Abstract
p63 is a transcription factor structurally related to the p53 tumor suppressor. The C-terminal region differs from p53's in that it contains a sterile alpha motif (SAM) domain and is subject to multiple alternative splicings. The N-terminal region is present in the transactivation (TA) and ΔN configurations, with the latter lacking the transcriptional activation domain 1. Single amino acid substitutions and frameshift mutations of p63 cause the human ankyloblepharon ectodermal dysplasia clefting (AEC) or ectrodactyly ectodermal dysplasia and facial clefting (EEC) syndromes. We have systematically compared the activities of the wild-type p63 isoforms and of the natural mutants in activation and repression assays on three promoters modulated by p53. We found that p63 proteins with an altered SAM domain or no SAM domain—the β isoforms, the EEC frameshift mutant, and the missense AEC mutations—all showed a distinctly higher level of activation of the MDM2 promoter and decreased repression on the HSP70 promoter. Fusion of SAM to the GAL4 DNA-binding domain repressed a heterologous promoter. A second activation domain, TA2, corresponding to exons 11 to 12, was uncovered by comparing the activation of ΔN isoforms on natural promoters and in GAL4 fusion systems. In colony formation assays, the AEC mutants, but not the EEC frameshift, were consistently less efficient in suppressing growth, in both the TA version and the ΔN version, with respect to their p63α counterparts. These data highlight the modularity of p63, identifying the SAM domain as a dominant transcriptional repression module and indicating that the AEC and EEC frameshift mutants are characterized by a subversion of the p63 transcriptional potential.
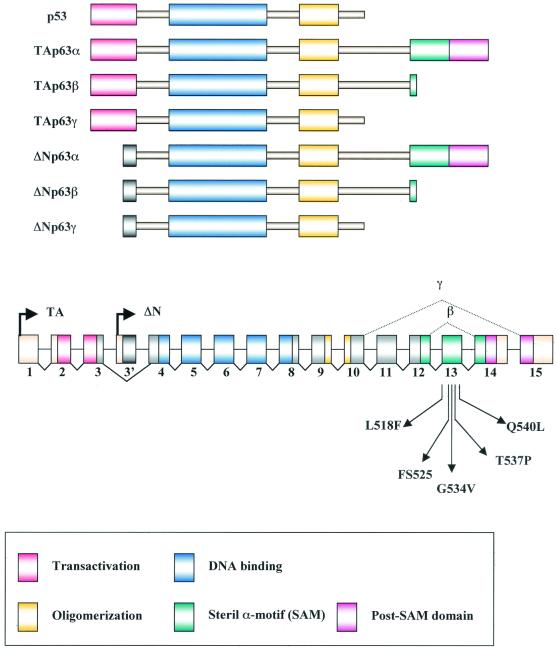
p53 is a sequence-specific transcription factor that plays a critical role in activating the expression of genes involved in cell cycle arrest and apoptosis. Under conditions of genotoxic stress, p53 stabilization leads to transcriptional activation and repression of different sets of genes (7). The MDM2, p21, and GADD45 genes are among those that are activated by p53 by direct binding to specific sites in their regulatory regions (32). Mutations in the p53 tumor suppressor gene are the most frequent somatic alterations found in human cancers, and hot spots of mutations are found within the DNA-binding subdomain (34, 47). Many genes are down-regulated at the transcriptional level by p53 with mechanisms that are less clearly defined, possibly involving direct interactions with components of the basal transcriptional apparatus (15, 22). More recently, two genes were uncovered, referred to as the p63 and p73 genes, encoding proteins that share significant amino acid identity with p53 in the transactivation, DNA-binding, and oligomerization domains (19, 20, 28, 37, 49). p63 and p73 are more similar to each other than to p53, and both can activate p53-responsive promoters (18, 26, 31, 48). An extended C-terminal coding region, not found in p53, is present in p63 and p73 and undergoes complex alternative splicing (21). Within this C-terminal extension, there is a sterile alpha motif (SAM) that is found in proteins that regulate mammalian development and is thought to be involved in protein-protein interactions (4, 44). In contrast to p53, p73 and p63 are rarely mutated in human cancers (34). Another notable difference is that p63 and p73 are present as two isoforms with respect to their N termini, with widely divergent biological properties: the transactivating (TA) isoforms, which resemble p53, are generated by the use of an upstream promoter; the ΔN isoforms, produced from an intronic promoter, contain the same DNA-binding and oligomerization domains as the TA but lack the transactivation domain (see Fig. 1). Both the TA and ΔN isoforms have three possible carboxyl termini, termed α, β, and γ. The β and γ isoforms lack the SAM domain.
FIG. 1.
Schematic representation of p63 structure. Intron-exon structure of the p53 and p63 genes, showing the p63 transcriptional start sites and the alternative splicing at the 3′ end that gives rise to the α, β, and γ isoforms, is shown. The change in the reading frame within exons 14 and 15 occurring in the β and γ isoforms is indicated. The positions of the EEC and AEC SAM domain mutations are indicated. All mutations fall within exon 13 and are predicted to affect only the α isoforms.
Inactivations of genes for p53-like proteins in mice have very different outcomes, implying that they play distinct roles in vivo. In fact, even though p53 modulates critical cellular processes and is ubiquitously expressed, p53 knockout mice are developmentally normal and show increased susceptibility to spontaneous tumorigenesis (13). Mice deficient for p73 exhibit profound defects, including hippocampal dysgenesis, hydrocephalus, chronic infections, and inflammations, as well as abnormalities in pheromone sensory pathways (50). Mice lacking p63 are born alive but die soon after birth, with several developmental defects, particularly in limb formation and in the skin compartment (35, 51). Important clues as to the physiological role of p63 came from the analysis of the molecular defects of patients with distinct autosomal dominant syndromes, caused by alteration of the p63 gene. (i) In ectrodactyly, ectodermal dysplasia and cleft lip/palate (EEC) syndrome, the phenotypes are similar to some of the features detected in p63-deficient mice (8). All but one of the almost 50 p63 mutations so far identified in EEC patients create amino acid substitutions that are predicted to abolish the DNA-binding capacity of p63. The remaining mutation introduces a frameshift within exon 13 of the p63 gene that affects p63α but not the p63β or γ isotype (46). (ii) Approximately 10% of the patients with isolated split hand-split foot malformation (SHFM) have p63 mutations, these being either amino acid substitutions in the DNA-binding domain or stop mutations at the C-terminal end of p63α (17, 46). (iii) For the Hay-Wells syndrome, also known as ankyloblepharon-ectodermal dysplasia-clefting (AEC), whose phenotype is similar to but clearly distinct from that of the EEC patients, all mutations identified so far are single amino acid substitutions encoded within exon 13 of the p63 gene (33).
