Abstract
Neurotrophins are key regulators of the fate and shape of neuronal cells and act as guidance cues for growth cones by remodeling the actin cytoskeleton. Actin dynamics is controlled by Rho GTPases. We identified a novel Rho GTPase-activating protein (Grit) for Rho/Rac/Cdc42 small GTPases. Grit was abundant in neuronal cells and directly interacted with TrkA, a high-affinity receptor for nerve growth factor (NGF). Another pool of Grit was recruited to the activated receptor tyrosine kinase through its binding to N-Shc and CrkL/Crk, adapter molecules downstream of activated receptor tyrosine kinases. Overexpression of the TrkA-binding region of Grit inhibited NGF-induced neurite elongation. Further, we found some tendency for neurite promotion in full-length Grit-overexpressing PC12 cells upon NGF stimulation. These results suggest that Grit, a novel TrkA-interacting protein, regulates neurite outgrowth by modulating the Rho family of small GTPases.
The Rho family of small GTPases (RhoA, Rac1, and Cdc42) control actin dynamics (6, 17, 55), whereas much less is known about the mechanism of the spatiotemporal regulation of their activities (16). Rho GTPases function as molecular switches, shuttling between a GDP-bound inactive state and a GTP-bound active state. Rho GTPases are regulated by the opposing effects of two classes of enzymes, guanine nucleotide exchange factors (GEFs) of the Dbl family and GTPase-activating proteins (GAPs). Dbl GEFs act to enhance the exchange of bound GDP for GTP and thus activate the Rho GTPases, whereas GAPs inhibit Rho family members by potentiating their intrinsic GTPase activity (55). In principle, local activation of GTPases in response to intra- or extracellular signals could occur by local activation of GEFs or inhibition of GAPs. The recent identification of novel Rho GTPase regulators (GEF/GAPs) associated with specific receptors (48, 52, 56) is beginning to unravel the molecular mechanism linking extracellular cues to highly localized changes in the actin cytoskeleton, underlining a variety of essential biological functions including cell division, adhesion, motility, and polarity.
Neuritogenesis can be seen as a particular form of cell motility driven by Rho GTPases; actin dynamics during growth cone navigation evolves into stabilization of the cytoskeleton and neurite elongation (13, 30). Rac1 and Cdc42 seem to be positive regulators, whereas RhoA seems to be a negative regulator, in process outgrowth from neuronal cells (26, 30). In neuronal morphogenesis, neurotrophins (NGF, brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5) are key regulators of the fate and shape of neuronal cells and have been shown to act as guidance cues for growth cones in vitro and in vivo (30, 39, 49). Trk tyrosine kinases, which are high-affinity receptors for neurotrophins, activate a variety of signaling cascades through their binding molecules composed of enzymes and adapters. Multiple effectors of Trk signalings, e.g., Ras, PI 3-kinase, and phospholipase Cγ, are coordinately regulated through their shared usage of adapter molecules—i.e., Shc family, IRS family, FRS-2, Grb2, Crk/CrkL, and Gab1/2, etc.—and they constitute dynamic networks (4, 24). Thus, to understand how neurotrophins regulate the growth of neuronal process, it is a critical issue to elucidate the mechanism by which localized cues of neurotrophins are transmitted to cause local reorganization of actin filaments through specific combinations in these networks.
Here we identified a novel neurally enriched Rho GTPase-activating protein (RhoGAP), Grit, which was constitutively associated with TrkA, a high-affinity receptor for NGF, and preferentially stimulated GTP hydrolysis of RhoA and Cdc42 over that of Rac1. Grit had additional partners, N-Shc and CrkL/Crk, both of which are adapter molecules in phosphotyrosine signaling downstream of activated receptor tyrosine kinases including Trk receptors (32, 34, 35). Through the binding of Grit to these adapters, Grit translocation was regulated by receptor stimulation. In PC12 cells, overexpression of the TrkA-binding region of Grit significantly inhibited NGF-induced neurite elongation. Further, we found some tendency for neurite promotion in full-length Grit-overexpressing PC12 cells upon NGF stimulation. These results indicate that Grit is one of the significant components in NGF-induced cytoskeletal changes leading to neurite outgrowth.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The bait plasmid pAS404-TrkA-NShc was designed to express a chimera between the cytoplasmic region of human TrkA (residues 433 to 790) and the phosphotyrosine binding (PTB) and CH1 domains of human p52N-Shc (residues 1 to 378) by using the pAS404 vector (47) derived from pAS1. Saccharomyces cerevisiae transformation and two-hybrid screening were carried out by using the Y190 strain, as described previously (20, 47). An 800-ml culture of Y190 cells containing pAS404-TrkA-NShc was transformed with 400 μg of a human adult brain cDNA library fused to the GAL4 transactivation domain (Clontech). The transformants were plated in selection medium (lacking Trp, Leu, and His) containing 25 mM 3-aminotriazole to select for histidine prototrophy. His-autotrophic colonies were lysed in liquid nitrogen and assayed for β-galactosidase activity on filters. A total of 1.2 × 107 transformants were screened, and 81 clones were isolated from the His-autotrophic and lacZ-positive colonies. The plasmids of positive clones were isolated, and their nucleotide sequences were determined with an ABI373S sequencer (Perkin-Elmer).
Cell culture and antibodies.
PC12 cells were grown in RPMI medium containing 10% horse serum and 5% fetal bovine serum (FBS). HeLa, 293, TIG1, COS-1, and Swiss 3T3 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% FBS. SH-SY5Y and SMS-KCN cells were grown in 10% FBS-containing RPMI medium. Two rabbit anti-Grit antibodies (PR205 and CT205) were raised against the purified recombinant Grit proteins expressed as histidine-tagged forms. Immunogens for PR205 and CT205 were residues 686 to 890 and 1534 to 1738 of human Grit, respectively. These antibodies were purified on affinity columns containing electrophoretically separated and eluted antigens. The specific reactivity of CT205 was further improved by removing cross-reactivity by adsorption to a column containing a Ni-Agarose binding fraction of bacterial lysates. Anti-T7 peptide antibody was obtained from Novagen, and anti-FLAG M2 antibody was obtained from Sigma. Antiphosphotyrosine antibody 4G10 was purchased from Upstate Biotechnology, and anti-Trk (C-14 and MCTrks), anti-CrkL, and anti-GST came from Santa Cruz. Anti-Cas, anti-Crk, and anti-epidermal growth factor receptor (anti-EGFR) (clone 13) were purchased from Transduction Laboratories. Anti-EGFR (6F1) was from MBL, and horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham-Pharmacia.
Expression plasmids.
cDNA for human Grit (KIAA0712) was obtained from Kazusa DNA Research Institute. Human TrkA and mouse TrkB were kindly provided by M. Barbacid. Rat TrkC was a gift from G. Yancopoulos. The wild type and all mutants of human Grit cDNA with the FLAG tag at the amino terminus were subcloned into pCI expression vector (Promega). The R58A mutant (from arginine 58 to alanine) of Grit RhoGAP construct was generated by PCR-mediated mutagenesis (Promega). A plasmid expressing the human TrkA protein in a hemagglutinin-tagged form was also subcloned into the pCI vector. Deletion mutants of TrkA were TrkAΔC1 (Δ710-790), TrkAΔC2 (Δ627-790), and TrkAΔC3 (Δ557-790). The T7-tagged N-Shc, Shc, and Sck constructs were previously described (33, 34).
Northern blot analysis and in situ hybridization.
Human multiple blots I and II (Clontech) were hybridized with a specific 32P-labeled cDNA probe of human Grit as described previously (34). Preparation of sections from Sprague-Dawley rats and in situ hybridization with a specific 35S-labeled cRNA probe of rat Grit were carried out as described previously (34).
RT-PCR.
Total RNAs from mock- and NGF-treated PC12 cells were extracted by using TRIZOL reagent (Invitrogen), and cDNA was synthesized from them by using the SuperScript First-Strand synthesis system (Invitrogen). The forward rat Grit primer was 5′-GCAAGCCATAGGCAGTTATGTGAG-3′; the reverse primer was 5′-CAACTCTATTGCTCCCGGGGCTC-3′. Rat Grit cDNA was amplified for 30 cycles with ExTaq polymerase (TaKaRa).
Immunofluorescence.
Cells were fixed in 3.7% formaldehyde and permeabilized with 0.2% Triton X-100. After having been soaked for 1 h in phosphate-buffered saline (PBS) containing 3% BSA and 0.1% Triton X-100, the samples were incubated overnight at 4°C with anti-Grit antibodies PR205 (5 μg/ml) or CT205 (8 μg/ml), washed with PBS, and then incubated for 30 min at room temperature with Alexa 488-conjugated anti-rabbit immunoglobulin G (IgG) (Molecular Probes). Staining of the same cells for TrkA, Cas, or EGFR was performed with anti-Trk (2 μg/ml; MCTrks), anti-Cas (2.5 μg/ml), or anti-EGFR (6F1; 5 μg/ml) antibody, respectively, followed by incubation with the combination of Alexa 488 anti-rabbit IgG and Alexa 594 anti-mouse IgG (Fig. 3E). The combination of Alexa 488 anti-mouse IgG and Alexa 594 anti-rabbit IgG was used in another experiment (Fig. 8E). The samples were then washed with PBS containing 0.2% Tween 20 and examined under a BX60 or IX71 microscope (Olympus) equipped with an MRC-1024 (Bio-Rad) or FV500 (Olympus) laser-scanning confocal imaging system.
