Abstract
Ethylene-responsive element binding factors (ERFs) are members of a novel family of transcription factors that are specific to plants. A highly conserved DNA binding domain known as the ERF domain is the unique feature of this protein family. To characterize in detail this family of transcription factors, we isolated Arabidopsis cDNAs encoding five different ERF proteins (AtERF1 to AtERF5) and analyzed their structure, DNA binding preference, transactivation ability, and mRNA expression profiles. The isolated AtERFs were placed into three classes based on amino acid identity within the ERF domain, although all five displayed GCC box–specific binding activity. AtERF1, AtERF2, and AtERF5 functioned as activators of GCC box–dependent transcription in Arabidopsis leaves. By contrast, AtERF3 and AtERF4 acted as repressors that downregulated not only basal transcription levels of a reporter gene but also the transactivation activity of other transcription factors. The AtERF genes were differentially regulated by ethylene and by abiotic stress conditions, such as wounding, cold, high salinity, or drought, via ETHYLENE-INSENSITIVE2 (EIN2)–dependent or –independent pathways. Cycloheximide, a protein synthesis inhibitor, also induced marked accumulation of AtERF mRNAs. Thus, we conclude that AtERFs are factors that respond to extracellular signals to modulate GCC box–mediated gene expression positively or negatively.
INTRODUCTION
Plants are exposed to a number of conditions that may adversely affect their growth and development. To adjust to changes in their environment, plants modulate the expression of specific sets of genes (O'Donnell et al., 1996, 1998; Ishitani et al., 1997; Yamaguchi-Shinozaki and Shinozaki, 1997; Lund et al., 1998; Xiong et al., 1999). A number of genes, including those that encode pathogenesis-related (PR) and antifungal proteins, are inducible by various forms of biotic and abiotic stress, such as pathogen attack, wounding, UV irradiation, high or low temperature, and drought, and by the gaseous hormone ethylene (Abeles et al., 1992; Ecker, 1995; O'Donnell et al., 1996, 1998; Penninckx et al., 1996, 1998). These forms of stress ultimately induce ethylene biosynthesis (Abeles et al., 1992; Ecker, 1995; O'Donnell et al., 1996), and ethylene has therefore been suggested to be a mediator of the stress response (Ecker, 1995; O'Donnell et al., 1996; Penninckx et al., 1996). Some defense-related genes that are induced by ethylene were found to contain a short cis-acting element known as the GCC box (AGCCGCC; Ohme-Takagi and Shinshi, 1990). This sequence was determined to be essential for the expression of several PR genes (Sessa et al., 1995; Shinshi et al., 1995; Sato et al., 1996) and to be the core sequence of the ethylene-responsive element (ERE) in tobacco (Ohme-Takagi and Shinshi, 1995).
ERE binding factor (ERF) proteins (formerly known as ERE binding proteins [EREBPs]) were isolated as GCC box binding proteins from tobacco (Ohme-Takagi and Shinshi, 1995), and their expression pattern was investigated in detail (Suzuki et al., 1998). They contain a highly conserved DNA binding domain (designated as the ERF domain; Hao et al., 1998) consisting of 58 or 59 amino acids (Ohme-Takagi and Shinshi, 1995). Recently, the solution structure of the ERF domain in complex with the GCC box has been determined (Allen et al., 1998). The ERF domain is novel both in its structure and in its mode of DNA recognition. It is composed of a β sheet and an α helix (β-α motif; see Figure 1), the β sheet interacting monomerically with the target DNA (Allen et al., 1998). The ERF domain binds to the GCC box with high affinity, the dissociation constant (Kd) for the ERF domain–GCC box interaction typically falling in the picomolar range (Hao et al., 1998). By contrast, basic leucine zipper proteins exhibit Kd values in the nanomolar range (Hurst, 1994).
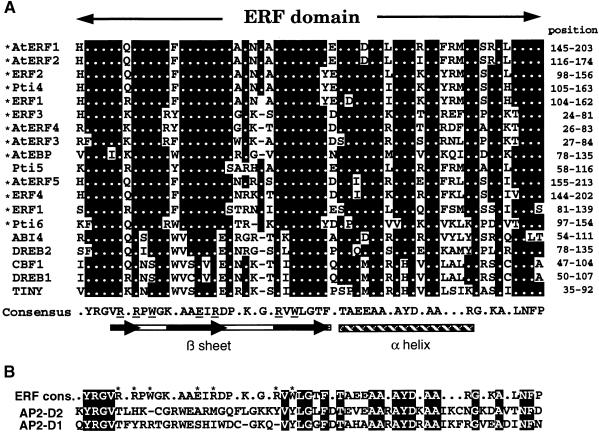
Figure 1.
Alignment of the ERF Domain from Various ERF Proteins.
(A) The amino acid sequences of the ERF domain from various ERF proteins are aligned. They include AtERF1 to AtERF5 (this article); tobacco ERF1 to ERF4 (Ohme-Takagi and Shinshi, 1995); Pti4, Pti5, and Pti6 (Zhou et al., 1997); TINY (Wilson et al., 1996); CBF1 (Stockinger et al., 1997); DREB1 and DREB2 (Liu et al., 1998); Arabidopsis ERF1 (Solano et al., 1998); AtEBP (Büttner and Singh, 1997); and ABI4 (Finkelstein et al., 1998). The asterisks represent those proteins that were shown to bind to the GCC box. Filled box areas with dots indicate amino acid identities; dashes indicate gaps introduced to maximize alignment. Numbers at right indicate the amino acid position of the ERF domain in each protein. Amino acid residues identified by nuclear magnetic resonance analysis (Allen et al., 1998) to interact with nucleotides within the GCC box are underlined in the ERF domain consensus sequence. The structure of the ERF domain is shown below the sequence; the bar and black arrows indicate the β sheet and the β strands within the β sheet, respectively. The cross-hatched box indicates the α helix (Allen et al., 1998).
(B) Comparison of the ERF domain consensus sequence with the AP2 domain. The consensus amino acid sequence of the ERF domain (ERF cons.) is compared with the D2 (AP2-D2) and D1 (AP2-D1) domains of APETALA2 (Jofuku et al., 1994). The amino acids with asterisks in the ERF consensus indicate residues that interact with nucleotides within the GCC box (Allen et al., 1998). Amino acids identical to the ERF consensus are boxed.
