Abstract
The photosystem II reaction center D1 protein is known to turn over frequently. This protein is prone to irreversible damage caused by reactive oxygen species that are formed in the light; the damaged, nonfunctional D1 protein is degraded and replaced by a new copy. However, the proteases responsible for D1 protein degradation remain unknown. In this study, we investigate the possible role of the FtsH protease, an ATP-dependent zinc metalloprotease, during this process. The primary light-induced cleavage product of the D1 protein, a 23-kD fragment, was found to be degraded in isolated thylakoids in the dark during a process dependent on ATP hydrolysis and divalent metal ions, suggesting the involvement of FtsH. Purified FtsH degraded the 23-kD D1 fragment present in isolated photosystem II core complexes, as well as that in thylakoid membranes depleted of endogenous FtsH. In this study, we definitively identify the chloroplast protease acting on the D1 protein during its light-induced turnover. Unlike previously identified membrane-bound substrates for FtsH in bacteria and mitochondria, the 23-kD D1 fragment represents a novel class of FtsH substrate—functionally assembled proteins that have undergone irreversible photooxidative damage and cleavage.
INTRODUCTION
Proteins that are rendered nonfunctional due to interactions with reactive oxygen species or free radicals undergo proteolysis and are replaced by newly synthesized copies. This is particularly significant in the chloroplast thylakoid membrane. Here, enzymes operate in a highly oxidizing environment and therefore are susceptible to impairment of structure and function. Within the thylakoid membrane, photosystem II (PSII) is the component most sensitive to oxidative damage. This sensitivity is partially due to its function in water splitting, a reaction that requires an oxidizing potential of 1.1 V, but it is also due to the intrinsic formation by PSII of toxic oxygen species (Andersson and Barber, 1994). PSII is a large multisubunit protein complex integral to the thylakoid membrane (Andersson and Barber, 1994). Its reaction center contains the homologous D1 and D2 proteins, PsbI, PsbW (in which Psb stands for PSII, and I and W denote specific subunits), and cytochrome b559. The D1/D2 heterodimer binds all of the chlorophylls, quinones, and metal ligands necessary to perform primary PSII photochemistry and electron transport. The structure of the PSII reaction center recently has been determined at a resolution of 8 Å (Rhee et al., 1998), and the structure of the dimeric PSII core complex has been set at a resolution of ∼9 Å (Hankamer et al., 1999).
Under conditions of high light intensity, electron transport within the complex is arrested, and consequently, the photosynthetic process is inactivated. This phenomenon is known as photoinhibition (Barber and Andersson, 1992; Prasil et al., 1992). The process is thought to occur via overreduction of the acceptor side of PSII, chlorophyll triplet formation, and production of toxic singlet oxygen (Vass et al., 1992; Telfer et al., 1994; Hideg et al., 1998). This reactive species in turn modifies the D1 protein, irreversibly impairing its function. To overcome permanent inhibition of photosynthetic electron transport, a highly coordinated repair process occurs within the chloroplast (Kyle et al., 1984; Prasil et al., 1992; Andersson and Aro, 1997; Melis, 1999). It includes transcription and translation of the plastid psbA gene, which encodes the D1 protein, insertion of the newly synthesized copy into the thylakoid membrane, binding of redox ligands, and reassembly of the PSII holocomplex. It is likely that repair is preceded by proteolytic removal of the damaged D1 protein; however, the identities of the proteases involved remain unknown despite intense research efforts.
After acceptor side photoinactivation, degradation of the D1 protein proceeds through two distinguishable steps (Andersson and Aro, 1997). The first is an endoproteolytic cleavage, which takes place in the stromal loop connecting the membrane-spanning helices D and E, yielding an N-terminal 23-kD fragment and a C-terminal 10-kD fragment. The protease responsible for catalyzing this cleavage reaction and the exact position at which it occurs have not been identified. The second step involves further digestion of the primary cleavage products.
The 23-kD fragment was detected in vivo by Greenberg et al. (1987) and also has been observed under in vitro conditions in isolated thylakoids and purified PSII complexes and reaction centers (Ohad et al., 1985; Salter et al., 1992; de Las Rivas et al., 1993). The primary cleavage recently was shown to be dependent on hydrolysis of membrane-bound GTP, whereas the addition of ATP had no effect (Spetea et al., 1999). In contrast, further degradation of the light-induced 23-kD D1 fragment was suggested to be mediated by an ATP-dependent zinc metalloprotease, because the steady state amount of this fragment was reduced by addition of ATP and zinc and increased by EDTA, a chelator of divalent metal ions (Spetea et al., 1999). Thus, at least two distinct proteases participate in degradation of the PSII D1 protein.
A few chloroplast homologs of bacterial proteases have been identified and characterized. These include the stromal ATP-dependent Clp protease (Shanklin et al., 1995; Adam, 1996; Sokolenko et al., 1998; Clarke, 1999), lumenal DegP (Itzhaki et al., 1998), and the thylakoid-bound FtsH protease (Lindahl et al., 1996; Ostersetzer and Adam, 1997). Bacterial FtsH is an ATP-dependent membrane-bound zinc metalloprotease (Tomoyasu et al., 1993a) and is the only essential energy-dependent protease in Escherichia coli (Gottesman, 1996). The FtsH proteases belong to the AAA family of ATPases. Members of this family have different functions but share a consensus sequence of ∼200 amino acids containing and flanking an ATP binding domain (Tomoyasu et al., 1993b).
Physiological substrates for FtsH in E. coli include the heat shock transcription factor σ32 (Herman et al., 1995; Tomoyasu et al., 1995), the phage λ CII protein (Shotland et al., 1997), and uncomplexed forms of the SecY protein (Kihara et al., 1995). A search of higher plant chloroplasts for homologs of this protease led to the finding of the thylakoid-bound FtsH (Lindahl et al., 1996). The ATP binding site and the catalytic zinc binding site, which are the functional domains of the bacterial enzyme, are both conserved in the plant FtsH. Moreover, topological studies have shown that FtsH is tightly bound to the thylakoid membrane and that the functional domains face the chloroplast stroma. FtsH is located exclusively in the stroma-exposed regions of the thylakoid membrane (Lindahl et al., 1996) and therefore is predicted to act on both soluble stromal and membrane-bound substrates. Consistent with this prediction, the FtsH protease has been implicated in the degradation of soluble and membrane-associated forms of unassembled Rieske Fe–S protein (Ostersetzer and Adam, 1997).
To date, FtsH is the only known ATP-dependent metalloprotease of higher plant chloroplasts. The finding that the steady state amount of the large 23-kD primary fragment of the damaged D1 protein is reduced by ATP and zinc (Spetea et al., 1999), together with the location of this fragment in the stroma-exposed thylakoid regions (Aro et al., 1993), suggests the involvement of FtsH in this process. To test this possibility, we overexpressed FtsH as a glutathione S-transferase (GST) fusion protein in E. coli and purified the recombinant protein by affinity chromatography. When introduced into isolated photoinactivated PSII core complexes in vitro, FtsH degraded the 23-kD D1 fragment, which was formed in the complexes during illumination. Moreover, mild treatment of thylakoid membranes with trypsin, which removed the extrinsic catalytic domain of FtsH, correlated with inhibition of proteolysis in situ. Degradation of the 23-kD D1 fragment was restored by adding purified FtsH to the membranes pretreated with trypsin, supporting a role for FtsH in the final degradation of oxidatively damaged D1 proteins. Thus, we have identified FtsH as a component of the complex PSII repair system that operates to overcome photoinhibition in plants.
