Abstract
Profilin is an actin monomer binding protein that, depending on the conditions, causes either polymerization or depolymerization of actin filaments. In plants, profilins are encoded by multigene families. In this study, an analysis of native and recombinant proteins from maize demonstrates the existence of two classes of functionally distinct profilin isoforms. Class II profilins, including native endosperm profilin and a new recombinant protein, ZmPRO5, have biochemical properties that differ from those of class I profilins. Class II profilins had higher affinity for poly-l-proline and sequestered more monomeric actin than did class I profilins. Conversely, a class I profilin inhibited hydrolysis of membrane phosphatidylinositol-4,5-bisphosphate by phospholipase C more strongly than did a class II profilin. These biochemical properties correlated with the ability of class II profilins to disrupt actin cytoplasmic architecture in live cells more rapidly than did class I profilins. The actin-sequestering activity of both maize profilin classes was found to be dependent on the concentration of free calcium. We propose a model in which profilin alters cellular concentrations of actin polymers in response to fluctuations in cytosolic calcium concentration. These results provide strong evidence that the maize profilin gene family consists of at least two classes, with distinct biochemical and live-cell properties, implying that the maize profilin isoforms perform distinct functions in the plant.
INTRODUCTION
Plant cells often respond to intracellular and extracellular cues by reorganizing their microtubule and actin microfilament cytoskeletons. Actin reorganization in particular is necessary for or coincident with a variety of environmentally influenced processes, including cell division, cell elongation, responses to wounding or pathogen attack, plastid positioning, and pollen germination and extension of the pollen tube (reviewed in Taylor and Hepler, 1997; Nick, 1999; Staiger, 2000). In all eukaryotic cells, actin reorganization is thought to be controlled by actin binding proteins that regulate the spatial and temporal polymerization and depolymerization of actin monomers (globular or G-actin) into filamentous actin (F-actin) and that also organize the cytoskeleton into macromolecular structures. Actin binding proteins can be placed into several broad groups, based on their functional characteristics in vitro. Many are sensitive to changes in calcium and pH, and some are thought to be regulated through signal transduction pathways by interacting with polyphosphoinositides or to act as downstream effectors of small G proteins (Schmidt and Hall, 1998).
Profilins are low molecular mass (12 to 15 kD), abundant, cytosolic actin monomer binding proteins that form a 1:1 complex with G-actin. In addition to actin, profilins also interact with poly-l-proline (PLP) and proline-rich proteins, membrane polyphosphoinositides, phosphatidylinositol-3-kinase, annexin, and several multiprotein complexes that are implicated in the regulation of actin nucleation and endocytosis (Machesky et al., 1994; Witke et al., 1998).
Profilin can have two opposite influences on the assembly of actin in vitro. By binding and sequestering G-actin, profilin causes the depolymerization of actin filaments. Under other conditions, however, profilin promotes actin polymerization. When a large pool of actin monomers is available, the profilin–actin complex can add to uncapped filament ends and stimulate polymerization (Pantaloni and Carlier, 1993; Kang et al., 1999). It also has been argued that profilin facilitates polymerization indirectly by stimulating the exchange of ADP for ATP on G-actin, because ATP-loaded G-actin adds onto filaments more readily (Goldschmidt-Clermont et al., 1992). However, as shown recently, even though Arabidopsis profilins are unable to enhance nucleotide exchange on vertebrate actin, they still promote polymerization in vitro (Perelroizen et al., 1996). The functional importance of stimulating actin nucleotide exchange is still a matter of debate (Korenbaum et al., 1998; Vinson et al., 1998) because a heterologous source of actin was used to test the effects of Arabidopsis profilin.
It is likely that the local cellular environment, especially the presence of other actin binding proteins and the stoichiometry of profilin to total actin, will determine the specific effect profilin has on actin polymerization. The ability of profilin to promote both polymerization and depolymerization has been confirmed genetically. In yeast and Dictyostelium, mutational analyses suggest that profilin has a sequestering function (Magdolen et al., 1993; Haugwitz et al., 1994), whereas Drosophila and Schizosaccharomyces pombe loss-of-function mutants indicate that profilin is necessary for actin polymerization (Cooley et al., 1992; Balasubramanian et al., 1994). Equivalent genetic studies to analyze plant profilin function have not been performed.
At least two profilin isoforms are expressed in several organisms. The amino acid sequence identity between the two isoforms from any given organism is between 54 and 83%, a diversity sometimes reflected in differences in their biochemical properties in vitro (Schlüter et al., 1997). Even small changes in amino acid sequence can alter the biochemical properties of profilin substantially. Several examples of single amino acid substitutions that change the association of profilin with one ligand without affecting binding to other ligands have been reported (Haarer et al., 1993; Sohn et al., 1995; Björkegren-Sjögren et al., 1997; Hájková et al., 1997; Gibbon et al., 1998; Korenbaum et al., 1998; Schlüter et al., 1998; Ostrander et al., 1999). Therefore, characterizing the biochemical properties of each profilin isoform is important for gaining insight about its potential cellular function.
Plant profilins, which originally were identified as a birch pollen allergen and plant panallergens (Valenta et al., 1991), exist in large gene families (Staiger et al., 1997; de Ruijter and Emons, 1999). As many as six isoforms are predicted to be expressed in both Arabidopsis and maize (Staiger et al., 1993; Christensen et al., 1996; Huang et al., 1996; Gibbon et al., 1998). An unresolved question is: why do multiple profilin isoforms exist in plants? Based on the differences among profilin isoforms from other kingdoms, plant profilin isoforms are also predicted to have biochemical and functional differences. Previous characterization of four recombinant maize profilin isoforms provided initial evidence that the isoforms indeed are not functionally equivalent (Gibbon et al., 1997, 1998, 1999a).
Here, we report the characterization of native maize profilins and compare them with recombinant maize profilin isoforms. The results demonstrate that the maize profilin multigene family comprises two functionally distinct classes. We also have identified a fifth maize profilin isoform, ZmPRO5, that is expressed predominantly in vegetative tissues but is also weakly detectable in pollen. Biochemical characterization shows that recombinant ZmPRO5 is similar to native profilin purified from a vegetative tissue (endosperm), whereas recombinant ZmPRO1 is similar to native profilin purified from a reproductive tissue (pollen). ZmPRO5 has much higher affinity for PLP and sequesters more G-actin than does ZmPRO1. Conversely, ZmPRO1 prevents phospholipase C from hydrolyzing phosphatidylinositol-4,5-bisphosphate (PtdIns[4,5]P2) more effectively than does ZmPRO5. These biochemical differences correlate with an increased ability of ZmPRO5 to disrupt actin arrays in live Tradescantia virginiana stamen hair cells. We also show that ZmPRO1 and ZmPRO5 slightly inhibit pollen actin nucleotide exchange, thereby implying that stimulation of nucleotide exchange is not important for plant profilin function. The native profilins isolated from endosperm and pollen are also functionally distinct. In agreement with the characteristics of the recombinant proteins, endosperm profilin is a better G-actin– sequestering and PLP binding protein and displaces the stamen hair cell nucleus markedly faster than does pollen profilin. These results demonstrate differences in expression pattern and functional properties between members of a monocot profilin gene family, differences that appear to delineate two distinct classes of isoforms.
RESULTS
ZmPRO5 Is a Plant Profilin
An expressed sequence tag from an illuminated maize leaf and sheath mRNA library was identified as a profilin by BLAST (Altschul et al., 1990) sequence comparison. The 703-nucleotide cDNA encoded an open reading frame of 396 nucleotides that contained consensus translation start and stop codons. The clone also contained 78 nucleotides of 5′ untranslated region and 229 nucleotides of 3′ untranslated region, the latter including a poly(A) tail of 17 nucleotides. Conceptual translation of the open reading frame produced a 131 amino acid protein, as shown in Figure 1, with a predicted mass of 14.1 kD and a pI of 4.39. The amino acid sequence of ZmPRO5 was very similar to that of ZmPRO4 (Gibbon et al., 1998), sharing 95% identity, but was less similar to the pollen profilins ZmPRO1, ZmPRO2, and ZmPRO3 (Staiger et al., 1993), with which it shared 80, 79, and 79% identity, respectively. The residues that are most highly conserved in profilins from different species are those implicated in PLP binding (Figure 1); they are present in the ZmPRO5 sequence. Residues thought to be involved in actin binding (Figure 1) are less well conserved among profilins from different species (Thorn et al., 1997).
Figure 1.
Deduced Amino Acid Sequence of ZmPRO5 Shown in Alignment with Other Plant Profilins.
Secondary structures, predicted according to crystallographic data from birch pollen profilin (Fedorov et al., 1997), are positioned above the primary amino acid sequence. Cylinders represent α helices, and arrows indicate β sheets. Conserved residues, which have been implicated in PLP binding (asterisks) and actin binding (pound signs), are indicated. The sequences used for the alignment, their sources, and GenBank accession numbers or original citations are as follows: AtPFN1, Arabidopsis thaliana profilin 1, U43325 (see also Huang et al., 1996); AtPFN2, Arabidopsis profilin 2, U43326 (see also Huang et al., 1996); AtPRF3, Arabidopsis profilin 3 (Huang et al., 1996); RcPRO1, Ricinus communis profilin 1, AF092547; ZmPRO4, Zea mays profilin 4, AF032370; ZmPRO5, maize profilin 5, AF201459 (this study); ZmPRO1, maize profilin 1, X73279; ZmPRO2, maize profilin 2, X73280; ZmPRO3, maize profilin 3, X73281; BvPRO1, Betula verrucosa profilin, M65179; AtPFN3, Arabidopsis profilin 3, U43323 (see also Huang et al., 1996); and AtPFN4, Arabidopsis profilin 4, U43324. The entire ZmPRO5 sequence is shown along with the differing residues from the other sequences. The sequence alignments were created by using the PileUp program from the Genetics Computer Group (Madison, WI; Devereux et al., 1984). Dashes indicate where gaps have been introduced to optimize the alignment.
ZmPRO5 Is Expressed Primarily in Vegetative Tissues
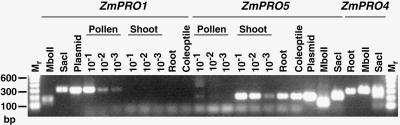
To examine maize tissues in which ZmPRO5 is expressed, we generated first-strand cDNA from ungerminated pollen, shoot, root, and coleoptile mRNA. The first-strand cDNA was used as template for polymerase chain reaction (PCR) amplification with gene-specific primers for ZmPRO1, ZmPRO4, and ZmPRO5, as shown in Figure 2. Qualitatively, the amplification product for ZmPRO5 was most abundant in vegetative tissues as determined by amplification from these tissues at 1:1000 dilution (in Figure 2, see Shoot; not shown for Coleoptile and Root). Only a minor product was produced from pollen first-strand cDNA at 1:10 dilution. In comparison, ZmPRO1 was abundant in pollen but was not detected in any vegetative tissue. This confirms our previous RNA gel blot analysis demonstrating that ZmPRO1 is expressed abundantly in pollen (Staiger et al., 1993). As also shown previously, with RNA gel blots and reverse transcription (RT)–PCR, ZmPRO4 transcripts are abundant in endosperm (Gibbon et al., 1998). ZmPRO5 and ZmPRO4 appear to be members of a predominantly vegetative profilin class, whereas ZmPRO1 is a member of a pollen-abundant class.