It has been argued that the ERF domain is closely related to the AP2 domain (Weigel, 1995; Okamuro et al., 1997). However, knowledge accumulated thus far has indicated that the AP2 and ERF domains belong to distinct families. Sequence alignment of extended family members between these two domains show <30% amino acid identity, whereas members of each family exhibit >60% sequence identity (Büttner and Singh, 1997; Hao et al., 1998; Riechmann and Meyerowitz, 1998; see Figure 1). The recent report of the solution structure of the ERF domain has contributed direct evidence that amino acid residues within the ERF domain that interact directly with the GCC box are not present in the AP2 domain (see Figure 1). Although detailed studies regarding this aspect have not been reported for the AP2 domain, the AP2 domain is likely to possess a mode of DNA recognition distinct from that of the ERF domain as well as a different DNA target sequence.
Sequences made available through the Arabidopsis genome project and expressed sequence tag collections indicate that numerous plant genes encode proteins that possess an ERF domain (referred to as ERF proteins in this work), representing a large, multigene family with many members in both dicots and monocots. They include the ERFs from tobacco; Pti4, Pti5, and Pti6 from tomato; and TINY, A. thaliana ERE binding protein (AtEBP), C-repeat/dehydration-responsive element (DRE) binding factor1 (CBF1), DRE binding protein1 (DREB1), DREB2, abscisic acid (ABA)–insensitive4 (ABI4), and ETHYLENE RESPONSIVE FACTOR1 (ERF1) from Arabidopsis (Ohme-Takagi and Shinshi, 1995; Wilson et al., 1996; Büttner and Singh, 1997; Stockinger et al., 1997; Zhou et al., 1997; Finkelstein et al., 1998; Liu et al., 1998; Solano et al., 1998). To date, genes encoding ERF proteins have been found only in higher plants and not in yeast or other fungi. It has been estimated that ∼125 genes in the Arabidopsis genome encode AP2/ERF proteins (Riechmann and Meyerowitz, 1998).
Several ERF proteins have been suggested to play a role in plant growth and development. Enhancement of TINY gene expression suppressed cell proliferation and resulted in a “tiny” phenotype (Wilson et al., 1996). Ectopic expression of CBF1 or DREB1 in Arabidopsis improved tolerance to freezing (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999). Pti4, Pti5, and Pti6 interact with the product of the Pto disease resistance gene (Zhou et al., 1997), supporting the idea that Pto may regulate the expression of defense-related genes by regulating the function of ERF proteins. AtEBP has been shown to interact with an octopine synthase (ocs) element binding protein (Büttner and Singh, 1997), suggesting that PR gene expression is regulated via a protein–protein interaction between an ERF protein and elicitor-responsive element binding factors. It has also been reported that ectopic expression of ERF1 in transgenic Arabidopsis results in a phenotype similar to that of the constitutive triple response1 mutant (Solano et al., 1998).
Although a number of ERF proteins have been shown to bind specifically to the GCC box (Ohme-Takagi and Shinshi, 1995; Büttner and Singh, 1997; Zhou et al., 1997; Solano et al., 1998), it is not known how ERF proteins possessing the same or similar binding specificity regulate transcription in plants. Genome projects are providing a wealth of information about sequences encoding ERF proteins in Arabidopsis, but information regarding physiological function and gene regulation of this family of transcription factors is still lacking. In this report, we isolated cDNAs encoding Arabidopsis ERF proteins—AtERF proteins—to investigate their role in transcriptional regulation in plants. We show that each AtERF is a factor that can act as either a transcriptional activator or a repressor for GCC box–dependent gene expression.
RESULTS
Isolation and Structure of AtERF cDNAs
We isolated cDNAs encoding AtERF proteins by screening a leaf cDNA library with the tobacco ERF cDNAs as probes. Five complete cDNAs, AtERF1 to AtERF5, which hybridized strongly with the tobacco ERF cDNAs, were further analyzed. Sequence analysis of the cDNAs demonstrated that AtERF1 to AtERF5 all contain the conserved ERF domain and that the cDNAs encode proteins of 266, 243, 225, 222, and 300 amino acids with predicted molecular masses of 28.9, 26.8, 25.2, 23.7, and 33.8 kD, respectively. Based on amino acid sequence identities within the ERF domain, each AtERF was categorized into one of three classes (Figures 2 and 3). AtERF1 and AtERF2 are 58% identical at the amino acid level and were grouped into class I. The class I AtERFs have a high degree of amino acid identity to ERF2 from tobacco and to Pti4 from tomato (Ohme-Takagi and Shinshi, 1995; Zhou et al., 1997) not only within the ERF domain but also within the acidic domain found in the N-terminal region. In addition, members of this group possess a region rich in basic amino acids (P/L-K-K/R-R-R) that could serve as a putative nuclear localization signal (Raikhel, 1992). Two cysteine residues flanking the ERF domain (Figure 3) were also conserved.
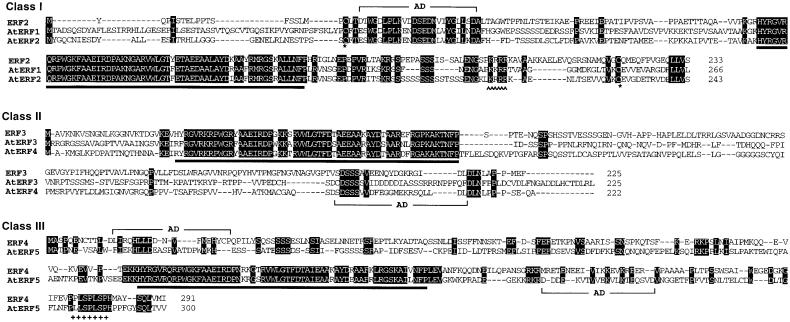
Figure 2.
AtERF cDNAs.
A schematic representation of the AtERF1 to AtERF5 cDNAs is shown. The AtERF proteins are grouped into three classes according to sequence similarity within the ERF domain. Boxes indicate the open reading frames, starting from the first ATG codon, and lines show putative untranslated regions. Black boxes indicate the ERF domain; hatched boxes represent acidic domains. Black boxes with a black triangle above them represent putative nuclear localization signals, and plus signs (+) indicate putative MAP kinase target sites. Numbers above the line indicate positions of amino acid residues; numbers below the line refer to nucleotide positions.
Figure 3.
Comparison of Deduced Amino Acid Sequences of AtERF Proteins.
Amino acid sequences grouped by class are aligned and compared with tobacco ERFs. Identity within the same class is indicated by shading. The filled bar below the sequences represents the ERF domain; AD indicates putative acidic domains. Carets represent putative nuclear localization signals. Plus signs (+) indicate putative MAP kinase target sites. Dashes indicate gaps used to optimize alignment. The GenBank, DDBJ, EMBL, and NCBI accession numbers of the nucleotide sequences of the AtERF cDNAs are AB008103 (AtERF1), AB008104 (AtERF2), AB008105 (AtERF3), AB008106 (AtERF4), and AB008107 (AtERF5).