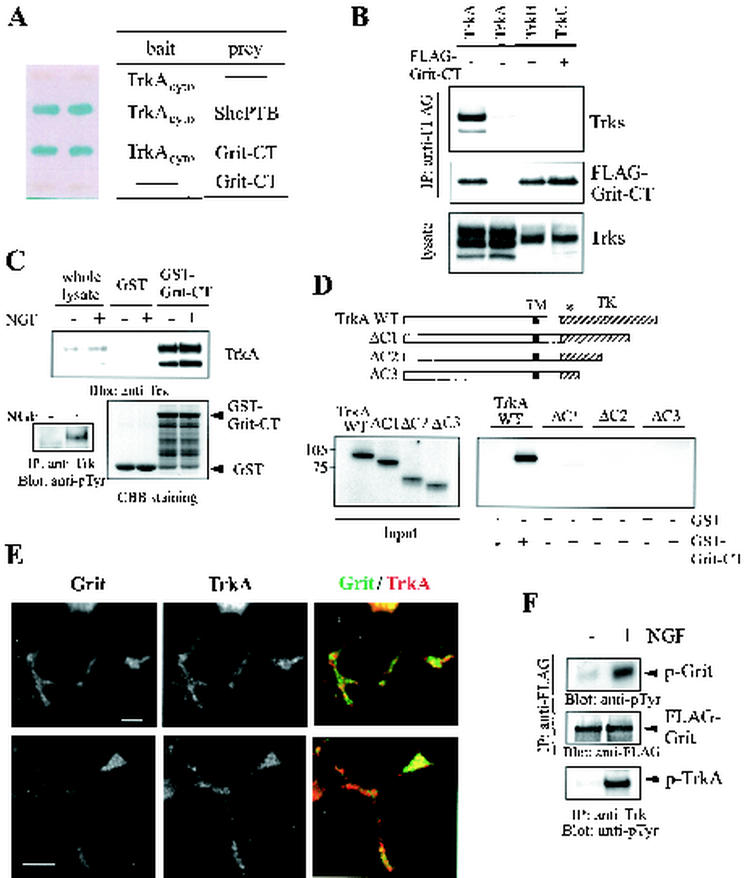
FIG. 3.
Interaction between Grit and TrkA. (A) The cytoplasmic region of human TrkA (not the TrkA/N-Shc chimera used in the initial screening) fused to the GAL4 DNA binding domain or the GAL4 DNA binding domain alone was coexpressed in the yeast two-hybrid assay with the GAL4 transactivation domain alone or with the GAL4 fusion to Shc PTB domain or Grit-CT region. Positive interactions were indicated by the induction of the lacZ reporter (blue color). (B) 293 cells were transfected with the FLAG-tagged Grit-CT construct and the indicated Trk cDNA. Anti-FLAG immunoprecipitates were probed with anti-pan Trk or anti-FLAG (for Grit-CT detection) antibody. Exposure times were adjusted so that comparable levels of signals were obtained in anti-Trk immunoblotting of cell lysates. (C) Lysates from PC12 cells with or without NGF treatment were incubated with either 25 μg of GST or GST fusion containing Grit-CT on glutathione-Sepharose. Whole-cell lysate (7% of starting material) and eluates from the affinity beads were analyzed by anti-Trk immunoblotting (top). Eluates were also subjected to SDS-PAGE and Coomassie staining (bottom right). For evaluation of NGF-induced Trk autophosphorylation, anti-Trk immunoprecipitates from mock- or NGF-treated PC12 cells were probed with antiphosphotyrosine antibody (bottom left). Abbreviations: IP, immunoprecipitation; CBB, Coomassie brilliant blue. (D) 35S-labeled in vitro-translated wild-type TrkA and a series of its mutants (top panel) were incubated with 10 μg of either GST or GST fusion containing Grit-CT protein. Inputs and eluates from the affinity beads were subjected to SDS-PAGE, and the labeled proteins were then detected by autoradiography. Abbreviations: TM, transmembrane domain; TK, tyrosine kinase domain; asterisk, ATP-binding site. (E) PC12 cells were treated with NGF for 2 days and costained as indicated above the panels (merged images at righ show Grit in green and TrkA in red). Scale bar, 5 μm. (F) 293 cells were transfected with FLAG-Grit cDNA, serum starved, and treated with NGF for 5 min. Anti-FLAG immunoprecipitates were probed with antiphosphotyrosine (top) or anti-FLAG (middle) antibody. NGF-dependent Trk autophosphorylation was confirmed as in Fig. 3C (bottom).
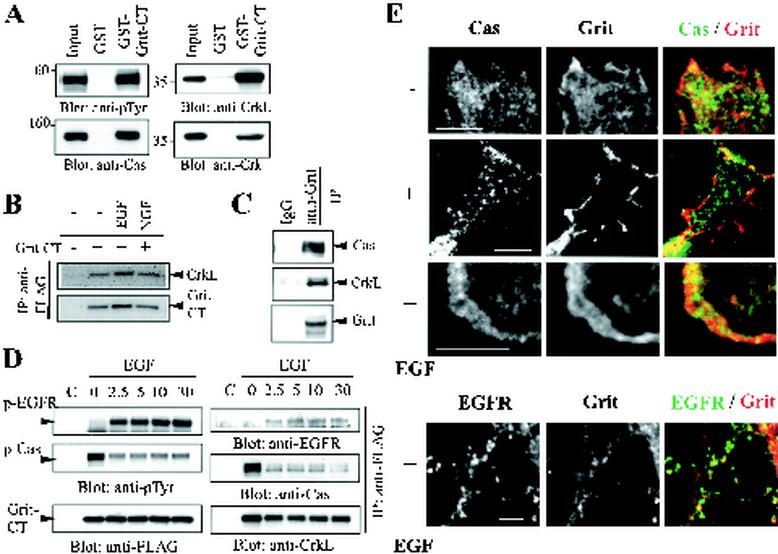
FIG. 8.
Association of Grit with CrkL/Crk and Cas. (A) PC12 cell lysates were incubated with either 25 μg of GST or GST fusion containing Grit-CT on glutathione-Sepharose. Cell lysates (7% of starting material) and eluates from the affinity beads were immunoblotted with the indicated antibodies. (B) PC12 cells were mock transfected or transfected with FLAG-Grit-CT plasmid, serum starved, and treated as indicated above the panels. Anti-FLAG immunoprecipitates were probed with anti-CrkL (top) or anti-FLAG (bottom) antibodies. (C) Grit-transfected COS-1 cells were serum-starved and lysed for immunoprecipitation with anti-Grit antibodies or control IgG. The immunoprecipitates were probed with anti-Cas, anti-CrkL, or anti-Grit antibodies. (D) COS-1 cells were transfected with FLAG-Grit-CT construct, deprived of serum, and stimulated for various lengths of time with EGF (0 to 30 min). Anti-FLAG immunoprecipitates were immunoblotted with antiphosphotyrosine, anti-FLAG (for Grit-CT detection), anti-EGFR, anti-Cas, or anti-CrkL antibodies. (E) Ligand-induced translocation of Grit in SH-SY5Y cells. SH-SY5Y cells were mock treated or treated with EGF for 5 min, and costained as indicated above the panels. Scale bars, 5 μm.
Immunoprecipitation and GST pull-down assay.
Cells were harvested in ice-cold lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1% Nonidet P-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 2 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride) containing protease inhibitors (Complete; Roche), and the lysates were then immunoprecipitated as described earlier (34). Immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with the desired antibodies. The glutathione S-transferase (GST) pull-down assay was performed as previously described (33). Cell lysates were incubated with 10 to 25 μg of GST fusion proteins on glutathione-Sepharose (Amersham-Pharmacia) for 2 h or overnight at 4°C. The washed beads were boiled in SDS-sample buffer, and the bound proteins were analyzed by immunoblotting.
RhoGAP assay.
Recombinant RhoA, Rac1, and Cdc42 were produced by using the pGEX-2T expression vector (Amersham-Pharmacia) as described (38). The proteins (1 μg, 40 pmol) were preloaded with [γ-32P]GTP (30 Ci/mmol; NEN) in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 10% glycerol, BSA (0.5 mg/ml), and 10 mM β-mercaptoethanol for 2 min at 37°C. For measuring the GAP activity of the recombinant Grit GAP domain, 30 ng of preloaded GTPase was diluted with a TM buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.5 mg/ml BSA) containing 2.5 mM GTP and the recombinant Grit GAP domain (residues 1 to 218) to a final volume of 20 μl, and then the mixture was incubated at 22°C. The reaction was stopped on ice at 5, 10, 15, or 20 min, and the samples were then filtered through 0.45-μm-pore-size nitrocellulose filers. The filters were washed three times with ice-cold wash buffer, and the radioactivity remaining as [γ-32P]GTP was determined by scintillation counting. For measuring the GAP activity of immunoprecipitated Grit protein or Grit GAP domain, the immune complex was washed with a TM buffer and resuspended in 20 μl of TM buffer containing 2.5 mM GTP. The resuspended immune complex was incubated with 6 ng of [γ-32P]GTP-loaded RhoA or Cdc42 at room temperature for 20 min (RhoA) or 5 min (Cdc42). The samples were then filtered and counted as above.
Microinjection.
Microinjection into Swiss 3T3 cells was performed as described elsewhere (38). Proteins were injected into the cytoplasm with fluorescein isothiocyanate (FITC)-dextran (5 mg/ml; Molecular Probes) to identify the microinjected cells. Cells were stimulated with lysophosphatidic acid (LPA) (20 ng/ml) for 30 min, platelet-derived growth factor (3 ng/ml) for 15 min, or bradykinin (100 ng/ml) for 5 min and then fixed for 30 min in 2% formaldehyde. For actin localization, cells were incubated with rhodamine-phalloidin (Molecular Probes) and examined with a Zeiss Axioscope microscope.
