Abstract
Arabidopsis plants have a system to specifically restrict the long-distance movement of tobacco etch potyvirus (TEV) without involving either hypersensitive cell death or systemic acquired resistance. At least two dominant genes, RTM1 and RTM2, are necessary for this restriction. Through a series of coinfection experiments with heterologous viruses, the RTM1/RTM2–mediated restriction was shown to be highly specific for TEV. The RTM2 gene was isolated by a map-based cloning strategy. Isolation of RTM2 was confirmed by transgenic complementation and sequence analysis of wild-type and mutant alleles. The RTM2 gene product is a multidomain protein containing an N-terminal region with high similarity to plant small heat shock proteins (HSPs). Phylogenetic analysis revealed that the RTM2 small HSP–like domain is evolutionarily distinct from each of the five known classes of plant small HSPs. Unlike most other plant genes encoding small HSPs, expression of the RTM2 gene was not induced by high temperature and did not contribute to thermotolerance of seedlings. The RTM2 gene product was also shown to contain a large C-terminal region with multiple repeating sequences.
INTRODUCTION
Systemic infection of plants by viruses generally requires genome replication in individual cells, cell-to-cell movement through plasmodesmata, and long-distance movement through the phloem (Carrington et al., 1996; Baker et al., 1997). A block at any of these steps, either by active defense responses or by incompatible combinations of viral and host factors, can lead to resistance. Several types of active resistance responses to viruses are known (Baker et al., 1997; Carrington and Whitham, 1998). Dominant or semidominant resistance (R) genes that trigger hypersensitive cell death or extreme resistance at initial infection sites, occurring most often in a strain-specific or “gene-for-gene” manner, are well known. The tobacco N gene and the potato Rx gene, which condition resistance against tobacco mosaic virus and potato virus X strains, respectively, belong to the nucleotide binding site, leucine-rich repeat (NBS-LRR) class of R genes (Whitham et al., 1994; Bendahmane et al., 1999). Whereas N confers a hypersensitive response concomitant with localized cell-to-cell movement in inoculated leaves, Rx confers an extreme resistance in initially infected cells (Bendahmane et al., 1995).
A second type of active resistance involves gene-silencing-like mechanisms. Virus-induced silencing is adaptive, meaning that a naive host can recognize viral nucleic acids and customize a sequence-specific response (Baulcombe, 1999; Grant, 1999). Thus, virus-induced silencing can limit virus accumulation, promote recovery (in some cases) from a systemic infection, and confer resistance to secondary infections with the same or homologous viruses (Ratcliff et al., 1997; Al-Kaff et al., 1998; Ratcliff et al., 1999). In contrast to resistance triggered by NBS-LRR–type R genes, resistance through silencing appears not to depend on a gene-for-gene recognition event. Passive resistance has been suggested to occur in cases in which the plant lacks one or more factors required for virus replication or movement. Several types of resistance conferred by recessive genes may be in this class (Callaway et al., 1996; Schaad and Carrington, 1996; Nicolas et al., 1997), although specific genes responsible for such resistance have not been isolated.
Resistance of Arabidopsis to tobacco etch potyvirus (TEV) does not fall into any of the known categories of resistance response. Resistance in Arabidopsis ecotypes such as Columbia-0 (Col-0) or Wassilewskija-2 (Ws-2) results from blockage of long-distance movement. Viral replication and cell-to-cell movement in inoculated leaves appear unaffected (Mahajan et al., 1998). Genetic analyses using natural ecotype variation and chemically induced mutations indicate that dominant genes at two loci, RTM1 and RTM2, are necessary for the resistance phenotype (Mahajan et al., 1998; Whitham et al., 1999). RTM1/RTM2–mediated resistance differs from typical R gene–mediated resistance in that hypersensitive cell death does not occur around infection foci, and moreover, mutants with defects in several genes known to be necessary for NBS-LRR–type resistance have no effect on TEV restriction. Markers associated with systemic acquired resistance are not induced after infection by TEV. Furthermore, depletion of salicylic acid through expression of salicylate hydroxylase in transgenic plants has no effect on restriction of TEV (Mahajan et al., 1998).
The mechanism whereby RTM1 and RTM2 cooperate to restrict long-distance movement of TEV is not clear, although several possibilities exist. The restriction could be the result of physical blockage of virus entry into, passage through, or exit from the phloem or involve the inhibition of a factor required for long-distance movement. The restriction could also be the result of recognition of TEV, followed by signaling to induce a TEV-restrictive state in phloem or in systemic tissue. Recently, the RTM1 gene was isolated and shown to encode a lectin-like protein with similarity to several proteins that have diverse roles in insect and pathogen defense (Chisholm et al., 2000). In this article, the specificity of the restriction system was investigated through a series of coinoculation experiments, and the RTM2 gene was isolated by a map-based cloning strategy. The isolated RTM2 gene encodes an unusual product consisting in part of a domain resembling small heat shock proteins (HSPs). Evidence is also presented to suggest that this domain has evolved a function distinct from those associated with other small HSPs.
RESULTS
Specificity of the Restricted Movement Phenotype
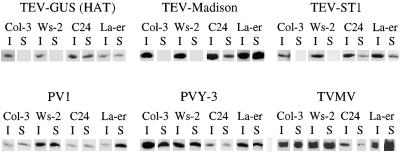
A qualitative survey of susceptibility of four Arabidopsis ecotypes to several TEV strains and other related potyviruses was conducted. Two of the ecotypes, Columbia-3 (Col-3) and Ws-2, contain dominant, restrictive alleles at RTM1 and RTM2, whereas ecotypes C24 and Landsberg erecta (Ler) contain restrictive RTM2 alleles and recessive rtm1 alleles that condition systemic susceptibility to TEV (HAT strain) (Mahajan et al., 1998; Whitham et al., 1999). Plants were inoculated with three strains of TEV, two strains of potato potyvirus Y (PVY), and one strain of tobacco vein mottling potyvirus (TVMV). Inoculated leaves and inflorescence tissue at 16 days postinoculation (DPI) were tested, using immunoblot analysis of normalized extracts, for capsid protein encoded by the respective viruses.
Capsid protein was detected in inoculated leaves of all ecotypes infected by each virus (Figure 1), indicating that each virus was able to establish primary infection sites. Each of the TEV strains moved to systemic sites in C24 and Ler but failed to move long distance in Col-3 and Ws-2. In contrast, all four ecotypes supported systemic infections by the PVY strains and TVMV (Figure 1). These four ecotypes also supported systemic infection by turnip mosaic potyvirus and cucumber mosaic cucumovirus (CMV) (data not shown). These data indicate that the RTM1/RTM2–mediated restriction systemic infection is relatively specific for TEV strains.
Figure 1.
Susceptibility of Arabidopsis Ecotypes to TEV, PVY, and TVMV Strains.
Plants were inoculated with three strains of TEV, two strains of PVY, and one strain of TVMV. Normalized samples from inoculated rosette (I) or systemic inflorescence (S) tissue at 15 DPI were subjected to immunoblot analysis by using anti–capsid protein serum specific for the respective viruses. The four ecotypes tested–Col-3, Ws-2, C24, and Ler (La-er)–each contain functional, dominant RTM2 alleles. Two ecotypes contain dominant, functional RTM1 alleles (Col-3 and Ws-2), and two ecotypes contain recessive rtm1 alleles (C24 and Ler).
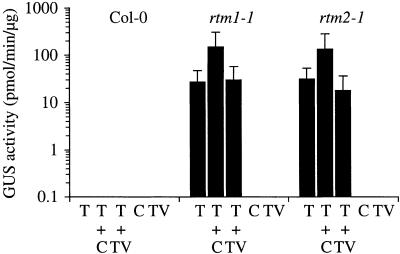
Two broad mechanisms could explain the specificity of restriction. First, the RTM1/RTM2 system could specifically recognize one or more TEV factors and elicit a TEV-specific or generalized defense response. Other viruses would not elicit the response, although they might have been sensitive to the response if activated. And second, the RTM1/RTM2 system could be potentially active against a broad range of viruses but be specifically suppressed by viruses other than TEV. To test these hypotheses, we conducted two sets of coinfection experiments. In the first set, parental Col-0, rtm1-1, and rtm2-1 plants were inoculated singly with TEV–β-glucuronidase (GUS), CMV, or TVMV or were inoculated doubly with TEV-GUS and CMV or TVMV. Inflorescence tissue was assayed for GUS activity at 16 DPI. If CMV and TVMV were able to infect plants containing a functional RTM1/RTM2 system by utilizing some defense suppression mechanism, then TEV-GUS was expected to be stimulated to infect Col-0 plants systemically in the presence of the heterologous viruses.