A number of studies have so far addressed the activation and growth suppression features of some of p63 isoforms (48). However, a comprehensive comparison of all known p63 isoforms on a natural rather than artificial construct is lacking. Thus, to better comprehend the biological complexity of p63, we undertook a systematic analysis of the transcriptional and growth-regulating activities of six splicing variants of p63 (TAp63α, -β, and -γ and ΔNp63α, -β, and -γ), of the exon 13 frameshift mutation isolated from an EEC patient, and of four missense mutations causing the AEC syndrome.
MATERIALS AND METHODS
Cell culture and transfections.
The Saos-2 cell line was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. For transfection, 80,000 cells were seeded into 24-well multiplates and on the following day were transfected with Lipofectamine 2000 (Gibco) under the conditions suggested by the manufacturer. The total amount of transfected DNA (1 μg) was kept constant using empty vector DNA when necessary. Thirty-six hours later, cells were collected for chloramphenicol acetyltransferase (CAT) assays (16). For RNA extraction, 800,000 Saos-2 cells were seeded into 60-mm-diameter dishes and, the day after, transfected with Lipofectamine 2000 with 10 μg of DNA (Gibco).
RNA preparation and analysis.
Total RNAs were extracted from Saos-2 cells by the Trizol reagent (Gibco) according to the instructions of the manufacturer. Fifteen micrograms of each sample was loaded on a 1% formaldehyde-containing agarose gel and transferred to a nitrocellulose membrane. The following fragments were used as probes: a 1.9-kb EcoRI fragment from the human MDM2 gene (5) and the rat glyceraldehyde-3-phosphate dehydrogenase fragment, used for normalization of RNA loading (40). Fragments were labeled with [α-32P]dCTP by the Random Primed DNA labeling kit (Boehringer).
Colony-forming efficiency assay.
Saos-2 cells were transfected with p63-containing plasmids, all carrying a neo-resistance gene. After 2 weeks of selection, the G418-resistant colonies were fixed with methanol-acetic acid (2:10 [vol/vol]) and visualized by staining with 2% (wt/vol) crystal violet for easy visualization.
Plasmids.
The p21/WAFCAT contains the 2.4-kb fragment from the p21/WAF promoter (6). The Bp100 CAT reporter contains two copies of the p53RE motif derived from the MDM2 intronic promoter (5). The Hsp70 CAT (23) and all p63 isoforms (wild type [wt] and mutants) were described previously (8, 33). The reporter GAL4 vectors, G5XΔN-CAT and G5-PB11CAT, were provided by B. Majello (29, 30).
GAL4 fusion proteins.
For construction of GAL4 fusion proteins, different oligonucleotide pairs were synthesized in order to specifically amplify exons 11, 12, and 14 from the wt TAα p63 cDNA and exon 13 from wt and AEC-mutated TAα p63 cDNAs. The sequences of the oligonucleotides were as follows: exon 11, 11 Forward (GGGGATCGATTCAGACCTCAATACAG 11) and 11 Reverse (CCCCCTCGAGTCAAATGTTGGCTCCCAT); exon 12, 12 Forward (CCCCATCGATTCCCATGATGGGCACC) and 12 Reverse (CCCCCTCGAGCTAGACAATGCTGCAATC); exon 13, 13 Forward (GGGGATCGATTAGTTTCTTAGCGAGG) and 13 Reverse (CCCCCTCGAGTCAATCCATGGAGTAATG); exon 14, 14 Forward (GGGGATCGATTGATCTGGCAAGTCTG) and 14 Reverse (CCCCCTCGAGTCACTCCCCCTCCTCTTT).
All oligonucleotides contained adequate restriction sites (underlined) to allow further cloning steps. The amplification products were purified after digestion with restriction enzymes and ligated into the pGAL4poly plasmid in frame with the GAL4 DNA-binding domain (27). All clones were authenticated by DNA sequencing.
Western immunoblot analysis.
Thirty-six hours after transfection, cells were lysed in loading buffer (2% sodium dodecyl sulfate, 30% glycerol, 300 mM β-mercaptoethanol, 100 mM Tris-HCl [pH 6.8]), and different volumes of total extracts were separated on SDS-8% polyacrylamide gels and transferred to a nitrocellulose membrane (Schleicher & Schuell). The blots were incubated with the following antibodies: p53 (FL 393, no. sc6243) c-myc (A14, no. sc789), and GAL4 (RK5C1, no. sc510) (Santa Cruz Biotechnology) and developed according to the manufacturer's instructions (Super Signal; Pierce).
RESULTS
Activation and inhibition of promoters by p63 isoforms.
p63 is present in multiple isoforms, resulting in part from differential splicing and in part from the use of two promoters that generate variations at the N-terminal end of the protein. Figure 1 shows a schematic representation of p63 complexity, as well as the positions of the mutations identified in EEC and AEC patients that were used in this study. Bona fide natural p63 gene targets have not been identified so far. To test the transcriptional activities of the different p63 isoforms, we used two target promoters that are activated (MDM2 and p21) and one promoter that is repressed (Hsp70) by p53. The Bp100-CAT and p21/WAFCAT plasmids contain the intronic MDM2 promoter and the upstream control sequences and promoter of p21, respectively, both harboring two DNA binding sites for p53 (5). The Hsp70-CAT plasmid contains a promoter with no known p53 binding sites that is efficiently down-regulated by p53 (1). In these experiments, we used the human osteosarcoma cell line Saos-2, which expresses very low levels of p73 and no p53 or p63 (6).