RESULTS
The Steady State Amount of the 23-kD D1 Fragment Is Reduced by ATP
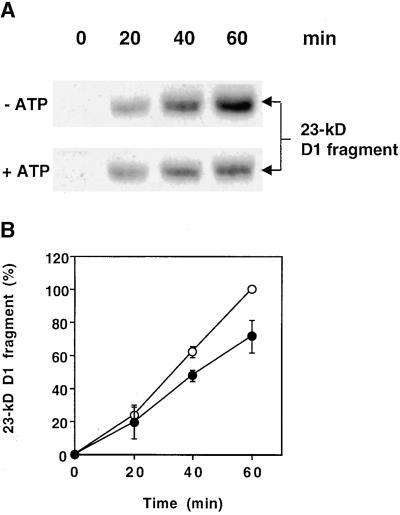
The amounts of the 23-kD D1 protein fragment present in thylakoid membranes during photoinhibitory illumination depend on the relative rates of primary cleavage of the D1 protein and further digestion of this 23-kD primary cleavage product. In Figure 1A, protein gel blot analysis reveals that photoinhibitory illumination of isolated thylakoids in the presence of ATP yielded a lesser amount of the 23-kD D1 protein fragment than illumination conducted in the absence of ATP. After 40 and 60 min in the light at an intensity of 1000 μmol m−2 sec−1, the amount of the 23-kD fragment was significantly lower in the presence of ATP (Figure 1B). This result is in agreement with a previous study of D1 protein degradation in isolated thylakoids. In this study, light-induced primary cleavage was shown to be independent of ATP, whereas the level of the 23-kD fragment was reduced in the presence of ATP (Spetea et al., 1999). Because the formation of this 23-kD fragment is unaffected by ATP, these observations suggest that its degradation is mediated by an ATP-dependent thylakoid membrane–bound protease.
Figure 1.
Effect of ATP on the Steady State Level of the 23-kD D1 Fragment during Photoinhibitory Illumination of Thylakoids.
(A) Thylakoids at a concentration of 0.4 mg of chlorophyll per mL were illuminated for up to 1 hr at an intensity of 1000 μmol m−2 sec−1 in the presence (+) or absence (−) of 2 mM ATP, and the amount of the 23-kD fragment of the D1 protein was examined by protein gel blot analysis.
(B) Quantification of the amount of the 23-kD fragment in the presence (filled circles) and absence (open circles) of ATP from three independent experiments. The amount of the fragment after 60 min illumination in the absence of ATP was defined as 100%. Means and the standard deviations are presented.
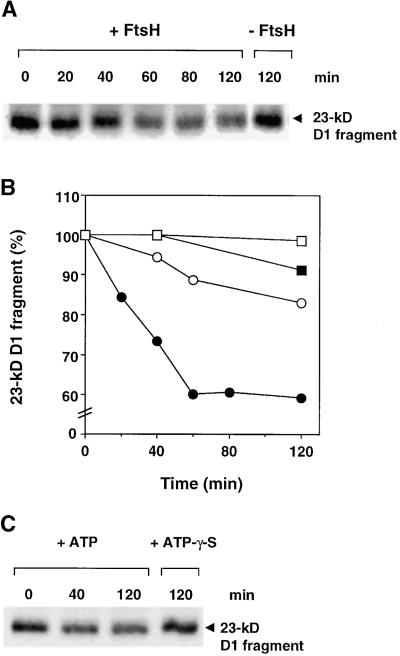
Degradation of the 23-kD D1 Fragment in Isolated Thylakoids Is Dependent on ATP Hydrolysis and Divalent Metal Ions
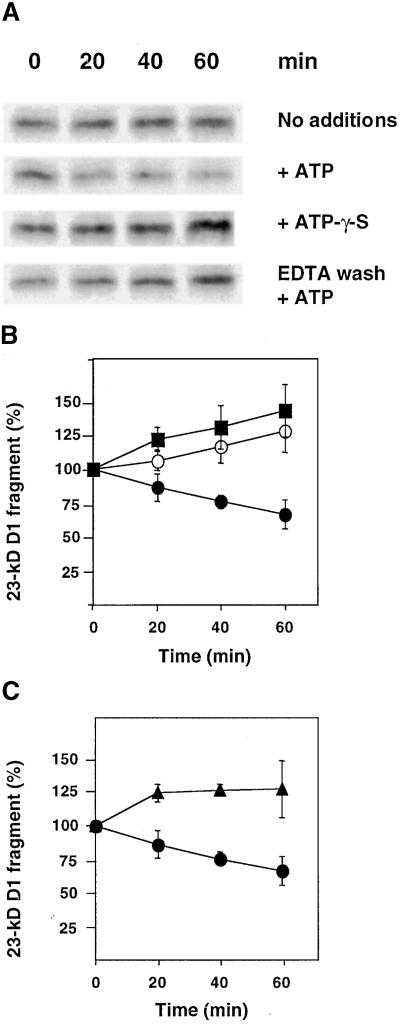
To study the possible involvement of FtsH in the degradation of the light-induced 23-kD D1 protein fragment, we required an experimental design separating primary and secondary proteolytic events. To this end, a period of illumination of thylakoids for generation of the 23-kD fragment was followed by a dark period for monitoring a putative net loss of this fragment. As shown in Figure 2, after a 45-min illumination of isolated thylakoids at an intensity of 800 μmol m−2 sec−1 (0 min), the 23-kD D1 fragment was readily detected by protein gel blot analysis. Subsequent incubation in the dark with no additions resulted in continued accumulation of this fragment, albeit at a slow rate (Figures 2A and 2B). This result is consistent with the notion that the primary cleavage occurs as a consequence of light-induced inactivation (Aro et al., 1993) and therefore that it continues after termination of the light treatment. The addition of ATP after transfer to the dark indeed caused a net loss of the 23-kD D1 fragment (Figures 2A and 2B). In contrast, the nonhydrolyzable analog ATP-γ-S did not support degradation of the 23-kD fragment (Figures 2A and 2B). Therefore, we concluded that proteolytic degradation of the light-induced 23-kD D1 fragment depends upon ATP hydrolysis. Note that although this experimental system allowed us to detect a net loss of the 23-kD fragment in the presence of ATP, the concomitant formation of this fragment masks the extent of its degradation. The actual loss therefore is considerably greater and can be calculated from the difference in the amounts of the 23-kD fragment in the presence and absence of ATP, respectively.
Figure 2.
Effects of ATP, ATP-γ-S, and Washing with EDTA on Degradation of the Light-Induced 23-kD D1 Fragment in Thylakoids.
(A) Thylakoids at a concentration of 0.4 mg chlorophyll per mL first were illuminated for 45 min at an intensity of 800 μmol m−2 sec−1 to induce fragmentation of the D1 protein. They then were incubated in the dark for up to 1 hr in the presence of either ATP or ATP-γ-S or with no additions. Some of the thylakoids were subjected to prewashing with 5 mM EDTA. The 23-kD D1 fragment was detected by protein gel blot analysis.
(B) Quantification of the amount of the 23-kD D1 fragment during dark incubation without addition of nucleotides (open circles) or with 2 mM ATP (filled circles) or 2 mM ATP-γ-S (filled squares).
(C) Quantification of the amount of the 23-kD fragment in thylakoids washed with EDTA and replenished with 5 mM MgCl2 in the presence of 2 mM ATP (filled triangles). Filled circles are as given in (B).
The values in (B) and (C) are the means of three independent experiments, and the standard deviations are presented by error bars.
Thylakoids that had been subjected to washing with EDTA after illumination and thereafter replenished with 5 mM MgCl2 did not exhibit degradation of the 23-kD D1 fragment during dark incubation in the presence of ATP (Figures 2A and 2C). This result indicates that the proteolytic reaction depends upon divalent metal ions other than Mg2+, and thus the responsible enzyme may be a metalloprotease.