AtERF3 and AtERF4 comprise class II and possess substantial amino acid identity within the DNA binding domain to the tobacco ERF3 protein (Figure 3). The characteristic features of class II ERFs are that the ERF domain is located close to the N terminus of the protein and that it consists of 58 amino acids, one residue shorter than that of class I ERFs. AtERF3 and AtERF4 do not exhibit pronounced amino acid sequence similarity outside of the ERF domain. However, class II AtERFs possess a similar cDNA structure with respect to the location of the ERF domain, length of the coding region, presence of the C-terminal acidic domain, and calculated pI.
AtERF5 is the only identified member of the third class, class III, possessing sequence similarity to ERF4 from tobacco. This class of ERF gene has the longest coding region among members of the ERF proteins isolated thus far. Acidic domains are located in both the N- and C-terminal regions flanking the ERF domain. In addition, a putative mitogen-activated protein (MAP) kinase phosphorylation site (PXXSPXSP, in which X represents any amino acid; Pearson and Kemp, 1991) is conserved in the C-terminal region of class III ERFs (Figure 3).
Database searches show that the nucleotide sequence of AtERF4 is almost identical to that of expressed sequence tag T45365, which defines the gene encoding RELATED TO APETALA2 PROTEIN5 (RAP2.5) (Okamuro et al., 1997), with the exception of a one-nucleotide insertion within the coding region. This insertion causes the reading frame to shift, provoking changes in the deduced amino acid sequence of the two genes. Because the nucleotide sequence of AtERF4 is identical to a chromosomal sequence (GenBank accession number AP000413), the extra nucleotide found in RAP2.5 may be attributed to intrinsic variation or to a sequencing error. Further database analyses revealed that AtERF3 and AtERF4 are found on chromosomes 1 and 3, respectively. AtERF2 and AtERF5 are located on chromosome 5, whereas AtERF1 resides on chromosome 4.
DNA Binding Preference of the AtERFs
To analyze whether the AtERFs have GCC box binding activity, we overexpressed the entire coding region of each AtERF as a maltose binding protein (MBP) fusion in Esche-richia coli and performed electrophoretic mobility shift assays. As shown in Figure 4, all MBP–AtERF fusion proteins were able to bind to a 16-bp oligonucleotide that contained the wild-type GCC box sequence (AGCCGCC), whereas binding activity was abolished when both G residues within the GCC box were replaced by T residues (ATCCTCC). This experiment indicated that all AtERFs have GCC box–specific binding activity.
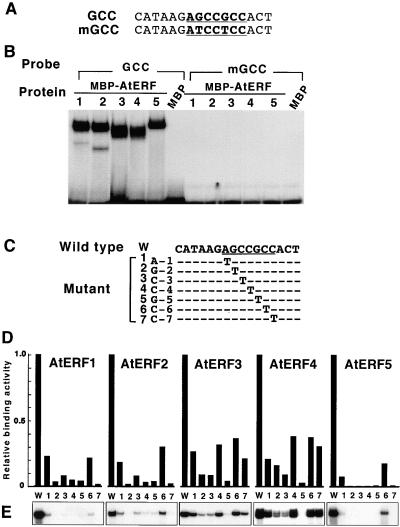
Figure 4.
Characterization of AtERF DNA Binding Affinity to the GCC Box.
(A) GCC box sequences used in this experiment. Underlining and boldface denote the GCC box. GCC, wild type; mGCC, a double mutant.
(B) AtERF possesses GCC box–specific binding activity. Electrophoretic mobility shift assays were performed using MBP–AtERF fusion proteins and a 16-bp wild-type GCC box fragment or a mutated version. MBP was used alone as a control. Numbers indicate MBP–AtERF1 to MBP–AtERF5, respectively.
(C) Sequence of the wild-type GCC box (W) and mutants (1 to 7) used for the DNA binding assays. The GCC box sequence, AGCCGCC, is underlined. Positions within the GCC box systematically substituted with a T residue are indicated as A-1, G-2, C-3, C-4, G-5, C-6, and C-7, respectively. Dashes indicate nucleotides identical to the wild-type sequence.
(D) Effect of single-base substitutions within the GCC box on DNA binding activity of the AtERFs. The binding activities of each AtERF to the mutated versions relative to the wild-type GCC box are shown graphically. W and the number (1 to 7) indicate the probes shown in (C).
(E) Autoradiographs of only the shifted bands from (D) are shown.
To investigate the DNA binding preference of each AtERF in more detail, we generated a series of oligonucleotides in which each nucleotide within the GCC box was separately substituted with a T nucleotide; these oligonucleotides were used to assess the relative binding activities of each AtERF in comparison to the wild-type GCC box sequence (Figures 4C to 4E). Results showed that any mutations within the GCC box reduced the binding ability of the AtERFs, indicating that GCC box is the optimal sequence for AtERF binding among the mutants investigated. The G residue at position 5 (G-5 in Figure 4C) appears to be essential for the GCC box sequence because no complex formation was detected between the AtERF proteins and the oligonucleotide carrying a mutation at this position. By contrast, positions A-1 and C-6 appeared to be less critical for binding, because all AtERFs could bind to some degree to the oligonucleotides carrying mutations at these positions. Substituting T for G-2, C-4, G-5, or C-7 reduced the binding activity of AtERF1, AtERF2, and AtERF5 to <10% of that on the wild-type GCC oligonucleotide, whereas AtERF3 and AtERF4 were able to bind to oligonucleotides mutated at the C-4 and C-7 positions with 30 to 40% efficiency. These data suggest that AtERF1, AtERF2, and AtERF5 are more sensitive to changes in the GCC box sequence whereas members of class II AtERFs appear to be more flexible in target sequence recognition than other AtERFs.
Transcriptional Regulation by the AtERFs
The ability of the AtERFs to regulate transcription in plant cells was tested by using transient assays (Figure 5). A luciferase (LUC)-encoding reporter gene, 4×HLS, which contains four copies of the GCC box sequence from the Arabidopsis HOOKLESS1 (HLS1) promoter (Lehman et al., 1996) fused to LUC, and an effector plasmid consisting of each AtERF under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Figure 5A) were delivered to Arabidopsis leaves by particle bombardment. LUC activity increased at least 12-fold when the reporter plasmid was coexpressed with AtERF1, AtERF2, or AtERF5 effector plasmids (Figure 5B). No such increase in LUC activity by the AtERFs was detected when a LUC reporter plasmid containing a mutated GCC box (ATCCTCC) was used (data not shown). These data indicate that AtERF1, AtERF2, and AtERF5 are able to function as GCC box sequence–specific transactivators in Arabidopsis leaves.