As expected, TEV-GUS moved long distances in singly inoculated rtm1-1 and rtm2-1 plants but not in parental Col-0 plants (Figure 2). No GUS activity was detected in any of the CMV or TVMV singly infected plants or in mock-inoculated plants. Importantly, no GUS activity was detected in TEV-GUS/CMV or TEV-GUS/TVMV dually infected Col-0 plants, suggesting that neither CMV nor TVMV was able to suppress the RTM1/RTM2 system during the coinfection. Similar results were obtained when plants were inoculated with one virus and then the other virus 3 days later, regardless of which virus was inoculated first (data not shown). In rtm1-1 and rtm2-1 plants, coinfection with TVMV failed to stimulate TEV-GUS to amounts any greater than that in TEV-GUS singly infected plants. In contrast, coinfection of the mutant plants with TEV-GUS and CMV resulted in stimulation of TEV-GUS to amounts approximately five- to 10-fold greater than in singly infected plants (Figure 2).
Figure 2.
Coinfection of Arabidopsis by TEV-GUS and CMV or TVMV.
Coinfection of wild-type Col-0 and rtm1-1 and rtm2-1 mutant plants was performed with TEV-GUS (T) and CMV (C) or TVMV (TV). Inflorescence tissue from singly infected or coinfected plants was analyzed by quantitative GUS assay at 16 DPI. Each bar represents the mean (±sd) GUS activity from 10 plants.
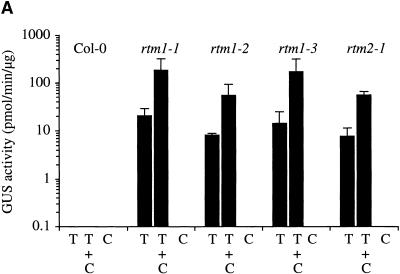
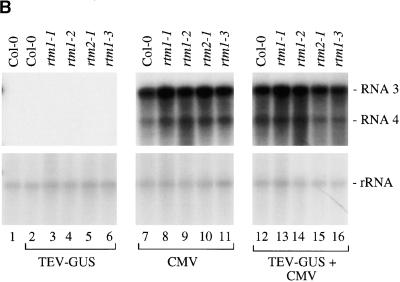
To investigate further the CMV-specific stimulation of TEV-GUS infection in rtm1 and rtm2 plants and to determine whether TEV-GUS induces a general antiviral response that affects heterologous viruses in Col-0 plants, we conducted a second set of coinfections. Inflorescence tissue from Col-0 and mutant plants infected singly or doubly with TEV-GUS and CMV was assayed for GUS activity at 24 DPI. In addition, accumulation of CMV was assayed by RNA gel blot analysis with a probe to detect RNA3 and RNA4. Mutant plants containing one of three rtm1 alleles or the rtm2-1 allele were tested. As in the previous experiments, coinfection with TEV-GUS and CMV resulted in stimulation of TEV-GUS in each mutant line but not in parental Col-0 plants (Figure 3 A). However, according to measurements of CMV RNA3 in systemic tissue, CMV accumulated to similar amounts in all singly and doubly infected Col-0 and mutant plants (Figure 3B and data not shown). These data indicate that the stimulation of TEV-GUS during coinfections of mutant plants with CMV is nonreciprocal. They also suggest that infection by TEV-GUS does not trigger a CMV-limiting response in Col-0 plants.
Figure 3.
Coinfection of Arabidopsis by TEV-GUS and CMV.
Coinfection of wild-type Col-0 and rtm1 and rtm2-1 mutant plants was performed with TEV-GUS (T) and CMV (C). Mutant plants containing rtm1-1, rtm1-2, and rtm1-3 alleles were tested.
(A) Quantitative GUS activity assays of inflorescence tissue from singly infected or coinfected plants sampled at 24 DPI. Each bar represents the mean (±sd) from three plants.
(B) RNA gel blot analysis of CMV RNA3, RNA4, and rRNA in inflorescence tissue from noninfected Col-0 (lane 1), TEV-GUS–infected (lanes 2 to 6), CMV-infected (lanes 7 to 11), and TEV-GUS/CMV–coinfected (lanes 12 to 16) wild-type Col-0 or mutant plants. Equal amounts of inflorescence tissue were pooled from each of three replicate plants before RNA extraction and blotting to nitrocellulose. The CMV RNA3, RNA4, and rRNA probes were labeled with phosphorus-32, and hybridization was detected by exposure to x-ray film.
Map-Based Cloning of RTM2
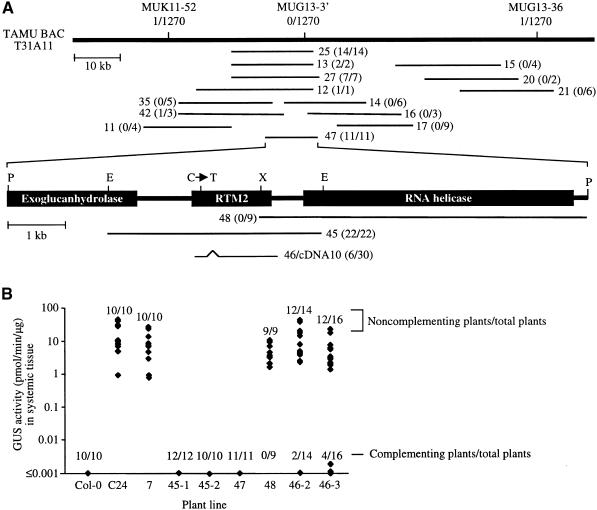
To further understand the basis for restriction of TEV movement, we isolated the RTM2 gene by using a map-based cloning strategy. The RTM2 locus had been previously mapped using a segregating population from a cross between an rtm2 mutant (rtm2-1 allele) of Col-0 ecotype and a Ws-2 ecotype (Whitham et al., 1999); the locus was mapped to an interval of ∼78 kb on chromosome 5 between the markers MUK11-52 and MUG13-36 (Figure 4A). Each marker identified a single, independent recombination event among 1270 chromosomes analyzed. No recombinants were detected between RTM2 and the marker MUG13-3′. The bacterial artificial chromosome (BAC) T31A11, identified from the TAMU BAC collection (Choi et al., 1995), was determined to contain the MUK11-52, MUG13-36, and MUG13-3′ markers (Figure 4A).
Figure 4.
Map-Based Cloning of RTM2.
(A) Map of TAMU BAC T31A11, cosmid subclones derived from T31A11 (thin horizontal lines), and putative ORFs around the RTM2 locus (expanded region). Indicated above the T31A11 diagram are the relative positions of markers MUK11-52, MUG13-3′, and MUG13-36 and the number of recombinants identified between each marker and RTM2 in a mapping population containing 1270 chromosomes. The code for each cosmid subclone (e.g., 25) is indicated adjacent to each diagram. The number of rtm2-1–complementing plants per total number of transgenic plants produced with each cosmid is indicated in parentheses. Clone 46 contains an insert from a cDNA library. The position of the C-to-T mutation in rtm2-1 plants is indicated above the RTM2 ORF diagram. E, EcoRI; P, PstI; X, XhoI.
(B) Quantitative GUS activity assays of systemic tissue from TEV-GUS–infected control or transgenic plants containing representative cosmid subclones. Each column contains data from nine to 16 individual transgenic plants or 10 Col-0 or C24 nontransformed control plants. Data from transgenic plants produced by using two independently constructed cosmids 45 and 46 are shown in separate columns. The numbers of susceptible (noncomplementing) and nonsusceptible (complementing) plants per total plants in each set are indicated. The transgenic plants labeled 7 contain an empty cloning vector.