FtsH Can Be Expressed in E. coli and Purified
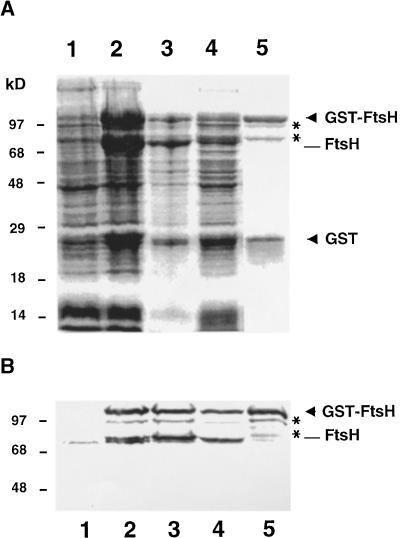
To allow direct examination of the putative role of FtsH during D1 protein degradation, we needed to purify FtsH. To this end, we constructed a GST–FtsH fusion protein that could be expressed in E. coli and purified in an active form. Purification was accomplished by using one-step affinity chromatography with immobilized glutathione to bind the GST moiety. Expression and purification were monitored by SDS-PAGE followed by staining with Coomassie blue and protein gel blot analysis by using antibodies raised against FtsH, as shown in Figure 3. In E. coli cells, before induction by isopropyl β-d-thiogalactopyranoside, a relatively small amount of endogenous bacterial FtsH (74 kD) was detected (Figure 3B, lane 1). Induction of expression of FtsH in strain A5039 of E. coli resulted in accumulation of the GST fusion protein (104 kD), as shown in Figures 3A and 3B (lanes 2). However, a fraction of this fusion protein was subject to processing between the GST moiety and the N terminus of FtsH, yielding large amounts of full-length FtsH (78 kD) and free GST (26 kD; Figures 3A and 3B, lanes 2). In fact, all other E. coli strains examined for expression of GST–FtsH accumulated only full-length FtsH and free GST: no fusion proteins remained intact (data not shown). In strain A5039, which was used in this study, typically half of the total recombinant GST–FtsH fusion protein remained intact after overnight expression at 20°C (Figures 3A and 3B, lanes 2).
Figure 3.
Overexpression and Purification of GST–FtsH.
E. coli cells were transformed with the plant cDNA encoding FtsH carried in pGEX-3X, and expression of GST–FtsH fusion protein was induced by 1 mM isopropyl β-d-thiogalactopyranoside.
(A) Bacterial proteins corresponding to 0.2 mL of cells at an OD600 of 1 were subjected to SDS-PAGE and stained with Coomassie blue.
(B) Bacterial proteins corresponding to 50 μL of cells at an OD600 of 1 were analyzed by protein gel blot analysis for FtsH.
In (A) and (B), lanes 1, uninduced E. coli cells; lanes 2, cells induced for expression; lanes 3, pellet after sonication and centrifugation; lanes 4, supernatant after sonication and centrifugation; and lanes 5, eluate after glutathione affinity chromatography. GST–FtsH and free GST are indicated by arrowheads, and small amounts of partially cleaved GST–FtsH (97 and 81 kD) are marked with asterisks.
After disruption of bacterial cells by sonication and centrifugation of the lysate, GST–FtsH as well as free FtsH partitioned between the pellet and supernatant (Figures 3A and 3B, lanes 3 and 4), but only the supernatant was used for further purification. The eluate from glutathione affinity chromatography contained GST–FtsH and free GST in approximately equal amounts (Figure 3A, lane 5). In addition, small amounts of two FtsH-derived protein species of 97 and 81 kD were detected (Figures 3A and 3B, lanes 5). Because they copurified with GST–FtsH and were recognized by FtsH antibodies, we concluded that they were intermediate breakdown products arising from cleavage of the fusion protein. The purification strategy involving glutathione affinity chromatography effectively excluded the possibility of endogenous bacterial FtsH contaminating the preparation, which indeed was found to be devoid of the 74-kD E. coli FtsH protein (Figure 3B, lane 5).
Purified Recombinant FtsH Is Proteolytically Active
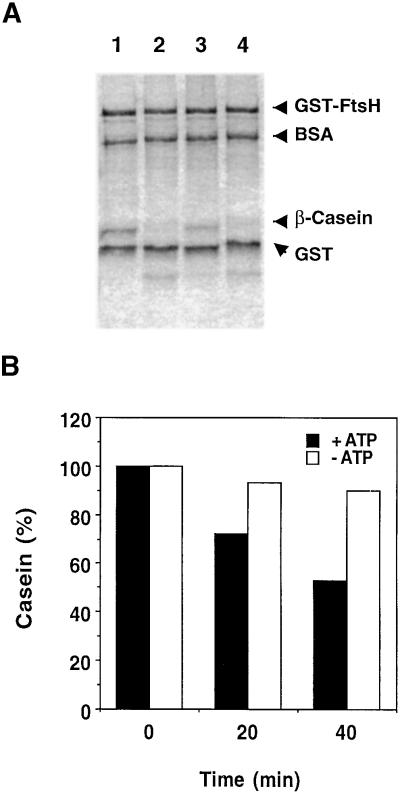
To ensure functionality of the purified recombinant protein, proteolytic activity of recombinant FtsH first was tested using β-casein as a substrate. Caseins are highly susceptible to proteolysis, probably due to their lack of defined secondary and tertiary structures, and therefore are commonly used for protease assays. Figure 4A shows that the FtsH-mediated degradation of β-casein in the presence of ATP was efficient and could be detected by Coomassie blue staining. In contrast, BSA and GST, which are compact globular proteins, remained stable throughout the experiment (Figure 4A). A cocktail of serine and cysteine protease inhibitors did not affect caseinolytic activity, whereas o-phenanthroline, a metalloprotease inhibitor, inhibited activity (Figure 4A). This confirms the identity of the protease, because zinc is an essential cofactor for proteolytic activity of FtsH. Degradation of β-casein also was found to depend on the presence of ATP (Figure 4B). Only after prolonged incubations could loss of β-casein be observed in the absence of ATP (data not shown).
Figure 4.
Degradation of β-Casein by Purified FtsH Fusion Protein.
GST–FtsH (0.96 μM) was incubated with β-casein (1.7 μM).
(A) The effect of protease inhibitors on β-casein degradation. Lane 1, a sample before incubation; lane 2, reaction mixture after 2 hr of incubation in the presence of 5 mM ATP; lane 3, as given for lane 2 with the addition of 10 mM o-phenanthroline; and lane 4, as given for lane 2 with the addition of a cocktail of serine and cysteine protease inhibitors. The proteins were resolved by SDS-PAGE and stained with Coomassie blue. Each lane contains 2 μg of FtsH fusion protein, and the initial amount of β-casein was 1 μg.
(B) Extent of β-casein degradation in the presence (+) or absence (−) of 5 mM ATP. Quantification is based on staining with Coomassie blue.
Purified Recombinant FtsH Degrades the Light-Induced 23-kD D1 Fragment in Isolated PSII Core Complexes
To test directly the ability of FtsH to degrade the 23-kD primary cleavage product of the D1 protein, purified recombinant FtsH was added to isolated PSII core complexes after photoinactivation. As shown in Figure 5, the addition of purified FtsH to the photoinactivated PSII cores containing large amounts of 23-kD D1 fragment resulted in degradation of this primary cleavage product. PSII core complexes alone, previously shown to be devoid of FtsH (Lindahl et al., 1996), did not display significant degradation of the 23-kD D1 fragment either in the presence or in the absence of ATP (Figures 5A and 5B). Maximum degradation of the 23-kD D1 fragment by FtsH added to the photoinactivated PSII cores was achieved in the presence of ATP. Under these conditions, ∼40% of the fragment was degraded during the first 60 min of the reaction (Figure 5B). In the absence of ATP, FtsH-mediated degradation was much slower, with <20% degraded after 2-hr incubation (Figure 5B). The addition of the nonhydrolyzable analog ATP-γ-S did not enhance degradation (Figure 5C), demonstrating that ATP hydrolysis is necessary for efficient proteolytic removal of the 23-kD D1 fragment.
Figure 5.
Degradation of the 23-kD D1 Fragment in Isolated PSII Core Complexes by Purified FtsH.
PSII cores were photoinactivated by illumination at 5000 μmol m−2 sec−1 for 60 min. The FtsH fusion protein (100 μg/mL) was incubated with 20 μg of chlorophyll per mL of PSII cores.