Neurite extension assays.
PC12 cells were grown in 35-mm-diameter glass-bottom dishes (Matsunami) coated with polyethylenimine (Sigma) and transfected with 1 μg of the indicated Grit construct and 0.1 μg of pEGFP-C1 (Clontech). Neurite outgrowth was stimulated with NGF (50 ng/ml; Calbiochem) and allowed to proceed for 58 h in RPMI medium containing 0.1% BSA. Then, the cells were fixed with 3.7% formaldehyde and examined by fluorescence microscopy to detect the presence of the green fluorescent cells that were expected to contain the exogenously expressed Grit proteins. Phase-contrast and fluorescent images were captured with a Zeiss Axiovert microscope. For each GFP-positive cell, lengths of cell body and neurite were measured by using NIH image 1.62. Adhesiveness of cells expressing the Grit GAP domain or its R58A mutant was evaluated by counting the number of GFP-positive cells for each sample by using CELLocate coverslips (Eppendorf).
Nucleotide sequence accession number.
The DNA sequence corresponding to a newly determined N terminus of the human Grit protein was deposited in GenBank/EMBL/DDBJ database under the accession number AB079856.
RESULTS
Identification of Grit, a novel member of the RhoGAP family, as a TrkA-interacting protein.
The yeast two-hybrid screen was used to search for proteins implicated in neurotrophin/Trk signaling in the course of our study on neuronal adapter N-Shc/ShcC. Our initial aim was to obtain candidate partners of N-Shc when it was phosphorylated by Trk receptors; thus, the bait consisted of a chimera between the TrkA cytoplasmic region having tyrosine kinase activity and the PTB-CH1 domains of N-Shc fused to the GAL4 DNA binding domain and was autophosphorylated when expressed in yeast (data not shown). We obtained 81 clones interacting with either phospho-TrkA or phospho-N-Shc upon screening a cDNA library from human adult brain. Fifteen of the clones encoded an uncharacterized member of RhoGAPs, which was identical to the human KIAA0712. We named this protein Grit, for GTPase regulator interacting with TrkA, because this protein was shown to interact with TrkA receptor in several binding assays (see below).
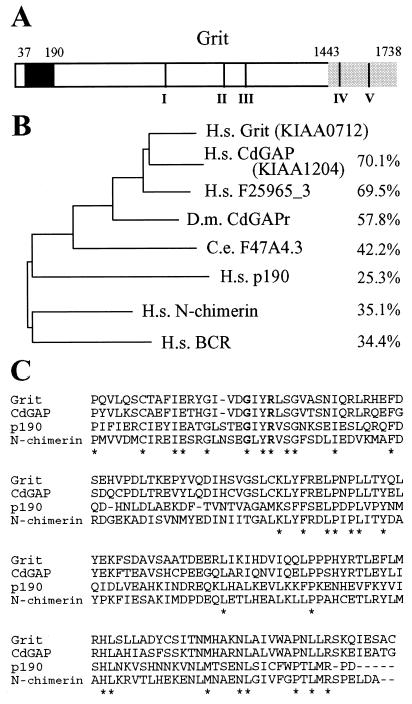
Grit was found to be a protein of 1,783 amino acids comprising an N-terminal RhoGAP domain, an internal proline-rich (PR) region, and a C-terminal (CT) TrkA-binding region (Fig. 1A). The internal PR region and the TrkA-binding region contained three and two potential SH3 binding motifs (61), respectively (Fig. 1A). Grit-related genes have been found in both invertebrates and vertebrates. The genomes of Drosophila melanogaster and Caenorhabditis elegans each contain a single gene with a high degree of homology to Grit within their RhoGAP domains. In mammals, there are at least two other genes that are highly homologous to Grit, i.e., the previously characterized RhoGAP member CdGAP (27) and one uncharacterized cDNA, F25965_3. We performed phylogenetic analysis of the RhoGAP domains of the Grit-related proteins and compared them with the RhoGAP domains of p190, N-chimerin, and Bcr (Fig. 1B and C). This analysis revealed that the GAP domains of Grit-related RhoGAPs were more closely related to each other than to the GAP domains of p190, N-chimerin, and Bcr and thus identified Grit-related RhoGAPs as a subfamily within the larger RhoGAP family (Fig. 1B). The CdGAP/Grit subfamily revealed a high degree of conservation within the N-terminal RhoGAP domain (Fig. 1B and C) and some similarity in succeeding sequences but little or no homology in the CT half (not shown).
FIG. 1.
Analysis of Grit amino acid sequence. (A) Schematic representation of human Grit (KIAA0712). The deposited KIAA0712 amino acid sequence (GenBank AB018255) was inferred to be truncated in the middle of the RhoGAP domain; we thus determined the corresponding nucleotide sequences of Grit cDNA clones obtained by 5′ rapid amplification of cDNA ends and added N-terminally 151 residues of Grit. RhoGAP domain, filled box; TrkA/adapter-binding CT region, hatched box. The amino acid positions of the domain boundaries are shown at top. Potential SH3 binding motifs I to V, predicted by the Scansite program, are shown by bars. (B) Phylogenetic analysis of the GAP domains of Grit and RhoGAP family proteins by use of the GENETYX program. The percent identities to human Grit are shown at the right. H.s., Homo sapiens; D.m., D. melanogaster; C.e., C. elegans. (C) Alignment of the GAP domain of Grit with that of other RhoGAP family members. Grit, residues 37 to 190; CdGAP, residues 35 to 188; p190, residues 1263 to 1409; N-chimerin, residues 282 to 434. Identical residues are indicated by asterisks. The critical glycine and arginine for GAP activity are in boldface type.
Grit expression is abundant in the nervous system.
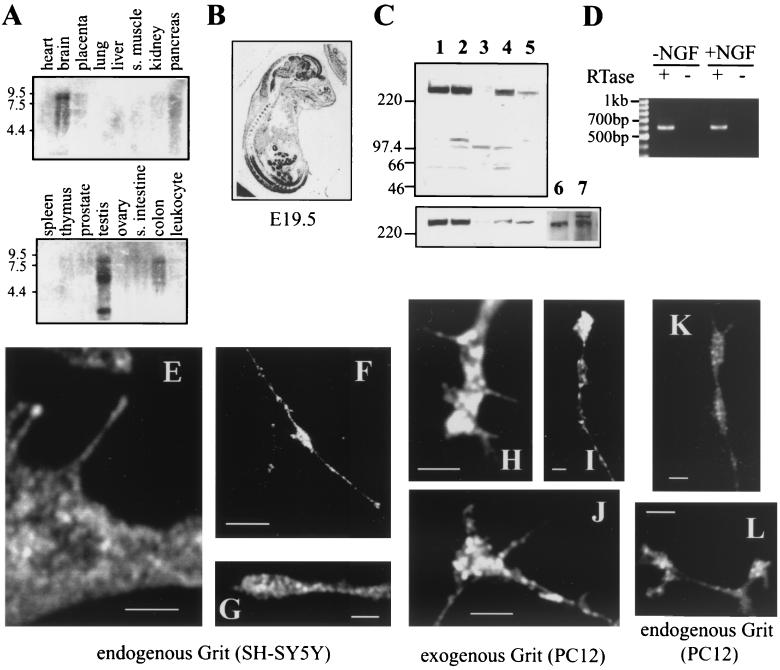
Northern blot analysis revealed three RNA transcripts of Grit at ∼9.0, 6.0, and 3.2 kb, with the highest levels of expression in the brain and testis and lower levels of transcripts in the pancreas and colon (Fig. 2A). Abundant expression of Grit mRNA in the embryonic day 19.5 rat was found by in situ hybridization to be restricted to the nervous system (both central and peripheral) and to a part of the gastrointestinal tract (Fig. 2B). We prepared two kinds of affinity-purified polyclonal antibodies (PR205 and CT205) against different regions of Grit. Although the predicted molecular mass of Grit protein was 191 kDa, either type of antibody reacted with a band of 250 kDa (Fig. 2C); therefore, we concluded that this 250-kDa band corresponded to endogenous Grit protein. Consistent with this, a band of 250 kDa was detected by both anti-FLAG and anti-Grit immunoblotting of the lysates of FLAG-tagged Grit-expressing COS-1 cells (data not shown). Grit protein was expressed at high levels in neuronal cells SH-SY5Y, SMS-KCN, and PC12 (Fig. 2C), as expected in light of its abundant mRNA expression in neural tissues. Further, comparable Grit expression was detected by RT-PCR in both naive and differentiated PC12 cells (Fig. 2D).
FIG. 2.