Figure 2.
RT-PCR Analysis of ZmPRO1 and ZmPRO5 in Four Maize Tissues.
The presence of profilin transcripts in different maize tissues was examined by RT-PCR detection of first-strand cDNA templates from pollen, shoot, root, and coleoptile tissue. Gene-specific primer pairs for ZmPRO1, ZmPRO4, and ZmPRO5 amplified products that were of identical size when plasmid DNA (not shown for ZmPRO4; see Gibbon et al., 1998) or maize tissue cDNAs were used as template. Serial dilutions of the cDNA templates demonstrated that ZmPRO5 transcript was considerably more abundant in vegetative tissues, whereas only a weak amplification product was apparent at the highest concentration of pollen template (10−1). Conversely, ZmPRO1 was detected at high concentrations in pollen, but no product was obtained from any vegetative tissue. The identity of the amplified products was confirmed by restriction digestion with MboII and SacI. For comparison with amplified product sizes, molecular standards (Mr) were loaded at left and right. Sizes (in base pairs) are shown at left.
Purification of Native Profilins from Maize Endosperm and Pollen
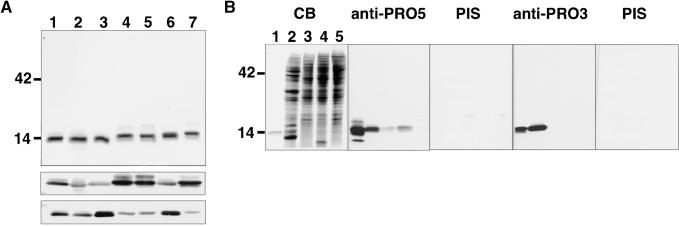
Profilin was purified from maize endosperm and pollen to investigate whether different plant tissues contained functionally distinct profilins. Purified, native endosperm profilin migrated more slowly on polyacrylamide gels than did native pollen profilin, as shown in Figure 3A. A similar difference was observed when recombinants ZmPRO4 and ZmPRO5 were compared with ZmPRO1, ZmPRO2, and ZmPRO3 (Figure 3A). To further test whether ZmPRO4 and ZmPRO5 were more similar to endosperm profilin than to pollen profilin, we raised an antiserum against recombinant ZmPRO5 (anti-ZmPRO5). Anti-ZmPRO5 recognized all five recombinant maize profilin isoforms and both native profilins, by protein gel blot analysis, as shown in Figure 3A (gel at center). Therefore, anti-ZmPRO5 is not isoform specific. However, anti-ZmPRO5 recognized recombinants ZmPRO4 and ZmPRO5 and native endosperm profilin more strongly than it did the other proteins tested. Conversely, an antibody raised against ZmPRO3 (anti-ZmPRO3; Figure 3A, bottom) recognized recombinants ZmPRO1, ZmPRO2, and ZmPRO3 and native pollen profilin most strongly. These results suggested that the major profilin isoforms present in pollen are ZmPRO1, ZmPRO2, or ZmPRO3, whereas native endosperm profilin is predominantly ZmPRO4 or ZmPRO5. To see whether differences in profilin isoforms extended to other plant tissues, we used the two antisera to probe blots of total protein extracts from pollen, root, coleoptile, and shoot tissues, as shown in Figure 3B. Anti-ZmPRO5 (second panel) recognized specifically a 14-kD band that was present in various amounts in all tissues examined, whereas anti-ZmPRO3 (fourth panel) recognized only a 14-kD band from pollen.
Figure 3.
Anti-ZmPRO5 and Anti-ZmPRO3 Recognize Complementary Sets of Profilin Isoforms.
(A) Purified profilins were separated by SDS-PAGE and stained with Coomassie blue (top) or were transferred to nitrocellulose and probed with antiserum raised against ZmPRO5 (anti-ZmPRO5; center) or ZmPRO3 (anti-ZmPRO3; bottom). Anti-ZmPRO5 recognized recombinant isoforms ZmPRO4 and ZmPRO5 (lanes 4 and 5) and native endosperm profilin (lane 7) most strongly. Anti-ZmPRO3 serum showed greatest reaction with recombinant isoforms ZmPRO1, ZmPRO2, and ZmPRO3 (lanes 1 to 3) and with native pollen profilin (lane 6). The positions of relevant molecular mass standards are shown at left in kilodaltons.
(B) Twenty micrograms of total protein extract from pollen (lane 2), roots (lane 3), coleoptiles (lane 4), and shoots (lane 5) along with 0.5 μg of recombinant ZmPRO5 (lane 1) was separated by SDS-PAGE. The resulting gels were stained with Coomassie blue (first gel) or transferred to nitrocellulose and probed with anti-ZmPRO5 (second gel), ZmPRO5 preimmune serum (third gel), anti-ZmPRO3 (fourth gel), or ZmPRO3 preimmune serum (fifth gel). Anti-ZmPRO5 recognized profilin from each maize tissue, whereas anti-ZmPRO3 detected profilin (more strongly than did anti-ZmPRO5) from pollen only. The positions of relevant molecular mass standards are shown at left in kilodaltons. CB, Coomassie blue; PIS, preimmune serum.
Microinjection of ZmPRO5 Results in Rapid Rearrangement of Cytoplasmic Architecture
To study the effects of increasing the cellular concentration of profilin, we developed a technique of microinjecting purified profilins into Tradescantia stamen hair cells (Staiger et al., 1994; Ren et al., 1997; Gibbon et al., 1997, 1998). Microinjection of profilin disrupts F-actin arrays and causes the nucleus to be displaced from a position at the center of the vacuole. By measuring the time it takes for the nucleus to move one nuclear diameter, we have been able to detect differences between recombinant maize profilin isoforms (Gibbon et al., 1998). In the studies reported here, 5 to 6 pL (∼10% of the cytoplasmic volume), at a needle concentration of 100 μM, was injected. This is estimated to increase the cytoplasmic profilin concentration from 6 to 16 μM (Staiger et al., 1994).
Previously, this laboratory had shown that recombinant ZmPRO4 displaces the nucleus of Tradescantia stamen hair cells more quickly than does recombinant ZmPRO1 (Gibbon et al., 1998). Because the amino acid sequence of ZmPRO5 is most similar to ZmPRO4, we expected that recombinant ZmPRO5 also would act more quickly than ZmPRO1. As shown in Table 1, this was indeed the case. The average time for nuclear displacement with ZmPRO5 (4.9 min) was almost identical to the average time for ZmPRO4 (4.7 min). Both ZmPRO5 and ZmPRO4 were significantly (P ⩽ 0.022) faster than ZmPRO1, which had an average displacement time of 7.0 min. Additionally, if endosperm and pollen profilins are composed of ZmPRO4- or ZmPRO5-like and ZmPRO1-like profilins, respectively, endosperm profilin should displace the stamen hair nucleus faster than pollen profilin does. Consistent with this prediction, endosperm profilin displaced the cell nucleus in an average time of 3.1 min, whereas pollen profilin averaged 6.2 min. For a negative control, we microinjected 1.4 mg/mL BSA; it caused no substantial movement of the nucleus during the 20-min assay.
Table 1.
Average Nuclear Displacement Times
| Injectate | Nuclear Displacement (min)aMean ±se (n) |
|---|---|
| BSAb | 19.5 ± 0.3 (22) |
| Cytochalasin D | 13.5 ± 1.1 (31)c |
| Latrunculin B | 2.1 ± 0.2 (31) |
| Recombinant | |
| ZmPRO1 | 7.0 ± 0.7 (45)d |
| ZmPRO4 | 4.7 ± 0.5 (31) |
| ZmPRO5 | 4.9 ± 0.6 (53) |
| Native | |
| Pollen profilin | 6.2 ± 0.7 (30) |
| Endosperm profilin | 3.1 ± 0.4 (32)e |
The average time required for nuclei to move outside the circumference of their original position was measured after Transcantia stamen hairs were injected with a 100 μM needle concentration of protein or inhibitor. Injected cells were monitored for a maximum period of 20 min.
To assess the effects of introducing foreign protein into the cytoplasm, we injected 1.4 mg/mL BSA as a negative control.
Cytochalasin D and latrunculin B were significantly different from each other (P ⩽ 0.001) and from all of the profilins (P ⩽ 0.0001 and P ⩽ 0.03, respectively) by the two-tailed t test.
ZmPRO1 was significantly different from ZmPRO4 and ZmPRO5 (P ⩽ 0.022).
Native endosperm profilin was significantly different from native pollen profilin (P = 0.0002).
For comparison with other known inhibitors of the actin cytoskeleton, we also microinjected cytochalasin D and latrunculin. Cytochalasin D, which binds to the barbed ends of actin filaments (Cooper, 1987), showed only a modest effect. It displaced the nucleus significantly (P ⩽ 0.0001) more slowly than did the profilins, with an average time of 13.5 min. Latrunculin B, which binds to plant G-actin with a high affinity (Kd of 74 nM; Gibbon et al., 1999b), displaced the nucleus significantly (P ⩽ 0.03) more quickly than did the profilins, with an average time of 2.1 min. These results demonstrated that molecules that bind to actin with different affinities, or have different modes of action, elicit distinct effects on the rate of nuclear movement in Tradescantia stamen hair cells.
Profilin Isoforms Differ in Affinity for PLP
To understand the biochemical basis underlying the isoform differences observed in the assay with live cells, we characterized the functional properties of recombinant and native profilins in vitro. The affinity of profilin for soluble PLP was measured by quantifying the increase in intrinsic tryptophan fluorescence when titrating a profilin solution with PLP. The average Kd value for ZmPRO5 was 164 μM proline residues, as shown in Table 2. These results were almost identical for ZmPRO4, which had a Kd of 167 μM. Both ZmPRO5 and ZmPRO4 were significantly (P < 0.0001) different from ZmPRO1, which had a Kd value of 289 μM. Endosperm profilin also had a significantly (P < 0.0001) lower average Kd value for PLP of 126 μM proline residues than for pollen profilin (Kd of 250 μM; Table 2). The Kd value of 320 μM that we obtained for recombinant human profilin I was similar to the previously reported value of 359 μM (Petrella et al., 1996).
Table 2.