(A) Protein gel blot of the 23-kD D1 fragment in photoinactivated PSII cores incubated in the dark, in the presence of 2 mM ATP, with (+) and without (−) FtsH.
(B) Quantification of the 23-kD D1 fragment in photoinactivated PSII core complexes. Core complexes were incubated with FtsH and ATP (filled circles), only with FtsH (open circles), only with ATP (filled squares), or in the absence of either FtsH or ATP (open squares).
(C) Protein gel blot of photoinactivated PSII cores incubated with FtsH in the presence (+) of 2 mM ATP or ATP-γ-S.
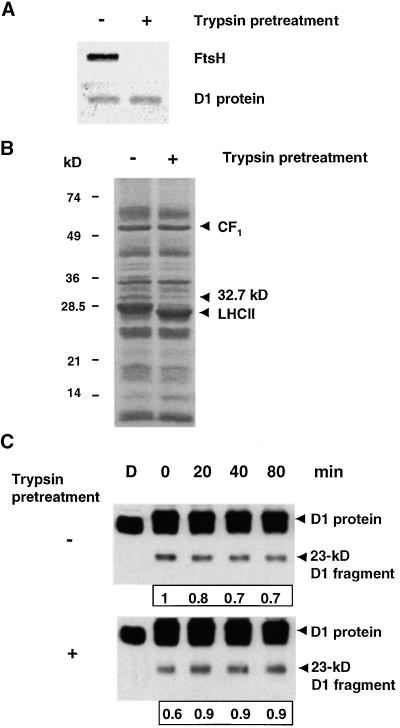
Removal of the Catalytic Domain of FtsH from Thylakoids Correlates with Loss of Degradation of the 23-kD D1 Fragment
In an attempt to produce FtsH-less thylakoids, we took advantage of the high sensitivity of FtsH to low concentrations of trypsin (Lindahl et al., 1996), generating thylakoids devoid of the catalytic domain of FtsH. Preliminary experiments showed that incubation of thylakoids with 1 μg/mL trypsin for 20 min on ice in the dark was the mildest and most efficient treatment for this purpose. Under these conditions, the extrinsic domain of FtsH containing the ATPase motif, the proteolytic active site, and the epitope for the antibody (Lindahl et al., 1996) were completely removed, as shown in Figure 6A. The stability of the D1 protein during the trypsin pretreatment was analyzed by protein gel blotting, and no loss was observed (Figure 6A). Also, no fragments of the D1 protein could be detected after trypsin pretreatment (Figure 6C, bottom, lane D). Apart from FtsH, only light-harvesting complex II and an unidentified 32.7-kD protein were sensitive to trypsin under these conditions, as determined by Coomassie blue staining (Figure 6B). In contrast to these proteins, the extrinsic stroma-exposed moiety of the ATP synthase, coupling factor CF1, remained unaffected by treatment with trypsin (Figure 6B).
Figure 6.
Removal of the Peripheral Catalytic Domain of FtsH from the Thylakoid Membrane and Its Effect on Degradation of the 23-kD D1 Fragment.
Isolated thylakoids at a concentration of 0.4 mg of chlorophyll per mL first were incubated on ice in the dark with 1 μg/mL trypsin for 20 min and then washed three times. A trypsin inhibitor, p-aminobenzamidine, was included at a concentration of 50 μM during subsequent treatments of both trypsin-pretreated and control membranes.
(A) Protein gel blot analysis of control (−) and trypsin-pretreated (+) thylakoids by using antibodies raised against FtsH and the D1 protein.
(B) Coomassie blue–stained gels of control (−) and trypsin-pretreated (+) thylakoids. The migration of molecular mass standards is indicated at left in kilodaltons, and the location of CF1, light-harvesting complex II (LHCII), and an unidentified protein (32.7 kD) are indicated by arrowheads.
(C) Control (−) and trypsin-pretreated (+) thylakoids were illuminated at 800 μmol m−2 sec−1 for 45 min and then incubated in the dark in the presence of 2 mM ATP. Protein gel blots are shown of the D1 protein and its 23-kD fragment in control and trypsin-pretreated thylakoids during dark incubation after illumination in the presence of 2 mM ATP. The boxed numbers under the blots represent the relative level of the 23-kD fragment in each sample. D denotes nonilluminated samples kept in the dark, and 0 represents the beginning of the dark period after the 45-min illumination.
Control and trypsin-pretreated thylakoids were illuminated for 45 min at 800 μmol m−2 sec−1 to generate fragments of the D1 protein. Before exposure to light, membranes were supplemented with p-aminobenzamidine to prevent possible residual trypsin activity. Trypsin-pretreated thylakoids were capable of producing the 23-kD fragment of the D1 protein when subjected to photoinhibitory light (Figure 6C, bottom), demonstrating that the proteolytic system responsible for primary cleavage remained intact during this treatment. After exposure to light, control and trypsin-pretreated thylakoids were kept in the dark in the presence of 2 mM ATP. Thylakoids used as control showed a net loss of the 23-kD fragment during dark incubation in the presence of ATP (Figure 6C, top), which is in agreement with the data shown in Figure 2. In contrast, thylakoids pretreated with trypsin continued to accumulate the fragment, yielding an increase in the amount of the fragment (Figure 6C, bottom). This result demonstrates that the endogenous ATP-dependent machinery responsible for degradation of the 23-kD D1 fragment indeed had been removed and implies that the loss of the degrading activity of the 23-kD fragment is correlated with the removal of FtsH.
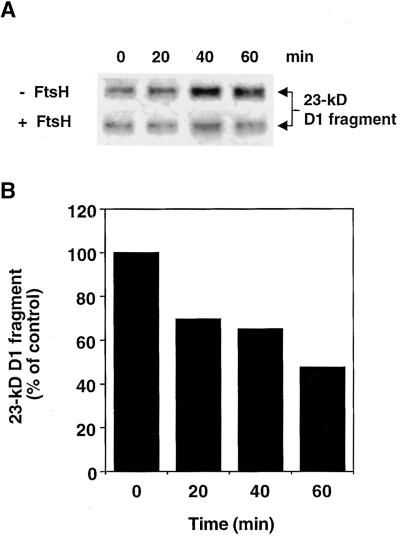
Purified FtsH Restores Degradation of the 23-kD D1 Fragment in Thylakoid Membranes Depleted of Endogenous FtsH
Because trypsin pretreatment could both remove the catalytic domain of FtsH and prevent degradation of the 23-kD fragment, we attempted to reconstitute proteolytic activity by adding purified FtsH to thylakoids that had been pretreated with trypsin. The trypsin-pretreated thylakoids were illuminated for 45 min at 800 μmol m−2 sec−1 and thereafter kept in the dark in the presence of ATP, with or without the addition of purified recombinant FtsH. As shown in Figure 7A (top), trypsin-pretreated membranes devoid of endogenous FtsH, without the addition of purified FtsH (control), showed continued accumulation of the 23-kD D1 fragment during dark incubation, which is in agreement with the results shown in Figure 6. However, the addition of purified FtsH abolished further accumulation of the fragment (Figure 7A, bottom), implying that this enzyme is responsible for the proteolytic removal of the 23-kD D1 fragment present in the thylakoid membranes after exposure to light. After a 60-min incubation in the dark in the presence of purified FtsH, the amount of the 23-kD D1 fragment was approximately half of that in control membranes without the addition of enzyme (Figure 7B). This reconstitution of activity further supports the notion that FtsH is involved in the degradation of the 23-kD fragment during the light-induced turnover of the PSII reaction center D1 protein.
Figure 7.
Restoration of Degradation of the 23-kD D1 Fragment by the Addition of Purified FtsH to Thylakoids Pretreated with Trypsin.
Thylakoids pretreated with trypsin at a concentration of 0.4 mg of chlorophyll per mL were illuminated at 800 μmol m−2 sec−1 for 45 min and then were incubated in the dark at 28°C in the presence of 5 mM ATP with or without 100 μg/mL purified FtsH.
