen. Genet. 250 305–315. [DOI] [PubMed] [Google Scholar]
- Pouteau, S., Huttner, E., Grandbastien, M.-A., and Caboche, M. (1991). Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 10 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, R.E., and Meyerowitz, E.M. (1986). Characterization of the genome of Arabidopsis thaliana. J. Mol. Biol. 187 169–183. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ronchi, A., Petroni, K., and Tonelli, C. (1995). The reduced expression of endogenous duplications (REED) in the maize R gene family is mediated by DNA methylation. EMBO J. 14 5318–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus, M.J., Galbiati, M., Ticknor, C., Chen, J., and Dellaporta, S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273 654–657. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., and Bennetzen, J.L. (1998). Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 81 37–44. [Google Scholar]
- SanMiguel, P. Tikhonov, A., Jin, Y.-K., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- Selker, E.U. (1999). Gene silencing: Repeats that count. Cell 97 157–160. [DOI] [PubMed] [Google Scholar]
- Surzycki, S.A., and Belknap, W.R. (1999). Characterization of repetitive DNA elements in Arabidopsis. J. Mol. Evol. 48 684–691. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Sugimoto, K., Otsuki, H., and Hirochika, H. (1998). Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 36 365–376. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Sugimoto, K., Otsuki, H., and Hirochika, H. (1999). A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18 383–393. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16 651–659. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., and Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260 1926–1928. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F., Cummings, M.P., Konieczny, A., Ausubel, F.M., and Rodermel, S.R. (1992). Copia-like retrotransposons are ubiquitous among plants. Proc. Natl. Acad. Sci. USA 89 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, C.P., Chaillet, J.R., and Bester, T.H. (1998). Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 20 116–117. [DOI] [PubMed] [Google Scholar]
- Wessler, S.R., Bureau, T.E., and White, S.E. (1995). LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5 814–821. [DOI] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., and Bester, T.H. (1997). Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13 335–340. [DOI] [PubMed] [Google Scholar]