Affinity of Maize Profilins for PLP
| Profilin | Kd (μM Proline Residues)aMean ±sd (n) |
|---|---|
| Recombinant | |
| Human profilin I | 320 ± 15 (8) |
| ZmPRO1 | 289 ± 32 (10)b |
| ZmPRO4 | 167 ± 25 (7) |
| ZmPRO5 | 164 ± 17 (13) |
| Native | |
| Pollen profilin | 250 ± 25 (6)c |
| Endosperm profilin | 126 ± 16 (5) |
The Kd values for recombinant and native profilins binding to PLP were determined by measuring an enhancement in intrinsic tryptophan fluorescence.
ZmPRO1 was significantly different from ZmPRO4 and ZmPRO5 by the two-tailed t test (P < 0.0001).
Native pollen profilin was significantly different from native endosperm profilin (P < 0.0001).
ZmPRO5 and Endosperm Profilin Inhibit Actin Polymerization Better than ZmPRO1 and Pollen Profilin
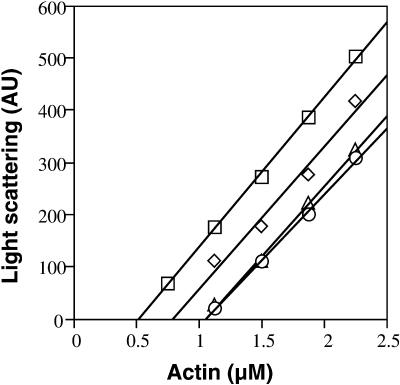
The G-actin–sequestering activity of the maize profilins was tested under steady state polymerizing conditions. An apparent Kd value was calculated by measuring the shift in the steady state critical concentration (Cc) for pollen actin assembly in the presence of profilin. The Cc is the minimum G-actin concentration at which polymerization will occur (Sheterline et al., 1998). The representative experiment shown in Figure 4 demonstrated a substantial difference in Cc values in the presence of ZmPRO5 when compared with ZmPRO1. The average apparent Kd values from several experiments were 1.2 μM for ZmPRO1, 0.3 μM for ZmPRO5, and 0.3 μM for ZmPRO4, as shown in Table 3. The values for ZmPRO5 and ZmPRO4 were significantly (P < 0.001) lower than those for ZmPRO1.
Figure 4.
ZmPRO5 Inhibits Actin Polymerization Better than ZmPRO1.
A representative experiment shows that ZmPRO5 and ZmPRO4 shift the steady state Cc values for actin assembly substantially more than does ZmPRO1. Increasing concentrations of pollen G-actin were polymerized in the absence (squares) or presence of 1.0 μM ZmPRO1 (diamonds), ZmPRO4 (circles), or ZmPRO5 (triangles). The Cc values (x axis intercept of each regression line) were 0.5 μM for actin alone and 0.8, 1.1, or 1.1 μM in the presence of ZmPRO1, ZmPRO4, or ZmPRO5, respectively. The resulting apparent Kd values were 1.2 μM for ZmPRO1, 0.3 μM for ZmPRO4, and 0.3 μM for ZmPRO5.
Table 3.
Apparent Kd Values for Pollen G-Actin
| Profilin | Kda |
|---|---|
| Recombinant | |
| Human profilin I | 0.2 ± 0.03 (5) |
| ZmPRO1 | 1.2 ± 0.6 (11)b |
| ZmPRO4 | 0.3 ± 0.3 (7) |
| ZmPRO5 | 0.3 ± 0.2 (11) |
| Native | |
| Pollen profilin | 0.7 ± 0.2 (5)c |
| Endosperm profilin | 0.3 ± 0.04 (3) |
The apparent Kd values for recombinant and native maize profilins binding to pollen actin under polymerizing conditions, in the presence of 55 μM Ca2+, were determined by measuring the shift in Cc values at steady state. All values, in μM, are reported as mean ±sd (n).
ZmPRO1 was significantly different from ZmPRO4 and ZmPRO5 by the two-tailed t test (P < 0.001).
Pollen profilin was significantly different from native endosperm profilin (P = 0.01).
Similarly, endosperm profilin sequestered significantly (P = 0.01) more pollen actin, with an average apparent Kd value of 0.3 μM, than did pollen profilin, with an average apparent Kd value of 0.7 μM (Table 3). Recombinant human profilin I also was found to have a high G-actin–sequestering activity under these conditions, with an apparent Kd value of 0.2 μM. Three additional types of qualitative assays were used to confirm that ZmPRO4 and ZmPRO5 have a greater association with G-actin than does ZmPRO1 (data not shown).
To test whether the observed differences between profilin isoforms were specific for maize pollen actin, we determined the apparent Kd values for binding to a heterologous source of actin. The affinity of ZmPRO5 and ZmPRO1 for rabbit skeletal muscle actin under polymerizing conditions gave average apparent Kd values of 0.3 ± 0.02 (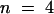 ) and 0.8 ± 0.08 μM (
) and 0.8 ± 0.08 μM (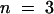 ), respectively—a significant difference (P < 0.0001). These results indicated that differences between profilin isoforms are not specific for maize pollen actin.
), respectively—a significant difference (P < 0.0001). These results indicated that differences between profilin isoforms are not specific for maize pollen actin.
Free [Ca2+] Affects Ability of Maize Profilins to Sequester G-Actin
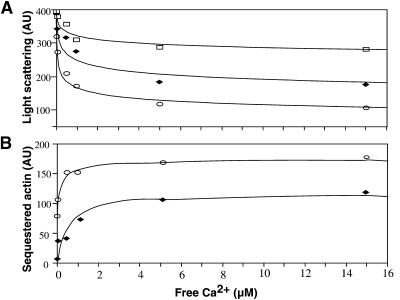
Free calcium concentration ([Ca2+]) has been shown to affect qualitatively the interaction between birch pollen profilin and muscle actin (Giehl et al., 1994) as well as the ability of profilin to promote polymerization (Perelroizen et al., 1996). Therefore, the G-actin–sequestering activity of ZmPRO1 and ZmPRO5 was tested over a range of [Ca2+], as shown in Figure 5A. As [Ca2+] increased, the amount of F-actin present at steady state was found to decrease. In the presence of either ZmPRO1 or ZmPRO5, the ability of profilin to sequester actin, and thereby further reduce F-actin levels at steady state, increased with the corresponding increase in [Ca2+]. By subtracting the extent of polymerization in the presence of either ZmPRO1 or ZmPRO5 from the extent of polymerized actin in the absence of profilin and plotting the difference versus [Ca2+], the effect of increasing [Ca2+] on actin alone was removed (Figure 5B). This shows that ZmPRO5 sequestered more G-actin than did ZmPRO1 at all [Ca2+] values tested. Maximum sequestering occurred at 5 μM Ca2+ for both profilin isoforms.
Figure 5.
Maize Profilins Sequester Pollen G-Actin in a Calcium-Dependent Manner.
(A) Pollen G-actin (2 μM) was polymerized in the absence (squares) or presence of 1.0 μM ZmPRO1 (diamonds) or ZmPRO5 (circles) with increasing concentrations of calcium. After reaching steady state, the amount of F-actin was measured by 90° light scattering. Plots of calcium concentration versus F-actin concentration revealed that both ZmPRO1 and ZmPRO5 sequestered more pollen G-actin at higher concentrations of calcium. The concentration of F-actin at steady state, in the absence of profilin, also decreases with increasing calcium.
(B) The plot of the F-actin concentration from the actin-only sample minus the F-actin concentration from either the actin + ZmPRO1 sample (diamonds) or the actin + ZmPRO5 sample (circles) versus calcium concentration clearly shows the sequestering effect of both profilins in the presence of increasing calcium. Maximum sequestering occurred at 5 μM Ca2+ for both isoforms, but ZmPRO5 sequestered more pollen G-actin than did ZmPRO1 at all calcium concentrations.
One possibility is that the affinity of the maize profilins for G-actin increases with increasing [Ca2+]. Alternatively, given that profilin adds Mg-ATP-actin but not Ca-ATP-actin subunits onto barbed ends (Perelroizen et al., 1996), the change in the amount of sequestered actin over a range of calcium concentrations could reflect a reduction in the ability of both ZmPRO1 and ZmPRO5 to induce polymerization as the actin subunits become Ca2+ loaded. In either case, at a given calcium concentration, a measured dissociation constant would reflect the amount of actin that is sequestered by profilin. Thus, apparent Kd values for native and recombinant profilins binding to G-actin were determined at the physiologically relevant Ca2+ concentrations of 25 nM and 1 μM. As shown in Table 4, the average Kd values were 2.6 and 1.8 μM for ZmPRO1, 1.7 and 0.6 μM for ZmPRO5, 2.9 and 0.8 μM for pollen profilin, and 2.5 and 0.4 μM for endosperm profilin, at 25 nM and 1 μM Ca2+, respectively. As was the case at 55 μM Ca2+, ZmPRO1 and pollen profilin acted significantly (P ⩽ 0.02) different from ZmPRO5 and endosperm profilin at 1 μM Ca2+. However, there was no significant difference between isoforms at 25 nM Ca2+. There was almost a threefold increase in ZmPRO5 sequestering activity at 1.0 μM Ca2+ compared with 25 nM Ca2+. ZmPRO1, on the other hand, had less than a 1.5-fold increase over the same Ca2+ range, indicating that ZmPRO5 was more sensitive to changes in [Ca2+] than was ZmPRO1.
Table 4.
Apparent Kd Values for Pollen G-Actin at Different Calcium Concentrationsa
| Profilin | 25 nM Ca2+ | 1.0 μM Ca2+ | 55 μM Ca2+ |
|---|---|---|---|
| Recombinant | |||
| ZmPRO1b | 2.6 ± 0.9 (3) | 1.8 ± 0.8 (7) | 1.2 ± 0.6 (11)c |
| ZmPRO5 | 1.7 ± 0.4 (4) | 0.6 ± 0.3 (4) | 0.3 ± 0.2 (11) |
| Native | |||
| Pollen profilin | 2.9 ± 1.0 (4) | 0.8 ± 0.1 (6) | 0.7 ± 0.2 (5) |
| Endosperm profilin | 2.5 ± 0.9 (3) | 0.4 ± 0.1 (4) | 0.3 ± 0.1 (3) |
The apparent Kd (μM, mean ±sd [n]) values for binding to pollen G-actin under physiological concentrations of Ca2+ were determined by measuring the shift in Cc at steady state.
ZmPRO1 and pollen profilin were significantly different from ZmPRO5 and endosperm profilin, respectively, at both 1.0 and 55 μM Ca2+ (P ⩽ 0.02) but not at 25 nM Ca2+. ZmPRO5 and endosperm profilin were significantly different at 25 nM from 1.0 μM and 1.0 μM from 55 μM Ca2+ (P ⩽ 0.02), whereas ZmPRO1 and pollen profilin were not significantly different at 1.0 μM from 55 μM Ca2+ (P > 0.09).
Data for 55 μM Ca2+ also were reported in Table 3.