(A) Protein gel blot of the 23-kD D1 fragment in trypsin-pretreated, light-treated thylakoids, during incubation in the dark in the presence (+) or absence (−) of purified recombinant FtsH. Zero (0) represents the beginning of the dark period after the light treatment.
(B) Quantification of the relative amount of the 23-kD D1 fragment in trypsin-pretreated light-treated thylakoids, after the addition of purified FtsH, based on the gel blot shown in (A). The values represent the ratio between the amounts of the fragment in the samples with and without FtsH for each time point.
DISCUSSION
The PSII D1 protein exhibits rapid turnover in response to high light intensity (Mattoo et al., 1984). Its high rate of synthesis and the corresponding high rate of degradation are related to the fact that this protein frequently undergoes light-induced, irreversible oxidative damage while catalyzing the light-driven electron transport from water to plastoquinone (Barber and Andersson, 1992). The transcription, translation, insertion, processing, and assembly of D1 protein have been studied thoroughly and characterized (see van Wijk et al., 1997). In contrast, the degradation mechanism remains an enigma, largely due to the lack of candidates for catalyzing primary and secondary cleavage events.
The discovery of FtsH in higher plant thylakoid membranes (Lindahl et al., 1996; Ostersetzer and Adam, 1997) and the finding that the steady state level of the light-induced 23-kD D1 fragment is reduced by ATP and zinc (Spetea et al., 1999) led us to hypothesize that FtsH plays a role in this proteolytic process. The final steps of D1 protein degradation take place in the nonappressed stroma-exposed thylakoid regions (Aro et al., 1993). FtsH is exclusively located in these membrane regions (Lindahl et al., 1996), thus fulfilling the spatial requirement for interaction with the putative protein substrate.
This study shows that recombinant FtsH is able to degrade the 23-kD D1 fragment present in purified photoinhibited PSII core complexes (Figure 5). Furthermore, degradation of this fragment in isolated thylakoid membranes (Figure 2) is abolished after removal of the catalytic domain of FtsH by mild pretreatment with trypsin (Figure 6) and can be restored by the addition of purified FtsH (Figure 7). Taken together, these results support a function of FtsH in the proteolytic removal of the 23-kD fragment of the D1 protein. In this study, we definitively have identified a chloroplast protease acting on the D1 protein during its light-induced turnover. Moreover, it provides biochemical evidence for the action of a particular protease on a specific physiological substrate in chloroplast protein degradation in general.
The rules governing protein stability are beginning to become unveiled. Proteases are known to be able to sense the folding state of potential substrates, thus selecting partially unfolded or misfolded proteins and exposing hydrophobic elements on their surface. A common group of targets for proteolysis are the unassembled proteins that normally constitute subunits of multiprotein complexes. The cause of their instability may well be incorrect folding together with enhanced accessibility in the absence of their neighboring subunits. Several studies of unassembled proteins in bacterial plasma membranes and mitochondrial inner membranes have shown that FtsH and its mitochondrial orthologs are responsible for their degradation (Pajic et al., 1994; Kihara et al., 1995; Nakai et al., 1995; Arlt et al., 1996; Guelin et al., 1996). Consistent with these observations, degradation of unassembled Rieske Fe–S protein in chloroplasts was suggested to be mediated by FtsH (Ostersetzer and Adam, 1997).
Unlike these previously identified membrane-bound FtsH substrates, the light-induced 23-kD D1 fragment is not present in excess amounts relative to the other subunits of the protein complex. On the contrary, it is present in substoichiometric amounts and resides in the center of the PSII complex. This location previously was demonstrated by first subjecting leaves to photoinhibitory illumination and thereafter isolating PSII reaction centers (Shipton and Barber, 1994). These PSII reaction centers indeed contained the 23-kD fragment, which was generated during the light treatment. Thus, the 23-kD D1 fragment represents an example of a new class of FtsH substrate–assembled proteins that have undergone irreversible damage.
The consideration of a possible mechanism of recognition between thylakoid FtsH and the 23-kD D1 fragment draws attention to the SsrA tagging system in E. coli. In this system, a C-terminal tail is added to translation products of incomplete mRNAs to tag these truncated polypeptides for degradation (Keiler et al., 1996). This tail (SsrA), encoded by the 10S stable RNA, consists of 11 amino acid residues, most of which are nonpolar. Herman et al. (1998) demonstrated that E. coli FtsH, as well as Clp proteases, is responsible for degradation of a cytosolic protein that has been modified to include SsrA or nonpolar pentapeptides at its C terminus. SsrA and other nonpolar C termini are thought to be recognized by proteases or chaperones (Levchenko et al., 1997). Thus, recognition of nonpolar residues at the C terminus of the 23-kD D1 fragment by thylakoid FtsH should be considered.
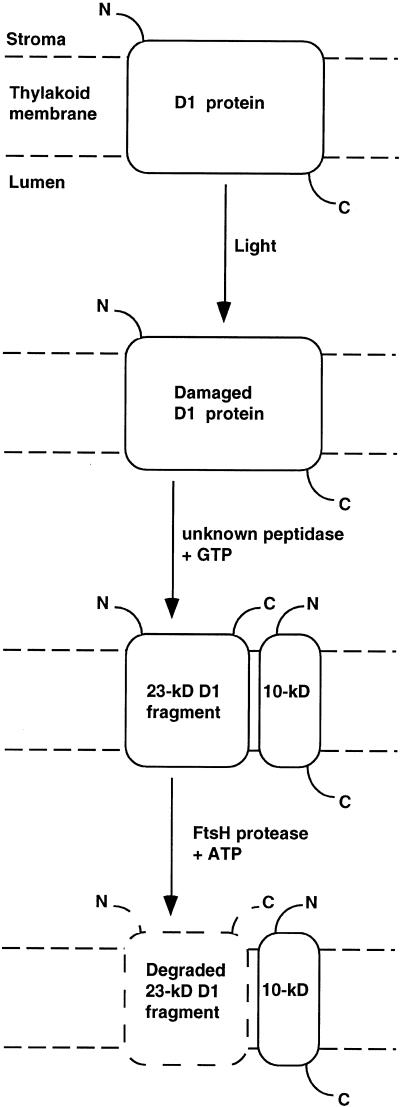
The light-induced 23-kD D1 fragment is generated by primary cleavage of the protein in the stromal loop between helices D and E (Greenberg et al., 1987; Shipton and Barber, 1994; see Figure 8). This cleavage is thought to be triggered by a structural change that occurs in the region around the QB quinone binding site as a consequence of oxidative damage (Jansen et al., 1993; Zer et al., 1994). The antibody used in this study to detect the 23-kD fragment was raised against residues 234 to 242. Nakajima et al. (1996) showed that the 23-kD fragment could be detected by an antibody raised against residues 239 to 247 of the D1 protein. This implies a primary cleavage after residue 247 of the protein. Between residue 247 and a predicted short α-helical structure (residues 252 to 261), peripheral to the stromal side of the thylakoid membrane, are four conserved nonpolar residues, Ile-Val-Ala-Ala (residues 248 to 251). A primary cleavage between alanine residue 251 and histidine residue 252 thus would yield a 23-kD D1 fragment with a nonpolar C terminus that could be recognized by binding domains of thylakoid FtsH. This mechanism would explain why the 23-kD fragment, but not the intact D1 protein, is a substrate for FtsH. In contrast to the C-terminal addition of an oligopeptide tagging a protein for degradation (Keiler et al., 1996), the novel mechanism suggested here involves a priming endoproteolytic cleavage yielding a new C terminus, which can be recognized by FtsH (Figure 8).
Figure 8.
Model Describing Stages in the Light-Induced Degradation of the PSII D1 Protein.
The schematic topology of the D1 protein in the thylakoid membrane is depicted. N and C denote the N and C termini, respectively. In the first stage, the D1 protein is photooxidatively damaged. The damaged protein is cleaved by an unknown mechanism requiring GTP. The resulting 23-kD fragment then is completely degraded by the FtsH protease.