The sequences of two P1 clones, MUK11 (GenBank accession number AB008271) and MUG13 (GenBank accession number AB005245), span the region corresponding to T31A11. The availability of sequence data enabled the construction of an initial series of 13 overlapping cosmid subclones derived from the interval between MUK11-52 and MUG13-36 in T31A11 (Figure 4A). The cosmids were transferred to Agrobacterium and used to transform the TEV-susceptible rtm2-1 mutant by using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transgenic seedlings containing each cosmid (T1 generation) were selected in soil and inoculated with TEV-GUS at ∼4 weeks of age. To test for systemic infection, we quantitatively assayed GUS activity in inflorescence tissue from the ends of two bolts at 15 and 22 DPI. Transgenic complementation with a clone containing the wild-type RTM2 allele was expected to result in restriction of TEV-GUS to inoculated leaves.
All rtm2-1 plants that were transformed with cosmids 12, 13, 25, and 27 exhibited the complementing phenotype of restricted TEV-GUS movement (Figure 4A). Transgenic complementation was also detected in one of the three plants transformed with cosmid 42. In all plants that lacked systemic infection, successful inoculation with TEV-GUS was confirmed by a colorimetric assay of GUS in inoculated leaves (data not shown). All plants transformed with each of the remaining eight cosmids (11, 14 to 17, 20, 21, and 34) were susceptible to TEV-GUS (Figure 4A).
Cosmid 25 was further subcloned, resulting in cosmids 45, 47, and 48, which were tested in transgenic complementation assays with rtm2-1 plants. Transgenic complementation was detected in each transformant containing cosmids 45 and 47 but not in those with cosmid 48 (Figures 4A and 4B). The nucleotide sequence corresponding to cosmid 47 (9.4 kb) contained three putative open reading frames (ORFs; Figure 4A). However, two of these putative ORFs (predicted to encode exoglucanhydrolase and RNA helicase) were represented only partially in complementing cosmid 45. The nucleotide sequence corresponding to the region represented in cosmid 47 from rtm2-1 plants was determined. A single mutation (C to T) was detected within the predicted ORF common to both cosmids 45 and 47 (Figure 4A). A 1234-bp cDNA clone (cDNA10) corresponding to this ORF was isolated from a wild-type Col-0 cDNA library (Newman et al., 1994) and sequenced (Figure 5). The rtm2-1 mutation resulted in conversion of the Gln-28 codon (CAA) to a stop codon (TAA). On the basis of the results of complementation and sequence analysis, the gene corresponding to cDNA10 was concluded to be RTM2.
Figure 5.
Nucleotide and Deduced Amino Acid Sequence of RTM2 cDNA.
Conceptual translation was started at the first ATG codon. The position of the C-to-T mutation in the rtm2-1 allele is indicated in reverse type. The inverted arrowhead indicates the position of the RTM2 intron in genomic DNA. The sequence with the thin underline is the α-crystallin–like domain. The boxed regions indicate the two types of repeated sequences. The italicized sequence corresponds to the region predicted by the Coils program (Lupas et al., 1991) to possess a continuous α-helical structure. The sequence with the thick underline is the predicted transmembrane sequence. The GenBank accession number for RTM2 is AF208051.
The cDNA10 sequence was transferred to a vector containing a constitutive 35S promoter and introduced into rtm2-1 plants for complementation analysis. Among 30 transgenic plants produced by using two independent fusion constructs (46-2 and 46-3), six restricted the TEV-GUS to inoculated leaves and 24 were susceptible (Figure 4B). These data suggest that the 35S-cDNA10 construct was partially functional in restoring RTM2 activity.
The RTM2 genomic sequence contains two exons separated by a 72-bp intron (Figures 4A and 5). The RTM2 gene encodes a predicted 366–amino acid protein with a deduced molecular mass of ∼41 kD.
The RTM2 Polypeptide—A Multidomain Structure with a Small HSP Sequence
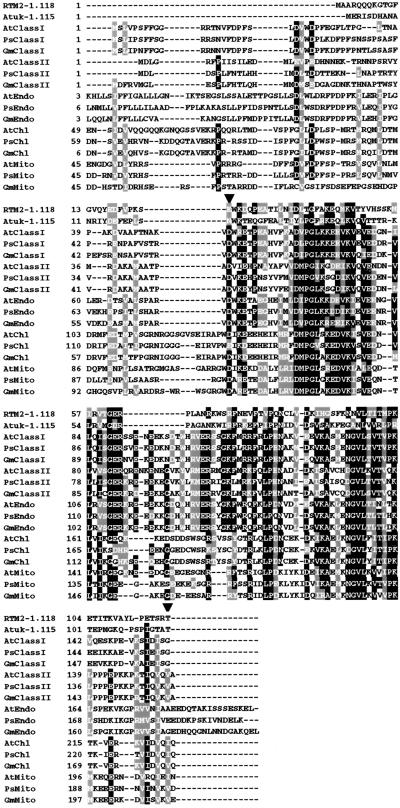
The most striking feature of the RTM2 protein, as revealed by BLAST searches (Altschul et al., 1990) and sequence alignments, is the presence of an N-terminal domain with high sequence similarity and predicted structural similarities to members of the small HSP family in plants (Figure 6). Plants contain five major classes of nuclear-encoded small HSPs, ranging in size from 16 to 27 kD, exclusive of signal peptides (Vierling, 1991; Waters et al., 1996). Two of the five classes of small HSPs are localized in the cytosol (classes I and II), and the remaining three are localized in the endoplasmic reticulum, the chloroplast, or the mitochondrion. These proteins form oligomeric complexes, possess chaperone activity, and contain a conserved region known as the α-crystallin domain (de Jong et al., 1998). The RTM2 sequence between residues 24 and 118 aligned well with the α-crystallin domain of members from each small HSP class from three selected species (Figure 6). No similarity was detected between RTM2 and small HSP sequences outside the α-crystallin–like domain.
Figure 6.
Alignment of RTM2 N-Terminal Sequence, α-Crystallin Domains of Plant Small HSPs, and the N-Terminal Region of Arabidopsis Atuk.
Gaps in the alignment are indicated by dashes. The α-crystallin domains are located between the arrowheads. Positions occupied by at least nine structurally conserved or identical residues are highlighted in gray or black, respectively. Representatives from each of the five major small HSP classes (cytosolic classes I and II, endomembrane [Endo], chloroplast [Chl], and mitochondrial [Mito]) from Arabidopsis (At), pea (Ps), and soybean (Gm) are shown. GenBank accession numbers are as follows: Atuk, AC005623; AtClassI, CAA35182.1; PsClassI, P19243; GmClassI, P05478; AtClassII, P29830; PsClassII, P19242; GmClassII, P05477; AtEndo, AAA19931.1; PsEndo, P19244; GmEndo, P30236; AtChl, AAA32818; PsChl, P09886; GmChl, P09887; AtMito, AAB38795; PsMito, P46254; and GmMito, Q39818.
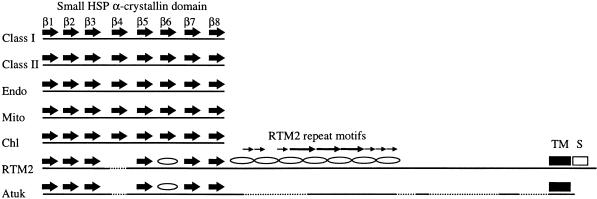
The α-crystallin domain of small HSPs forms an eight-stranded β-sandwich structure (de Jong et al., 1998; Kim et al., 1998). Secondary structure predictions made with the PHDsec program (Rost and Sander, 1994) and sequence alignments with small HSPs revealed that the α-crystallin–like domain of RTM2 probably possesses a similar structure, with two notable exceptions: not only is β strand 4 lacking from the RTM2 polypeptide, resulting in a 12-residue gap in the alignment (Figures 6 and 7), but the sequence corresponding to β strand 6 was predicted to form an α helix in RTM2.