Maize Profilins Do Not Enhance the Rate of Actin Nucleotide Exchange
Profilins from diverse eukaryotic organisms have been shown to increase the rate of G-actin nucleotide exchange on muscle actin (Schlüter et al., 1997). However, Arabidopsis profilins did not enhance nucleotide exchange on vertebrate actin (Perelroizen et al., 1996). We examined the possibility that plant profilins would increase the rate of nucleotide exchange with a relevant source of actin by measuring the increase in fluorescence emission when pollen G-actin incorporates the ATP analog ε-ATP. These reactions were performed in the presence of 50 mM KCl, 0.5 mM MgCl2, and 2 μM Ca2+ to simulate the polymerizing conditions shown to facilitate formation of profilin–actin complexes.
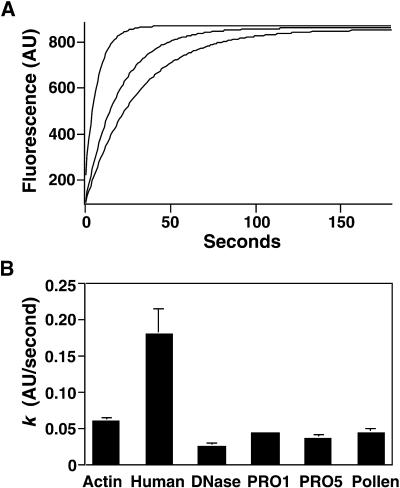
As shown in Figure 6A, human profilin I enhances, whereas the maize profilins slightly inhibit, nucleotide exchange. The average turnover rate (k) for 0.5 μM pollen actin alone was 0.059 arbitrary fluorescence units (AU)/sec (Figure 6B). A substoichiometric amount (0.1 μM) of human profilin I, which stimulates vertebrate actin nucleotide exchange (Korenbaum et al., 1998), increased the rate of pollen actin nucleotide exchange to 0.18 AU/sec. In contrast, equal stoichiometric amounts of either ZmPRO1 or ZmPRO5 reduced the rate of pollen actin nucleotide exchange to 0.043 and 0.036 AU/sec, respectively—rates significantly (P < 0.001) slower than that of pollen actin alone. A known inhibitor of actin nucleotide exchange, DNase I, reduced the rate of pollen actin nucleotide exchange to 0.025 AU/sec. Similar results were obtained by using saturating amounts of the maize profilins under physiologic and low ionic strength conditions and by measuring the dissociation of ε-ATP (data not shown). These results demonstrate that the maize profilins do not stimulate actin nucleotide exchange and actually may inhibit the process.
Figure 6.
Maize Profilins Inhibit Nucleotide Exchange on Pollen Actin.
(A) The incorporation of ε-ATP by 0.5 μM pollen actin in the absence (curve at center) or presence of 0.1 μM human profilin I (curve at top) or 0.5 μM ZmPRO5 (curve at bottom) was monitored over time. Human profilin I dramatically enhances the initial nucleotide exchange rate, whereas ZmPRO5 inhibits the rate slightly.
(B) The rate of incorporation of ε-ATP (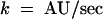 ) was determined by fitting the data with a single exponential function as shown in (A). For several experiments, the average rate for actin alone was 0.059 ± 0.0045 (
) was determined by fitting the data with a single exponential function as shown in (A). For several experiments, the average rate for actin alone was 0.059 ± 0.0045 (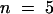 ; column 1). The average rates in the presence of 0.1 μM human profilin I, 0.5 μM DNase I, 0.5 μM ZmPRO1, 0.5 μM ZmPRO5, or 0.5 μM native pollen profilin were 0.18 ± 0.035 (
; column 1). The average rates in the presence of 0.1 μM human profilin I, 0.5 μM DNase I, 0.5 μM ZmPRO1, 0.5 μM ZmPRO5, or 0.5 μM native pollen profilin were 0.18 ± 0.035 (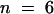 ), 0.025 ± 0.0048 (
), 0.025 ± 0.0048 (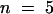 ), 0.043 ± 0.0032 (
), 0.043 ± 0.0032 (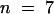 ), 0.036 ± 0.0052 (
), 0.036 ± 0.0052 (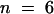 ), and 0.043 ± 0.0058 (
), and 0.043 ± 0.0058 (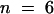 ), respectively. The rate of ε-ATP incorporation by pollen actin alone was significantly different by the two-tailed t test from incorporation of ε-ATP by pollen actin in the presence of the profilins or DNase I (
), respectively. The rate of ε-ATP incorporation by pollen actin alone was significantly different by the two-tailed t test from incorporation of ε-ATP by pollen actin in the presence of the profilins or DNase I (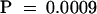 ). Error bars represent sd.
). Error bars represent sd.
Maize Profilins Differ in Ability to Inhibit Hydrolysis of PtdIns(4,5)P2 by Phospholipase C
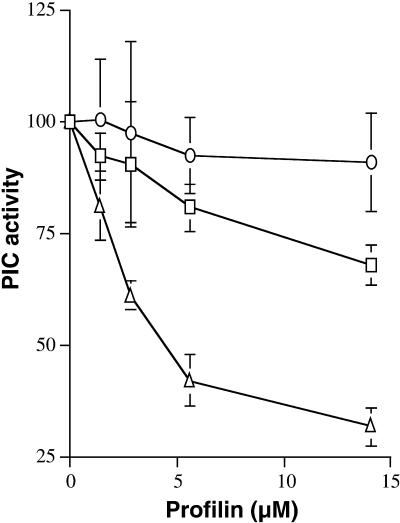
To measure the interaction of the maize profilins with membrane phospholipids, we assayed ZmPRO1 and ZmPRO5 for the ability to inhibit the hydrolysis of PtdIns(4,5)P2 by phosphoinositide-specific phospholipase C from bean membrane. As shown in Figure 7, ZmPRO1 was found to inhibit phosphoinositidase activity more effectively than did ZmPRO5. Moreover, the positive control, recombinant human profilin I, inhibited phosphoinositidase activity more effectively than did either maize profilin. These results imply that ZmPRO1 binds to PtdIns(4,5)P2 with a greater affinity than does ZmPRO5. However, the possibility that ZmPRO1 and ZmPRO5 bind to PtdIns(4,5)P2 with similar affinities but in different manners cannot be ruled out.
Figure 7.
Inhibition of Plant Polyphosphoinositide-Specific Phospholipase C.
The hydrolysis of PtdIns(4,5)P2 by phospholipase C (PIC) was measured in the presence of increasing concentrations of ZmPRO1 (squares), ZmPRO5 (circles), or human profilin I (triangles). Each data point represents the average (±sd) of four independent determinations made with three individual batches of profilin. The variation between experiments and profilin batches in all cases was <10%. Phospholipase C activity in the absence of profilin was set to 100%. The phospholipase C kcat was 14.3 nmol mg−1 min−1, based on 10-min incubations. ZmPRO1 and ZmPRO5 are significantly different at 14.1 μM (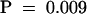 ).
).
DISCUSSION
We have demonstrated that the maize profilin multigene family contains two distinct classes that differ in their expression patterns, biochemical properties, and live-cell effects. Furthermore, we show that native profilins isolated from pollen and endosperm mimic the differences seen between recombinant ZmPRO1 and ZmPRO5, respectively. An overview of the properties of class I (ZmPRO1, ZmPRO2, ZmPRO3, and pollen) and class II (ZmPRO4, ZmPRO5, and endosperm) maize profilins is shown in Table 5. These classes also are compared with other plant profilins and human profilin I. When microinjected into Tradescantia stamen hair cells, class II profilins disrupt cytoplasmic architecture significantly more rapidly than do class I profilins. The biochemical basis for enhanced potency in the live cell may be threefold, because complementary associations with three known profilin ligands have been demonstrated. Class II profilins have a higher affinity for PLP and sequester more G-actin than do class I profilins. Conversely, ZmPRO1 (class I) inhibits hydrolysis of PtdIns(4,5)P2 by phosphoinositidase more strongly than does ZmPRO5 (class II), providing indirect evidence that ZmPRO1 binds to membrane phosphoinositides with greater affinity than does ZmPRO5. This combination of complementary association with three profilin ligands has not been demonstrated for profilin isoforms from any other organism. The results indicate that the maize profilin family contains at least two classes with specific patterns of expression and biochemical activity; the results also support the possibility that individual isoforms perform distinct functions in the plant.
Table 5.
Overview of the Properties of Plant Profilin
|
Kd Valuesa
|
|||||
|---|---|---|---|---|---|
| Profilin Source | Actin | PLP | PtdIns(4,5)P2 | Live Cellb | References |
| Human profilin I | 0.1–0.5c 0.2d |
359 305–320 |
0.2 +++e |
3.0–5.0 |
Gieselmann et al., 1995; Sohn et al., 1995; Petrella et al., 1996; Hájková et al., 1997 Gibbon et al., 1997, 1998; this study |
| Zea mays | Gibbon et al., 1997, 1998, 1999a; this study | ||||
| Class I | |||||
| PRO1 | 1.1–1.2d | 275–289 | +e | 6.0–8.5 | |
| PRO2 | NDf | 262 | 6.0–8.5 | ||
| PRO3 | ND | 251 | 6.0–8.5 | ||
| Native pollen | 0.7 | 249–250 | 6.0–8.5 | ||
| Class II | |||||
| PRO4 | 0.3–0.4 | 167–173 | 3.0–5.0 | ||
| PRO5 | 0.3 | 164 | −e | 3.0–5.0 | |
| Native endosperm | 0.3 | 126 | 3.0–5.0 | ||
| B. verrucosa | 1.8–6.5c | 150 | 24 | ND |
Drøbak et al., 1994; Giehl et al., 1994; Domke et al., 1997; Ballweber et al., 1998 |
| Arabidopsis | 1.8–2.3c | ND | ND | ND | Perelroizen et al., 1996 |
| P. rhoeas | ND | 130 | ND | 5.7 | Clarke et al., 1998 |
| R. communis | 0.07d | 121 | ND | 3.0–5.0 | Schobert et al., 2000 |
Kd values are reported in μM.
Time (min) for Tradescantia stamen hair nuclear displacement upon injection of 100 μM profilin. Range of times accounts for variations in injection conditions that cannot be held constant between studies.
Kd values for binding to rabbit skeletal muscle actin at steady state.
Apparent Kd values for binding to maize pollen actin at steady state.
Relative ability to inhibit plant phospholipase C.
ND, not determined.
With the discovery that many cytoskeletal proteins are encoded by multigene families, researchers have asked the intriguing question of whether the resulting isoforms have unique functional properties or simply ensure redundancy (for plants, see Meagher, 1991; Meagher et al., 1999). The distinct temporal and spatial expression patterns of cytoskeletal gene family members indicate that a complex regulation of expression has evolved, but the necessity for these differences remains to be answered. Perhaps protein isoforms with unique functions are required at different times or locations. Because the expression patterns of individual gene family members overlap, functionally distinct isoforms also could be required at the same time in the same tissue or cell. Evidence that cytoskeletal protein family members have unique functions is accumulating. Transient expression of cytoplasmic actin isoforms, but not muscle actin isoforms, in cultured cells causes dramatic morphological changes, which suggests that cytoplasmic and muscle actins have different biochemical characteristics (von Arx et al., 1995; Mounier et al., 1997). In Drosophila, only one actin isoform is expressed in the indirect flight muscle. Flies lacking indirect flight muscle actin have severe defects when complemented by a nonmuscle actin isoform, indicating that the fly actin isoforms are not functionally equivalent (Fyrberg et al., 1998).