Chaperone-like activity was demonstrated to be performed by the isolated ATP binding domain of the i-AAA protease, a mitochondrial ortholog of FtsH (Leonhard et al., 1999). Leonhard et al. (1996) suggested that the mitochondrial FtsH-like m-AAA protease participates in the partitioning of a membrane-bound model substrate to the catalytic site by virtue of its chaperone activity, thereby allowing cleavage between residues that normally are embedded in the lipid bilayer. This possibility was further supported by a recent study by Kihara et al. (1999) showing that extracytoplasmic domains of membrane-bound substrates were degraded by E. coli FtsH. Therefore, it is possible that FtsH actively pulls the 23-kD D1 fragment out of the membrane during the process of degradation. The previously reported endoproteolytic cleavages in lumen-exposed loops of the D1 protein should facilitate complete digestion (Andersson and Aro, 1997), and in vivo degradation may be aided by lumenal proteases, such as DegP (Itzhaki et al., 1998).
Our studies show that degradation of the 23-kD D1 fragment by FtsH in vitro is stimulated severalfold by ATP (Figure 5B). The lack of effect of the nonhydrolyzable analog ATP-γ-S implies that the reaction depends on ATP hydrolysis (Figure 5C). Similarly, degradation of the 23-kD fragment in situ in intact thylakoid membranes was shown to be dependent on ATP hydrolysis (Figures 2A and 2B). These observations are consistent with simultaneous chaperone and proteolytic activities. An additional role for ATP has been implied in promoting the formation of an active oligomeric complex of the FtsH-like m-AAA protease (Arlt et al., 1996) and another ATP-dependent protease, ClpAP (Hoskins et al., 1998). Such complex formation, however, does not require hydrolysis of ATP. Yet, another ATP-dependent step is the transfer of a substrate polypeptide chain from the initial binding site to the proteolytic site within the Clp protease complex (Hoskins et al., 1998). This feeding mechanism operates at the expense of ATP hydrolysis. There are no available data supporting a substrate-feeding mechanism for FtsH-like proteases, and the quaternary structure of thylakoid FtsH is not known. Therefore, it remains to be determined at which steps ATP binding and hydrolysis are necessary for FtsH-mediated degradation of the 23-kD D1 fragment.
FtsH-mediated membrane protein degradation has been studied in several systems with different degrees of complexity. Unassembled proteins that are degraded in wild-type organisms have been shown to become stable in deletion or site-directed mutants of FtsH (Pajic et al., 1994; Kihara et al., 1995; Nakai et al., 1995; Arlt et al., 1996; Guelin et al., 1996). Purified SecY (Akiyama et al., 1996a; Kihara et al., 1996) and YccA, a membrane protein of unknown function (Kihara et al., 1998), are degraded by purified E. coli FtsH in vitro. In addition, SecY, as well as subunit a of the ATPase Fo sector, is degraded in crude detergent-solubilized E. coli plasma membranes after the addition of purified FtsH (Akiyama et al., 1996a, 1996b). Our study of FtsH-mediated degradation of the 23-kD D1 fragment shows FtsH-dependent protein degradation in vitro of a membrane protein substrate present in a purified and assembled protein complex that has been damaged. We also demonstrate FtsH-dependent proteolysis in situ in isolated membranes. An important notion is that FtsH may not be the only thylakoid protease removed by trypsin pretreatment and that other unidentified enzymes may be discovered as participants in the highly complex process of D1 protein turnover.
We demonstrate here that the FtsH protease is involved in proteolytic removal of an oxidatively damaged and cleaved membrane protein, the PSII reaction center D1 protein (Figure 8). However, the details of this process are far from clear. The site of primary GTP-dependent cleavage and the enzyme responsible for catalyzing this event need to be identified. The fate of other reported fragments, such as the 10-kD C-terminal D1 fragment, should be clarified. The possible participation of other proteases in the degradation of the D1 protein also needs to be examined. Within the larger context of membrane protein degradation, elucidating the mechanism for partitioning of hydrophobic transmembrane regions to the proteolytic site and the coupled energy conversion remains a challenge.
METHODS
Subcloning and Overexpression of FtsH
The cDNA clone encoding the Arabidopsis thaliana FtsH precursor protein (Lindahl et al., 1996) was used as a template for polymerase chain reaction amplification of a 1.96-kb fragment encoding the mature FtsH protein. The oligonucleotides used were the 5′ end primer 5′-GCGGATCCCCAATTCGCCATTCTC-3′ and the 3′ end primer 5′-CGGAATTCTTAACCGTCAATAAAGAGAC-3′. This fragment was cloned into the BamHI-EcoRI sites of pGEX-3X (Pharmacia) to generate a glutathione S-transferase–FtsH (GST–FtsH) fusion protein, with the GST moiety linked to the N terminus of the FtsH protein. The accuracy of the construct was verified by DNA sequencing. Recombinant FtsH was expressed in strain A5039 of Escherichia coli with LacZ LacIq::(Kanr). For each expression event, freshly transformed cells were used. Liquid cultures (300 mL) were grown in Luria-Bertani broth in the presence of 50 μg/mL ampicillin at 30°C. Expression was induced by adding 1 mM isopropyl β-d-thiogalactopyranoside (final concentration) to cell cultures at an OD600 of 0.4, and the cultures were transferred to 20°C and grown overnight. The final OD600 was typically 1.5. Optical density of cell cultures was determined using a Uvikon 930 spectrophotometer (Kontron Instruments, Zurich, Switzerland).
Purification of GST–FtsH Fusion Protein
E. coli cells from a 300-mL culture were harvested by centrifugation at 1000g for 10 min and washed once in 30 mL of PBS (Biological Industries, Beit Haemek, Israel) adjusted to pH 8.0 with NaOH. The pellet was resuspended in 1 mL of PBS, pH 8.0, and stored at −70°C until use. Cells were thawed on ice and resuspended to 6.5 mL in PBS, pH 8.0. Reduced DTT was added to a 20 mM final concentration, and Complete, EDTA-free (Boehringer Mannheim), a cocktail of inhibitors of serine and cysteine proteases, was added to the cell suspension. Cells were disrupted by sonication on ice at 12 μm for 5 × 10 sec with 30-sec cooling intervals. The lysate was centrifuged at 10,000g for 15 min, and the resulting supernatant was mixed with 300 μL of settled glutathione–agarose (Sigma). This affinity matrix was preequilibrated with PBS, pH 8.0, and 20 mM DTT. Binding was performed at 4°C overnight under gentle agitation. The glutathione–agarose beads then were allowed to sediment on ice, and the supernatant containing unbound proteins was discarded. The beads were resuspended in PBS, pH 8.0, to 10 mL, allowed to resettle, and separated from the supernatant. For additional washes, the beads were transferred to a 1.5-mL microcentrifuge tube, resuspended to 1.5 mL in PBS, pH 8.0, and centrifuged at 2000g for 10 sec. This washing procedure was repeated five times. Finally, GST–FtsH was eluted in a batch procedure by addition of 150 μL of 50 mM Tris-HCl, pH 8.0, containing 20 mM reduced glutathione followed by gentle agitation at 4°C for 2 hr. Protein concentration of the eluate was measured using the assay of Bradford (Bio-Rad) with BSA as a standard. The relative amounts of intact GST–FtsH and free GST in the eluate were determined by SDS-PAGE, Coomassie Brilliant Blue R 250 staining, and subsequent quantification of the respective bands by using the National Institutes of Health Image program.