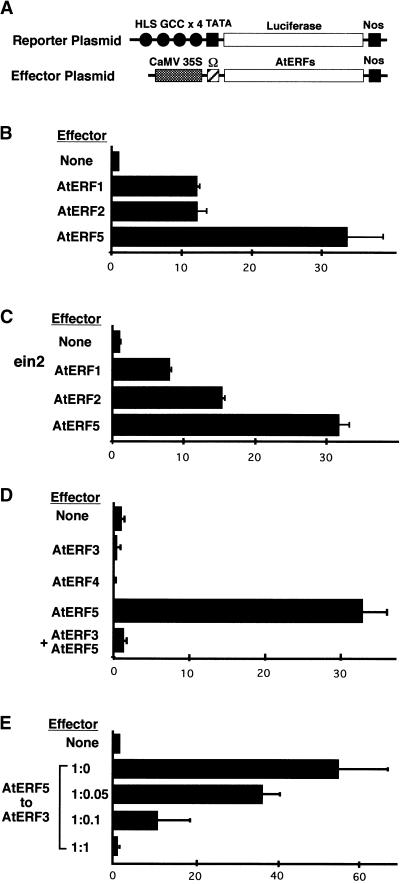
Figure 5.
AtERFs Can Transactivate or Repress GCC-Mediated Gene Expression in Arabidopsis Leaves.
(A) Schematic diagram of the reporter and effector plasmids used in transient assays. A region of the Arabidopsis HLS1 gene, which contains the GCC box (filled circles), was fused to a minimal TATA box and a firefly LUC gene. Effector plasmids were under the control of the CaMV 35S promoter. Nos denotes the terminator signal of the gene for nopaline synthase. Ω indicates translational enhancer of tobacco mosaic virus.
(B) Transactivation of the 4×HLS GCC box–LUC reporter gene by AtERF1, AtERF2, and AtERF5.
(C) Transactivation of the reporter gene by AtERF1, AtERF2, and AtERF5 in the ethylene-insensitive mutant ein2.
(D) Repression of reporter gene activity by AtERF3 and AtERF4 and suppression of AtERF5-mediated transactivation by AtERF3.
(E) Dependence of repression on the amount of AtERF3 effector plasmid. Activation of the reporter gene by AtERF5 is reduced by AtERF3 in a dose-dependent manner. Different concentrations of AtERF3 effector were co-bombarded with AtERF5 effector (from a ratio of 1:0 to 1:1; AtERF3 to AtERF5) and the reporter plasmid.
Values shown are averages of results from three independent experiments. Error bars indicate standard deviation. All LUC activities are expressed relative to the reporter construct alone (value set at 1).
Some genes containing the GCC box in their promoter region are known to be upregulated by ethylene in Arabidopsis (Chen and Bleecker, 1995; Lehman et al., 1996; Penninckx et al., 1996). We investigated whether components of the ethylene signaling pathway affect the activation of GCC box–mediated transcription by the AtERFs. The 4×HLS reporter gene and the AtERF effector constructs were introduced into leaves of the ethylene-insensitive mutant ein2 (Guzman and Ecker, 1990). LUC activity increased when AtERF1, AtERF2, or AtERF5 were coexpressed in ein2 leaves, approaching levels observed in the wild type (Figure 5C). These data suggest that transactivation activity of the AtERFs is not affected by a mutation in the EIN2 ethylene signal transducer.
By contrast, AtERF3 and AtERF4 (class II ERFs) did not activate transcription but appeared to repress reporter gene activity. Coexpression of AtERF3 or AtERF4 with the 4×HLS reporter construct resulted in a 50% reduction in the basal LUC activity. Furthermore, coexpression of AtERF3, AtERF5, and the 4×HLS reporter resulted in a level of LUC activity that was close to the levels obtained with the reporter construct alone (Figure 5D), indicating that AtERF3 repressed not only basal activity of a reporter gene but also the activity of another transcriptional activator. AtERF3 suppression of AtERF5 activation was found to be dose dependent, with the addition of increasing amounts of AtERF3 effector progressively reducing the transactivation ability of AtERF5 (Figure 5E). Thus, this effect is unlikely to result from nonspecific squelching.
To determine whether class II AtERFs are active repressors that possess an intrinsic repression activity that is distinct from their DNA binding activity, we constructed a reporter gene (5GAL4GCC) containing two different cis-elements in the promoter region, the yeast GAL4 sequence and the GCC box. The purpose of this construct was to examine whether a repressor can suppress the activity of an activator without competition for the same DNA binding site. The 5GAL4GCC reporter gene and the effector plasmid containing the GAL4 DNA binding domain fused to the transcriptional activation domain of viral protein 16 (VP16) from herpes simplex virus (Triezenberg et al., 1988) were cobombarded with or without the AtERF3 effector plasmid (Figure 6). Reporter gene activity increased by a minimum of 20-fold when the VP16 effector alone was bombarded. The activation by the VP16 activator was reduced to ∼50% when AtERF3 was coexpressed with the VP16 plasmid (Figure 6B). These results indicate that class II AtERFs are active repressors that can suppress the activity of other transcriptional activators without competing for DNA binding sites. Additionally, AtERF3 and AtERF4 could repress the activity of a GAL4 reporter gene when fused to the GAL4 DNA binding domain (data not shown), suggesting the presence of a discrete repression domain in class II AtERF proteins.
Figure 6.
AtERF3 Suppresses Transactivation without Competing for the Same DNA Binding Site.
(A) Schematic diagram of the reporter and effector plasmids used. The GAL4 binding site (patterned hatched boxes) and GCC box sequence (filled circles) were fused to a minimal TATA box and the LUC gene. The AtERF3 effector and VP16 effector plasmid containing the GAL4 DNA binding domain (GAL4DBD) fused upstream of the VP16 activation domain (VP16 AD) were under the control of the CaMV 35S promoter. Nos denotes the terminator signal of the gene for nopaline synthase. Ω indicates translational enhancer of tobacco mosaic virus.
(B) AtERF3 represses VP16-mediated transactivation without competing for the same DNA binding site. Values shown are averages of results from three independent experiments. Error bars indicate standard deviation. All LUC activities are expressed relative to the reporter construct alone (value set at 1).