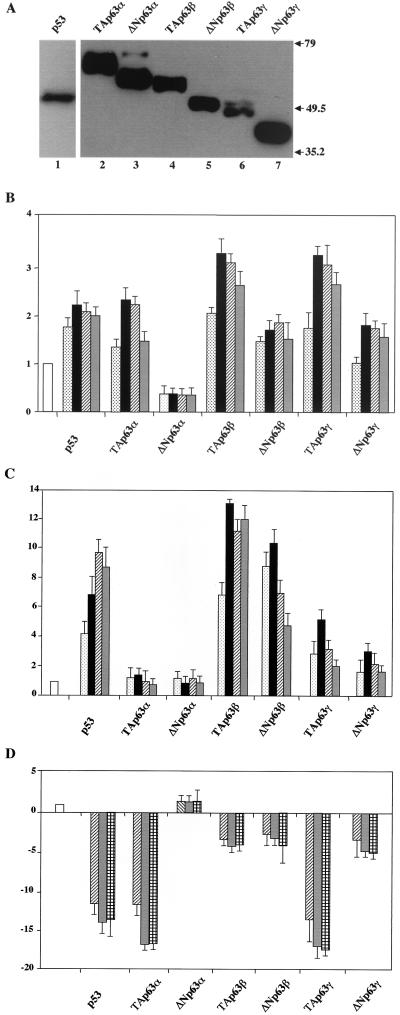
Initially, we verified the expression levels of the transfected p63 isoforms. Figure 2A shows that all proteins were expressed from the pcDNA vectors and their abundance varies to some extent, with the α isoforms being the more abundant and the γ being the less abundant, in agreement with published results (5); for this reason, different amounts of total cell lysates were used: 25 μl for TAp63β and TAp63γ (Fig. 2A, lanes 2 and 6) and 10 μl for other p63 isoforms and p53. To take on these differences, which could possibly alter transactivation potentials, we systematically transfected different doses of plasmids in the subsequent experiments. Results in Fig. 2B show the transcriptional activities of dose responses of all p63 isoforms on the p21 promoter: as expected, p53, as well as TAp63α and TAp63γ, activate transcription, albeit modestly, whereas ΔNp63α does not. ΤAp63β and, surprisingly, ΔNp63γ and ΔNp63β also activate p21 over a wide range of transfected DNA. A different activation profile emerges from the analysis of the MDM2 reporter (Fig. 2C). Both the TAp63α and ΔNp63α isoforms were inefficient as transcriptional activators. The TAp63γ and ΔNp63γ isoforms activated transcription modestly, fourfold on average (Fig. 2C). On the other hand, the TAp63β and ΔNp63β isoforms activated transcription very efficiently, 10- to 15-fold, even at low levels of transfected plasmids, with TAp63β being a stronger activator than p53. This is remarkable, since the protein levels of the β isoforms are consistently lower by a factor of three than those of the α isoforms (Fig. 2A); thus, we are possibly underestimating the TA and ΔNp63β activation potential. Taken together, these results are surprising, since (i) the robust activation obtained on the MDM2 promoter with ΔNp63β is effective despite the lack of the canonical TA activation domain, suggesting that an additional transcriptional activation function(s) is contained within p63, and (ii) the lack of activation of the TAp63α isoform is most likely due to sequences at the C-terminal end, possibly suggesting that a repressive domain is present in exons 13 to 14, which are absent in the β isoform.
FIG. 2.
Transactivation potential of p63 isoforms. (A) The transfected proteins are correctly expressed. Saos-2 cells were transiently transfected with 0.5 μg of the indicated plasmids and lysed in 100 μl of lysis buffer 36 h after transfection. Different volumes of total cell extracts were immunoblotted with anti-myc antibodies: p53 (10 μl) (lane 1), TAp63α (10 μl) (lane 2), ΔNp63α (10 μl) (lane 3), TAp63β (25 μl) (lane 4), ΔNp63β (10 μl) (lane 5), TAp63γ (25 μl) (lane 6), and ΔNp63γ (10 μl) (lane 7). (B) Saos-2 cells were transfected with the p21/WAFCAT reporter plasmid (0.5 μg) (open bar). Different amounts of expression plasmids for p53 and p63 isoforms were cotransfected (5, 25, 100, and 300 ng; dotted, black, dashed, and gray bars, respectively). After 36 h, cells were harvested and CAT activity was determined. (C) Saos-2 cells were transfected with the pBP100 reporter plasmid (0.5 μg) (open bar) and with expression plasmids as in panel B. (D) Saos-2 cells were transfected with the Hsp70 reporter plasmid (0.25 μg) (open bar) and with expression plasmids (100, 300, and 500 ng; dashed, gray and squared bars, respectively). The basal activity of the reporters was set to 1. Data are presented as fold activation or repression relative to the sample without effector. Each histogram bar represents the mean of three independent transfection duplicates. Standard deviations are indicated.
Several promoters were shown to be inhibited by p53 (15). Repression assays were performed with p63α and γ isoforms but not with p63β (36). We wished to verify whether p63 isoforms have repression capacity by cotransfecting increasing amounts of p63 plasmids with the Hsp70 promoter, a p53 target which possesses a high level of intrinsic activity even without activation by heat shock. Indeed, promoter activity is very efficiently repressed by p53 (10- to 15-fold; see Fig. 2D), a result entirely consistent with previous reports (1). TAp63α and TAp63γ potently inhibited transcription, whereas the corresponding ΔN isoforms showed a clear reduction of the inhibition capacity. On the other hand, both the TA and ΔNp63β isoforms modestly inhibited Hsp70 promoter function. These data suggest that two domains influence the repressing activity: the TA domain, whose presence in the α and γ isoforms guarantees stronger repression, and the C terminus of the protein, the absence of which decreases repression.
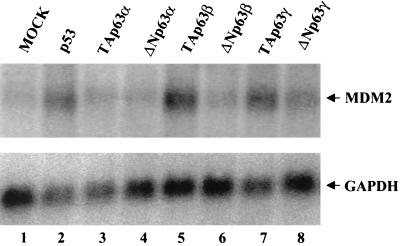
No correlation has been shown so far between transient coexpression with transfected p63 isoforms and the actual modulation of endogenous genes. For this reason, we felt it was important to validate the results obtained with the MDM2-CAT reporter constructs by checking the expression of the endogenous MDM2 gene upon cotransfection of the different p63 isoforms described above. Thirty-six hours after the addition of p63 cDNAs, total RNA was extracted and used for Northern blot analysis. As shown in Fig. 3, we observed a fivefold increase with p53 and an even higher induction with the TAp63β and TAp63γ isoforms (Fig. 3A, compare lane 1 with lanes 2, 5, and 7). The TA and ΔN p63α isoforms were completely ineffective (Fig. 3A, lanes 3 and 4), while the ΔNβ and ΔNγ isoforms were very modestly active (Fig. 3A, lanes 6 and 8). These results are in line with those previously reported for p53 (52) and largely confirmatory of the data obtained with the reporter assays, with the exception of ΔNp63β, which is much more efficient in the latter experimental setup.
FIG. 3.
The endogenous MDM2 mRNA is induced upon p63 transfection. Northern analysis of total RNA extracted from Saos-2 cells transiently transfected with the indicated plasmids is shown. MDM2 RNA was barely detectable in mock-transfected cells (lane 1). As expected, p53 transfection induced MDM2 RNA expression (lane 2); an even stronger induction was observed in the TAβ transfectants (lane 5); transfection of the TAγ isoform resulted in a good induction of MDM2 RNA (lane 7). Transfection of the TAp63α, ΔNp63α, and ΔNp63β isoforms did not induce MDM2 RNA expression (lanes 3, 4, and 6). The same filter was hybridized to a rat glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe for normalization of RNA loading.
Dissection of C-terminal repression and activation subdomains.