Figure 7.
Diagrammatic Representation of Known or Predicted Structural Features of Plant Small HSPs, RTM2, and Atuk.
The relative positions of the eight β strands in the small HSP α-crystallin domain are indicated by large arrows. The predicted α-helical structures in the α-crystallin–like domain of RTM2 and Atuk are shown by ovals. The repeated sequences in the central region of RTM2 are indicated by the two sets of small arrows. The region predicted to form a long α-helical structure by the Coils program (Lupas et al., 1991) is indicated by the intertwining diagram. Gaps to maintain structural alignment are indicated by the dotted lines. Chl, chloroplast; Endo, endomembrane; Mito, mitochondrial; S, serine-rich sequence; TM, predicted transmembrane sequence.
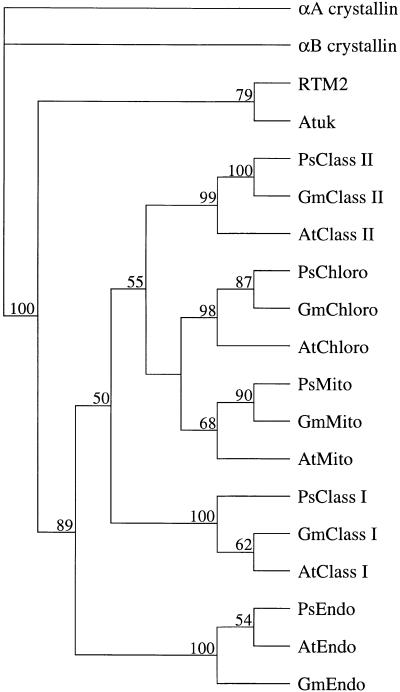
The evolutionary relatedness of the RTM2 small HSP domain with Arabidopsis, pea, and soybean small HSPs was inferred from cladistic analysis with the PAUPsearch and PAUPdisplay programs (Wisconsin Package version 10.0; Genetics Computer Group, Madison, WI). As shown by others (Waters et al., 1996; de Jong et al., 1998), phylogenetic analysis reveals that the members within each class of dicot small HSPs share a common lineage (Figure 8). RTM2 was clearly placed in a branch (supported in 89% of bootstrap replicates) that was distinct from each of the five small HSP classes (Figure 8). These groupings were reinforced by phylogram analysis using the neighbor-joining method (data not shown).
Figure 8.
Cladogram of RTM2, Atuk, and Small HSPs.
The α-crystallin–like domains were aligned as in Figure 6 and assembled into a single tree by a heuristic search using bootstrap analysis (100 replicates) with simple sequence addition, tree bisection-reconnection branch swapping, steepest descent off, and MULPARS on in PAUPsearch and PAUPdisplay. The human αA-crystallin and αB-crystallin sequences were assigned to the outgroup. The numbers above branch points indicate bootstrap percentage values. Abbreviations and the GenBank accession number for each sequence other than αA-crystallin (P02489) and αB-crystallin (P02511) are given in the legend for Figure 6.
No significant sequence similarities were identified between the remainder of RTM2 and any other protein of known function. However, several features of the central and C-terminal regions of RTM2 were identified through sequence analysis and secondary structure predictions with programs using the PredictProtein server (Rost, 1996). Amino acid residues 129 to 225 contained two types of repeating sequences. The five–amino acid sequence LEE(S,K,R)(E,R,K) occurs six times (Figure 5). Three repeats of this motif flank both sides of a second type of repeated sequence consisting of three tandemly arranged motifs of (I,M,A)(R,K)KLQEEAKAKE(E,K)(L,A) (Figure 5). Both of these repeat elements represent highly charged regions within RTM2. The sequence containing residues 119 to 223 was also predicted with the Coils program (Lupas et al., 1991) to form an uninterrupted α helix, although the highly charged nature of this region can lead to inaccurate assignment of such structures. Amino acid residues 327 to 344 were predicted, in the PHDsec program (Rost et al., 1995), to form a transmembrane helix (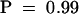 ). An N-terminal signal peptide for entry into the secretory pathway was not identified, suggesting that association with a membrane, if it occurs, might happen post-translationally. In addition, amino acid residues 345 to 362 compose a serine-rich serine region in which 16 of 17 are serine residues (Figure 5).
). An N-terminal signal peptide for entry into the secretory pathway was not identified, suggesting that association with a membrane, if it occurs, might happen post-translationally. In addition, amino acid residues 345 to 362 compose a serine-rich serine region in which 16 of 17 are serine residues (Figure 5).
An RTM2-like Gene in Arabidopsis
The protein most similar to RTM2 in the sequence databases was a deduced protein (designated Atuk) of unknown function encoded on Arabidopsis chromosome 2 (GenBank accession number AC005623). The Atuk sequence clearly contained an α-crystallin–like domain (Figure 6). Phylogenetic analysis suggested that the α-crystallin–like domain of Atuk was most closely related to that of RTM2 (Figure 8). Aside from sequence similarity, RTM2 and Atuk contained several predicted structural and organizational features in common. Both had a relatively short sequence to the N-terminal side of the α-crystallin–like domain. Both were predicted to lack the sequence corresponding to β strand 4 and to form α helices in place of β strand 6 within the α-crystallin–like region (Figures 6 and 7). Both proteins also contained domains beyond the small HSP–like sequence, although the Atuk sequence in this region was considerably shorter than that of RTM2 (Figure 7). Despite sequence similarity in the region that had been predicted to form an extended α helix in RTM2, secondary structure predictions indicated that an α-helical configuration in this region of Atuk was unlikely. Both proteins were predicted to contain a transmembrane α helix near the C terminus (Figure 7). No Atuk sequence comparable to the serine-rich C-terminal region of RTM2 was identified, although the actual C terminus of the Atuk polypeptide could not be deduced solely from the genomic DNA sequence.
Analysis of RTM2 Gene Expression
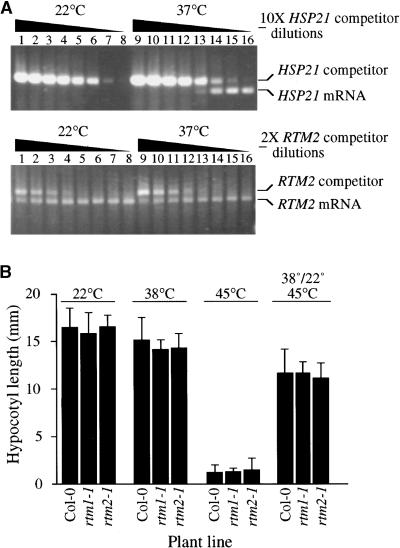
The presence of a small HSP–like domain in RTM2 suggested that the RTM2 gene might be regulated by heat stress. Transcription of most genes encoding small HSPs is rapidly induced by at least 1000-fold during heat shock. To determine whether the expression of RTM2 was heat inducible, competitive quantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed with Col-0 RNA isolated from plants treated at 22 or 37°C. As a control, expression of ATHSP21 mRNA encoding a small HSP of Arabidopsis chloroplasts (Chen and Vierling, 1991) was analyzed at the same time. The quantitative RT-PCR technique involved comparison of PCR product intensities from reactions containing normalized concentrations of cDNA and successively dilute concentrations of competitor DNA corresponding to genomic (intron-containing) DNA (Becker-Andre and Hahlbrock, 1989; Gilliand et al., 1990; Siebert and Larrick, 1992). Expression of ATHSP21 mRNA was not detected in plants maintained at 22°C, even at the lowest concentration of competitor DNA tested (Figure 9A, top, lanes 1 to 8). Expression of ATHSP21 mRNA was induced by treatment of plants at 37°C. Product corresponding to ATHSP21 mRNA was detected first in reactions containing competitor diluted to a concentration of 10−4 (Figure 9A, top, lane 13) relative to the initial competitor concentration (lane 9).
Figure 9.
RTM2 Expression Is Not Heat Inducible and Does Not Affect Thermotolerance.