Assay for Proteolytic Activity of FtsH against β-Casein
Reaction mixtures contained 0.96 μM GST–FtsH (100 μg/mL) and 1.7 μM β-casein (50 μg/mL). Thus, 2 μg of purified GST–FtsH and 1 μg of β-casein (Sigma) were added per a 20-μL reaction volume. These proteins were incubated in a buffer composed of 50 mM Tris-acetate, pH 8.0, 5 mM Mg-acetate, 12.5 μM Zn-acetate, 80 mM NaCl, 100 μg/mL BSA, and 1.4 mM β-mercaptoethanol (in vitro buffer) at 38°C for up to 2 hr. When indicated, the reaction was supplemented with 5 mM ATP. Protease inhibitors used were 10 mM o-phenanthroline (Sigma) and Complete, EDTA-free (Boehringer Mannheim). At the end of each incubation, Laemmli solubilizing buffer (Laemmli, 1970) was added, and the samples were stored on ice. Samples were subjected to SDS-PAGE by using 12% acrylamide gels. The relative amounts of β-casein were determined using Coomassie Brilliant Blue R 250 staining followed by quantification using the National Institutes of Health Image program.
Degradation of the 23-kD D1 Protein Fragment in Isolated Thylakoids
Thylakoids were isolated from 10- to 12-day-old pea (Pisum sativum) leaves, as previously described (Andersson et al., 1976). For removal of the extrinsic domain of FtsH, thylakoids were resuspended in a medium containing 10 mM sodium phosphate, pH 7.4, 100 mM sucrose, 5 mM NaCl, and 5 mM MgCl2 to a concentration of 0.4 mg of chlorophyll per mL. Trypsin (Boehringer Mannhein) was added to a concentration of 1 μg/mL, and the mixture was incubated in the dark on ice for 20 min. Trypsin was removed by centrifugation at 3000g for 5 min at 4°C, and the thylakoid pellet was washed three times in the same medium. Possible residual trypsin activity was inhibited by including 50 μM p-aminobenzamidine (Sigma) in subsequent treatments. Control thylakoids were subjected to identical treatments without trypsin.
For photoinhibitory illumination, control and trypsinated thylakoids were resuspended in 50 mM Hepes-NaOH, pH 7.4, 0.4 M sucrose, 15 mM NaCl, and 5 mM MgCl2 to a concentration of 0.4 mg of chlorophyll per mL. Illumination was performed at 22°C and at the indicated light intensities. Subsequent to the light treatment, degradation and accumulation of the light-induced 23-kD D1 protein fragment was followed in the dark at 25°C in the presence or absence of 2 mM ATP or ATP-γ-S.
Washing thylakoids with EDTA to remove bound zinc was performed after illumination and before incubation in the dark. Thylakoids were kept on ice in 50 mM Hepes-NaOH, pH 7.4, 0.4 M sucrose, 15 mM NaCl, and 5 mM MgCl2 at a concentration of 0.4 mg of chlorophyll per mL. EDTA was added to 5 mM final concentration; after centrifugation at 5000g for 5 min at 4°C, thylakoids were resuspended in the same medium without EDTA, thereby replenishing the membranes with Mg2+.
Degradation of the 23-kD D1 protein fragment in trypsin-pretreated thylakoid membranes was restored by adding 100 μg/mL purified GST–FtsH to thylakoids at a concentration of 80 μg of chlorophyll per mL. Reaction mixtures were incubated in the dark in the presence of 5 mM ATP at 28°C. The amount of the 23-kD D1 fragment was analyzed by protein gel blotting as described below.
Degradation of the 23-kD D1 Protein Fragment in Purified Photosystem II Core Complexes
Photosystem II (PSII) core complexes were isolated as previously described (Roobol-Boza and Andersson, 1996) and resuspended to a concentration of 0.2 mg of chlorophyll per mL in a buffer containing 50 mM Hepes-NaOH, pH 7.4, 0.4 M sucrose, 15 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, and 0.01% (w/v) dodecyl maltoside. Photoinactivation of PSII core complexes was performed by illumination at 5000 μmol m−2 sec−1 in the presence of 0.2 mM GTP at 22°C for 60 min, causing >90% loss of oxygen evolution. PSII core complexes also were kept in the dark as control material. Control and photoinhibited PSII cores were frozen in liquid N2 and stored at −80°C until use. The reactions assaying proteolytic activity of FtsH toward the 23-kD D1 fragment were performed in a medium consisting of 50 mM Tris-acetate, pH 8.0, 5 mM Mg-acetate, 12.5 μM Zn-acetate, 80 mM NaCl, and 1.4 mM β-mercaptoethanol. The concentration of GST–FtsH was 100 μg/mL, and PSII cores were added to a concentration of 20 μg of chlorophyll per mL. When indicated, 2 mM ATP was included. Reactions were incubated at 35°C in the dark, and aliquots were withdrawn at the times indicated, solubilized in Laemmli sample buffer, and stored on ice until SDS-PAGE analysis. The 23-kD D1 fragment was detected and quantified by gel blotting, as described below.
Protein Gel Blot Analysis
Quantitative immunoblotting of the D1 protein and the 23-kD fragment was performed using a polyclonal antibody raised against a polypeptide corresponding to amino acids 234 to 242 (DE loop) of the spinach D1 protein (Spetea et al., 1999). For quantification of the D1 protein, 0.25 μg of chlorophyll thylakoid membranes was loaded per lane, and the antibody was used at a dilution of 1:5000. For quantification of the 23-kD D1 protein fragment, 1 μg of chlorophyll thylakoid membranes or 0.4 μg of chlorophyll PSII cores was loaded per lane, and the antibody was used at a dilution of 1:3000. Immune complexes were detected by enhanced chemiluminescence.
FtsH was detected using a polyclonal antibody raised against a 16–amino acid polypeptide corresponding to the second region of homology of the Arabidopsis FtsH protein (Lindahl et al., 1996) at a dilution of 1:1000. Thylakoid membranes (3 μg of chlorophyll) or 50 μL of bacterial cells at an OD600 of 1 was loaded per lane. The alkaline phosphatase color development reaction (nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate) was used to detect signals.
Acknowledgments
This research was supported by grants from the Israel Science Foundation, founded by the Israel Academy of Sciences and Humanities–Charles H. Revson Foundation (to Z.A.), and the Swedish Natural Science Research Council and the Swedish Foundation for Strategic Research (to B.A.). M.L. is a recipient of a Marie Curie Training and Mobility of Researchers postdoctoral fellowship from the European Commission. We thank Itzhak Ohad and Nir Keren of Hebrew University for many insightful discussions. The valuable technical advice of Dinah Teff is gratefully acknowledged.
References
- Adam, Z. (1996). Protein stability and degradation in chloroplasts. Plant Mol. Biol. 32 773–783. [DOI] [PubMed] [Google Scholar]
- Akiyama, Y., Kihara, A., and Ito, K. (1996. a). Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 399 26–28. [DOI] [PubMed] [Google Scholar]
- Akiyama, Y., Kihara, A., Tokuda, H., and Ito, K. (1996. b). FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271 31196–31201. [DOI] [PubMed] [Google Scholar]
- Andersson, B., and Aro, E.-M. (1997). Proteolytic activities and proteases of plant chloroplasts. Physiol. Plant. 100 780–793. [Google Scholar]
- Andersson, B., and Barber, J. (1994). Composition, organization and dynamics of thylakoid membranes. In Advances in Molecular and Cell Biology, E.E. Bittar, ed (Greenwich, UK: Jai Press Inc.), pp. 1–53.