Increasing evidence suggests that profilin isoforms have different affinities for their known ligands and distinct subcellular localizations (reviewed in Schlüter et al., 1997). Acanthamoeba profilin isoforms I and II have similar affinities for both G-actin and PLP, but isoform II has at least a 10-fold greater affinity for PtdIns(4,5)P2 (Kaiser et al., 1986; Machesky et al., 1990; Petrella et al., 1996). As a result, isoform II localizes preferentially to the plasma membrane (Bubb et al., 1998). Complementary affinities for two ligands have been detected between the bovine profilin isoforms (Lambrechts et al., 1995, 1997). In the presence of capped barbed ends, there are no differences in actin binding. Bovine profilin II has a greater affinity for PLP, whereas bovine profilin I has a greater affinity for PtdIns(4,5)P2. The functional importance of PLP binding is indicated by additional experiments. Bovine profilin II, but not profilin I, in the presence of a proline-rich peptide that mimics the intracellular profilin ligand vasodilator-stimulated phosphoprotein (VASP), is able to promote actin filament nucleation (Jonckheere et al., 1999). However, vertebrate profilin I binds more strongly to the proline-rich protein N-WASP than does vertebrate profilin II, demonstrating the importance of characterizing the affinity of profilins for their actual protein binding partners (Suetsugu et al., 1998).
At present, cDNAs for five maize profilin genes are known and, on the basis of amino acid sequence identity, can be placed into two groups (Staiger et al., 1993; Gibbon et al., 1998; this study). Expression of ZmPRO1, ZmPRO2, and ZmPRO3 has been detected only in pollen by using RNA gel blot analysis or RT-PCR. ZmPRO4 and ZmPRO5 appear to be expressed predominantly in vegetative tissues, but they are also present at lower amounts in pollen. ZmPRO4 is also quite abundant in endosperm tissue (Gibbon et al., 1998). Although antisera raised against either purified ZmPRO3 or ZmPRO5 recognize each recombinant and native maize profilin, anti-ZmPRO5 reacts more strongly with ZmPRO4, ZmPRO5, and endosperm profilin, whereas anti-ZMPRO3 reacts more strongly with ZmPRO1, ZMPRO2, ZmPRO3, and pollen profilin.
To examine whether functionally distinct profilin isoforms are expressed in plants, we assayed native maize profilins in the live cell for the ability to alter actin cytoarchitecture. Microinjection into Tradescantia stamen hair cells showed that endosperm profilin caused nuclear displacement in half the time required for pollen profilin. If excess profilin acts as a sequestering protein in these interphase cells (Staiger et al., 1997), the simple explanation for our observations would be that endosperm profilin is a better G-actin–sequestering protein than pollen profilin. In support of this hypothesis, under steady state polymerizing conditions in vitro, endosperm profilin sequestered more G-actin than did pollen profilin. The recombinant profilin isoforms mimic the live-cell and biochemical differences discovered during investigation of the native profilins. ZmPRO4 and ZmPRO5 sequester more G-actin than does ZmPRO1, which correlates with their increased ability to disrupt actin cytoarchitecture in the live-cell assay.
The ability of the maize profilins to prevent actin polymerization in vitro depends on [Ca2+]. As [Ca2+] increases, both class I and II profilins sequester more actin monomers, which results in less polymerized actin at steady state. Even though profilin is distributed uniformly in the cytoplasm of pollen tubes (Grote et al., 1995; Vidali and Hepler, 1997), its activity could be modified by local changes in [Ca2+]. This is relevant because pollen tubes have a fluctuating, tip-high, Ca2+ gradient (Rathore et al., 1991; Miller et al., 1992; Holdaway-Clarke et al., 1997; Messerli and Robinson, 1997). The shank of the pollen tube has [Ca2+] of 100 to 500 nM, whereas the extreme apex may see pulses of [Ca2+] that reach 1 to 10 μM (Holdaway-Clarke et al., 1997; Messerli and Robinson, 1997). In addition, during stimulus-mediated cessation of pollen tube growth, a Ca2+ wave >1.5 μM floods the shank of the tube, and the tip-high gradient dissipates (Franklin-Tong et al., 1993, 1997).
Because sequestering of G-actin by native pollen profilin is dependent on [Ca2+], we propose a model whereby profilin acts as an efficient sensor that alters actin organization in response to Ca2+ fluxes. The apparent Kd value of native pollen profilin for pollen G-actin is 2.9 μM at 25 nM Ca2+ and 0.8 μM at 1.0 μM Ca2+. These values allow us to calculate whether changes in profilin sequestering activity can account for fluxes in actin polymerization. The concentrations of profilin and total actin are equal in maize pollen (125 μM; Gibbon et al., 1999b). At 25 nM Ca2+, from the equation and assumptions of Gibbon et al. (1999b), the calculated profilin–actin complex, G-actin (Cc), and F-actin concentrations would be 107, 0.4, and 17.6 μM, respectively. At 1 μM Ca2+, the calculated profilin–actin complex, G-actin, and F-actin concentrations would be 115, 0.4, and 9.6 μM, respectively. This 45% reduction in the concentration of F-actin is likely to be significant, especially given recent demonstrations that even small reductions in F-actin levels caused by latrunculin B can inhibit pollen tube growth (Gibbon et al., 1999b).
Moreover, our data predict that increased profilin sequestering activity will contribute importantly to local changes in polymer levels in response to fluctuations in calcium concentration, such as the decreased amounts of F-actin at the extreme apex of actively growing pollen tubes (Miller et al., 1996; Kost et al., 1998). We also predict that the flood of Ca2+ during stimulus-mediated cessation of pollen tube growth may induce profilin sequestering activity and thus result in a reduction of F-actin (B. Snowman, V.E. Franklin-Tong, and C.J. Staiger, unpublished data). When [Ca2+] in lily pollen tubes is forced to concentrations >10 μM experimentally, actin filaments are fragmented and cytoplasmic streaming stops (Kohno and Shimmen, 1987, 1988). It would be interesting to determine whether the altered appearance of actin also correlates with a measurable reduction in F-actin.
Actin depolymerizing-factor (ADF)/cofilin proteins are able to rapidly depolymerize F-actin by increasing the loss of ADP-loaded actin subunits from filament ends (Carlier et al., 1997). Because vertebrate and Acanthamoeba profilins enhance nucleotide exchange (Blanchoin and Pollard, 1998), it has been suggested that profilin uses this pool of ADP-loaded G-actin to stimulate polymerization of ATP-loaded G-actin. However, Arabidopsis profilins have no effect on the rate of nucleotide exchange yet still can induce polymerization of rabbit muscle actin (Perelroizen et al., 1996), and birch pollen profilin has little effect on the nucleotide exchange rate of yeast actin (Eads et al., 1998). Therefore, either the ability to enhance G-actin nucleotide exchange may not be important for all profilins or profilins may need to be tested with physiologically relevant sources of actin to demonstrate this property (Korenbaum et al., 1998; Vinson et al., 1998). We observed that two classes of maize profilins slightly inhibited nucleotide exchange on maize pollen actin. Thus, plant profilins appear not to enhance nucleotide exchange. Our data, therefore, are in agreement with a modified model in which profilin synergizes with ADF/cofilin to increase filament turnover without profilin-induced nucleotide exchange (Didry et al., 1998).
In addition to G-actin, profilins also interact with PLP, with proteins that contain proline-rich regions, and with PtdIns(4,5)P2. The maize profilins were found to have complementary differences in their interaction with both PtdIns(4,5)P2 and PLP. Class II profilins are better PLP binding proteins than are class I profilins, whereas compared with ZmPRO5, ZmPRO1 has an enhanced association with PtdIns(4,5)P2 (Table 5). These combinations of biochemical properties are unique among eukaryotic profilin isoforms. For instance, human profilin I is a good PtdIns(4,5)P2 and actin binding protein but has a low affinity for PLP. This suggests that the plant profilins bind to unique endogenous ligands or that actin arrays specific to plant cells may require actin binding proteins with properties that are different from their nonplant counterparts.
Two families of proline-rich proteins, VASP and VASP-like proteins (Reinhard et al., 1995) and the formin-homology proteins (reviewed in Frazier and Field, 1997), have been identified as partners of profilin from nonplant systems. Originally, profilin–proline-rich protein interactions were thought to target profilin to subcellular domains in which actin polymerization is regulated. A recent study, however, demonstrated that interaction with a proline-rich peptide from VASP alters the effect of profilin on actin dynamics (Jonckheere et al., 1999). The importance of PLP binding for maize profilin function has been tested by a single amino acid substitution mutant (Y6F), which selectively increases the affinity for PLP without affecting actin binding (Gibbon et al., 1998, 1999a). ZmPRO1-Y6F has an affinity for PLP similar to that of ZmPRO4 and ZmPRO5 and displaces the stamen hair cell nucleus just as quickly. To further substantiate these results, investigators should test profilin mutants having less affinity for PLP but unaltered in actin binding properties.
Cell signaling through membrane phospholipids also affects profilin regulation, because profilin bound to PtdIns(4,5)P2 is unable to interact with G-actin (reviewed in Schlüter et al., 1997). Phosphorylated but not unphosphorylated phospholipase C can hydrolyze profilin-bound PtdIns(4,5)P2, releasing profilin as well as the second messengers diacyglycerol and inositol 1,4,5-trisphosphate. Released profilin then would be available to regulate actin polymerization. When injected into Tradescantia stamen hair cells, a profilin with a high affinity for PtdIns(4,5)P2 might be sequestered at the plasma membrane and thus be unable to affect actin dynamics. This presumably would lead to a lessened ability to disrupt actin cytoarchitecture and a slower nuclear displacement time, compared with a profilin with a lower affinity for PtdIns(4,5)P2.
It has been suggested that the angiosperm profilin isoforms represent two ancient classes, nonreproductive (vegetative) and reproductive, based on amino acid sequence and expression patterns (Huang et al., 1996; Meagher et al., 1999). Such a divergence of the two classes, it is thought, would predate the split between monocots and dicots. Accordingly, a vegetative maize profilin would be predicted to be more similar to a vegetative Arabidopsis profilin than to a reproductive maize profilin. With the increase in plant profilin sequences in current databases, a reexamination of this hypothesis would be timely, but it is too early to state with confidence that the biochemical differences observed between the maize profilin classes can be extended to all plant profilins.