(A) Effect of heat treatment on expression of Arabidopsis HSP21 (top) and RTM2 (bottom) genes. Competitive quantitative RT-PCR assays using successive 10-fold (10× HSP21) or twofold (2× RTM2) dilutions of competitor DNA (as indicated by the triangles) were performed with mRNA isolated from plants treated at 22 or 37°C for 1 hr. cDNA prepared from each set of treated plants was used for both HSP21 and RTM2 reactions. The competitor DNA was derived from intron-containing genomic DNA for both HSP21 and RTM2.
(B) Thermotolerance assay with Arabidopsis seedlings. Hypocotyl lengths (mean ±sd) were measured 2 days after temperature treatment in plants grown in the dark on solid medium. Plants were treated for 1 hr at the temperatures indicated above the graph. The temperature shift experiment involved treatments at 38°C for 1 hr, 22°C for 1 hr, and 45°C for 1.5 hr.
Preliminary quantitative RT-PCR using 10-fold dilutions of competitor DNA revealed that RTM2 was expressed at similar levels at both 22 and 37°C (data not shown). To discern more subtle differences in expression levels after the two treatments, we performed quantitative RT-PCR by using twofold dilutions of competitor DNA, starting at a competitor DNA concentration at which RTM2 mRNA–derived products were first detected in the preliminary reactions. The dilution of competitor DNA at which the ratio of PCR products from competitor and RTM2 mRNA templates was equivalent was twofold higher when using mRNA from plants at 22°C than that at 37°C (Figure 9A, bottom, lanes 2 and 11), indicating that RTM2 mRNA was not induced by heat shock. This result is consistent with the observation that the RTM2 promoter region lacks canonical heat shock elements (5′-aGAAgcTTCtaGAAg-3′; Barros et al., 1992; Schoffl et al., 1998).
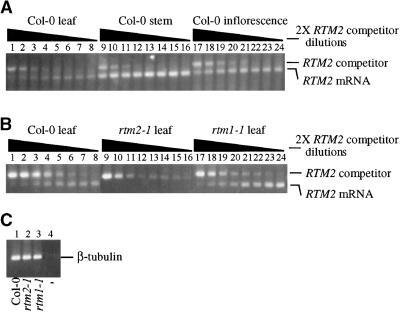
The expression of RTM2 was analyzed further in three tissue types in TEV-restrictive Col-0 plants and in leaves of susceptible rtm2-1 and rtm1-1 plants. Competitive quantitative RT-PCR was first used to compare the amounts of RTM2 mRNA in rosette leaves, stems, and inflorescence clusters from 6-week-old plants. In three independent experiments, of which one is shown in Figure 10A, the competitor DNA concentration at which competitor- and cDNA-derived PCR products were most similar was clearly higher when stem mRNA was used. This suggests that RTM2 mRNA accumulates to the highest levels as a percentage of total mRNA, in stem tissue. The relative abundance of RTM2 mRNA in rosette leaves of Col-0 and rtm1-1 plants was similar (Figure 10B, lanes 1 to 8 and 17 to 25), suggesting that alteration of RTM1 function has little or no effect on expression of RTM2. However, rtm2-1 mRNA was not detected, even at the most dilute concentration of competitor DNA tested (Figure 10B, lanes 9 to 16). Using the same cDNA reactions from Col-0, rtm2-1, and rtm1-1 plants, β-tubulin mRNA–derived PCR product was detected in each case (Figure 10C), indicating that the failure to detect rtm2-1 mRNA was not attributable to a lack of cDNA synthesis or the presence of an inhibitor. These results suggest that the rtm2-1 mutation leads to destabilization of the mRNA.
Figure 10.
Expression of RTM2 and rtm2-1.
Competitive quantitative RT-PCR analysis of RTM2 mRNA in wild-type Arabidopsis Col-0 leaf, stem, and inflorescence tissue. The cDNA concentration used for each set of reactions was normalized to allow direct comparisons between the three samples. The triangles indicate successive twofold (2×) dilutions of competitor DNA in each reaction.
(A) and (B) Competitive quantitative RT-PCR analysis of RTM2 or rtm2-1 mRNA in leaves of wild-type Col-0, rtm2-1, or rtm1-1 plants. The rtm1-1 plants contained the wild-type RTM2 allele. The cDNA concentration used for each set of reactions in (A) was normalized to allow direct comparisons among the three samples but differed from that used in the reactions in (B).
(C) RT-PCR reactions to amplify β-tubulin from the cDNA preparations used in (B). PCR reaction with no cDNA template (–) is also shown.
RTM2 Does Not Contribute to Thermotolerance
A major role of small HSPs is to protect cells from heat and other stresses. Overexpression of small HSPs in homologous or heterologous organisms has been shown to confer enhanced thermotolerance (Schoffl et al., 1998). Although expression of RTM2 mRNA is not regulated by the heat shock response, it was still possible that constitutive expression contributes to thermotolerance.
To determine whether RTM2 contributes to thermotolerance in Arabidopsis, we tested wild-type Col-0 and rtm2-1 plants in hypocotyl elongation assays under different temperature conditions. Plants containing the rtm1-1 allele with wild-type RTM2 were also tested. We hypothesized that if RTM2 contributes to thermotolerance, plants containing the rtm2-1/rtm2-1 genotype should exhibit greater sensitivity to heat stress. Seedlings were germinated in dishes on solid medium at 22°C and subjected to one of four treatments: (1) continuous 22°C; (2) 38°C for 1 hr; (3) 45°C for 1 hr; or (4) a temperature shift series of 38°C (1 hr), 22°C (1 hr), and 45°C (1.5 hr). Plates were then incubated at 22°C for 2 days, and hypocotyl lengths were measured. Growth of wild-type and both mutant plants after the 22 and 38°C treatments was similar (Figure 9B). The 45°C heat treatment, without the adaptive preincubation at 38°C, severely inhibited growth of the wild-type and mutant plants (Figure 9B). Incubating the plants at 38°C (which induces a protective heat shock response) before performing the 45°C treatment resulted in thermotolerance of wild-type and mutant plants (Figure 9B). However, no statistically significant differences were detected between hypocotyl lengths of wild-type and mutant plants in any of the temperature treatments (P > 0.75), including the temperature shift treatment, indicating that rtm2-1 plants were not compromised in responding to high temperatures.
DISCUSSION
Specificity of TEV Restriction in Arabidopsis
Analysis of systemic susceptibility of Arabidopsis ecotypes, which have functionally different RTM1 or rtm1 alleles, revealed that the RTM1/RTM2–mediated restriction of long-distance movement is relatively specific for TEV. The basis for specificity was investigated through a series of coinfection experiments with TVMV and CMV. We tested whether heterologous viruses systemically suppress the restriction system and whether TEV specifically elicits a general, systemic antiviral response. There is considerable precedence for systemic antiviral responses that are either elicited or suppressed by viruses (Baker et al., 1997; Carrington and Whitham, 1998). Coinfections of RTM1/RTM2–competent plants (Col-0) with TEV-GUS and TVMV or CMV failed to stimulate TEV-GUS to move long distances. The most direct interpretation of these results is that the restriction system is still functional in the presence of TVMV or CMV, which argues against the systemic suppression hypothesis.
The coinfection data also cast doubt on the hypothesis that TEV strains specifically activate a general, systemic defense response. Such a model would require that TEV strains, but not other viruses tested, encode an elicitor that triggers the response. CMV accumulated to similar extents in RTM1/RTM2 active and inactive plants, regardless of whether or not it was coinoculated with TEV-GUS. These data are most easily interpreted to mean that the restriction system, even if activated by TEV-GUS, has little or no activity against CMV. A potential weakness of all of the coinfection experiments, however, is that they may not be sufficiently sensitive to detect local effects. For example, the system may restrict TEV because of a general response that is active only in infected cells associated with the vasculature. If TEV-GUS and CMV infect different cells within the same plant, long-distance movement and systemic infection by CMV may not be restricted. The coinfection experiments as designed address only potential responses that might occur systemically.