Expression pattern of Grit mRNA and protein. (A) Northern blot analysis. Blots of mRNA from adult human tissues were probed with a fragment of human Grit corresponding to the CT region. Size markers are indicated on the left in kilobases. Abbreviations: s. muscle, skeletal muscle; s. intestine, small intestine. (B) In situ hybridization of Grit mRNA. A sagittal section from an embryonic day 19.5 rat embryo was hybridized with an antisense probe of the internal region of rat Grit. The adjacent section hybridized with a sense control probe gave no specific signals (not shown). (C) Western blot analysis of Grit was carried out by using two kinds of anti-Grit polyclonal antibodies (top, PR205; bottom, CT205) and lysates from various cell lines. Lanes 1 and 6, SH-SY5Y; lane 2, SMS-KCN; lane 3, HeLa; lane 4, 293; lane 5, TIG1; lane 7, PC12. (D) Agarose gel showing Grit RT-PCR products amplified from total RNA isolated from mock- or NGF-treated PC12 cells with (+) or without (−) reverse transcriptase (RTase). (E to G) SH-SY5Y cells incubated with retinoic acid for 2 days were stained with the anti-Grit antibody PR205. Scale bars, 2 μm (E and G) or 10 μm (F). (H to L) PC12 cells transfected (H to J) or not (K and L) with human Grit cDNA were treated with NGF for 2 days, and stained with anti-Grit antibody, PR205 (H to J) or CT205 (K and L). Scale bars: 2 μm (H to J) or 5 μm (K and L). Specificity of anti-Grit staining was checked as follows. (i) Control staining of SH-SY5Y and PC12 cells without primary antibody gave no significant signals at the same dilution of second antibody used here. (ii) Cell staining with a serial dilution of anti-Grit antibodies showed a proportional decrease in specific signals as shown in Fig. 2E-L. (iii) When COS-1 cells were transfected with FLAG-tagged Grit cDNA and costained by anti-Grit PR205 and anti-FLAG antibodies, both staining patterns were identical.
The subcellular localization of Grit was examined in neurite-bearing neuronal cells. In SH-SY5Y human neuroblastoma cells differentiated with retinoic acid, Grit gave a punctate pattern of staining throughout the cytoplasmic compartment with anti-Grit antibody PR205 (Fig. 2E to G). In particular, Grit was also detected at the cell periphery (Fig. 2E) and at the tips of neurites (Fig. 2F and G). To examine Grit localization in PC12 rat pheochromocytoma cells expressing the TrkA receptor, we first transfected PC12 cells with human Grit cDNA and stained them with anti-Grit antibody PR205, because the anti-Grit PR205, with excellent specificity for human Grit (Fig. 2C), was largely unreactive with rodent Grit. The exogenous Grit protein in NGF-treated PC12 cells presented a similar punctate staining (Fig. 2H to J) as the endogenous one in SH-SY5Y cells. Anti-Grit-CT205, having less reactivity than PR205, could recognize rodent Grit (Fig. 2C, lane 7), and the distribution of endogenous Grit in NGF-treated PC12 cells stained with anti-Grit-CT205 (Fig. 2K and L) was essentially the same as that of the exogenous one (Fig. 2H-J).
Interaction between Grit and TrkA receptor.
The cytoplasmic region of TrkA directly interacted with the Grit-CT region (residues 1443 to 1738) in the yeast two-hybrid assay (Fig. 3A). The shortest Grit fragment binding to TrkA in yeast consisted of the CT 53 amino acids, although its affinity for TrkA was considerably lower than that of Grit-CT (not shown). The specific interaction between Grit and TrkA, when the two were coexpressed in mammalian cells, was examined in 293 cells. As shown in Fig. 3B, the interaction between TrkA and the Grit-CT region was readily detected in a coimmunoprecipitation experiment, whereas neither of two other neurotrophin receptors, TrkB and TrkC, bound to the Grit-CT region. We carried out an extensive database search for a sequence homologous to this TrkA-binding CT region but did not find significant homology to any sequence.
Grit bound to TrkA independently of phosphorylation of the receptor. Lysates prepared from mock- or NGF-treated PC12 cells were incubated with GST alone or with GST fusion protein containing Grit-CT. Comparable levels of TrkA were recovered from both cell lysates with GST-Grit-CT, although TrkA did not bind to GST alone (Fig. 3C, top). Tyrosine-phosphorylation of TrkA upon NGF treatment was confirmed (bottom-left).
Grit bound to the C terminus of the TrkA receptor. In vitro-translated wild-type and deletion mutants of TrkA were detected at comparable levels (Fig. 3D, left) and tested for their interaction with Grit by use of the GST pull-down assay. TrkA receptors lacking their most CT part (TrkAΔC1) failed to bind to GST-Grit-CT (Fig. 3D, right).
We investigated by double labeling whether there was overlap in the distribution of endogenous Grit with TrkA in neurite-bearing PC12 cells treated with NGF. As shown in Fig. 2E to L, Grit showed punctate staining, and a substantial pool of Grit colocalized with TrkA along and at the tips of the neurites (Fig. 3E).
It is well-known that the TrkA receptor activated upon NGF stimulation phosphorylates many of its binding molecules (4, 24). Anti-FLAG immunoprecipitates from mock- or NGF-treated 293 cells cotransfected with FLAG-Grit and TrkA were analyzed by antiphosphotyrosine immunoblotting. Grit was clearly tyrosine phosphorylated following NGF stimulation (Fig. 3F).
Grit is a GAP for Rho/Rac/Cdc42 and preferentially stimulates GTP hydrolysis of Rho and Cdc42 over that of Rac.
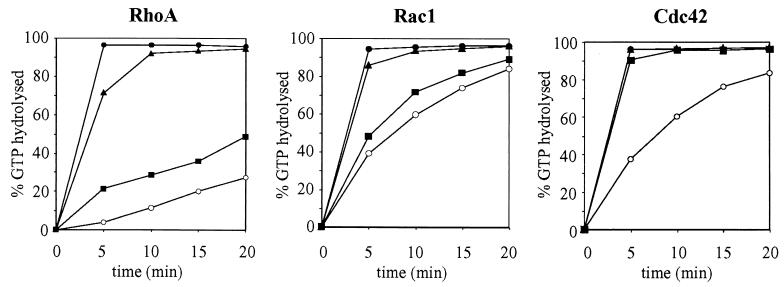
The recombinant RhoGAP domain of Grit was incubated with RhoA, Rac1, or Cdc42 preloaded with [γ-32P]GTP, and the amount of hydrolyzed GTP was measured (Fig. 4). Grit had an in vitro GAP activity toward Rho/Rac/Cdc42; the Grit GAP domain efficiently stimulated GTP hydrolysis of RhoA and Cdc42, whereas the effect on Rac1 was a moderate one (compare the GTP hydrolysis stimulated by 2 nM GAP for 5 min with the corresponding intrinsic GTPase activity).
FIG. 4.
Characterization of in vitro RhoGAP activity of Grit protein. GTPase activation of RhoA, Rac1, and Cdc42 (40 nM each) stimulated by various amounts of recombinant Grit GAP domain (▪, 2 nM; ▴, 20 nM; •, 200 nM RhoGAP) and the intrinsic GTPase activity (○) of the GTPases were measured at 5-min intervals. The percentage γPi associated with the GTPase at each time point was subtracted from 100% to give the percentage of GTP hydrolyzed.
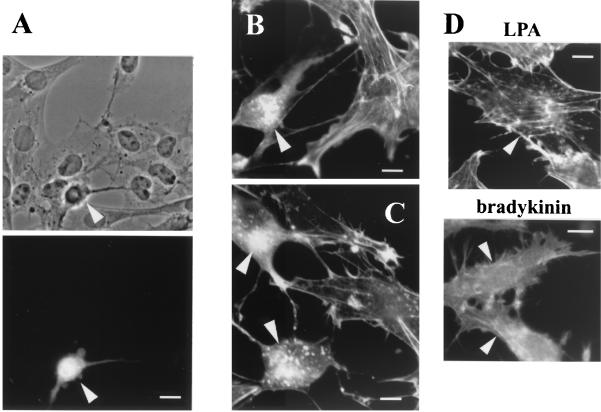
We further examined the effect of the Grit RhoGAP domain on the morphology of Swiss 3T3 fibroblasts, in which RhoA, Rac1, and Cdc42 were shown earlier to elicit distinct morphological changes (36). When the Grit GAP domain was overexpressed, the Swiss 3T3 cells rounded up and extended very long and beaded processes (Fig. 5A, arrowhead); this change closely resembled the morphological change of fibroblasts transfected with p190 RhoGAP (51), which was shown to preferentially regulate GTP hydrolysis of RhoA (43). Next, the recombinant Grit GAP domain was microinjected into serum-starved Swiss 3T3 cells, and its effect on morphological changes induced by extracellular agonists was examined. Rho-dependent, LPA-induced stress fiber formation (Fig. 5B) and formation of Cdc42-dependent, bradykinin-induced filopodia (Fig. 5C) were completely abolished by the Grit GAP microinjection. Although Rac-dependent, platelet-derived growth factor-induced membrane ruffling was also inhibited by the Grit GAP microinjection, a larger amount of RhoGAP was required to inhibit the Rac-dependent actin change than to inhibit RhoA-dependent or Cdc42-dependent structural changes (data not shown); this was in line with the result of the in vitro GAP assay (Fig. 4). Mock injection had no effect on the formation of Rho-dependent, LPA-induced stress fibers or Cdc42-dependent, bradykinin-induced filopodia (Fig. 5D). These results indicate that the Grit GAP domain worked toward Rho/Rac/Cdc42 and preferentially stimulated GTP hydrolysis of RhoA and Cdc42 over that of Rac1. Also, full-length Grit protein exhibited GAP activity for RhoA and Cdc42 (see below, Fig. 7F).
FIG. 5.
Characterization of in vivo RhoGAP activity of Grit protein. (A) Swiss 3T3 cells were transfected with the pIRES-EGFP-Grit GAP domain construct. After 2 days of incubation, phase-contrast (top) and fluorescent (bottom) images were captured with a Zeiss Axiovert microscope. Scale bar, 10 μm. (B and C) Serum-starved Swiss 3T3 cells were microinjected with the purified Grit RhoGAP domain before stimulation with LPA (B) or bradykinin (C). To identify the microinjected cells, we coinjected FITC-dextran along with RhoGAP protein (shown by arrowheads). Actin filaments were visualized by staining with rhodamine-phalloidin. Scale bars, 10 μm. (D) For controls, starved Swiss 3T3 cells were microinjected with FITC-dextran alone (shown by arrowheads) and stimulated with LPA (top) or bradykinin (bottom). Actin filaments were visualized as above. Scale bars, 10 μm.