The actin binding properties of vegetative (AtPFN1) and reproductive (AtPFN3) Arabidopsis profilins have been reported (Perelroizen et al., 1996). Under conditions of low ionic strength, AtPFN1 and AtPFN3 bind vertebrate actin almost equally, in the 2.0-μM range. Under steady state polymerizing conditions with capped barbed ends, AtPFN1 has a slightly greater affinity (Kd of 1.8 μM) for vertebrate G-actin than does AtPFN3 (Kd of 2.6 μM). Before declaring that functionally distinct classes I and II are a general feature of plant profilin families and that these differences predate the ancient split between monocots and dicots, a thorough analysis of the biochemical and live-cell properties of multiple profilin isoforms from a dicotyledonous plant must be performed. The presence of tissue-specific classes of profilins indicates that actin organization within different maize tissues might require actin binding proteins with unique functional properties.
The evidence reported here suggests that the maize profilin gene family contains isoforms that have unique patterns of expression and biochemical properties. Because members of both classes are present in mature maize pollen, we propose that individual isoforms play distinct roles, or have unique subcellular distributions, or both. The generation of isoform-specific antibodies and GFP–profilin fusion proteins and the identification of class-specific mutants should help address these possibilities. Further studies also will focus on the effect of maize profilins on actin polymerization, the physiologic relevance of interactions with PLP and PtdIns(4,5)P2, and the influence of other actin binding proteins on profilin function.
METHODS
Nucleotide Sequence Analysis
The cDNA for an expressed sequence tag (GenBank accession number AA054790), identified as a profilin by a basic local alignment search tool (BLAST; Altschul et al., 1990) search, was kindly provided by T.A. Musket (University of Missouri–Columbia Probe Center). The original library was prepared from mRNA isolated from illuminated leaves and sheaths of 5-week-old maize plants (C. Baysdorfer, California State University, Hayward). Both strands of the 703-bp EcoRI-XhoI insert in the pBluescript II SK− vector (Stratagene, La Jolla, CA) were sequenced by automated fluorescence sequencing (ALF Express; Pharmacia Biotech, Piscataway, NJ). This clone is referred to as ZmPRO5, and the full nucleotide sequence information has GenBank accession number AF201459.
Polymerase Chain Reaction Amplification
Total RNA was isolated from ungerminated pollen, roots, shoots, or coleoptiles of the maize inbred line A188 with TRIzol reagent (Gibco BRL), according to the manufacturer's instructions. Poly(A)+ mRNA was isolated with a Quick-Prep Micro mRNA purification kit (Pharmacia Biotech) and reverse transcribed with a first-strand cDNA synthesis kit (Pharmacia Biotech). The polymerase chain reaction (PCR) was performed with gene-specific primers (5′-TAAGTTTGTCATAATGC-3′ and 5′-CGAGATATGAGGAGATGG-3′) that were designed to amplify a 190-bp fragment (nucleotides 472 to 662) in the 3′ untranslated region of the ZmPRO5 transcript. Primers designed to amplify a portion of the 3′ untranslated region from ZmPRO1 and ZmPRO4 have been described previously (Staiger et al., 1993; Gibbon et al., 1998). PCR was performed with annealing temperatures of 50, 57, and 40°C for ZmPRO1, ZmPRO4, and ZmPRO5, respectively. Purified plasmid DNA for ZmPRO1 and ZmPRO5 served as controls. The identity of the gene-specific amplification products was confirmed by digestion of gel-purified PCR products with specific restriction enzymes (MboII and SacI; Promega). The 190-base-long ZmPRO5 gene-specific product was cut by MboII (108 and 82 bp) but not by SacI, whereas the 285-base-long ZmPRO4 gene-specific product was cut by SacI (209 and 77 bp) but not by MboII. As expected, the ZmPRO1 gene-specific product also was cut by MboII but not by SacI. However, the gene-specific primers for ZmPRO5 and ZmPRO1 did not amplify cDNA from ZmPRO1 and ZmPRO5 plasmid DNA, respectively (data not shown).
Protein Purification
The ZmPRO5 coding region was amplified with VentR DNA polymerase (New England Biolabs, Beverly, MA). The 5′ primer (5′-CCCATATGTCGTGGCAGGCGTACG-3′) mimics the first six codons and introduces an NdeI restriction site (underlined). The 3′ primer (5′-GGGGATCCTTAGAAGCCCTGTGCGATCAG-3′) mimics the last six codons, including the stop translation codon, and introduces a BamHI restriction site (underlined). After PCR amplification, the 400-bp product was ligated into pGEM-T (Promega). Plasmid DNA from transformants was digested with NdeI and BamHI, and the insert DNA was subcloned into pET-23a (Novagen, Madison WI) digested with the same enzymes. The fidelity of the cloned product was verified by sequencing. The resulting pET-23a–ZmPRO5 construct was transformed into strain BL21(DE3) of Escherichia coli, and protein expression was induced by the addition of 0.4 mM isopropyl β-d-thiogalactopyranoside. Recombinant ZmPRO5 protein was purified on poly-l-proline (PLP)–Sepharose, according to methods described previously (Karakesisoglou et al., 1996), with the addition of a 3 M urea wash step before the elution with 7 M urea. Yields of purified ZmPRO5 were ∼7.5 mg/L bacteria.
Native pollen profilin was purified on PLP–Sepharose as described previously (Gibbon et al., 1997), also with the addition of a 3 M urea wash before the 7 M urea elution. Native profilin was purified from endosperm 16 days after pollination by grinding in liquid nitrogen and resuspending in buffer I (20 mM Tris-HCl, 150 mM KCl, and 0.2 mM DTT, pH 7.5) supplemented with 0.1% Triton X-100, 1:200 protease inhibitor cocktail (Ren et al., 1997), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). The extract then was sonicated in five 30-sec bursts. The sonicate was clarified with two consecutive centrifugations of 30,000 and 46,000g and was passed through a Sepharose CL-4B column (Sigma) to remove lipids. The flow-through liquid was loaded onto a PLP–Sepharose column, which then was washed with three volumes each of buffer I, buffer I + 2 M urea, and buffer I + 3 M urea. Profilin was eluted with 7 M urea, and the protein-containing fractions were pooled and concentrated in a 10,000 D cutoff Centricon (Millipore, Bedford, MA). The average yield (±sd) from five purifications was 540 ± 105 μg of profilin from 100 g of endosperm.
Maize pollen actin was purified as described previously (Ren et al., 1997), with a few modifications. A 5- to 10-g sample of frozen pollen was ground with a mortar and pestle for 25 min in 50 mL of buffer A (10 mM Tris-HCl, pH 8.5, 0.5 mM CaCl2, 0.01% NaN3, 50 mM NaF, 30 mM NaPPi, 0.4 mM ATP, 0.5 mM DTT, 0.5 mM PMSF, and 1:200 protease inhibitor cocktail). During the grinding, a total of 25 mg of recombinant human profilin I was added in three steps. After sonication (five 30-sec bursts) and two clarification steps (Ren et al., 1997), the extract was supplemented with 0.4 mM ATP, but not the 1:200 protease inhibitor cocktail or 0.5 mM PMSF. The actin that was eluted with 1 M KCl was used, after a cycle of assembly–disassembly, for experiments in this study.
Rabbit skeletal muscle actin (99% pure) was purchased from Cytoskeleton Inc. (Denver, CO) and prepared with one cycle of polymerization and depolymerization. Polymerization was induced by supplementing the manufacturer's buffer with 100 mM KCl and 5 mM MgCl2. Filamentous (F)-actin was collected by centrifugation and dialyzed against buffer G (containing 5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.01% NaN3, 0.5 mM DTT, and 0.2 mM ATP), as described for maize pollen actin (Ren et al., 1997).
Profilin concentrations were determined with the Bradford assay (Bio-Rad, Hercules, CA), with BSA as a standard. An extinction coefficient (ε280 =16,000 M−1 cm−1) for the maize profilins was determined (Gill and von Hippel, 1989) and gave calculated protein concentrations within 5% of the Bradford assay results. The concentration of pollen and rabbit skeletal muscle actin was determined from an A290 of 0.63 for a 1 mg/mL solution (Houk and Ue, 1974). Pollen actin and rabbit skeletal muscle actin at equivalent concentrations, as determined by A290, were indistinguishable when separated by SDS-PAGE and stained with Coomassie Brilliant Blue R (data not shown).
Protein Gel Blots
Antisera against recombinant ZmPRO5 (anti-ZmPRO5) were raised in a New Zealand white rabbit, as described previously (Karakesisoglou et al., 1996), by staff of the Purdue University Cancer Center Antibody Production Facility. High-titer antisera (1:1000) were found for each weekly bleed, beginning after the fourth injection. Bleed 3, at 1:1000 dilution, was used for this study.
Pollen was collected from field-grown plants. Roots were collected from 1-week-old etiolated seedlings grown at room temperature between moist paper towels. Shoot and coleoptile tissues were collected from 3-day-old etiolated seedlings grown at 30°C. Total protein extracts were prepared by freezing the tissues in liquid nitrogen and then grinding with a mortar and pestle. Tissue powders were resuspended in buffer I, supplemented with a 1:200 dilution of protease inhibitors from a stock solution, and sonicated. Frozen pollen was resuspended in buffer I directly. The extracts were clarified by centrifugation at 4°C and 2000g for 5 min, and protein concentration was determined by the Bradford assay. Extracts were boiled in protein sample buffer, separated by PAGE along with purified recombinant and native pollen maize profilins as standards, and blotted onto nitrocellulose (Schleicher and Schuell). Filters were probed with anti-ZmPRO5 or anti-ZmPRO3 antisera (Karakesisoglou et al., 1996), incubated with a horseradish peroxidase–conjugated secondary antibody (Sigma), and developed as described previously (Karakesisoglou et al., 1996).
Nuclear Displacement Assay
Tradescantia virginiana stamen hair cells, collected from freshly opened flowers, were microinjected with recombinant and native profilins, cytochalasin D (Sigma), or latrunculin B (Calbiochem, San Diego, CA), at 100-μM needle concentration as described previously (Staiger et al., 1994; Karakesisoglou et al., 1996; Gibbon et al., 1997, 1998). For each protein or inhibitor, at least 30 injections were performed, using three independent batches of each profilin isoform. BSA (Bio-Rad), at an equivalent protein concentration of 1.4 mg/mL, was injected as a control.
PLP and Monomeric-Actin Binding
The Kd value for profilin binding to PLP was determined by measuring the increase of intrinsic tryptophan fluorescence on complex formation (Perelroizen et al., 1994; Petrella et al., 1996), as described in detail previously (Gibbon et al., 1997, 1998). At least three independent batches of each profilin isoform were used to determine Kd values for PLP binding.