Expression of the AtERF mRNAs
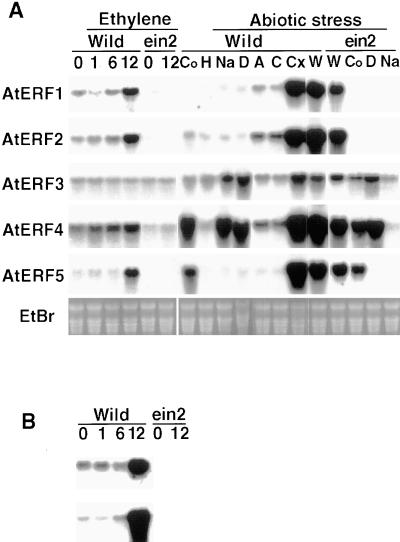
The expression of the AtERF genes was investigated by RNA gel blot analysis with total RNA isolated from Arabidopsis leaves (Figure 7). mRNAs corresponding to each of the AtERFs could be detected in air-grown, healthy Arabidopsis plants, although the basal levels of each AtERF transcript differed. Because some GCC box–containing genes, such as that encoding chitinase or PDF1.2, are known to be induced by ethylene (Chen and Bleecker, 1995; Penninckx et al., 1996), we exposed Arabidopsis plants to ethylene for 1, 6, and 12 hr and monitored expression of the AtERFs. Expression of genes encoding chitinase and PDF1.2 was induced 12 hr after ethylene treatment. Correspondingly, amounts of AtERF1, AtERF2, and AtERF5 transcripts increased two- to threefold 12 hr after ethylene treatment, whereas AtERF3 or AtERF4 transcript amounts did not increase. In the ein2 mutant, the expression of the AtERFs was not induced by ethylene treatment, suggesting that ethylene induction of the AtERFs is regulated under the ethylene signaling pathway, as is the case for the genes that encode chitinase and PDF1.2 (Figure 7). We observed that the mRNA levels of all of the AtERFs except AtERF3 were consistently lower in ein2 plants compared with the wild type.
Figure 7.
Induction of AtERF mRNA Expression.
(A) Expression pattern of AtERF mRNAs after exposure to ethylene and abiotic stress in the wild type (wild) or an ethylene-insensitive mutant, ein2 (ein2). Total RNA was prepared from Arabidopsis leaves treated with ethylene gas for 0, 1, 6, and 12 hr, respectively, or treated with cold (Co), heat (H), NaCl (Na), drought (D), abscisic acid (A), CHX (Cx), or wounded (W) for 6 hr. C, the control for CHX treatment; EtBr, ethidium bromide staining.
(B) Expression patterns of the genes encoding PDF1.2 (top) and chitinase (bottom) in wild-type (Wild) and ein2 (ein2) Arabidopsis plants treated with ethylene for 0, 1, 6, and 12 hr.
Ten micrograms of RNA was loaded and hybridized with the cDNA encoding AtERF, chitinase (Chen and Bleecker, 1995), or PDF1.2 (Penninckx et al., 1996).
Next, we investigated whether abiotic stresses such as wounding, cold, heat, high NaCl concentration, or drought induced AtERF expression (Figure 7). In these experiments, plants were exposed to stress conditions for only 6 hr to avoid possible secondary effects. Wounding induced marked accumulation of mRNAs for all of the AtERFs except AtERF3. AtERF4 was also induced by cold, high NaCl concentration, and drought stress, whereas AtERF5 expression was upregulated by cold stress. AtERF3 expression was moderately induced by salinity and drought stress. Cold and drought stresses are known to promote ABA biosynthesis (Zeevaart and Creelman, 1988; Chandler and Robertson, 1994); however, exogenous ABA did not induce AtERF expression. Similarly, high temperature stress did not affect the expression of any AtERF gene.
The induction of the AtERFs by wounding, cold, and drought stress was observed in ein2 mutants as well as in wild-type plants, although the fold induction was slightly lower in ein2 than it was in the wild type. In contrast, the induction of AtERF3 and AtERF4 by high salinity was not observed in ein2 (Figure 7), indicating that the ethylene signaling pathway mediated by EIN2 may regulate the salt stress–mediated induction of AtERF3 and AtERF4, even though ethylene itself did not induce expression of these AtERF genes.
By contrast, treatment of Arabidopsis plants with cycloheximide (CHX), a protein synthesis inhibitor, resulted in the marked accumulation of AtERF mRNAs, indicating that de novo protein synthesis may not be required for AtERF expression (Figure 7). By 6 hr after CHX treatment, the mRNA levels of AtERF1, AtERF2, AtERF4, and AtERF5 increased 20- to 100-fold compared with the control, whereas AtERF3 mRNA increased only approximately fivefold. The observed accumulation of mRNA after CHX treatment may be due to the inhibition of de novo production of labile transcriptional repressors and/or mRNA-degrading enzymes (Berberich and Kusano, 1997) and suggests that transcriptional or post-transcriptional regulatory mechanisms may be involved in AtERF expression.
DISCUSSION
Structural Features
The five AtERFs isolated during the course of this work were grouped into three classes based on their amino acid sequence identities within the ERF domain, although other regions of AtERF1 and AtERF2 also share extensive amino acid similarity. Nevertheless, AtERF1 and AtERF2 are not thought to have evolved simply by gene duplication because the genes that encode them are located on different chromosomes. In contrast, members of the CBF/DREB family most likely arose after gene duplication events because the proteins are highly similar. Moreover, the corresponding genes are closely linked and have the same transcriptional orientation (Gilmour et al., 1998; Shinwari et al., 1998; Medina et al., 1999). AtERF1 and AtERF2 share high amino acid sequence identities with Pti4, which interacts with the product of the Pto disease resistance gene (Zhou et al., 1997). It is possible, then, that class I AtERFs are involved in pathogen signal transduction in Arabidopsis via protein–protein interactions.
By contrast with AtERF1 and AtERF2, the class II proteins AtERF3 and AtERF4 do not share much amino acid sequence identity except for residues within the ERF domain. Nevertheless, the overall structure and the function of these two proteins as transcriptional repressors are conserved, indicating that AtERFs in this class may have diverged from a single ancestral origin.
The involvement of protein kinases has been reported in ethylene signal transduction and in the transactivation of GCC box–dependent transcription (Kieber et al., 1993; Raz and Fluhr, 1993; Sessa et al., 1996; Suzuki et al., 1998). Therefore, phosphorylation may regulate some aspects of AtERF function, such as DNA binding activity, nuclear localization, or transactivation. The fact that a putative site for MAP kinase–mediated phosphorylation is found in the class III protein AtERF5, although such sites appear not to be present in representatives of the other two classes, suggests that phosphorylation may play an important role in the function of class III ERFs.