FIG. 7.
Association of Grit with N-Shc adapter molecule. (A) COS-1 cells were transfected with the FLAG-Grit-CT construct and the indicated T7-tagged Shc family cDNA, serum-starved, and treated with EGF. Anti-FLAG immunoprecipitates were probed with anti-T7 (top) or anti-FLAG (middle, for Grit-CT detection) antibody. Anti-T7 immunoblotting of cell lysates showed comparable levels of expression of Shc family members (bottom). (B) COS-1 cells were cotransfected with T7-tagged wild-type or mutant N-Shc constructs and FLAG-Grit-CT plasmid. Cell lysates and anti-FLAG immunoprecipitates were probed with anti-T7 antibody. (C) Lysates from COS-1 cells transfected with Grit cDNA were incubated with either 20 μg of GST or GST fusion with N-Shc SH2 domain on glutathione-Sepharose. Cell lysates and eluates from the affinity beads were probed with anti-Grit (top) or anti-GST (bottom) antibodies. (D) FLAG-Grit cDNA with or without N-Shc construct was used to transfect COS-1 cells. The transfected cells were deprived of serum and then mock treated or treated with EGF. Anti-T7 immunoprecipitates were probed with anti-Grit (top) or anti-T7 (middle, for N-Shc detection) antibodies. (E) COS-1 cells were transfected with FLAG-Grit cDNA along with N-Shc or N-Shc PTB-CH1 construct as indicated above the panels. The transfected cells were serum starved and treated or not with EGF. Anti-FLAG immunoprecipitates were probed with antiphosphotyrosine or anti-Grit antibodies. Phosphorylated proteins in the top panel were identified by immunoblotting with their respective antibodies (see Fig. 8). Anti-T7 blot of cell lysates confirmed the expression of N-Shc constructs (bottom). (F) COS-1 cells were mock transfected or transfected with FLAG-Grit and N-Shc constructs or FLAG-tagged Grit GAP domain construct, serum starved, and treated with or without EGF. [γ-32P]GTP-loaded RhoA or Cdc42 was incubated with anti-FLAG immunoprecipitates from lysates of the transfected cells, and the hydrolyzed GTP was measured as in Fig. 4.
Effect of wild-type and mutant Grits on NGF-induced neurite outgrowth in PC12 cells.
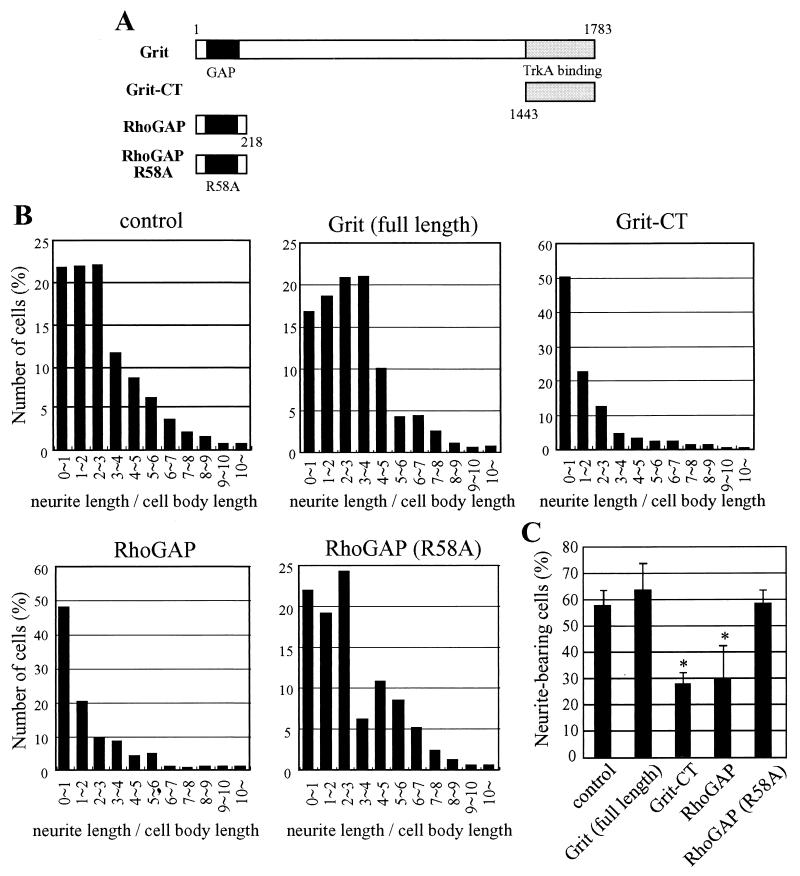
Within a wide range of effects of NGF/TrkA signaling, neurite outgrowth is most closely linked to actin remodeling controlled by Rho GTPases. The PC12 cell has been extensively used for studies on neurite extension due to its ability to undergo differentiation to a neuron-like phenotype in response to NGF. To investigate the function of Grit in neurite outgrowth, we examined the effect of expression of FLAG-tagged wild-type Grit or its mutants (Fig. 6A) in PC12 cells. By anti-FLAG immunoblotting, we estimated that the expression level of full-length Grit was 1/10 to 1/20 of the levels of the three mutant products, possibly due to their different sizes (not shown), and thus the effect of Grit on neurite outgrowth was assessed relative to that in the control cells (transfected with GFP alone). Naive PC12 cells did not show significant neurite outgrowth by the transfection with wild-type or mutant Grits only (not shown). The cells transfected with GFP alone developed neurites by 2.5 days of treatment with NGF (Fig. 6B, control). In contrast, transient overexpression of the Grit-CT region and RhoGAP domain strongly inhibited neurite extension induced by NGF (Fig. 6B). The proportions of neurite-bearing cells were calculated (Fig. 6C). In GFP-transfected cells (control), the proportion of neurite-bearing cells was 58% in the presence of NGF. However, only 28% of the Grit-CT-expressing cells bore neurites even in the presence of NGF (Fig. 6C). These results suggest that Grit-CT, i.e., the TrkA-binding region of Grit, significantly inhibited NGF-induced neurite outgrowth and acted in a dominant-negative manner in NGF signaling. In contrast, the cells overexpressing full-length Grit efficiently developed neurite in response to NGF (Fig. 6B). The proportion of neurite-bearing cells in full-length Grit-expressing cells was 64% (Fig. 6C), indicating that Grit overexpression promoted neurite outgrowth, although not at a significant level. PC12 cells were also transfected with the R58A mutant of full-length Grit (Grit R58A), which was expected to lack most of its GAP activity, and examined in the same way. The proportion of neurite-bearing cells in the Grit R58A-expressing cells (55%; not shown) was comparable to that in the control cells (58% in Fig. 6C). The reason why the dominant-negative effect was observed for the Grit-CT expression but not for the Grit R58A expression could be that the expression level of the former was 10- to 20-fold higher than that of the latter, due to their different sizes. However, at least we can say that the tendency for neurite-promotion in the full-length Grit overexpressing cells upon NGF treatment disappeared in the Grit R58A-expressing cells. In this assay, the Grit GAP domain inhibited neurite extension (neurite bearing, 30%), and this inhibitory effect was not detected in the cells transfected with the R58A mutant of RhoGAP domain (neurite bearing: 59%). Considering the different effects between the Grit GAP domain and full-length Grit, the RhoGAP activity of Grit appears to be regulated by an elaborate mechanism (see Discussion). Taken together, these results suggest that Grit is one of the significant components in NGF-induced neurite outgrowth.
FIG. 6.
Neurite extension assay of wild-type and mutant Grit proteins. Plasmids expressing FLAG-tagged wild-type or mutants of Grit were transfected into PC12 cells, along with pEGFP-C1. The cells were cultured with NGF for 58 h and fixed for microscopy. The neurite extension assay was carried out as described in Materials and Methods. More than three separate experiments were done for each construct, and at least 50 cells were counted for each experiment. (A) Diagrams of FLAG-tagged wild-type and mutants of Grit. (B) The ratio of neurite length/cell body length was divided into 11 segments as indicated under the horizontal axes. For each construct, a histogram plotting the percentage of cell number in each segment is presented. (C) Cells having neurites whose lengths were twofold longer than their cell body lengths were scored as neurite bearing. Results are expressed as the mean percentage of neurite-bearing cells with the standard deviation (error bars). Symbols indicate the results of t test analysis; *, P < 0.01 compared with the control.
Other partners of Grit, N-Shc and CrkL/Crk, are adapter molecules in phosphotyrosine signaling.