The ability of profilin to sequester maize pollen globular (G)-actin under polymerizing conditions was determined by monitoring a shift in the critical concentration (Cc) at steady state, as described previously (Gibbon et al., 1998), with important modifications. Errors were found in the previously reported methods used to measure the actin-sequestering ability of profilin (Gibbon et al., 1998). A correction appeared in Plant Cell (Gibbon et al., 1999a), and new methods and criteria for data analysis (see below) have been established. Actin polymerization was measured by the amount of 90° scatter of 450-nm light (Cooper and Pollard, 1982). Briefly, in a 1.8-mL reaction volume, 1 μM profilin (instead of 2 μM; Gibbon et al., 1998) was incubated with five different concentrations of actin (from 0.75 to 2.25 μM) in LSF buffer (5 mM Hepes, 0.5 mM DTT, 0.2 mM ATP, 50 μM CaCl2, and 0.1% [w/v] NaN3, pH 7.0). To keep the Ca2+ concentration (55 μM) equal, we added actin from a 50-μM stock solution in buffer G and diluted it to a constant volume of 108 μL with buffer G. Background light scattering was recorded, and polymerization was induced by the addition of 5 mM MgCl2 and 100 mM KCl. After 16 hr, the final extent of polymerization was measured by light scattering, and the Cc value for actin was determined by plotting the difference between the final and initial light scattering values for each sample versus actin concentration. Only curves that contained at least four data points in the linear range and had slopes that differed by <20% from the slope of the actin-only curve were used to determine apparent Kd values. In this report, eight of 86 profilin curves (9%) did not meet the defined criteria and were discarded. Assuming that the profilin–actin complex forms in a 1:1 molar ratio and that profilin does not facilitate subunit addition at the barbed end, the apparent Kd value is equal to the concentration of unbound profilin, multiplied by the Cc for actin polymerization in the absence of profilin, and divided by the concentration of profilin–actin complex. At least four independent batches of each recombinant profilin and three independent batches of each native profilin were assayed.
Apparent Kd values also were determined at 25 nM Ca2+ and 1 μM Ca2+ by supplementing 2 × LSF buffer with 2 mM and 168.4 μM EGTA, respectively. All Ca2+ concentrations in this report were computed with the CalCalc software program (Jan Schmid, Massey University, New Zealand, and Wilhelmus Schreurs, Colorado State University, personal communication). Experiments at 25 nM Ca2+ were performed in the presence of 2 μM profilin to increase the shift in Cc. The average Cc for pollen actin alone was 0.33 μM ± 0.05 (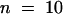 ), 0.34 ± 0.1 (
), 0.34 ± 0.1 (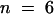 ), and 0.40 ± 0.12 (
), and 0.40 ± 0.12 (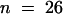 ) in the presence of 25 nM, 1.0 μM, and 55 μM Ca2+, respectively.
) in the presence of 25 nM, 1.0 μM, and 55 μM Ca2+, respectively.
The effect of a range of Ca2+ concentrations on the ability of ZmPRO1 and ZmPRO5 to sequester actin monomers at a single stoichiometry (2 μM actin to 1 μM profilin) was tested by adding increasing amounts of EGTA from a 50-mM stock in LSF buffer. The difference between the final and initial light scattering values for each sample was plotted versus Ca2+ concentration. Because Ca2+ decreases the extent of actin polymerization at steady state, the effect of varying [Ca2+] on profilin sequestering activity was expressed after the values for ZmPRO1 and ZmPRO5 were subtracted from those of actin alone. These data were plotted versus Ca2+ concentration.
Nucleotide Exchange Analysis
The rate of nucleotide exchange on pollen G-actin, in the absence or presence of 0.1 μM human profilin I, or 0.5 μM ZmPRO1, ZmPRO5, native pollen profilin, or DNase I (Sigma), was determined by measuring the increase in fluorescence on incorporation of 1,N6-ethenoadenosine 5′-triphosphate (ε-ATP; Sigma; Goldschmidt-Clermont et al., 1992). The ε-ATP (50 μM) and profilin or DNase I (in a constant volume of 115 μL) were mixed with 2 × buffer (4 mM Tris-HCl, pH 7.5, 1.0 mM DTT, 100 mM KCl, and 1.0 mM MgCl2) and brought to a final reaction volume of 1.485 mL with water. The initial fluorescence was determined in a spectrofluorimeter with excitation at 360 nm and emission at 410 nm. The reaction was initiated by adding 0.5 μM pollen G-actin, from 50 μM stock reagent in buffer G, and monitored for 300 sec. The final concentration of free Ca2+ under these conditions was 2.0 μM. The rate of incorporation (Δfluorescence [arbitrary units]/sec) was determined by fitting the data for the first 180 sec with a single exponential function. At least two batches of each profilin and two batches of actin were used for these analyses.
Inhibition of Phospholipase C Activity
The inhibition of bean (Vicia faba) plasma membrane phosphoinositide phospholipase C activity by the maize profilins was measured as described previously (Drøbak et al., 1994). Briefly, phosphoinositidase activity was assayed by incubating bean plasma membranes at 25°C in 50 μL of buffer E (50 mM Tris/malate, pH 6.0, and 10 μM CaCl2) with 50 μM phosphatidylinositol-4,5-bisphosphate (PtdIns[4,5]P2) and 0.86 kBq 3H-PtdIns(4,5)P2, in the presence of 0.0, 1.4, 2.8, 5.7, or 14.2 μM recombinant ZmPRO1, ZmPRO5, or human profilin I. Reactions were stopped by the addition of 1 mL of chloroform/methanol (2:1 [v/v]). After a 5-min incubation on ice and the addition of 250 μL of 0.6 M HCl, the samples were vortex-mixed and centrifuged at 14,000g for 2 min. Four hundred microliters of the top phase was removed, and its radioactivity was determined by liquid scintillation spectrometry (model 1410; LKB-Wellac, Turku, Finland) after addition of scintillation fluid (Hionic-Fluor; Hewlett-Packard, UK).
Acknowledgments
We thank Peter Watkins (John Innes Centre) for preparation of bean plasma membranes, Milos Tanurdzic for preparing first-strand cDNA, and Dan Szymanski, David Asai, Joann Otto, David Wilkes, and Bryan Gibbon for helpful comments and discussions. This research was funded by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the Purdue Research Foundation to C.J.S.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M.K., Hirani, B.R., Burke, J.D., and Gould, K.L. (1994). The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 125, 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballweber, E., Giehl, K., Hannappel, E., Huff, T., Jockusch, B.M., and Mannherz, H.G. (1998). Plant profilin induces actin polymerization from actin:β-thymosin complexes and competes directly with β-thymosins and with negative co-operativity with DNaseI for binding to actin. FEBS Lett. 425, 251–255. [DOI] [PubMed] [Google Scholar]
- Björkegren-Sjögren, C., Korenbaum, E., Nordberg, P., Lindberg, U., and Karlsson, R. (1997). Isolation and characterization of two mutants of human profilin I that do not bind poly(l-proline). FEBS Lett. 418, 258–264. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., and Pollard, T.D. (1998). Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 273, 25106–25111. [DOI] [PubMed] [Google Scholar]
- Bubb, M.R., Baines, I.C., and Korn, E.D. (1998). Localization of actobindin, profilin I, profilin II, and phosphatidylinositol-4,5-bisphosphate (PIP2) in Acanthamoeba castellanii. Cell Motil. Cytoskeleton 39, 134–146. [DOI] [PubMed] [Google Scholar]
- Carlier, M.-F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G.-X., Hong, Y., Chua, N.-H., and Pantaloni, D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 136, 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, H.E.M., Ramachandran, S., Tan, C.T., Surana, U., Dong, C.H., and Chua, N.-H. (1996). Arabidopsis profilins are functionally similar to yeast profilins: Identification of a vascular bundle–specific profilin and a pollen-specific profilin. Plant J. 10, 269–279. [DOI] [PubMed] [Google Scholar]
- Clarke, S.R., Staiger, C.J., Gibbon, B.C., and Franklin-Tong, V.E. (1998). A potential signaling role for profilin in pollen of Papaver rhoeas. Plant Cell 10, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, L., Verheyen, E., and Ayers, K. (1992). chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69, 173–184. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A. (1987). Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J.A., and Pollard, T.D. (1982). Methods to measure actin polymerization. Methods Enzymol. 85, 182–210. [DOI] [PubMed] [Google Scholar]
- de Ruijter, N.C.A., and Emons, A.M.C. (1999). Actin-binding proteins in plant cells. Plant Biol. 1, 26–35. [Google Scholar]
- Devereux, J., Haeberli, P., and Smithies, O. (1984). A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didry, D., Carlier, M.-F., and Pantaloni, D. (1998). Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273, 25602–25611. [DOI] [PubMed] [Google Scholar]
- Domke, T., Federau, T., Schlüter, K., Giehl, K., Valenta, R., Schomburg, D., and Jockush, B.M. (1997). Birch pollen profilin: Structural organization and interaction with poly-(l-proline) peptides as revealed by NMR. FEBS Lett. 411, 291–295. [DOI] [PubMed] [Google Scholar]
- Drøbak, B.K., Watkins, P.A.C., Valenta, R., Dove, S.K., Lloyd, C.W., and Staiger, C.J. (1994). Inhibition of plant plasma membrane phosphoinositide phospholipase C by the actin-binding protein, profilin. Plant J. 6, 389–400. [Google Scholar]
- Eads, J.C., Mahoney, N.M., Vorobiev, S., Bresnick, A.R., Wen, K.K., Rubenstein, P.A., Haarer, B.K., and Almo, S.C. (1998). Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry 37, 11171–11181. [DOI] [PubMed] [Google Scholar]
- Fedorov, A.A., Ball, T., Mahoney, N.M., Valenta, R., and Almo, S.C. (1997). The molecular basis for allergen cross-reactivity: Crystal structure and IgE-epitope mapping of birch pollen profilin. Structure 5, 33–45. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong, V.E., Ride, J.P., Read, N.D., Trewavas, A.J., and Franklin, F.C.H. (1993). The self-incompatibility response in Papaver rhoeas is mediated by cytosolic free calcium. Plant J. 4, 163–177. [Google Scholar]
- Franklin-Tong, V.E., Hackett, G., and Hepler, P.K. (1997). Ratio-imaging of Ca2+i in the self-incompatibility response in pollen tubes of Papaver rhoeas. Plant J. 12, 1375–1386. [Google Scholar]
- Frazier, J.A., and Field, C.M. (1997). Actin cytoskeleton: Are FH proteins local organizers? Curr. Biol. 7, R414–R417. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E.A., Fyrberg, C.C., Biggs, J.R., Saville, D., Beall, C.J., and Ketchum, A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36, 271–287. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C., Ren, H., and Staiger, C.J. (1997). Characterization of maize (Zea mays) pollen profilin function in vitro and in live cells. Biochem. J. 327, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon, B.C., Zonia, L.E., Kovar, D.R., Hussey, P.J., and Staiger, C.J. (1998). Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell 10, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon, B.C., Zonia, L.E., Kovar, D.R., Hussey, P.J., and Staiger, C.J. (1999. a). Correction: Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell 11, 1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999. b). Latrunculin B has different effects on maize pollen germination and tube growth. Plant Cell 11, 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl, K., Valenta, R., Rothkegel, M., Ronsiek, M., Mannherz, H.-G., and Jockusch, B.M. (1994). Interaction of plant profilin with mammalian actin. Eur. J. Biochem. 226, 681–689. [DOI] [PubMed] [Google Scholar]