- Andersson, B., Åkerlund, H.E., and Albertsson, P.Å. (1976). Separation of subchloroplast membrane particles by counter-current distribution. Biochim. Biophys. Acta 423 122–132. [DOI] [PubMed] [Google Scholar]
- Arlt, H., Tauer, R., Feldmann, H., Neupert, W., and Langer, T. (1996). The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85 875–885. [DOI] [PubMed] [Google Scholar]
- Aro, E.-M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17 61–66. [DOI] [PubMed] [Google Scholar]
- Clarke, A.K. (1999). ATP-dependent Clp proteases in photosynthetic organisms—A cut above the rest! Ann. Bot. 83 593–599. [Google Scholar]
- de Las Rivas, J., Shipton, C.A., Ponticos, M., and Barber, J. (1993). Acceptor side mechanism of photoinduced proteolysis of the D1 protein in photosystem II reaction centers. Biochemistry 32 6944–6950. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. (1996). Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30 465–506. [DOI] [PubMed] [Google Scholar]
- Greenberg, B.M., Gaba, V., Mattoo, A.K., and Edelman, M. (1987). Identification of a primary in vivo degradation product of the rapidly turning-over 32 kD protein of photosystem II. EMBO J. 6 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelin, E., Rep, M., and Grivell, L.A. (1996). Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 381 42–46. [DOI] [PubMed] [Google Scholar]
- Hankamer, B., Morris, E.P., and Barber, J. (1999). Revealing the structure of the oxygen-evolving core dimer of photosystem II by cryoelectron crystallography. Nat. Struct. Biol. 6 560–564. [DOI] [PubMed] [Google Scholar]
- Herman, C., Thevenet, D., Dari, R., and Bouloc, P. (1995). Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92 3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, C., Thevenet, D., Bouloc, P., Walker, G.C., and D'Ari, R. (1998). Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg, E., Kalai, T., Hideg, K., and Vass, I. (1998). Photoinhibition of photosynthesis in vivo results in singlet oxygen production detection via nitroxide-induced fluorescence quenching in broad bean leaves. Biochemistry 37 11405–11411. [DOI] [PubMed] [Google Scholar]
- Hoskins, J.R., Pak, M., Maurizi, M.R., and Wickner, S. (1998). The role of the ClpA chaperone in proteolysis by ClpAP. Proc. Natl. Acad. Sci. USA 95 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki, H., Naveh, L., Lindahl, M., Cook, M., and Adam, Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273 7094–7098. [DOI] [PubMed] [Google Scholar]
- Jansen, M.A., Depka, B., Trebst, A., and Edelman, M. (1993). Engagement of specific sites in the plastoquinone niche regulates degradation of the D1 protein in photosystem II. J. Biol. Chem. 268 21246–21252. [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R.H., and Sauer, R.T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271 990–993. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1995). FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. USA 92 4532–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1996). A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15 6122–6131. [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1998). Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: An implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol. 279 175–188. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1999). Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18 2970–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle, D.J., Ohad, I., and Arntzen, C.J. (1984). Membrane protein damage and repair: Selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl. Acad. Sci. USA 81 4070–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leonhard, K., Herrmann, J.M., Stuart, R.A., Mannhaupt, G., Neupert, W., and Langer, T. (1996). AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15 4218–4229. [PMC free article] [PubMed] [Google Scholar]
- Leonhard, K., Stiegler, A., Neupert, W., and Langer, T. (1999). Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 398 348–351. [DOI] [PubMed] [Google Scholar]
- Levchenko, I., Smith, C.K., Walsh, N.P., Sauer, R.T., and Baker, T.A. (1997). PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell 91 939–947. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Tabak, S., Cseke, L., Pichersky, E., Andersson, B., and Adam, Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271 29329–29334. [DOI] [PubMed] [Google Scholar]
- Mattoo, A.K., Hoffman-Falk, H., Marder, J.B., and Edelman, M. (1984). Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc. Natl. Acad. Sci. USA 81 1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis, A. (1999). Photosystem-II damage and repair cycle in chloroplasts: What modulates the rate of photodamage? Trends Plant Sci. 4 130–135. [DOI] [PubMed] [Google Scholar]
- Nakai, T., Yasuhara, T., Fujiki, Y., and Ohashi, A. (1995). Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol. Cell. Biol. 15 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, Y., Yoshida, S., Inoue, Y., and Ono, T. (1996). Occupation of the Q(B)-binding pocket by a photosystem II inhibitor triggers dark cleavage of the D1 protein subjected to brief preillumination. J. Biol. Chem. 271 17383–17389. [DOI] [PubMed] [Google Scholar]
- Ohad, I., Kyle, D.J., and Hirschberg, J. (1985). Light-dependent degradation of the QB-protein in isolated pea thylakoids. EMBO J. 4 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer, O., and Adam, Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: The possible role of the FtsH protease. Plant Cell 9 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic, A., Tauer, R., Feldmann, H., Neupert, W., and Langer, T. (1994). Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 353 201–206. [DOI] [PubMed] [Google Scholar]
- Prasil, O., Adir, N., and Ohad, I. (1992). Dynamics of photosystem II: Mechanism of photoinhibition and recovery processes. In Topics in Photosynthesis (Amsterdam: Elsevier Science Publishers), pp. 295–348.
- Rhee, K.H., Morris, E.P., Barber, J., and Kuhlbrandt, W. (1998). Three-dimensional structure of the plant photosystem II reaction centre at 8 Å resolution. Nature 396 283–286. [DOI] [PubMed] [Google Scholar]
- Roobol-Boza, M., and Andersson, B. (1996). Isolation of hydrophobic membrane proteins by perfusion chromatography—Purification of photosystem II reaction centers from spinach chloroplasts. Anal. Biochem. 235 127–133. [DOI] [PubMed] [Google Scholar]
- Salter, A.H., Virgin, I., Hagman, A., and Andersson, B. (1992). On the molecular mechanism of light-induced D1 protein degradation in PSII core particles. Biochemistry 31 3990–3998. [DOI] [PubMed] [Google Scholar]
- Shanklin, J., Dewitt, N.D., and Flanagan, J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipton, C.A., and Barber, J. (1994). In vivo and in vitro photoinhibition reactions generate similar degradation fragments of D1 and D2 photosystem-II reaction-centre proteins. Eur. J. Biochem. 220 801–808. [DOI] [PubMed] [Google Scholar]
- Shotland, Y., Koby, S., Teff, D., Mansur, N., Oren, D.A., Tatematsu, K., Tomoyasu, T., Kessel, M., Bukau, B., Ogura, T., and Oppenheim, A.B. (1997). Proteolysis of the phage lambda CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol. 24 1303–1310. [DOI] [PubMed] [Google Scholar]
- Sokolenko, A., Lerbs-Mache, S., Altschmied, L., and Herrmann, R.G. (1998). Clp protease complexes and their diversity in chloroplasts. Planta 207 286–295. [DOI] [PubMed] [Google Scholar]
- Spetea, C., Hundal, T., Lohmann, F., and Andersson, B. (1999). GTP bound to the chloroplast thylakoid membranes is required for light-induced multi-enzyme degradation of photosystem II D1 protein. Proc. Natl. Acad. Sci. USA 96 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., Bishop, S.M., Phillips, D., and Barber, J. (1994). Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J. Biol. Chem. 269 13244–13253. [PubMed] [Google Scholar]
- Tomoyasu, T., Yamanaka, K., Murata, K., Suzaki, T., Bouloc, P., Kato, A., Niki, H., Hiraga, S., and Ogura, T. (1993. a). Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175 1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu, T., Yuki, T., Morimura, S., Mori, H., Yamanaka, K., Niki, H., Hiraga, S., and Ogura, T. (1993. b). The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J. Bacteriol. 175 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu, T., Gamer, J., Bukau, B., Kanemori, M., Mori, H., Rutman, A.J., Oppenheim, A.B., Yura, T., Yamanaka, K., Niki, H., Hiraga, S., and Ogura, T. (1995). Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14 2551–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk, K.J., Roobol-Boza, M., Kettunen, R., Andersson, B., and Aro, E.-M. (1997). Synthesis and assembly of the D1 protein into photosystem II: Processing of the C terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36 6178–6186. [DOI] [PubMed] [Google Scholar]
- Vass, I., Styring, S., Hundal, T., Koivuniemi, A., Aro, E.-M., and Andersson, B. (1992). Reversible and irreversible intermediates during photoinhibition of photosystem II: Stable reduces QA species promote chlorophyll triplet formation. Proc. Natl. Acad. Sci. USA 89 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zer, H., Prasil, O., and Ohad, I. (1994). Role of plastoquinol oxidoreduction in regulation of photochemical reaction center II D1 protein turnover in vivo. J. Biol. Chem. 269 17670–17676. [PubMed] [Google Scholar]