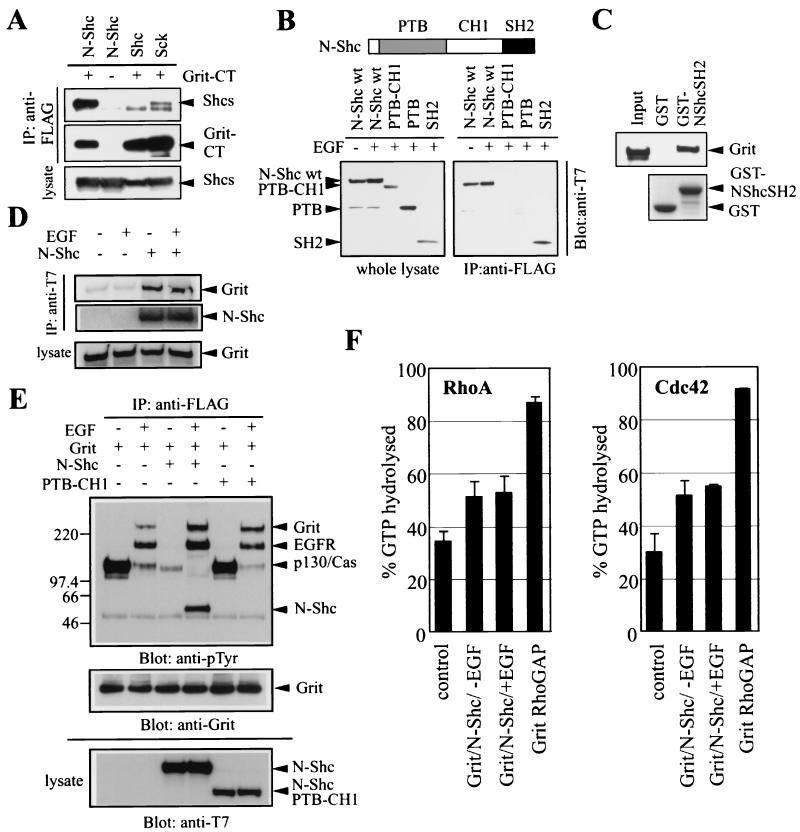
Grit was expressed throughout the nervous system, including the brain (Fig. 2), whereas few neurons in the brain express TrkA receptor in contrast to the predominant expression of TrkA in peripheral neurons (4). The question thus arises as to whether there are any binding partners of Grit in TrkA-negative neurons. To address this issue, we studied the interaction between Grit and members of Shc adapter family (Shc, N-Shc, and Sck). This family has been implicated in neurotrophin signaling in both central and peripheral neurons (11, 33, 34, 44, 50) as well as in many other phosphotyrosine-mediated signalings including the EGF pathway (7, 41). By using EGF-treated COS-1 cells overexpressing FLAG-tagged Grit-CT and T7-tagged Shc family members, we found that N-Shc, a central-neuron specific adapter (34), bound efficiently to Grit-CT (Fig. 7A).
The interaction between Grit and N-Shc occurred through the N-Shc SH2 domain. Wild-type and deletion mutants of N-Shc were expressed at comparable levels in COS-1 cells (Fig. 7B, left) and tested for their interaction with Grit-CT by coimmunoprecipitation. The N-Shc SH2 domain interacted with Grit-CT as effectively as did the wild-type N-Shc, whereas N-Shc mutants lacking the SH2 domain did not bind to Grit-CT (Fig. 7B, right). This association between Grit and the N-Shc SH2 domain was confirmed by the GST pull-down assay (Fig. 7C). Although the SH2 domain is generally thought to be a phosphotyrosine-binding module, the interaction between full-length Grit and N-Shc was detected in both the absence and presence of EGF (Fig. 7D), indicating that this interaction was constitutive, as reported for other SH2-mediated interactions (12, 46).
Since N-Shc was shown to bind to the phosphorylated EGF receptor via its PTB domain (37), the Grit-N-Shc complex may relocate to the activated receptor upon ligand stimulation. To assess this possibility, we transfected COS-1 cells with the FLAG-Grit construct with or without N-Shc constructs. The transfected cells were serum-starved and then stimulated with EGF. Anti-FLAG immunoprecipitates were analyzed by antiphosphotyrosine immunoblotting. Even in the absence of N-Shc, a moderate association between Grit and EGFR was observed following EGF stimulation (lanes 1 and 2 in Fig. 7E); however, as we expected, the amount of coimmunoprecipitated EGFR was significantly increased in the presence of N-Shc (compare lanes 2 and 4). This increase was not detected in the presence of the N-Shc PTB-CH1 mutant, which did not bind to Grit (lane 6). Also, a trace level of Grit tyrosine-phosphorylation was observed upon EGF stimulation (lane 2), and its level was clearly increased in the presence of N-Shc (lane 4). These results indicate that Grit was recruited to the activated EGF receptor and tyrosine-phosphorylated following ligand stimulation, at least in part, through its interaction with N-Shc.
Next, we measured the RhoGAP activity of full-length Grit protein and examined the effect of Grit tyrosine-phosphorylation on its GAP activity. To increase Grit phosphorylation, we cotransfected COS-1 cells with FLAG-Grit and N-Shc cDNAs as in Fig. 7E. The transfected cells were lysed for immunoprecipitation with anti-FLAG antibody. The immunoprecipitated Grit was incubated with [γ-32P]GTP-loaded RhoA or Cdc42, and the GTP hydrolysis was measured. Similarly, the transfected FLAG-tagged Grit GAP domain was recovered, and its GAP activity was measured. The amount of Grit protein was estimated to be 1/10 to 1/20 of that of Grit GAP domain by anti-FLAG immunoblotting (not shown). As shown in the left panel of Fig. 7F, the immunoprecipitated Grit GAP domain exhibited a 2.6-fold increase in RhoA-directed GAP activity (lane 4) compared with the mock immunoprecipitate from untransfected cells (lane 1). Grit proteins from EGF-treated and untreated cells each exhibited an approximately 50% increase (lanes 2 and 3). Likewise, when a 3.1-fold increase in Cdc42-directed GAP activity was detected for the Grit GAP domain, the Grit protein displayed a 70 to 80% increase in the Cdc42GAP activity with no marked difference between mock- and EGF-treated conditions (Fig. 7F, right). These results indicate that full-length Grit could stimulate GTP hydrolysis of Rho GTPases and that there was no evidence for the effect of Grit phosphorylation on its GAP activity.
Figure 7E shows that Grit was associated directly or indirectly with a highly phosphorylated protein (p130) in unstimulated cells and that the binding was abruptly decreased either by stimulation of the cells with EGF or by expression of N-Shc. We found that this p130 protein was Cas. Coimmunoprecipitation using Grit deletion mutants revealed that p130 interacted with the Grit-CT region (data not shown); this was confirmed by the GST pull-down assay (Fig. 8A, top left). Two-hybrid screening using Grit-CT as a bait revealed Crk, an SH2-SH3 containing adapter (32), as a candidate partner of Grit-CT (data not shown). This is consistent with the existence of SH3 binding motifs in the Grit-CT region (Fig. 1A). Among Crk SH2-binding molecules, Cas is a tyrosine-phosphorylated protein of 130 kDa (45). Therefore, we presumed a Grit/Crk/Cas complex formation, and so we examined this presumption by conducting a pull-down assay using GST-Grit-CT affinity beads. As shown in Fig. 8A, the Grit-CT-interacting p130 was Cas (left panels). Crk and its cousin CrkL were also recovered on GST-Grit-CT beads from PC12 cell lysates, although Grit-CrkL binding was severalfold tighter than that of Grit-Crk (right). In PC12 cells, constitutive interaction between the transfected Grit-CT protein and CrkL was observed (Fig. 8B), indicating that separate pools of Grit bound to TrkA and CrkL/Crk adapter in TrkA-positive cells. The presence of full-length Grit/CrkL/Cas complex in mammalian cells was confirmed by immunoprecipitation of lysates from serum-starved COS-1 cells transfected with Grit cDNA (Fig. 8C); furthermore, a prominent increase in Cas was observed in the anti-Grit immunoprecipitate when CrkL was overexpressed with Grit in COS-1 cells (data not shown). Since EGF induced the binding of Crk/CrkL to EGFR (15, 19), Fig. 7E suggests that the Grit/CrkL/Cas complex might be replaced by the Grit/CrkL/EGFR complex following EGF stimulation; in fact, a similar exchange of Crk's partner upon ligand stimulation was previously observed (25).
To examine the possible partner exchange of Grit-CrkL upon EGF stimulation, we transfected COS-1 cells with FLAG-Grit-CT, serum-starved them, and then treated them with EGF for the periods indicated in Fig. 8D. EGF-induced association between Grit and phospho-EGFR (top panels) and a concurrent decrease in Grit-phospho-Cas binding (middle panels) were observed. Grit-CrkL binding remained at a significant level during this period (bottom right). Because N-Shc bound to the Grit-CT region (Fig. 7A and B), and CrkL/Crk bound to the same region of Grit (Fig. 8A, B and D), N-Shc-Grit as well as CrkL/Crk-Grit interactions were independent and competed with each other; and this could account for the large decrease in CrkL/Crk-binding Cas protein in the Grit-containing complex by N-Shc expression (Fig. 7E).
Many SH2-containing signaling molecules, including Crk/CrkL, are localized mostly in the cytoplasm and are translocated to the plasma membrane upon growth factor stimulation (40); thus, the partner exchange of Grit-CrkL upon ligand stimulation could lead to translocation to the cell periphery. This possibility was examined by double labeling of Grit and Cas in mock- or EGF-treated SH-SY5Y cells. In the absence of EGF, considerable overlap in the distribution of Grit and Cas was observed (Fig. 8E, top). Upon EGF treatment, Grit was clearly translocated to the edge of the cell periphery, whereas Cas mostly remained in the cytoplasm (the second panels). Likewise, a substantial Grit relocation occurred in membrane ruffles, and the Grit concentration into the ruffles was more prominent than that of Cas (the third panels). Upon EGF treatment, the relocated Grit colocalized with a subset of EGFR in the cell periphery (Fig. 8E, bottom panels), and the accumulation of phosphorylated Crk at membrane ruffles following EGF stimulation was previously reported (19); therefore, the Grit translocation could occur, at least in part, through Grit-CrkL/Crk complex and its partner exchange from Cas to EGFR.