In rtm1 and rtm2 plants, coinoculation by TEV-GUS and CMV resulted in nonreciprocal stimulation of TEV-GUS. If TEV-GUS stimulation by CMV also occurred in Col-0 plants, it was insufficient to overcome the active RTM1/RTM2 system. Nonreciprocal stimulation of one virus by another has been well documented in plants and is frequently associated with synergistic disease syndromes (Matthews, 1991). Interestingly, TEV stimulates systemic accumulation of CMV in a nonreciprocal manner in Nicotiana spp, a phenomenon that likely involves suppression of gene silencing responses by TEV-encoded HC-Pro (Pruss et al., 1997; Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998). CMV also encodes a suppressor of gene silencing (the 2b protein), but it may function to inhibit a component of the silencing pathway that differs from that suppressed by HC-Pro (Beclin et al., 1998; Brigneti et al., 1998). The CMV 2b protein may be relatively effective in suppressing gene silencing in Arabidopsis, whereas TEV-encoded HC-Pro may be relatively effective in Nicotiana spp. Such species specificity of silencing suppression may explain the basis for the host range of many viruses. However, because coinfection with CMV was insufficient to overcome the block to TEV in Col-0 plants, the RTM1/RTM2 system and any host responses (such as silencing) that are suppressed by CMV are suggested to be independent.
RTM2
A map-based cloning strategy was used to isolate the RTM2 gene. Isolation of RTM2 was confirmed by transgenic complementation in the TEV-susceptible rtm2-1 mutant and by sequence analysis of the wild-type and rtm2-1 loci. The mutant locus was predicted to encode a protein truncated beyond residue 28, indicating that the rtm2-1 mutation likely resulted in a loss-of-function allele.
The most notable feature of the RTM2 polypeptide is the similarity of the N-terminal region with small HSPs. The small HSP superfamily is widely distributed throughout prokaryotes and eukaryotes and is defined by a conserved 100– to 110–amino acid motif known as the α-crystallin domain (de Jong et al., 1998). The amino acid sequences flanking this domain in small HSPs are highly variable. Most small HSPs do not possess extensive C-terminal extensions. In contrast, RTM2 has a predicted molecular mass > 41 kD, and its C terminus extends 243 residues beyond the α-crystallin domain. However, there are few clues to the function of the RTM2 C-terminal region, because no proteins of known function with sequence similarity to this region were identified in database searches.
A common feature of small HSPs is that they assemble into water-soluble, oligomeric complexes that possess chaperone activity in vitro. Chaperone activity has been demonstrated for small HSPs by showing that they prevent unfolding and aggregation of model proteins in a way that lacks substrate specificity (de Jong et al., 1998). The crystal structure of a small HSP from Methanococcus janschii was recently solved at atomic resolution (Kim et al., 1998). The α-crystallin domain, which comprises eight β strands, provides the scaffold on which the oligomeric structure is assembled.
Several lines of evidence suggest that although the RTM2 N-terminal domain is related to small HSPs, RTM2 is unlikely to possess typical chaperone functions. Unlike members of the five major classes of plant small HSPs, the RTM2 gene was not heat inducible under conditions that stimulate the heat shock response and appears not to affect thermotolerance. Sequence analysis of genomic DNA indicated that the RTM2 promoter lacks cis-regulatory elements required for heat-inducible transcriptional activation. Alignment of the RTM2 sequence with sequences of plant small HSPs further revealed that two of the eight α–crystallin domain β strands (strands 4 and 6) are either missing or unlikely to form within RTM2. Given that β strand 6 of one monomer interacts with β strand 1 of a different monomer in the small HSP oligomeric structure (Kim et al., 1998), RTM2 may form tertiary and quaternary structures that differ markedly from those of small HSPs. On the basis of phylogenetic analysis, the α-crystallin–like domain of RTM2 may define a distinct class of plant small HSPs. This class may also include the deduced Atuk protein because RTM2 and α-crystallin–like domains are consistently clustered within a distinct branch in phylogenetic trees. Taken together, these data suggest either that a heat-inducible chaperone role for RTM2 was lost through evolutionary processes or that RTM2 is derived from a small HSP–like progenitor that lacked such temperature-related functions. By analogy with other HSP-related proteins that are expressed at normal temperatures, perhaps RTM2 is most appropriately referred to as a small heat shock cognate protein.
The similarities of RTM1 and RTM2 to lectin-like (Chisholm et al., 2000) and small HSP–like proteins, respectively, suggest that these proteins provide structural rather than catalytic roles. Several models that incorporate structural roles for RTM1 and RTM2 in TEV-specific restriction are currently viable. Because the restriction requires dominant RTM1 and RTM2 alleles, inhibition of TEV probably involves a mechanism requiring active RTM1 and RTM2 proteins. In a direct inhibition model, RTM1 or RTM2 or both could interact with, or inactivate, one or more viral proteins necessary for long-distance movement. Several TEV proteins, including capsid protein, CI helicase, HC-Pro, and NIa, provide functions required for long-distance movement (Dolja et al., 1994; Cronin et al., 1995; Kasschau et al., 1997; Schaad et al., 1997; Carrington et al., 1998), and interference with one or more of these could restrict systemic infection. Components of the RTM1/RTM2 system may also interact with cellular structures necessary for long-distance movement. Such an interaction may specifically occlude interaction with TEV-encoded movement factors, although not with comparable proteins encoded by other viruses. It is also possible that the inhibition of long-distance movement is the result of direct inhibition of viral or cellular factors necessary for virus replication. In any direct inhibition model, the relevant interactions must be postulated to occur only in cells associated with the vasculature, given the lack of evidence to suggest that RTM1 or RTM2 affects viral functions in epidermal or mesophyll cells (Mahajan et al., 1998). The relative abundance of RTM2 mRNA in stem tissue may indicate that RTM2 is expressed in greater amounts in cells in or around the vasculature, although this remains to be determined through more rigorous localization experiments.
Several indirect models are also plausible. RTM1 and RTM2 could stimulate or promote the activity of an anti-TEV or movement-restricting factor in inoculated leaves. RTM2, for example, could promote stability of a complex that has TEV-restricting activity. In fact, many types of multisubunit complexes are stabilized by interaction with heat shock cognate proteins (Boston et al., 1996). Such a complex might be necessary for production of a localized antiviral response that restricts movement out of initially infected leaves. An alternative indirect model states that RTM1 and RTM2 are necessary to produce, transport, or perceive a signal that results in establishment of a TEV-restrictive state in systemic tissue. Such a signal would be induced specifically by TEV infection in inoculated leaves, and the restrictive state would either prohibit movement of virus out of the vasculature or suppress replication in cells after exit from the phloem. In such a case, the signal or antiviral effector would have to be highly specific for TEV.
METHODS
Plant and Virus Propagation
Arabidopsis thaliana ecotypes Columbia-0 (Col-0), Columbia-3 (Col-3), C24, Landsberg erecta (Ler), Wassilewskija-2 (Ws-2), and tobacco etch potyvirus (TEV)–susceptible Col-0 mutants rtm1-1, rtm1-2, rtm1-3, and rtm2-1 were grown in a greenhouse with a 14-hr photoperiod at ∼22°C, unless indicated otherwise.
All viruses were propagated in Nicotiana tabacum cv Xanthi-nc, and except for the cucumber mosaic cucumovirus (CMV), inocula for infectivity tests were prepared as described (Whitham et al., 1999). CMV inoculum was prepared by homogenizing symptomatic tissue in 20 mM potassium phosphate buffer, pH 7.0, at a 1:1 (w/v) tissue/buffer ratio.
Virus Infectivity and Coinoculation Assays
Rosette leaves in Arabidopsis plants were inoculated with TEV, potato potyvirus Y (PVY), or tobacco vein mottling potyvirus (TVMV) strains at 3.5 to 4 weeks postgermination by the artist airbrush method (Whitham et al., 1999). Infection of inoculated leaves was tested at 15 days postinoculation (DPI) by immunoblot assay with capsid protein antiserum specific for each virus. Pools of three leaves were homogenized in protein dissociation buffer. Immunoblot assays using anti–rabbit IgG conjugated to horseradish peroxidase (Amersham) and an enhanced chemiluminescence detection system (Amersham) were performed as described (Whitham et al., 1999). Systemic infection by each virus was tested using immunoblot assay with pools of two inflorescence clusters from independent bolts of each plant at 15 DPI.