AtERFs Are GCC Box Binding Factors
Each of the ERFs isolated in this study displayed GCC box–specific binding activity, and a number of previously identified ERF proteins have also been shown to possess GCC box binding activity (see Figure 1). Binding assays revealed that the GCC box (AGCCGCC) appears to be the optimal target site for the AtERF proteins and that the G-5 nucleotide within this sequence is essential for AtERF binding (Figure 4). However, results also showed that each AtERF possessed a distinct DNA binding preference. AtERF1, AtERF2, and AtERF5 appear to be most sensitive to the single-nucleotide substitutions within the GCC box sequence because these AtERF proteins were hardly able to bind to five of the seven single mutated oligonucleotides that were tested.
By contrast, AtERF3 and AtERF4 appear to be more flexible than other AtERF proteins with respect to their target sequence preferences because they were able to bind to several mutated oligonucleotides to which other AtERF proteins did not bind. The binding flexibility of the class II AtERF proteins implies that these proteins might interact with genes with which other classes of AtERF proteins cannot. Nevertheless, it is important to analyze whether the DNA binding activities obtained in vitro correlate with the transcriptional activities of the AtERFs in vivo.
It is difficult to explain at this stage where differences in binding preferences between class II AtERFs and other AtERFs might arise because the amino acid residues that have been shown to interact with DNA (Allen et al., 1998) are conserved in all ERF domains (Figure 1). The slight sequence variations across the ERF domains may result in conformational changes that affect DNA binding specificity; however, although we used the entire AtERF coding region in our DNA binding assays, our results are consistent with those in which only the minimum DNA binding domains were tested (Hao et al., 1998). This suggests that the regions flanking the DNA binding domain are not critical for mediating the protein–DNA interaction in vitro.
Transcriptional Regulation
We showed that AtERF1, AtERF2, and AtERF5 act as transcriptional activators for GCC box–dependent transcription in Arabidopsis leaves. Similarly, other ERF proteins, such as CBF1 and DREB, have been shown to function as transactivators in Arabidopsis or yeast (Stockinger et al., 1997; Liu et al., 1998). Additionally, ERF1 (Solano et al., 1998) has been shown to activate a subset of ethylene-inducible genes, such as PDF1.2 and the gene encoding chitinase, when overexpressed. The AtERFs were capable of activating transcription of a reporter gene in ein2 mutant as well as in wild-type leaves in our transient expression system, indicating that AtERF proteins may not be regulated post-transcriptionally by the ethylene signaling pathway. Similar results have been reported in transgenic plants overexpressing ERF1 (Solano et al., 1998).
One of our most interesting findings was that AtERF3 and AtERF4 can act as transcriptional repressors in Arabidopsis leaves. This suggests that a dynamic system utilizing antagonistic mechanisms for controlling GCC box–dependent transcription operates in plants. Repression can be categorized broadly into two modes: passive and active (Cowell, 1994; Hanna-Rose and Hansen, 1996). In plants, the maize Dof2 protein (Yanagisawa and Sheen, 1998) appears to be a passive repressor that downregulates the transcriptional activity of Dof1 by competing for the same DNA binding site; it does not possess intrinsic repressive activity. On the other hand, active repression involves obstruction of transcription initiation by proteins that contain intrinsic repression activity, as distinct from DNA binding activity (Cowell et al., 1992; Cowell, 1994; Hanna-Rose and Hansen, 1996). In plants, soybean G box Binding Factor2 is thought to be an active repressor (Liu et al., 1997).
Our results demonstrate that class II AtERFs utilize an active repression mechanism because they suppress the transactivation activity of other transcription factors without competing for the same DNA binding site (Figure 6). Several possible mechanisms of active repression have been proposed (Cowell, 1994; Hanna-Rose and Hansen, 1996; Pazin and Kadonaga, 1997). One is that repressors interfere with transcriptional activators that interact with the basal transcriptional machinery, such as TFIID. Alternatively, repressors might recruit co-repressors such as histone deacetylase, which modifies chromatin structure and prevents other transcriptional activators from binding to their target cis-elements (Pazin and Kadonaga, 1997). Analyses of factors that interact with the repression domain of AtERF would demonstrate the mechanism by which class II AtERFs function as transcriptional repressors.
Because all AtERFs can bind to the same target sequence, one point of regulation of transactivation or repression lies in DNA binding affinity. Because lower amounts of AtERF3 effector can suppress transactivation by AtERF5 (Figure 5E) and because the dissociation constant (DNA binding affinity) of the class II ERF domain to the GCC box sequence was comparable to that of the class I or class III ERF domains (Hao et al., 1998; D. Hao and M. Ohme-Takagi, unpublished results), the repression activity displayed by class II AtERFs appears to be stronger than the transactivation activities of class I or III AtERFs. As GCC box–mediated transcription is upregulated in plant cells in response to extracellular signals, mechanisms that downregulate the repression activity of class II AtERFs, such as protein degradation or regulation at the mRNA level, should be operating.
AtERF Proteins Are Stress Signal–Responsive Factors
AtERF genes respond differently not only to ethylene but also to various forms of abiotic stress (Figure 7). AtERF1, AtERF2, and AtERF5 were induced by ethylene 12 hr after treatment, whereas the ethylene response of ERF1 is observed within 2 hr (Solano et al., 1998). Thus, the AtERFs may function later in the induction of ethylene-inducible genes. In contrast, responses of AtERF genes to abiotic stress occur much more quickly than do those to ethylene, suggesting that the AtERFs are involved in regulating responses to abiotic stresses in addition to ethylene. This idea is supported by our recent database survey (M. Ohta, S. Fujimoto, and M. Ohme-Takagi, unpublished results). This survey indicates that several stress-inducible genes, such as ARSK1 and a dehydrin gene, both of which are induced by ABA, NaCl, cold, and/or wounding (Hwang and Goodman, 1995; Rouse et al., 1996), possess the GCC box sequence in their 5′ upstream regions. Moreover, the GCC box has been recently shown to act as an elicitor-responsive element in tobacco cells (Yamamoto et al., 1999). These findings suggest that the GCC box may act as a cis-regulatory element for biotic and abiotic stress signal transduction in addition to its role as an ERE, again implying that the AtERFs may act as transcription factors for stress-responsive genes.