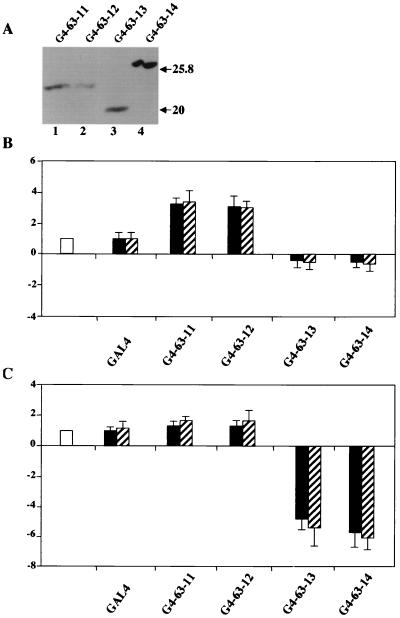
To dissect the function(s) of the p63 C-terminal region, we used the GAL4 recruitment assay with Saos-2 cells. We generated constructs containing exon 11, 12, 13, or 14 fused to the yeast GAL4 DNA-binding domain. The activities of the resulting constructs, termed hereafter G4-63-11, G4-63-12, G4-63-13, and G4-63-14, were checked with two reporter plasmids: the first, the G5XΔN construct, which contains five GAL4 binding sites fused to the human myc promoter sequences spanning from −93 to +54 of the P2 transcription start site, has low intrinsic activity and was used to study activation (30). The second one, the G5-PB11 plasmid, a construct that harbors a minimal simian virus 40 promoter with 6 Sp1 binding site and a TATA box, driving high basal activity, was used to score for repression (29). Initially, we verified that the proteins were correctly expressed upon transfection into Saos-2 cells (Fig. 4A). As shown in Fig. 4B, the G5XΔN was activated three- to fivefold by both the G4-63-11 and the G4-63-12 fusion proteins. A construct in which both exons 11 and 12 were fused to GAL4 DNA-binding domain did not increase transcription further (data not shown). The G4-63-13 and G4-63-14 proteins had a slight inhibitory effect on the very low level of transcription from this reporter. On the other hand, when the latter were used with the G5-PB11 reporter plasmid, which has high intrinsic activity, they repressed transcription four- to fivefold (Fig. 4C). These data uncover two new functions located at the C terminus of p63: (i) an activation domain, termed hereafter TA2, encoded within exons 11 and 12 that is most likely responsible for the transcriptional activation observed with the ΔN isoforms in transient reporter assays; and (ii) a repressive domain, encoded by sequences within exons 13 and 14.
FIG. 4.
Transcriptional activation and repression by Gal4-p63 exons. (A) Saos-2 cells were transiently transfected with 0.5 μg of the indicated plasmids and lysed in 100 μl of lysis buffer 36 h after transfection. Ten microliters of total cell extracts were immunoblotted with anti-GAL4 antibodies. (B) G5XΔN reporter (0.5 μg) (open bar) was cotransfected into Saos-2 cells together with indicated GAL4 vectors (black and dashed bars; 0.3 and 0.5 μg, respectively). (C) PB11 reporter (0.5 μg) (open bar) was cotransfected into Saos-2 cells together with indicated GAL4 vectors (black and dashed bars; 0.3 and 0.5 μg, respectively). Data are presented as fold activation or repression relative to the sample without effector. Each histogram bar represents the mean of three independent transfection duplicates. Standard deviations are indicated.
Analysis of transcriptional activity of the EEC and AEC p63 mutants.
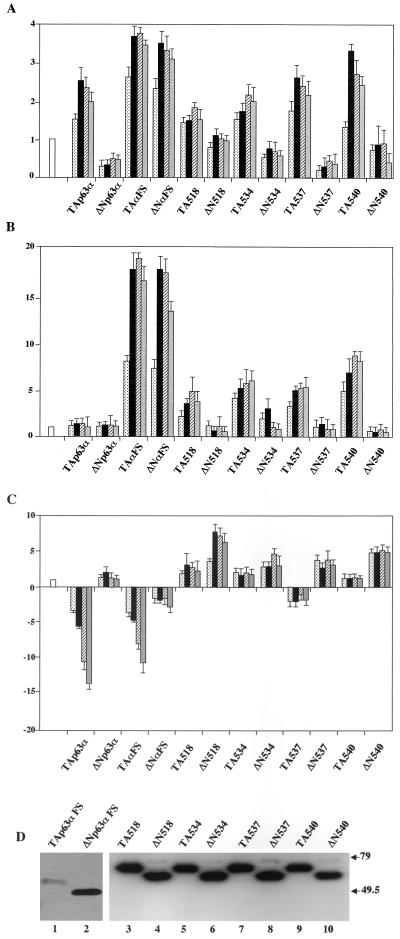
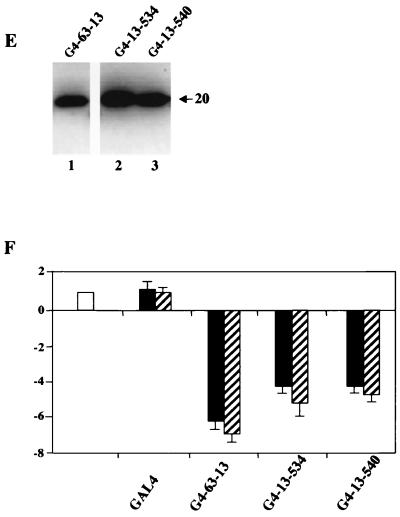
The delineation of exon 13 sequences as a transcriptional repressing part of p63 prompted us to examine the activities of mutants causative of the human EEC and AEC syndromes (8, 33). p63αFS contains a mutation found in an EEC syndrome patient containing an A insertion that generates a frameshift at amino acid 525 encoded within exon 13, leading to a predicted premature stop; p63 AEC missense mutants L518F, G534V, T537P, and Q540L are also encoded within exon 13 in the α1 and α3 helices and L2 of the SAM domain (33). Because of the alternative splicing at the C terminus (Fig. 1), these mutations are present only in the α isoforms. We performed the reporter assays outlined above with expression vectors coding for these mutants in the TA and ΔN configurations. The TA and ΔN p63αFS mutants showed similar behavior, activating the p21 and MDM2 promoters (Fig. 5A and B): both were among the strongest activators, unlike their wt counterparts, which were ineffective in MDM2 activation. With the AEC mutants, all the ΔN isoforms were unable to activate p21 or the MDM2 promoter, while the TA isoforms were active on MDM2 but not on p21. The gain of transcription function of TA540 is particularly striking, for it is the strongest among the activators tested on MDM2. In repression assays, the frameshift mutants showed no differences with respect to the wt isoforms (Fig. 5C), whereas the TA AEC mutants completely lost the capacity to repress Hsp70 transcription. Indeed, all ΔN AEC constructs, whose ΔN wt counterpart has no repression capacity, showed a robust activation of this promoter. The mutants were checked for protein expression in immunoblot analysis with specific anti-myc tag antibodies, and all were expressed at equivalent levels (Fig. 5D). From this analysis, we conclude that single amino acid substitutions within the SAM domain turn an ineffective activator, such as TAp63α, into a powerful one (Fig. 5B) and at the same time render the proteins incapable of repression (Fig. 5C). Furthermore, the transcriptional properties/behavior of p63αFS is clearly distinct from that of the AEC mutants. To further clarify whether the AEC mutations alter the exon 13 repression domain identified above (Fig. 4C), we generated constructs containing the AEC G534V and Q540L exon 13 mutants fused to the yeast GAL4 DNA-binding domain (G4-13-534 and G4-13-540). The activities of the resulting constructs were assayed with the reporter used above (Fig. 4C). The proteins were correctly expressed upon transfection into Saos-2 cells (Fig. 5E). As shown in Fig. 5F, when assayed with the G5-PB11 reporter, the repression capacities of G4-13-534 and G4-13-540 were modestly reduced compared to that of G4-63-13. These data suggest that the AEC mutations do not disrupt the exon 13 repression domain, but rather, in the entire p63 protein, AEC mutations could prevent (i) the association of a corepressor and/or (ii) interactions of the SAM domain with the TA1 of p63.