- Gieselmann, R., Kwiatkowski, D.J., Janmey, P.A., and Witke, W. (1995). Distinct biochemical characteristics of two human profilin isoforms. Eur. J. Biochem. 229, 621–628. [DOI] [PubMed] [Google Scholar]
- Gill, S.C., and von Hippel, P.H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, P.J., Furman, M.I., Wachsstock, D., Safer, D., Nachmias, V.T., and Pollard, T.D. (1992). The control of actin nucleotide exchange by thymosin β4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell 3, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote, M., Swoboda, I., Meagher, R.B., and Valenta, R. (1995). Localization of profilin- and actin-like immunoreactivity in in vitro–germinated tobacco pollen tubes by electron microscopy after special water-free fixation techniques. Sex. Plant Reprod. 8, 180–186. [Google Scholar]
- Haarer, B.K., Petzold, A.S., and Brown, S.S. (1993). Mutational analysis of yeast profilin. Mol. Cell. Biol. 13, 7864–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hájková, L., Sjögren, C.B., Korenbaum, E., Nordberg, P., and Karlsson, R. (1997). Characterization of a mutant profilin with reduced actin-binding capacity: Effects in vitro and in vivo. Exp. Cell Res. 234, 66–77. [DOI] [PubMed] [Google Scholar]
- Haugwitz, M., Noegel, A.A., Karakesisoglou, J., and Schleicher, M. (1994). Dictyostelium amoeba that lack G-actin–sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell 79, 303–314. [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke, T.L., Feijo, J.A., Hackett, G.R., Kunkel, J.G., and Hepler, P.K. (1997). Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9, 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk, T.W., Jr., and Ue, K. (1974). The measurement of actin concentration in solution: A comparison of methods. Anal. Biochem. 62, 66–74. [DOI] [PubMed] [Google Scholar]
- Huang, S.R., McDowell, J.M., Weise, M.J., and Meagher, R.B. (1996). The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 111, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere, V., Lambrechts, A., Vandekerckhove, J., and Ampe, C. (1999). Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin β4. FEBS Lett. 447, 257–263. [DOI] [PubMed] [Google Scholar]
- Kaiser, D.A., Sato, M., Ebert, R.F., and Pollard, T.D. (1986). Purification and characterization of two isoforms of Acanthamoeba profilin. J. Cell Biol. 102, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, F., Purich, D.L., and Southwick, F.S. (1999). Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 274, 36963–36972. [DOI] [PubMed] [Google Scholar]
- Karakesisoglou, I., Schleicher, M., Gibbon, B.C., and Staiger, C.J. (1996). Plant profilins rescue the aberrant phenotype of profilin-deficient Dictyostelium cells. Cell Motil. Cytoskeleton 34, 36–47. [DOI] [PubMed] [Google Scholar]
- Kohno, T., and Shimmen, T. (1987). Ca-induced fragmentation of actin filaments in pollen tubes. Protoplasma 141, 177–179. [Google Scholar]
- Kohno, T., and Shimmen, T. (1988). Mechanism of Ca2+ inhibition of cytoplasmic streaming in lily pollen tubes. J. Cell Sci. 91, 501–509. [Google Scholar]
- Korenbaum, E., Nordberg, P., Björkegren-Sjögren, C., Schutt, C.E., Lindberg, U., and Karlsson, R. (1998). The role of profilin in actin polymerization and nucleotide exchange. Biochemistry 37, 9274–9283. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Lambrechts, A., Vandamme, J., Goethals, M., Vandekerckhove, J., and Ampe, C. (1995). Purification and characterization of bovine profilin II. Actin, poly(l-proline) and inositolphospholipid binding. Eur. J. Biochem. 230, 281–286. [PubMed] [Google Scholar]
- Lambrechts, A., Verschelde, J.L., Jonckheere, V., Goethals, M., Vandekerckhove, J., and Ampe, C. (1997). The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 16, 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., Goldschmidt-Clermont, P.J., and Pollard, T.D. (1990). The affinities of human platelet and Acanthamoeba profilin isoforms for polyphosphoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., Atkinson, S.J., Ampe, C., Vandekerckhove, J., and Pollard, T.D. (1994). Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen, V., Drubin, D.G., Mages, G., and Bandlow, W. (1993). High levels of profilin suppress the lethality caused by overproduction of actin in yeast cells. FEBS Lett. 316, 41–47. [DOI] [PubMed] [Google Scholar]
- Meagher, R.B. (1991). Divergence and differential expression of actin gene families in higher plants. Int. Rev. Cytol. 125, 139–163. [DOI] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasamy, M.K. (1999). Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 11, 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli, M., and Robinson, K.R. (1997). Tip localized Ca2+ pulses are coincident with peak pulsatile growth rates in pollen tubes of Lilium longiflorum. J. Cell Sci. 110, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Miller, D.D., Callaham, D.A., Gross, D.J., and Hepler, P.K. (1992). Free Ca2+ gradient in growing pollen tubes of Lilium. J. Cell Sci. 101, 7–12. [Google Scholar]
- Miller, D.D., Lancelle, S.A., and Hepler, P.K. (1996). Actin microfilaments do not form a dense meshwork in Lilium longiflorum pollen tube tips. Protoplasma 195, 123–132. [Google Scholar]
- Mounier, N., Perriard, J.-C., Gabbiani, G., and Chaponnier, C. (1997). Transfected muscle and nonmuscle actins differentially sorted by cultured smooth muscle and nonmuscle cells. J. Cell Sci. 110, 839–846. [DOI] [PubMed] [Google Scholar]
- Nick, P. (1999). Signals, motors, morphogenesis: The cytoskeleton in plant development. Plant Biol. 1, 169–179. [Google Scholar]
- Ostrander, D.B., Ernst, E.G., Lavoie, T.B., and Gorman, J.A. (1999). Polyproline binding is an essential function of human profilin in yeast. Eur. J. Biochem. 262, 26–35. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., and Carlier, M.-F. (1993). How profilin promotes actin filament assembly in the presence of thymosin β4. Cell 75, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Perelroizen, I., Marchand, J.-B., Blanchoin, L., Didry, D., and Carlier, M.-F. (1994). Interaction of profilin with G-actin and poly(l-proline). Biochemistry 33, 8472–8478. [DOI] [PubMed] [Google Scholar]
- Perelroizen, I., Didry, D., Christensen, H., Chua, N.-H., and Carlier, M.-F. (1996). Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 271, 12302–12309. [DOI] [PubMed] [Google Scholar]
- Petrella, E.C., Machesky, L.M., Kaiser, D.A., and Pollard, T.D. (1996). Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35, 16535–16543. [DOI] [PubMed] [Google Scholar]
- Rathore, K.S., Cork, R.J., and Robinson, K.R. (1991). A cytoplasmic gradient of Ca2+ is correlated with the growth of lily pollen tubes. Dev. Biol. 148, 612–619. [DOI] [PubMed] [Google Scholar]
- Reinhard, M., Giehl, K., Abel, K., Haffner, C., Jarchau, T., Hoppe, V., Jockusch, B.M., and Walter, U. (1995). The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 14, 1583–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H., Gibbon, B.C., Ashworth, S.L., Sherman, D.M., Yuan, M., and Staiger, C.J. (1997). Actin purified from maize pollen functions in living plant cells. Plant Cell 9, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter, K., Jockusch, B.M., and Rothkegel, M. (1997). Profilins as regulators of actin dynamics. Biochim. Biophys. Acta 1359, 97–109. [DOI] [PubMed] [Google Scholar]
- Schlüter, K., Schleicher, M., and Jockusch, B.M. (1998). Effects of single amino acid substitutions in the actin-binding site on the biological activity of bovine profilin I. J. Cell Sci. 111, 3261–3273. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., and Hall, M.N. (1998). Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14, 305–338. [DOI] [PubMed] [Google Scholar]
- Schobert, D., Gottschalk, M., Kovar, D.R., Staiger, C.J., Yoo, B.-C., and Lucas, W.J. (2000). Characterization of Ricinus communis phloem profilin, RcPRO1. Plant Mol. Biol., in press. [DOI] [PubMed]
- Sheterline, P., Clayton, J., and Sparrow, J.C. (1998). Actin. Protein Profile 4, 1–272. [PubMed] [Google Scholar]
- Sohn, R.H., Chen, J., Koblan, K.S., Bray, P.F., and Goldschmidt-Clermont, P.J. (1995). Localization of a binding site for phosphatidylinositol-4,5-bisphosphate on human profilin. J. Biol. Chem. 270, 21114–21120. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J. (2000). Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 257–288. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Goodbody, K.C., Hussey, P.J., Valenta, R., Drøbak, B.K., and Lloyd, C.W. (1993). The profilin multigene family of maize: Differential expression of three isoforms. Plant J. 4, 631–641. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Yuan, M., Valenta, R., Shaw, P.J., Warn, R., and Lloyd, C.W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4, 215–219. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Gibbon, B.C., Kovar, D.R., and Zonia, L.E. (1997). Profilin and actin depolymerizing factor: Modulators of actin organization in plants. Trends Plant Sci. 2, 275–281. [Google Scholar]
- Suetsugu, S., Miki, H., and Takenawa, T. (1998). The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 17, 6516–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L.P., and Hepler, P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491. [DOI] [PubMed] [Google Scholar]
- Thorn, K.S., Christensen, H.E.M., Shigeta, R., Huddler, D., Shalaby, L., Lindberg, U., Chua, N.H., and Schutt, C.E. (1997). The crystal structure of a major allergen from plants. Structure 5, 19–32. [DOI] [PubMed] [Google Scholar]
- Valenta, R., Duchêne, M., Pettenburger, K., Sillaber, C., Valent, P., Bettelheim, P., Breitenbach, M., Rumpold, H., Kraft, D., and Scheiner, O. (1991). Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 253, 557–560. [DOI] [PubMed] [Google Scholar]
- Vidali, L., and Hepler, P.K. (1997). Characterization and localization of profilin in pollen grains and tubes of Lilium longiflorum. Cell Motil. Cytoskeleton 36, 323–338. [DOI] [PubMed] [Google Scholar]
- Vinson, V.K., Delacruz, E.M., Higgs, H.N., and Pollard, T.D. (1998). Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880. [DOI] [PubMed] [Google Scholar]
- von Arx, P., Bantle, S., Soldati, T., and Perriard, J. (1995). Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J. Cell Biol. 131, 1759–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke, W., Podtelejnikov, A.V., Di Nardo, A., Sutherland, J.D., Gurniak, C.B., Dotti, C., and Mann, M. (1998). In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 17, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]