Induction of the AtERFs by wounding, cold, and drought appears to be independent from the ethylene signaling pathway because responses to these abiotic stresses were observed in ein2 plants. By contrast, induction of the genes encoding AtERF3 and AtERF4 by high salinity stress seems to be regulated by the ethylene signaling pathway mediated by EIN2, although the expression of these class II AtERFs is not induced by ethylene. Thus, AtERF expression appears to be controlled by a complex pathway that is independent from or dependent on ethylene signal transduction.
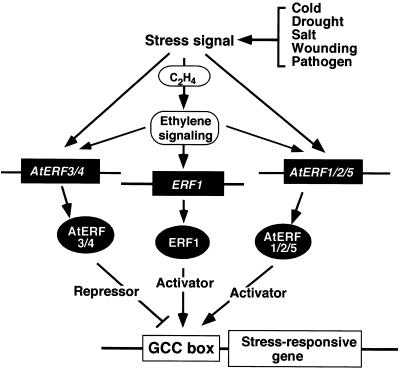
A model for the regulation of GCC box–dependent transcription mediated by ERF proteins is shown in Figure 8. Several possible roles for ERF proteins in the regulation of GCC box–dependent transcription are presented. As the AtERF1 to AtERF5 genes are expressed in response to different stimuli, the AtERF proteins probably function as signal- and sequence-specific transcription factors by activating or repressing the expression of GCC box–containing stress-responsive genes dependent on or independent of the ethylene signal. Other proteins, such as ERF1 (Solano et al., 1998), probably act as activators of GCC box–containing stress- and/or ethylene-responsive genes by positively modulating gene expression in response to external or internal ethylene. This model suggests that the AtERFs are regulated differentially at both the transcriptional and post-transcriptional levels and that subsets of GCC box–containing genes may be regulated by different ERF proteins.
Figure 8.
A Model for GCC Box–Mediated Stress Signal—Dependent Transcription by ERF Proteins in Arabidopsis.
After reception of a stress signal by plants, ERF genes may be upregulated via an ethylene-dependent pathway or an ethylene-independent pathway. ERF genes such as ERF1 (Solano et al., 1998) are induced by ethylene and can interact with GCC box–containing stress response genes. The AtERFs are able to interact with the GCC box sequence in an ethylene-independent manner. AtERF1, AtERF2, or AtERF5 may activate a specific subset of GCC box–containing genes, whereas AtERF3 and AtERF4 may repress the expression of these genes. These data suggest that the ERF proteins may act as factors responsive to extracellular signals and that they are involved in regulating a subset of GCC box–containing stress response genes.
METHODS
Plant Material and Treatments
Wild-type (Arabidopsis thaliana ecotype C24) and ein2 plants were grown in a 1:1 mixture of Soil Mix (Sakata Co., Kyoto, Japan) and vermiculite in growth chambers at 22°C with a 16-hr light/dark cycle for 20 to 30 days. Treatments were performed at 22°C unless otherwise noted and were as follows: ethylene—plants were placed in an ethylene flow chamber (100 ppm C2H4, at 5 mL/min); abscisic acid (ABA)—0.1 mM ABA was sprayed on plants; temperature stress—plants were incubated at either 37 or 4°C; drought—whole plants were placed on filter paper and allowed to desiccate; high salt—plants in pots were placed directly into a container of 300 mM NaCl; wounding—rosette leaves were detached from the plant, cut into ∼5-mm strips, and floated on 50 mM phosphate buffer, pH 7.0; and cycloheximide (CHX)—the root systems of whole plants were washed gently with water to remove soil and then the plants were incubated in 10 μg/mL CHX in 0.1% (v/v) DMSO or 0.1% (v/v) DMSO as a control. Aerial tissues were excised after treatment and immediately frozen in liquid nitrogen.
cDNA Screening
An Arabidopsis λgt10 cDNA library prepared from air-grown plant leaves was screened with the tobacco ethylene-responsive element binding factor (ERF) cDNA coding regions as probes by using standard procedures (Sambrook et al., 1989).
Electrophoretic Mobility Shift Assay
The coding region of each AtERF was cloned into the pMAL vector (New England Biolabs, Beverly, MA) and expressed in Escherichia coli BL21 cells. Maltose binding protein (MBP)–AtERF fusion proteins were purified using amylose resin, as specified by the manufacturer.
Both strands of the following oligonucleotides were synthesized: wild-type GCC box (W: CATAAGAGCCGCCACT) and double mutant GCC box (mGCC: CATAAGATCCTCCACT). In addition, each single mutant GCC box oligonucleotide in which each of the nucleotides in the GCC box was systematically replaced with T (Figure 4C; Hao et al., 1998) was synthesized. Electrophoretic mobility shift assays were performed in a 10-μL volume that contained 0.1 μg of MBP–AtERF fusion protein, 1 μg of poly(dA-dT), 4 fmol of a 16-bp oligonucleotide end-labeled with phosphorus-32, and DNA binding buffer (25 mM Hepes-KOH, pH 7.5, 5% [v/v] glycerol, 40 mM KCl, 0.5 mM EDTA, and 1 mM DTT). After a 15-min incubation at room temperature, the reaction mixture was subjected to 5% nondenaturing PAGE in 0.25 × TBE (1 × TBE is 22.5 mM Tris-borate, pH 8.0, and 0.25 mM EDTA). Binding was quantified on a BAS-2000 system (Fuji, Tokyo, Japan).
Transient Expression
For a reporter gene construct, a 29-bp region containing the GCC box sequence from the Arabidopsis HLS1 gene (ATGAGTTAACGCAGACATAGCCGCCATTT) was multimerized four times, placed upstream of the minimal −42 to +8 TATA box from the cauliflower mosaic virus (CaMV) 35S promoter, and fused transcriptionally to the firefly luciferase (LUC) gene (Promega). For the reporter construct with two cis-elements, the GAL4 binding site repeated in tandem five times and the GCC box sequence (GATCAGCCGCCGGATC) multimerized four times were placed upstream of the reporter gene containing the minimal TATA box. For effector plasmids, the β-glucuronidase (GUS) gene in pBI221 (Clonetech, Palo Alto, CA) was replaced by the coding region of each AtERF. The tobacco mosaic virus Ω sequence (Gallie et al., 1987) was inserted downstream of the CaMV 35S promoter in pBI221 to enhance the efficiency of translation. For the viral protein 16 (VP16) activator plasmid, the GAL4 DNA binding domain (amino acid positions 1 to 147; Ma and Ptashne, 1987) was fused upstream of the VP16 activation domain (positions 413 to 490; Triezenberg et al., 1988).