FIG. 5.
Transcriptional regulation by EEC and AEC mutation. (A) Saos-2 cells were transfected with the p21/WAFCAT reporter plasmid (0.5 μg) (open bar). Different amounts of expression plasmids for p63 isoforms were cotransfected (5, 25, 100, and 300 ng; dotted, black, dashed, and gray bars, respectively). After 36 h, cells were harvested and CAT activity was determined. (B) Saos-2 cells were transfected with the pBp100 reporter plasmid (0.5 μg) (open bar) and cotransfected as in panel A. (C) Saos-2 cells were transfected with the Hsp70 reporter plasmid (0.25 μg) (open bar) and cotransfected with expression plasmids (5, 25, 100, 300 and ng; dotted, black, dashed, and gray bars, respectively). (D and E) Saos-2 cells were transiently transfected with 0.5 μg of the indicated plasmids and lysed in 100 μl of lysis buffer 36 h after transfection. Twenty microliters of total cell extracts were immunoblotted with anti-myc (D) and anti-GAL4 (E) antibodies. (F) G5-PB11 reporter (0.5 μg, open bar) was cotransfected into Saos-2 cells together with indicated GAL4 vectors (black and dashed bars, 0.3 and 0.5 μg, respectively). Data are presented as fold activation or repression relative to the sample without effector. Each histogram bar represents the mean of three independent transfection duplicates. Standard deviations are indicated.
Growth suppression by wt and mutant p63 isoforms.
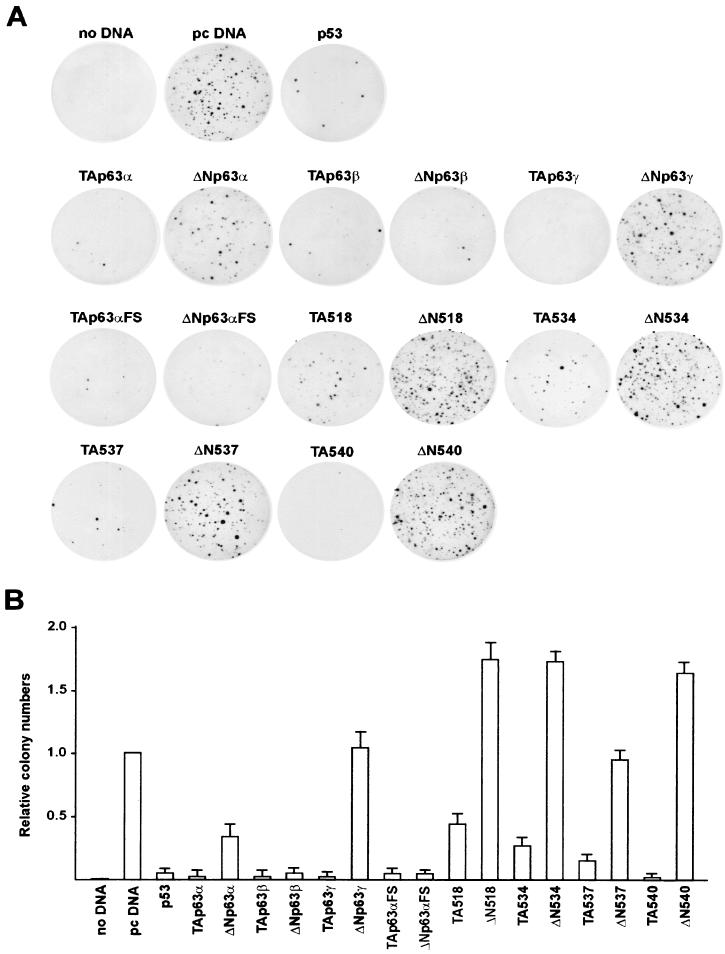
p53 is known to inhibit growth and some of p63 isoforms have been already checked for growth suppression in colony formation assays, known to reveal cell cycle blocking and proapoptosis functions (37). We performed such experiments with Saos-2 cells stably transfected with the constructs used above, which harbor a neomycin selection marker. The numbers of colonies scored after 2 weeks of addition of G418 in the medium are shown in Fig. 6A. As expected, p53 induced a dramatic decrease (50-fold) in colony numbers. In line with previous reports, essentially the same picture was evident with all the TAp63 isoforms. Among the mutants, TAp63αFS and TA540 behaved like the wt p63α isoform, whereas TA518, TA534, and to a lesser degree TA537 yielded five- to sevenfold more colonies than TAp63α yet clearly fewer than the pcDNA control. As for the ΔN isoforms, p63α and, more profoundly, p63β inhibited growth, whereas p63γ and AEC mutants were essentially negative in this assay: in fact, ΔN518, ΔN534, and ΔN540 generated a consistently higher number of colonies. On the other hand, p63αFS behaved essentially like the p63β isoform, being the strongest repressor of cell growth. Taken together, these data highlight remarkable differences in terms of growth suppression between (i) the TA and ΔN isoforms, (ii) the α/γ and β isoforms, and (iii) the EEC and AEC mutants.
FIG. 6.
Growth suppression by expression of wt and mutated p63 isoforms. (A) p53, p63 isoforms, and pcDNA expression vectors (0.3 μg each) were transfected into Saos-2 cells. After 2 weeks of selection, colonies were fixed and stained to demonstrate suppression of colony formation. (B) The graph represents the number of colonies obtained with the indicated plasmids relative to that detected on the pcDNA transfected plates, which was set to 1. The experiment was repeated three times in duplicate. Standard deviations are indicated.