DISCUSSION
The control of activities of individual Rho GTPases is crucial to neurotrophin-induced neurite outgrowth (3, 58, 60). Our results suggest that Grit, a novel TrkA-interacting protein, regulates neurite outgrowth through modulating Rho GTPases. In PC12 cells, NGF oppositely regulated the activities of Rac1 and RhoA, activating Rac1 and suppressing RhoA (58). Sos1 has been implicated in NGF-induced Rac activation, and the Sos1/E3b1/eps8 complex, endowed with Rac-specific GEF activity, was proposed to be located somewhat distantly from the activated receptors (21). In contrast, RhoA was shown to interact constitutively with p75NTR, a low-affinity neurotrophin receptor (59). While p75NTR-mediated down-regulation of RhoA upon neurotrophin stimulation was previously reported (59), we assume a possible involvement of TrkA/Grit in the NGF-induced RhoA suppression to explain the result of our neurite extension assay (Fig. 6), because RhoA was one of preferred targets of Grit GAP activity (Fig. 4 and 5). The association between Trk and p75NTR was demonstrated previously, and neurotrophin was suggested to reinforce the Trk-p75NTR interaction (5). Based on this finding, it is possible that TrkA/Grit could be prompted to associate with p75/RhoA complex upon NGF stimulation; Grit could thereby down-regulate RhoA in close proximity to the two neurotrophin receptors and positively contribute to NGF-induced neurite outgrowth.
The remarkable difference between the positive effect of the full-length Grit protein and the negative one of the Grit GAP domain on NGF-induced neurite outgrowth supports the idea that RhoGAP activity of the Grit GAP domain should be properly regulated spatiotemporally by other domains including the TrkA-binding region. As in the case of p190, overexpression of Rho-specific GAP could promote neurite outgrowth (9). However, Grit had a GAP activity toward Rho/Rac/Cdc42; thus, it is not surprising that overexpression of the Grit GAP domain throughout the cell (free from the spatiotemporal control by other domains of Grit) did not accelerate neurite extension, because down-regulation of Rho could increase neurite production, whereas down-regulation of Rac/Cdc42 could decrease neurite outgrowth (26, 30). During the 2.5-day incubation in the neurite extension assay, a large excess of Grit GAP domain markedly reduced cell adhesion; the number of Grit GAP domain-expressing cells (mean ± standard deviation) that remained on the substrate was 47.5% ± 6.0% relative to that of controls (100%), whereas the RhoGAP R58A expression did not affect cell adhesiveness (104.0% ± 26.3%). Because the neurite growth rate is changed depending on the strength of cell adhesion (29), the decrease in neurite outgrowth in the Grit GAP domain-expressing cells could be largely attributed to the decreased adhesiveness.
Another pool of Grit was associated with phosphotyrosine adapter molecules, N-Shc and CrkL/Crk, and thereby could be relocated to a close vicinity of activated receptor tyrosine kinases. Membrane relocation has been described for other Rho regulators, e.g., Tiam1 (10), KIAA0380 (53), and p190 RhoGAP (8). However, to our knowledge, the present study provides the first demonstration of a relocation mechanism through a direct adapter binding of a RhoGAP. The relocated Grit would down-regulate Rho/Rac/Cdc42 depending on which GTPase was positioned close to the activated receptors. The N-Shc-Grit interaction and the CrkL/Crk-Grit interaction were independent, and thus Grit relocation was separately mediated by N-Shc and CrkL/Crk. In the case of neurotrophin stimulation, both N-Shc and Crk/CrkL can recruit their partners to the Trk-containing complex (34, 54, 57). Thus, we suppose that Grit is recruited to this complex and may turn off RhoA also relocated to this complex through the induced Trk-p75 interaction, thereby contributing to neurotrophin-induced neurite outgrowth.
GAP activity itself can be modulated, and the regulation of RhoGAP activity has been classified into two major mechanisms, i.e., RhoGAP phosphorylation and protein-protein interaction. Tyrosine phosphorylation of p190 RhoGAP by Src or focal adhesion kinase has been proposed to regulate its GAP activity (2, 14, 42). A similar regulation was reported for other GEF/GAPs (18, 28). However, in the present study, neither an increase nor a decrease in Grit GAP activity was detected following its tyrosine phosphorylation, and thus the functional significance of Grit phosphorylation remains to be clarified. The regulation of GAP activity via protein-protein interaction was recently demonstrated for CdGAP (23), another member of the CdGAP/Grit family. The CdGAP sequence containing five SH3 binding motifs was responsible for down-regulation of GAP activity by the SH3-containing protein intersectin, and showed a partial homology to the corresponding Grit sequence having three SH3 binding motifs. We have obtained several candidate partners containing SH3 domains by the two-hybrid screening using this region of Grit as a bait (T. Nakamura and N. Mori, unpublished data). Modulation of Grit GAP activity by these candidate binders will be a subject of future research.
Accumulating evidence indicates the critical roles of Rho GTPases in neuronal morphogenesis, including axon growth and guidance (19, 22, 31), dendrite elaboration (31, 62), migration, polarity, and plasticity (reviewed in reference 30). A variety of effects of different Rho GTPases in different neuronal compartments indicate that functions and regulations of Rho GTPases may be more complex in neurons than in fibroblasts (30). Further, the expression of constitutive-active and dominant-negative Rho GTPase mutants often produced similar phenotypes (1, 31, 62), indicating that Rho GTPase signaling pathway has a cyclic mode of action, particularly prominent in neuronal cells, e.g., a rapid cycling between extension and retraction of filopodia during axonal growth. These views make it increasingly important to elucidate the elaborate mechanism of spatiotemporal control of Rho GTPase activity in individual neuronal compartments. Recently identified Rho GTPase regulators (GEF/GAPs) associated with specific receptors (48, 52, 56), including Grit, should advance our understanding of these signaling pathways linking extracellular cues, receptors, and molecular switches (GTPases).
Acknowledgments
We are grateful to Kimiko Watanabe, Itsuko Nakano, and Yumiko Kadokawa for their technical assistance; to Seisuke Hattori for valuable advice on the GAP assay; to Michiyuki Matsuda for directional advice; and to M. Barbacid and G. Yancopoulos for providing plasmids.
This work was supported by a grant from CREST, JST, and also, in part, by the Strategic Promotion System for Brain Science from STA, the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim, and Funds for Comprehensive Research on Aging and Health from the Ministry of Health and Welfare (to N.M.).
REFERENCES
- 1.Albertinazzi, C., D. Gilardelli, S. Paris, R. Longhi, and I. de Curtis. 1998. Overexpression of a neural-specific rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons. J. Cell Biol. 142:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, W. T., L. A. Petch, and K. Burridge. 2000. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr. Biol. 10:719-722. [DOI] [PubMed] [Google Scholar]
- 3.Banzai, Y., H. Miki, H. Yamaguchi, and T. Takenawa. 1992. 2000. Essential role of neural Wiskott-Aldrich syndrome protein in neurite extension in PC12 cells and rat hippocampal primary culture cells. J. Biol. Chem. 275:11987-11991. [DOI] [PubMed] [Google Scholar]
- 4.Bibel, M., and Y. A. Barde. 2000. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 14:2919-2937. [DOI] [PubMed] [Google Scholar]
- 5.Bibel, M., E. Hoppe, and Y. A. Barde. 1999. Biochemical and functional interactions between the neurotrophin receptors trk and p75NTR. EMBO J. 18:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 7.Bonfini, L., E. Migliaccio, G. Pelicci, L. Lanfrancone, and P. G. Pelicci. 1996. Not all Shc's roads lead to Ras. Trends Biol. Sci. 21:257-261. [PubMed] [Google Scholar]
- 8.Brouns, M. R., S. F. Matheson, K. Q. Hu, I. Delalle, V. S. Caviness, J. Silver, R. T. Bronson, and J. Settleman. 2000. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development 127:4891-4903. [DOI] [PubMed] [Google Scholar]
- 9.Brouns, M. R., S. F. Matheson, and J. Settleman. 2001. p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat. Cell Biol. 3:361-367. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, F. G., C. M. Elliot, M. Gibbs, and J. H. Exton. 2000. Translocation of the Rac1 guanine nucleotide exchange factor Tiam1 induced by platelet-derived growth factor and lysophosphatidic acid. J. Biol. Chem. 275:9742-9748. [DOI] [PubMed] [Google Scholar]
- 11.Cattaneo, E., and P. G. Pelicci. 1998. Emerging roles for SH2/PTB-containing Shc adaptor proteins in the developing mammalian brain. Trends Neurosci. 21:476-481. [DOI] [PubMed] [Google Scholar]
- 12.De Sepulveda, P., S. Ilangumaran, and R. Rottapel. 2000. Suppressor of cytokine signaling-1 inhibits VAV function through protein degradation. J. Biol. Chem. 275:14005-14008. [DOI] [PubMed] [Google Scholar]
- 13.Dickson, B. J. 2001. Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 11:103-110. [DOI] [PubMed] [Google Scholar]