Transient assays were performed by using the particle gun bombardment method, as follows: 510 μg of 1-μm gold biolistic particles (Bio-Rad) were coated with 1.6 μg of reporter plasmid and a total amount of 1.2 μg of effector plasmid or blank plasmid. As a reference, the Renilla LUC gene (Promega) under the control of the CaMV 35S promoter was used at a reference gene–reporter gene ratio of 1:5. Bombardment was done with a Bio-Rad PDS-1000/He machine. After bombardment, the samples were floated on 50 mM phosphate buffer, pH 7.0, incubated in a plant growth chamber at 22°C for 6 hr, frozen in liquid nitrogen, and quantified for LUC activity.
RNA Analyses
Total RNA was extracted as described previously (Fukuda et al., 1991). Aliquots of 10 μg of total RNA were denatured, separated on 1.2% formaldehyde–agarose gels, transferred to GeneScreen Plus (NEN Life Science Products, Boston, MA), and allowed to hybridize with a 32P-labeled probe. The intensity of the hybridization signal was analyzed by using the BAS-2000 system (Fuji). Equal loading of samples was confirmed by visualization of rRNA after ethidium bromide staining.
Acknowledgments
We are grateful to Dr. Kazuko Yamaguchi-Shinozaki for the Arabidopsis cDNA library and to the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the C-24 and ein2 Arabidopsis seed. We also thank Kyoko Matsui for technical assistance. S.Y.F. and M.O. were recipients of Science and Technology Agency Fellowships from the Japan Science and Technology Corporation. S.Y.F. would also like to thank the National Science Foundation for its support.
References
- Abeles, F.B., Morgan, P.W., and Saltveit, M.E., Jr. (1992). Ethylene in Plant Biology, 2nd ed. (San Diego, CA: Academic Press).
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich, T., and Kusano, T. (1997). Cycloheximide induces a subset of low temperature–inducible genes in maize. Mol. Gen. Genet. 254 275–283. [DOI] [PubMed] [Google Scholar]
- Büttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., and Robertson, M. (1994). Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 113–141. [Google Scholar]
- Chen, Q.G., and Bleecker, A.B. (1995). Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 108 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell, I.G. (1994). Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 19 38–42. [DOI] [PubMed] [Google Scholar]
- Cowell, I.G., Skinner, A., and Hurst, H.C. (1992). Transcriptional repression by a novel member of the bZip family of transcription factors. Mol. Cell. Biol. 12 3070–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker, J.R. (1995). The ethylene signal transduction pathway in plants. Science 268 667–675. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, Y., Ohme, M., and Shinshi, H. (1991). Gene structure and expression of a tobacco endochitinase gene in suspension-cultured cells. Plant Mol. Biol. 16 1–10. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R., Sleat, D.E., Watts, J.W., Turner, P.C., and Wilson, T.M.A. (1987). A comparison of eukaryotic viral 5′ leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 15 8693–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16 433–442. [DOI] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12 229–234. [DOI] [PubMed] [Google Scholar]
- Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plants. J. Biol. Chem. 273 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hurst, H.C. (1994). Protein Profile: Transcription Factors 1: bZip Proteins. (London: Academic Press). [PubMed]
- Hwang, I., and Goodman, H.M. (1995). An Arabidopsis thaliana root-specific kinase homologue is induced by dehydration, ABA, and NaCl. Plant J. 8 37–43. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interaction and convergence of abscisic acid–dependent and abscisic acid–independent pathways. Plant Cell 9 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G.W., Van Montagu, M., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnol. 17 287–291. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain, separate two cellular signal transduction pathways in drought- and low temperature–responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Hagen, G., and Guilfoyle, T.J. (1997). A G-box-binding protein from soybean binds to the E1 auxin-responsive element in the soybean GF3 promoter and contains a proline-rich repression domain. Plant Physiol. 115 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, A.A., Blum, P.H., Bhattramakki, D., and Elthon, T.E. (1998). Heat-stress response of maize mitochondria. Plant Physiol. 116 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Ptashne, M. (1987). Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48 847–853. [DOI] [PubMed] [Google Scholar]
- Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain–containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Truesdale, M.R., Calvert, C.M., Dorans, A., Roberts, M.R., and Bowles, D.J. (1998). A novel tomato gene that rapidly responds to wound-and pathogen-related signals. Plant J. 14 137–142. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1990). Structure and expression of a tobacco β-1,3-glucanase gene. Plant Mol. Biol. 15 941–946. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D.. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin, M.J., and Kadonaga, J.T. (1997). What's up and down with histone deacetylation and transcription? Cell 89 325–328. [DOI] [PubMed] [Google Scholar]
- Pearson, R.B., and Kemp, B.E. (1991). Protein kinase phosphorylation site sequences and consensus specificity motifs: Tabulations. Methods Enzymol. 200 62–81. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Metraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel, N.V. (1992). Nuclear targeting in plants. Plant Physiol. 100 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz, V., and Fluhr, R. (1993). Ethylene signal is transduced via protein phosphorylation events in plants. Plant Cell 5 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Rouse, D.T., Marotta, R., and Parish, R.W. (1996). Promoter and expression studies on an Arabidopsis thaliana dehydrin gene. FEBS Lett. 381 252–256. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sato, F., Kitajima, S., Koyama, T., and Yamada, Y. (1996). Ethylene-induced gene expression of osmotin-like protein, a neutral isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol. 37 249–255. [DOI] [PubMed] [Google Scholar]
- Sessa, G., Meller, Y., and Fluhr, R. (1995). A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol. Biol. 28 145–153. [DOI] [PubMed] [Google Scholar]
- Sessa, G., Raz, V., Savaldi, S., and Fluhr, R. (1996). PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell 8 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi, H., Usami, S., and Ohme-Takagi, M. (1995). Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol. Biol. 27 923–932. [DOI] [PubMed] [Google Scholar]
- Shinwari, Z.K., Nakashima, K., Miura, S., Kasuga, M., Seki, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250 161–170. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain–containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15 657–665. [DOI] [PubMed] [Google Scholar]
- Triezenberg, S.J., Kingsbury, R.C., and McKnight, S.L. (1988). Functional dissection of VP16, the transactivator of Herpes simplex virus immediate early gene expression. Genes Dev. 2 718–729. [DOI] [PubMed] [Google Scholar]
- Weigel, D. (1995). The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Ishitani, M., and Zhu, J.-K. (1999). Interaction of osmotic stress, temperature and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 119 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., Suzuki, K., and Shinshi, H. (1999). Elicitor-responsive, ethylene-independent activation of GCC box–mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J. 20 571–579. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., and Sheen, J. (1998). Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 439–473. [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]