DISCUSSION
We have performed the first comprehensive and systematic investigation of the transcriptional and growth inhibition activities of the p63 isoforms identified so far: the results are summarized in Table 1. We uncovered a novel activation domain, TA2, encoded within exons 11 and 12 and a repression function encoded by exons 13 and 14. Most importantly, single amino acid substitutions in the SAM domain completely abolish inhibition function and, within the TA isoforms, acquire activation potential. The biological importance of the SAM domain was further detailed in colony assays using AEC mutants. We conclude that the SAM domain is a dominant transcriptional repression module crucial for the function of the α isoform.
TABLE 1.
Summary of the transcriptional and growth suppression activities of wild-type and mutated p63a
| Protein | Activation | Repression | Growth suppr. |
|---|---|---|---|
| p53 | +++ | +++ | +++ |
| TAp63α | + | +++ | +++ |
| ΔNp63α | − | − | + |
| TAp63β | ++++ | + | +++ |
| ΔNp63β | +++ | + | +++ |
| TAp63γ | ++ | +++ | +++ |
| ΔNp63γ | + | ++ | − |
| TAp63FS | ++++ | +++ | +++ |
| ΔNp63FS | ++++ | + | +++ |
| TAα518 | ++ | −− | + |
| ΔNα518 | − | −− | −− |
| TAα534 | ++ | −− | ++ |
| ΔNα534 | − | −− | −− |
| TAα537 | ++ | + | ++ |
| ΔNα537 | − | −− | − |
| TAα540 | +++ | − | +++ |
| ΔNα540 | − | −− | −− |
Data obtained concerning transcriptional activation, transcriptional repression, and growth suppression (suppr.) capacities of p63 are shown. The strength of the response indicated by the + and − signs is relative to that obtained with p53, which was set as +++ in all cases.
p63 TA2 activation domain.
For all p53 family members, the very N terminus of the protein harbors a transcriptional activation domain. Although far less conserved than the DNA-binding subdomain, this highly acidic stretch shows some degree of similarity among the three family members. The β isoform in the ΔN configuration has powerful activation potential, suggesting that a second activation function was present in p63. The analysis of ΔNγ, lacking the SAM domain and less active in transcription, suggested that sequences encoded by exons 11 and 12 are important for activation. That a second activation domain, TA2, is positioned between amino acids 410 and 512 is indeed documented by the GAL4 fusion experiments (Fig. 4B). Several additional indications are in keeping with the importance of TA2: (i) it is confined to discrete exon-intron boundaries, as is often the case for functionally defined domains, and its genomic organization, differential splicing, and primary amino acid sequence are conserved between p63 and p73. (ii) The analysis of the transcriptional effects of p63α on natural promoters with TA and ΔNp63 inducible cell lines showed that one of the p53 potential targets, GADD45, was activated exclusively by ΔNp63α, a result well in line with the requirement of a second activation domain (12). (iii) p73 was shown to contain an activation function active in yeast and in mammalian cells between amino acids 380 and 499, a region that corresponds to p63 gene exons 11 and 12 (38, 45); moreover, one of the two p73 missense mutations identified in human neuroblastomas involves a proline, P425L, that is conserved in p63: the mutation indeed affects activation function in GAL4 assays (43). (iv) This region has a high proline content that is found in the activation domains of many other transcription factors (25). Interestingly, the Blandino and colleagues have recently shown that Yes-associated protein, a WW domain adaptor, binds to a stretch of p73 that is conserved within exon 12 of p63 and coactivates p73- and p63-mediated transcription (42).
Finally, the residual activity of γ isoforms observed in our assays could well be due to 26 amino acids at the very N-terminal end of the protein outside TA1 which are encoded by additional splicing variants (3). A further activation function was recently mapped to this region (12). In general, we provide evidence that an endogenous gene is activated by the panel of transfected p63 isoforms. It should be mentioned, however, that we were unable to activate the endogenous MDM2 gene by overexpressing ΔNp63β in the absence of TA1, unlike the robust activation observed with the TA versions. This result is remindful of the p73 case, for which activation in reporter assays by overexpression was not completely paralleled by stimulation of endogenous gene expression (24). Several explanations can be invoked to explain this behavior, since (i) the reporter genes are not recapitulating the whole regulation and (ii) the endogenous gene is constrained by chromatin structures.
The p63 SAM repression domain.
The repressive activity elicited by the SAM domain encoded by exon 13 was characterized in more detail, revealing the following. (i) Removal of this region by alternative splicing (β versus α) dramatically increases the activation potential of p63 on MDM2. (ii) Fusion of this domain to a GAL4 DNA-binding domain repressed an SV40 promoter reporter construct. (iii) In repression assays with the HSP70 promoter, the AEC mutants led to a complete subversion of the repression behavior within the TAα configuration, turning these mutants into activators, albeit weak ones. Results obtained with p73 support this idea: a C-terminal deletion to amino acid 427, including the SAM domain, has been shown to yield a more active protein (43, 45). A clear difference between the α and β isoforms within the TA configuration was reported on the MDM2 and GADD45 promoters but not on the p21 promoter (24). We also found that a C-terminal deletion augmented activity.
The three-dimensional structure of the p73 SAM domain between amino acids 491 to 550 has been recently solved by nuclear magnetic resonance: it is composed of five α-helices that are structurally very similar to the prototype SAM domain of the ephrin B2 receptor (9). Interestingly, the AEC mutations studied here are clustered and are predicted to be either modifying the overall structure and stability of the helices or causing subtle differences in the solvent-accessible surface of α3 (33). The β isoforms, in addition to splicing of the SAM, also are subject to a change in the frame and loss of the amino acids encoded by exon 14. Interestingly, mutations causing premature stop codons in exon 14 have been associated with limb mammary syndrome and SHFM syndrome, pointing to a relevant role of this exon (46).
What are the mechanisms by which the α isoforms inhibit transcription? The interpretation must consider the fact that the DNA-binding domain is intact and presumably capable of targeting the protein to the various promoters. The SAM domain is thought to be a protein-protein interacting module that mediates homo- and heterodimerization (44). Self-association of p63 and binding to p73 are not elicited by the SAM domain (10). Rather, it is tempting to speculate that a corepressor protein is normally bound to this part of p63α isoforms, possibly interfering with DNA binding and rendering p63 transcriptionally inactive: alternatively spliced β and γ isoforms or mutations would prevent the association of such a corepressor and hence unmask the activation domains.
AEC/EEC and p63 transcriptional control.