- 14.Dumenil, G., P. Sansonetti, and V. N. Tran. 2000. Src tyrosine kinase activity down-regulates Rho-dependent responses during Shigella entry into epithelial cells and stress fibre formation. J. Cell Sci. 113:71-80. [DOI] [PubMed] [Google Scholar]
- 15.Fukazawa, T., S. Miyake, V. Band, and H. Band. 1996. Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins. J. Biol. Chem. 271:14554-14559. [DOI] [PubMed] [Google Scholar]
- 16.Gulli, M. P., and M. Peter. 2001. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 15:365-379. [DOI] [PubMed] [Google Scholar]
- 17.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 18.Han, J., B. Das, W. Wei, L. Van Aelst, R. D. Mosteller, R. Khosravi-Far, J. K. Westwick, C. J. Der, and D. Broek. 1997. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 17:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto, Y., H. Katayama, E. Kiyokawa, S. Ota, T. Kurata, N. Gotoh, N. Otsuka, M. Shibata, and M. Matsuda. 1998. Phosphorylation of CrkII adaptor protein at tyrosine 221 by epidermal growth factor receptor. J. Biol. Chem. 273:17186-17191. [DOI] [PubMed] [Google Scholar]
- 20.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Innocenti, M., P. Tenca, E. Frittoli, M. Faretta, A. Tocchetti, P. P. Di Fiore, and G. Scita. 2002. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 156:125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jalink, K., E. J. van Corven, T. Hengeveld, N. Morii, S. Narumiya, and W. H. Moolenaar. 1994. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J. Cell Biol. 126:801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenna, S., N. K. Hussain, E. I. Danek, I. Triki, S. Wasiak, P. S. McPherson, and N. Lamarche-Vane. 2002. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J. Biol. Chem. 277:6366-6373. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan, D. R., and F. D. Miller. 2000. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 10:381-391. [DOI] [PubMed] [Google Scholar]
- 25.Khwaja, A., B. Hallberg, P. H. Warne, and J. Downward. 1996. Networks of interaction of p120cbl and p130cas with Crk and Grb2 adaptor proteins. Oncogene 12:2491-2498. [PubMed] [Google Scholar]
- 26.Kozma, R., S. Sarner, S. Ahmed, and L. Lim. 1997. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamarche-Vane, N., and A. Hall. 1998. CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J. Biol. Chem. 273:29172-29177. [DOI] [PubMed] [Google Scholar]
- 28.Lanzetti, L., V. Rybin, M. G. Malabarba, S. Christoforidis, G. Scita, M. Zerial, and P. P. Di Fiore. 2000. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408:374-377. [DOI] [PubMed] [Google Scholar]
- 29.Lockerbie, R. O. 1987. The neuronal growth cone: a review of its locomotory, navigational and target recognition capabilities. Neuroscience 20:719-729. [DOI] [PubMed] [Google Scholar]
- 30.Luo, L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173-180. [DOI] [PubMed] [Google Scholar]
- 31.Luo, L., Y. J. Liao, L. Y. Jan, and Y. N. Jan. 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8:1787-1802. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda, M., and T. Kurata. 1996. Emerging components of the Crk oncogene product: the first identified adaptor protein. Cell Signal. 8:335-340. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, T., M. Komiya, N. Gotoh, S. Koizumi, M. Shibuya, and N. Mori. 2002. Discrimination between phosphotyrosine-mediated signaling properties of conventional and neuronal Shc adapter molecules. Oncogene 21:22-31. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, T., S. Muraoka, R. Sanokawa, and N. Mori. 1998. N-Shc and Sck, two neuronally expressed Shc adapter homologs. Their differential regional expression in the brain and roles in neurotrophin and Src signaling. J. Biol. Chem. 273:6960-6967. [DOI] [PubMed] [Google Scholar]
- 35.Nakazawa, T., I. Nakano, M. Sato, T. Nakamura, M. Tamai, and N. Mori. 2002. Comparative expression profiles of Trk receptors and Shc-related phosphotyrosine adapters during retinal development: potential roles of N-Shc/ShcC in brain-derived neurotrophic factor signal transdcution and modulation. J. Neurosci. Res. 68:668-680. [DOI] [PubMed] [Google Scholar]
- 36.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 37.O'Bryan, J. P., Z. Songyang, L. Cantley, C. J. Der, and T. Pawson. 1996. A mammalian adaptor protein with conserved Src homology 2 and phosphotyrosine-binding domains is related to Shc and is specifically expressed in the brain. Proc. Natl. Acad. Sci. USA 93:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta, Y., N. Suzuki, S. Nakamura, J. H. Hartwig, and T. P. Stossel. 1999. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. USA 96:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, T. D., A. Jackman, F. L. Rice, J. Kucera, and W. D. Snider. 2000. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron 25:345-357. [DOI] [PubMed] [Google Scholar]
- 40.Pawson, T., and G. D. Gish. 1992. SH2 and SH3 domains: from structure to function. Cell 71:359-362. [DOI] [PubMed] [Google Scholar]
- 41.Ravichandran, K. S. 2001. Signaling via Shc family adapter proteins. Oncogene 20:6322-6330. [DOI] [PubMed] [Google Scholar]
- 42.Ren, X. D., W. B. Kiosses, D. J. Sieg, C. A. Otey, D. D. Schlaepfer, and M. A. Schwartz. 2000. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J. Cell Sci. 113:3673-3678. [DOI] [PubMed] [Google Scholar]
- 43.Ridley, A. J., A. J. Self, F. Kasmi, H. F. Paterson, A. Hall, C. J. Marshall, and C. Ellis. 1993. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozakis-Adcock, M., J. McGlade, G. Mbamalu, G. Pelicci, R. Daly, W. Li, A. Batzer, S. Thomas, J. Brugge, P. G. Pelicci, J. Schlessinger, and T. Pawson. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360:689-692. [DOI] [PubMed] [Google Scholar]
- 45.Sakai, R., A. Iwamatsu, N. Hirano, S. Ogawa, T. Tanaka, H. Mano, Y. Yazaki, and H. Hirai. 1994. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 13:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmandt, R., S. K. Liu, and C. J. McGlade. 1999. Cloning and characterization of mPAL, a novel Shc SH2 domain-binding protein expressed in proliferating cells. Oncogene 18:1867-1879. [DOI] [PubMed] [Google Scholar]
- 47.Sekiguchi, T., E. Hirose, N. Nakashima, M. Ii, and T. Nishimoto. 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276:7246-7257. [DOI] [PubMed] [Google Scholar]
- 48.Shamah, S. M., M. Z. Lin, J. L. Goldberg, S. Estrach, M. Sahin, L. Hu, M. Bazalakova, R. L. Neve, G. Corfas, A. Debant, and M. E. Greenberg. 2001. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 105:233-244. [DOI] [PubMed] [Google Scholar]
- 49.Song, H. J., and M. M. Poo. 1999. Signal transduction underlying growth cone guidance by diffusible factors. Curr. Opin. Neurobiol. 9:355-363. [DOI] [PubMed] [Google Scholar]
- 50.Stephens, R. M., D. M. Loeb, T. D. Copeland, T. Pawson, L. A. Greene, and D. R. Kaplan. 1994. Trk receptors use redundant signal transduction pathways involving SHC and PLC-γ1 to mediate NGF responses. Neuron 12:691-705. [DOI] [PubMed] [Google Scholar]
- 51.Tatsis, N., D. A. Lannigan, and I. G. Macara. 1998. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J. Biol. Chem. 273:34631-34638. [DOI] [PubMed] [Google Scholar]
- 52.Taya, S., N. Inagaki, H. Sengiku, H. Makino, A. Iwamatsu, I. Urakawa, K. Nagao, S. Kataoka, and K. Kaibuchi. 2001. Direct interaction of insulin-like growth factor-1 receptor with leukemia-associated RhoGEF. J. Cell Biol. 155:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Togashi, H., K. Nagata, M. Takagishi, N. Saitoh, and M. Inagaki. 2000. Functions of a rho-specific guanine nucleotide exchange factor in neurite retraction. Possible role of a proline-rich motif of KIAA0380 in localization. J. Biol. Chem. 275:29570-29578. [DOI] [PubMed] [Google Scholar]
- 54.Torres, M., and E. Bogenmann. 1996. Nerve growth factor induces a multimeric TrkA receptor complex in neuronal cells that includes Crk, SHC and PLC-γ1 but excludes p130(CAS). Oncogene 12:77-86. [PubMed] [Google Scholar]
- 55.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 56.Wong, K., X. R. Ren, Y. Z. Huang, Y. Xie, G. F. Liu, H. Saito, H. Tang, L. Wen, S. M. Brady-Kalnay, L. Mei, J. Y. Wu, W. C. Xiong, and Y. Rao. 2001. Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107:209-221. [DOI] [PubMed] [Google Scholar]
- 57.Wu, C., C. F. Lai, and W. C. Mobley. 2001. Nerve growth factor activates persistent Rap1 signaling in endosomes. J. Neurosci. 21:5406-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaguchi, Y., H. Katoh, H. Yasui, K. Mori, and M. Negishi. 2001. RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J. Biol. Chem. 276:18977-18983. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita, T., K. L. Tucker, and Y. A. Barde. 1999. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron 24:585-593. [DOI] [PubMed] [Google Scholar]
- 60.Yasui, H., H. Katoh, Y. Yamaguchi, J. Aoki, H. Fujita, K. Mori, and M. Negishi. 2001. Differential responses to nerve growth factor and epidermal growth factor in neurite outgrowth of PC12 cells are determined by Rac1 activation systems. J. Biol. Chem. 276:15298-15305. [DOI] [PubMed] [Google Scholar]
- 61.Yu, H. T., J. K. Chen, S. B. Feng, D. C. Dalgarno, A. W. Brauer, and S. L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933-945. [DOI] [PubMed] [Google Scholar]
- 62.Zipkin, I. D., R. M. Kindt, and C. J. Kenyon. 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90:883-894. [DOI] [PubMed] [Google Scholar]