Coinoculation of Arabidopsis Col-0 or mutant plants with TEV–β-glucuronidase (GUS) and CMV (Y strain) was performed in two steps. Plants were first dusted with carborundum and inoculated with CMV by using a cotton tip applicator. Plants were then inoculated with TEV-GUS by the artist airbrush method. Coinoculation of Arabidopsis with TEV-GUS and TVMV by the artist airbrush method was done after mixing the inocula together in a 1:1 (v/v) ratio. TEV-GUS was measured in inflorescence tissue at 16 or 24 DPI by quantitative GUS activity assay as described (Mahajan et al., 1998). TVMV was detected in inflorescence tissue of each coinoculated plant by immunoblot assay. CMV was measured in inflorescence tissue at 16 DPI by an RNA gel blot assay. The RNA was extracted from pools of two inflorescence clusters from independent bolts as described (Chomczynski and Sacchi, 1987). Total RNA (1 μg) from each of three independent plants was pooled to minimize plant-to-plant variation. The RNA was denatured in the presence of glyoxal and DMSO, separated by electrophoresis in a 1% agarose gel, and blotted to a nylon membrane as described (Ausubel et al., 1995). Blots were hybridized with a 32P-labeled probe specific for CMV RNA3 and RNA4. After exposure to x-ray film and quantification of radioactivity corresponding to RNA3 with a phosphorimager, blots were stripped and reanalyzed with a radiolabeled probe to detect rRNA. Hybridization to rRNA was detected by exposure to x-ray film and quantified with a phosphorimager.
Cosmid Subclones and Arabidopsis Transformation
The TAMU bacterial artificial chromosome (BAC) T31A11 was digested partially with HindIII and subcloned into the binary cosmid vector pSLJ755I5 (Jones et al., 1992). Partially digested products were size selected by electrophoresis in an 0.8% agarose gel, and fragments longer than 15 kb were recovered by using Gene Clean III (Bio 101, Vista, CA). The fragments were ligated to HindIII-digested and dephosphorylated pSLJ755I5. Ligation products were packaged with Gigapack III XL (Stratagene, La Jolla, CA) extract, transfected into Escherichia coli DH10B cells, and plated on medium containing 12.5 μg/mL tetracycline, isopropyl-β-d-thiogalactopyranoside, and X-gal (Inalco, Milan, Italy). A set of 13 overlapping cosmid clones that spanned the RTM2 locus (Figure 4A) was identified by screening 360 clones with nine polymerase chain reaction (PCR) markers (sequence available on request) spaced at ∼10-kb intervals throughout T31A11. Selected clones were subjected to fingerprint analysis with HindIII.
Cosmid clones were transferred to Agrobacterium tumefaciens strain GV3101 and used to transform rtm2-1 plants by the vacuum infiltration method (Bechtold and Pelletier, 1998). Transformants were selected on soil that had been pre-wetted with 25 mg/L glufosinate ammonium (Finale; AgrEvo, Montvale, NJ).
Cosmid 25, one of four cosmids that complemented the rtm2-1 mutation, was further fragmented and subcloned into pSLJ755I5. Subclones containing a 6.5-kb XhoI-PstI fragment, a 3.1-kb EcoRI fragment, and a 9.4-kb PstI fragment (Figure 4A) were produced. Subclones were introduced into E. coli DH10b by electroporation and transferred to Agrobacterium by conjugation. Transformed plants were produced and selected as described.
Isolation of RTM2 cDNA
The BAC clone T31A11 was used as a probe to screen ∼4 × 105 plaque-forming units from an Arabidopsis Col-0 cDNA library (Newman et al., 1994). A total of 135 clones were obtained and converted to plasmids by in vivo excision. These clones were further screened with a 10-kb PCR product known on the basis of complementation data to span the RTM2 locus (Figure 1A). A single clone (cDNA10) was identified and sequenced. The RTM2 cDNA was amplified by using Pfu polymerase (Stratagene) and primers that added XhoI and BamHI sites to the 5′ and 3′ ends, respectively. The PCR product was digested with XhoI and BamHI and then inserted into the expression vector pRTL2 (Restrepo et al., 1990), which contains an enhanced CMV 35S promoter and terminator. The expression cassette was digested with PstI and inserted into pSLJ755I5, generating clones 46-2 and 46-3. These clones were mobilized into Agrobacterium and used to transform Arabidopsis rtm2-1 plants as described above.
Infectivity Assays in Transgenic Arabidopsis
Rosette leaves of Arabidopsis plants were inoculated with TEV-GUS as described (Whitham et al., 1999). GUS activity in inflorescence tissue at the ends of dominant bolts was assayed at 15 and 22 DPI (Mahajan et al., 1998). All plants that lacked GUS activity in upper, noninoculated tissues were tested to confirm that they were successfully infected in inoculated leaves. Rosette leaves exposed to inoculum were infiltrated with the colorimetric substrate X-gluc (Inalco), which allowed the infection foci to be scored as multicellular blue spots (Mahajan et al., 1998).
Sequence Analysis of the rtm2-1 Locus
Three primer pairs were used to amplify a 9.4-kb interval of Arabidopsis rtm2-1 genomic DNA corresponding to the sequence in complementing cosmid 47. To reduce the possibility of errors in the sequence from PCR artifacts, products from eight independent PCR reactions were pooled and then purified by gel electrophoresis. The PCR products were subjected to a cycle sequencing protocol (Perkin-Elmer), and reactions were analyzed on an ABI 377 sequencing apparatus (Applied Biosystems, Foster City, CA). Sequences were assembled by using Genetics Computer Group software (Wisconsin Package version 10.0) and aligned with the wild-type genomic and cDNA10 sequences by using PileUp and CLUSTAL W programs (Thompson et al., 1994).
Database and Phylogenetic Analyses
The sequence of RTM2 was used to search the GenBank database using the BLAST program (Altschul et al., 1990). Sequences of small heat shock proteins (HSPs) and the RTM2 small HSP–like region were aligned with the assistance of the CLUSTAL W program. The secondary structures of the RTM2 and Atuk proteins were predicted with the PredictProtein server (http://dodo.cpmc.columbia.edu/predictprotein/predictprotein.html [Rost, 1996]).
The phylogenetic relationships of the α-crystallin domains of representative plant small HSPs, human αA- and αB-crystallin, and the α-crystallin–like domains of RTM2 and Atuk were analyzed with PAUPsearch and PAUPdisplay (Wisconsin Package version 10.0). A heuristic search was performed by tree bisection-reconnection branch swapping with MULPARS on and steepest descent off. Bootstrap analysis was done for 100 replicates with simple sequence addition.
mRNA Extraction and Quantitative Reverse Transcription–PCR
RNA was extracted from Arabidopsis tissues as described (Chomczynski and Sacchi, 1987). Contaminating genomic DNA was removed by digestion with RNase-free DNase I (Ambion, Austin, TX). The RNA was extracted with phenol:chloroform (1:1) and then chloroform, precipitated with ethanol, and resuspended in sterile deionized water. Poly(A)+ RNA was isolated by using Oligotex resin (Qiagen, Chatsworth, CA). Before cDNA synthesis, the samples were incubated at 70°C for 10 min, chilled on ice, and then prewarmed at 42°C. First-strand cDNA was synthesized for 1 hr at 42°C in reactions (10 μL) containing ∼200 ng of poly(A)+ RNA, 0.5 μg/mL oligo dT primer, 5 pmol/μL RTM2-RTR primer (5′-CTCCTCTAGCTTCGCCGCCTTCTC-3′), buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl, and 3 mM MgCl2), 20 mM DTT, 1 mM each deoxynucleotide triphosphate, 2 μCi 32P-dATP (3000 Ci/mM; Amersham), and 200 units of Superscript II reverse transcriptase (Gibco BRL). Unincorporated nucleotides were removed by filtration on Sephadex G50 centrifuge columns (Boehringer Mannheim). Radioincorporation was measured, and cDNA concentrations were normalized for concomitant PCR reactions.