Both AEC and EEC syndromes are dominant traits caused by the dominant-negative activities of the mutated p63 isoforms. The EEC single amino acid mutations described so far concern the DNA-binding subdomain of p63 and affect all isoforms (2, 8): many correspond to residues in p53, such as R204, R279, R280, and R304 in p63, which are the equivalents of p53 mutational hot spots in innumerable human tumors (34, 41). Their effects on artificial p53 reporters have been observed: they were unable to activate when transfected alone and were unable to repress cotransfected p53 or TAp63γ (8, 33, 48). The equivalent p53 mutants are known to behave in very much the same way: indeed, some of the p63 missense mutations in EEC syndrome, such as R204 and R304, which are shared between EEC and SHFM (46), behaved very much like the p53 equivalents in transfections assays (8). Biochemical work indicated that heterotetramers between p63 and p73 with p53 are not easily observed in vitro and in vivo (10). However, it was recently found that p53 DNA-binding mutants are specifically capable of heteromerizing with p63 and p73, hence lending support to a model whereby the function of these two proteins would also be influenced in a negative way (14).
A fundamentally different scenario is evident for the AEC missense mutants. First, none of the mutations identified so far involves the DNA-binding domain; second, unlike the missense EEC, they are present only in the α isoforms. These mutants showed no differences in the activation of an artificial reporter containing 13 p53 binding sites in front of a minimal promoter, compared to the wt TAp63α counterpart (33): this behavior is indeed confirmed in our study of the p21 promoter, but it is clearly different from the pattern of activation observed on MDM2, suggesting that AEC mutants affect only a subset of p63 targets. The results obtained with AEC mutations in the GAL4 context (Fig. 5F) suggest that rather than disrupting the exon 13 repression domain, AEC mutations prevent association with a corepressor necessary to induce the down-modulation normally seen in the TAp63α protein. This suggests that the role of the repression domain is dominant, a notion that is consistent with the results obtained with exon 13-lacking β and γ isoforms. It is even possible that the negative role of the SAM domain affects the TA1 activation function.
AEC patients have a peculiar phenotype of skin lesions, in particular severe scalp dermatitis (46). Skin biopsies of AEC and EEC syndrome patients documented p63 staining in the differentiating cells of the suprabasal layer, where p63 is normally absent (33). The TA and ΔN isoforms are known to be differentially expressed in keratinocyte differentiation systems: the former increases and the latter conversely decreases upon loss of growth capacity and differentiation (11, 39). Growth suppression assays used in our study are the results of both activation of proapoptosis genes and repression of cell cycle promoting genes. In general, the colony assays of the AEC TA and ΔN isoforms suggest a more complex picture that might involve an altered capacity in activating growth-stimulating genes and in repressing proapoptosis genes. The dramatic increase in growth inhibition in the ΔNβ and ΔNFS isoforms possibly indicates that proapoptosis genes are not repressed. Conversely, the importance of TA2 is highlighted by the comparison between the ΔNβ and γ isoforms, in which it is evident that loss of exons 11 and 12 leads to lack of activation of proapoptosis genes. Thus, the severe skin phenotype of these patients might be a complex combination of altered cell renewal and lack of expression of highly specific differentiation genes. This interpretation requires the exact knowledge of p63 isoforms present in the basal layer in vivo, at present unknown, and the identification of p63 endogenous targets, some of which are emerging (11). Identification of bona fide p63 targets in vivo is clearly mandatory in order to fully understand the activation and repression functions of this transcription factor.
Acknowledgments
We thank Francesco Blasi for critical reading of the manuscript and for continuous support to L.G. We also thank Romina Colombo for help with some of the experiments and Simone Sabbioneda for computer assistance.
This work was supported by a grant from MURST to L.G.
Footnotes
Luisa dedicates this work to the memory of Giusi.
REFERENCES
- 1.Agoff, S. N., J. Hou, D. I. Linzer, and B. Wu. 1993. Regulation of the human hsp70 promoter by p53. Science 259:84-87. [DOI] [PubMed] [Google Scholar]
- 2.Andrew, S. E. 2001. When p63 goes awry: SAM domain mutations. Clin. Genet. 60:11-12. [Google Scholar]
- 3.Bamberger, C., and H. Schmale. 2001. Identification and tissue distribution of novel KET/p63 splice variants. FEBS Lett. 501:121-126. [DOI] [PubMed] [Google Scholar]
- 4.Bork, P., and E. V. Koonin. 1998. Predicting functions from protein sequences; where are the bottlenecks? Nat. Genet. 18:313-318. [DOI] [PubMed] [Google Scholar]
- 5.Calabrò, V., G. Mansueto, T. Parisi, M. Vivo, R. Calogero, and G. La Mantia. 2002. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homologue p63. J. Biol. Chem. 277:2674-2681. [DOI] [PubMed] [Google Scholar]
- 6.Calabrò, V., G. Mansueto, T. Parisi, M. Vivo, R. Calogero, and G. La Mantia. 2002. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J. Biol. Chem. 22:196-206. [DOI] [PubMed] [Google Scholar]
- 7.Caspari, T. 2000. Checkpoints: how to activate p53. Curr. Biol. 10:R315-R317. [DOI] [PubMed] [Google Scholar]
- 8.Celli, J., P. Duijf, B. C. J. Hamel, M. Bamshad, B. Kramer, A. P. T. Smits, R. Newbury-Ecob, R. C. M. Hennekam, G. Van Buggenhout, A. van Haeringen, C. G. Woods, A. J. van Essen, R. de Waal, G. Vriend, D. A. Haber, A. Yang, F. McKeon, H. G. Brunner, and H. van Bokhoven. 1999. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143-153. [DOI] [PubMed] [Google Scholar]
- 9.Chi, S.-W., A. Ayed, and C. H. Arrowsmith. 1999. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 18:4438-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274:18709-18714. [DOI] [PubMed] [Google Scholar]
- 11.De Laurenzi, V., A. Rossi, A. Terrinoni, D. Barcaroli, M. Levrero, A. Costanzo, R. A. Knight, P. Guerrieri, and G. Melino. 2000. p63 and p73 transactivate differentiation gene promoters in human keratinocyte. Biochem. Biophys. Res. Commun. 273:342-346. [DOI] [PubMed] [Google Scholar]
- 12.Dohn, M., S. Zhang, and X. Chen. 2001. p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20:3193-3205. [DOI] [PubMed] [Google Scholar]
- 13.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 14.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginsberg, D., F. Mechta, M. Yaniv, and M. Oren. 1991. Wild-type p53 can down-modulate the activity of various promoters. Proc. Natl. Acad. Sci. USA 88:9979-9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, S. K., L. Guerrini, and F. Blasi. 1994. Differential DNA sequence specificity and regulation of HIV-1 enhancer activity by cRel-RelA transcription factor. J. Biol. Chem. 269:22230-22237. [PubMed] [Google Scholar]
- 17.Ianakiev, P., M. W. Kilpatrick, I. Toudjarska, D. Basel, P. Beighton, and P. Tsipouras. 2000. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 67:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin, M. S., and W. G. Kaelin. 2001. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 12:337-349. [PubMed] [Google Scholar]
- 19.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. C. Biscan, A. Valent, A. Minty, P. Chalon, J. M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 20.Katoh, I., K. Aisaki, S. Kurata, S. Ikawa, and Y. Ikawa. 2000. p51A (TAp63γ), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene 19:3126-3130. [DOI] [PubMed] [Google Scholar]
- 21.Kong, X. T., V. A. Valentine, S. T. Rowe, M. B. Valentine, S. T. Ragsdale, B. G. Jones, D. A. Wilkinson, G. M. Brodeur, S. L. Cohn, and A. T. Look. 1999. Lack of homozygously inactivated p73 in single-copy MYCN primary neuroblastomas and neuroblastoma cell lines. Neoplasia 1:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause, K., M. Wasner, W. Reinhard, U. Haugwitz, C. L. Dohna, J. Mossner, and K. Engeland. 2000. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 28:4410-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landsberger, N., and A. P. Wolffe. 1995. Role of chromatin and Xenopus laevis heat shock transcription factor in regulation of transcription from X. laevis hsp70 promoter in vivo. Mol. Cell. Biol. 15:6013-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, C. W., and N. B. La Thangue. 1999. Promoter specificity and stability control of the p53-related protein p73. Oncogene 18:4171-4181. [DOI] [PubMed] [Google Scholar]
- 25.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 26.Levrero, M., V. De Laurenzi, A. Costanzo, S. Sabatini, J. Gong, J. Y. J. Wang, and G. Melino. 2000. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113:1661-1670. [DOI] [PubMed] [Google Scholar]
- 27.Liberati, C., A. di Silvio, S. Ottolenghi, and R. Mantovani. 1999. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J. Mol. Biol. 285:1441-1455. [DOI] [PubMed] [Google Scholar]
- 28.Lohrum, M. A. E., and K. H. Vousden. 2000. Regulation and function of the p53-related proteins: same family, different rules. Trends Cell Biol. 10:197-202. [DOI] [PubMed] [Google Scholar]
- 29.Majello, B., P. De Luca, G. Hagen, G. Suske, and L. Lania. 1994. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 22:4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majello, B., P. De Luca, and L. Lania. 1997. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 272:4021-4026. [DOI] [PubMed] [Google Scholar]
- 31.Marin, M. C., and W. G. Kaelin, Jr. 2000. p63 and p73: old members of a new family. Biochim. Biophys. Acta 1470:M93-M100. [DOI] [PubMed] [Google Scholar]
- 32.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18:7621-7636. [DOI] [PubMed] [Google Scholar]
- 33.McGrath, J. A., P. H. G. Duijf, V. Doetsch, A. D. Irvine, R. de Waal, K. R. J. Vanmolkot, V. Wessagowit, A. Kelly, D. J. Atherton, W. A. D. Griffiths, S. J. Orlow, A. van Haeringen, M. G. E. M. Ausems, A. Yang, F. McKeon, M. A. Bamshad, H. G. Brunner, B. C. J. Hamel, and H. van Bokhoven. 2001. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum. Mol. Genet. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 34.Michael, D., and M. Oren. 2002. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 12:53-59. [DOI] [PubMed] [Google Scholar]
- 35.Mills, A. A., B. Zheng, X.-J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 36.Nishi, H., M. Senoo, K. H. Nishi, B. Murphy, T. Rikiyama, Y. Matsumura, S. Habu, and A. C. Johnson. 2001. p53 homologue p63 represses epidermal growth factor receptor expression. J. Biol. Chem. 276:41717-41724. [DOI] [PubMed] [Google Scholar]
- 37.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 38.Ozaki, T., M. Naka, N. Takada, M. Tada, S. Sakiyama, and A. Nakagawara. 1999. Deletion of the COOH-terminal region of p73α enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res. 59:5902-5907. [PubMed] [Google Scholar]
- 39.Pellegrini, G., E. Dellambra, O. Golisano, E. Martinelli, I. Fantozzi, S. Bondanza, D. Ponzin, F. McKeon, and M. De Luca. 2001. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 98:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piechaczyk, M., J. M. Blanchard, L. Marty, C. Dani, F. Panabieres, S. El Sabouty, P. Fort, and P. Jeanteur. 1984. Post-transcriptional regulation of glyceraldehyde-3-phosphate-dehydrogenase gene expression in rat tissues. Nucleic Acids Res. 25:6951-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raycroft, L., J. R. Schmidt, K. Yoas, M. Hao, and G. Lozano. 1991. Analysis of p53 mutants for transcriptional activity. Mol. Cell. Biol. 11:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strano, S., E. Munarriz, M. Rossi, L. Castagnoli, Y. Shaul, A. Sacchi, M. Oren, M. Sudol, G. Cesareni, and G. Blandino. 2001. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276:15164-15173. [DOI] [PubMed] [Google Scholar]
- 43.Takada, N., T. Ozaki, S. Ichimiya, S. Todo, and A. Nakagawara. 1999. Identification of a transactivation activity in the COOH-terminal region of p73 which is impaired in the naturally occurring mutants found in human neuroblastomas. Cancer Res. 59:2810-2814. [PubMed] [Google Scholar]
- 44.Thanos, C. D., and J. U. Bowie. 1999. p53 family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 8:1708-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueda, Y., M. Hijikata, S. Takagi, T. Chiba, and K. Shimotohno. 1999. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene 18:4993-4998. [DOI] [PubMed] [Google Scholar]
- 46.van Bokhoven, H., B. C. J. Hamel, M. Bamshad, E. Sangiorgi, F. Gurrieri, P. H. G. Duijf, K. R. J. Vanmolkot, E. van Beusekom, S. E. C. van Beersum, J. Celli, G. F. M. Merkx, R. Tenconi, J. P. Fryns, A. Verloes, R. A. Newbury-Ecob, A. Raas-Rotschild, F. Majewski, F. A. Beemer, A. Janecke, D. Chitayat, G. Crisponi, H. Kayserili, J. R. W. Yates, G. Neri, and H. G. Brunner. 2001. p63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am. J. Hum. Gen. 69:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X. W., and C. C. Harris. 1997. p53 tumor-suppressor gene: clues to molecular carcinogenesis. J. Cell. Physiol. 173:247-255. [DOI] [PubMed] [Google Scholar]
- 48.Yang, A., and F. McKeon. 2000. p63 and p73: p53 mimics, menaces and more. Mol. Cell. Biol. 1:199-207. [DOI] [PubMed] [Google Scholar]
- 49.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 50.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 51.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 52.Zauberman, A., D. Flusberg, Y. Haupt, Y. Barak, and M. Oren. 1995. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 23:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]