Competitive PCR reactions (Becker-Andre and Hahlbrock, 1989; Gilliand et al., 1990; Siebert and Larrick, 1992) were performed with normalized cDNA, competitor DNA at various concentrations as high as 100 amol/μL, buffer (20 mM Tris-HCl, pH 8.8, 10 mM KCl, 10 mM [NH4]2SO4, 0.1% Triton X-100, 0.1 mg/mL BSA), 200 μM each deoxynucleotide triphosphate, 5 pmol/L each primer, and 1.0 units of Pfu Turbo polymerase (Stratagene). After an initial incubation at 94°C for 1 min, 30 PCR cycles were performed at 94°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec. Control PCR reactions were performed without competitor to show that no genomic DNA contaminated the reactions or without cDNA to show that reaction components were not contaminated with cDNA or genomic DNA (data not shown). The RTM2 sequence was amplified with the RTM2-RTR and RTM2-RTF (5′-TGGAAAGATCAACCTGAAGCC-3′) primers. The ATHSP21 sequence was amplified with the ATHSP21F (5′-GCTGCATCGGCTCTATGTTC-3′) and ATHSP21R (5′-GATCGAGTCCTACTGAAT-CTGG-3′) primers. Competitor DNA for both genes was amplified from Col-0 genomic DNA, gel-purified, and quantified by measurements of A260 and A280. The RTM2 and ATHSP21 PCR products were analyzed by electrophoresis in 2 and 1% agarose gels, respectively, followed by staining with ethidium bromide.
Seedling Thermotolerance Assay
Wild-type Col-0, rtm2-1, and rtm1-1 seeds were sterilized and plated on PNS medium: 5 mM KNO3, 2 mM MgSO4, 2 mM Ca(NO3)2, 50 mM FeEDTA, 2.5 mM KPO4, pH 5.5, 70 μM H3BO3, 14 μM MnCl2, 0.5 μM CuSO4, 1 μM ZnSO4, 0.2 μM Na2MoO4, 10 μM NaCl, 0.01 μM CoCl2, 0.5% sucrose, and 0.8% agar. Plates were wrapped in foil and kept at 4°C for 2 days and then were transferred to 22°C for 2 days. Plates were subjected to one of four temperature shift treatments: continuous incubation at 22°C; 38°C for 1 hr; 45°C for 1 hr; or a temperature series of 38°C for 1 hr, 22°C for 1 hr, and then 45°C for 1.5 hr. All plates were incubated at 22°C after the heat treatments. The experiment was conducted in triplicate, with 10 to 16 plants of all three genotypes included on each plate. The hypocotyl length of all seedlings was measured 2 days after the heat treatment. The data for each temperature treatment were evaluated by analysis of variance.
Acknowledgments
We thank Elizabeth Vierling for helpful discussions and advice about small HSPs and seedling thermotolerance assays and Jonathan Jones for supplying pSLJ755I5. We also thank Judy Schnurr for assistance with immunoblot analysis. This work was supported by grants from the National Institutes of Health (Nos. AI43288 and AI27832 to J.C.C. and No. GM18529 to S.A.W.) and the United States Department of Agriculture (National Research Initiative 98-35303-6485).
References
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279, 2113–2115. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Short Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant–microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Barros, M.D., Czarnecka, E., and Gurley, W.B. (1992). Mutational analysis of a plant heat shock element. Plant Mol. Biol. 19, 665–675. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (1999). Viruses and gene silencing in plants. Arch. Virol. 15 (suppl.), 189.–201. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Becker-Andre, M., and Hahlbrock, K. (1989). Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res. 17, 9437–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252, 313–317. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kohn, B.A., Dedi, C., and Baulcombe, D.C. (1995). The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 8, 933–941. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston, R.S., Viitanen, P.V., and Vierling, E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32, 191–222. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Wan-Xiang, L., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing. EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Callaway, A., Liu, W., Andrianov, V., Stenzler, L., Zhao, J., Wettlaufer, S., Jayakumar, P., and Howell, S.H. (1996). Characterization of cauliflower mosaic virus (CaMV) resistance in virus-resistant ecotypes of Arabidopsis. Mol. Plant-Microbe Interact. 9, 810–818. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Whitham, S.A. (1998). Viral invasion and host defense: Strategies and counter-strategies. Curr. Opin. Plant Biol. 1, 336–341. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance movement of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Jensen, P.E., and Schaad, M.C. (1998). Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Chen, Q., and Vierling, E. (1991). Analysis of conserved domains identifies a unique structural feature of a chloroplast heat shock protein. Mol. Gen. Genet. 226, 425–431. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Mahajan, S.K., Whitham, S.A., Yamamoto, M.L., and Carrington, J.C. (2000). Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc. Natl. Acad. Sci. USA 97, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. (1995). Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World 2, 17–20. [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Cronin, S., Verchot, J., Haldeman-Cahill, R., Schaad, M.C., and Carrington, J.C. (1995). Long-distance movement factor: A transport function of the potyvirus helper component-proteinase. Plant Cell 7, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, W.W., Caspers, G.J., and Leunissen, J.A. (1998). Geneology of the α-crystallin small heat-shock protein superfamily. Int. J. Biol. Macromol. 22, 151–162. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V., Haldeman, R., Robertson, N.L., Dougherty, W.G., and Carrington, J.C. (1994). Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13, 1482–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliand, G., Perrin, S., Blanchard, K., and Bunn, H.F. (1990). Analysis of cytokine mRNA and DNA: Detection and quantification by competitive polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, S.R. (1999). Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell 96, 303–306. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counter-defensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Cronin, S., and Carrington, J.C. (1997). Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228, 251–262. [DOI] [PubMed] [Google Scholar]
- Kim, K.K., Kim, R., and Kim, S.H. (1998). Crystal structure of a small heat-shock protein. Nature 394, 595–599. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Mahajan, S.K., Chisholm, S.T., Whitham, S., and Carrington, J.C. (1998). Identification and characterization of a locus (RTM1) in Arabidopsis thaliana that restricts long-distance movement of tobacco etch virus. Plant J. 14, 177–186. [DOI] [PubMed] [Google Scholar]
- Matthews, R.E.F. (1991). Plant Virology, 3rd ed. (San Diego, CA: Academic Press).
- Newman, T., de Bruijin, F.J., Green, P., Keegstra, K., Kende, H., McIntosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., Thomashow, M., Retzel, E., and Somerville, C. (1994). Genes galore: A summary of methods for assessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, O., Dunnington, S.W., Gotow, L.F., Pirone, T.P., and Hellmann, G.M. (1997). Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237, 452–459. [DOI] [PubMed] [Google Scholar]
- Pruss, G., Ge, X., Shi, X.M., Carrington, J.C., and Vance, V.B. (1997). Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F.G., MacFarlane, S.A., and Baulcombe, D.C. (1999). Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B. (1996). PHD: Predicting one-dimensional protein structure by profile based on neural networks. Methods Enzymol. 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Rost, B., and Sander, C. (1994). Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19, 55–72. [DOI] [PubMed] [Google Scholar]
- Rost, B., Casadio, R., Fariselli, P., and Sander, C. (1995). Transmembrane helices predicted with 95% accuracy. Protein Sci. 4, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C., and Carrington, J.C. (1996). Suppression of long-distance movement of tobacco etch virus in a nonsusceptible host. J. Virol. 70, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C., Lellis, A.D., and Carrington, J.C. (1997). VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71, 8624–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffl, F., Prandl, R., and Reindl, A. (1998). Regulation of the heat-shock response. Plant Physiol. 117, 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, P.D., and Larrick, J.W. (1992). Competitive PCR. Nature 359, 557–558. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling, E. (1991). The heat shock response in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 579–620. [Google Scholar]
- Waters, E.R., Lee, G.J., and Vierling, E. (1996). Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 47, 325–338. [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Whitham, S., Yamamoto, M., and Carrington, J.C. (1999). Selectable viruses and altered susceptibility mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]