Abstract
snm1 mutants of Saccharomyces cerevisiae have been shown to be specifically sensitive to DNA interstrand crosslinking agents but not sensitive to monofunctional alkylating agents, UV, or ionizing radiation. Five homologs of SNM1 have been identified in the mammalian genome and are termed SNM1, SNM1B, Artemis, ELAC2, and CPSF73. To explore the functional role of human Snm1 in response to DNA damage, we characterized the cellular distribution and dynamics of human Snm1 before and after exposure to DNA-damaging agents. Human Snm1 was found to localize to the cell nucleus in three distinct patterns. A particular cell showed diffuse nuclear staining, multiple nuclear foci, or one or two larger bodies confined to the nucleus. Upon exposure to ionizing radiation or an interstrand crosslinking agent, the number of cells exhibiting Snm1 bodies was reduced, while the population of cells with foci increased dramatically. Indirect immunofluorescence studies also indicated that the human Snm1 protein colocalized with 53BP1 before and after exposure to ionizing radiation, and a physical interaction was confirmed by coimmunoprecipitation assays. Furthermore, human Snm1 foci formed after ionizing radiation were largely coincident with foci formed by human Mre11 and to a lesser extent with those formed by BRCA1, but not with those formed by human Rad51. Finally, we mapped a region of human Snm1 of approximately 220 amino acids that was sufficient for focus formation when attached to a nuclear localization signal. Our results indicate a novel function for human Snm1 in the cellular response to double-strand breaks formed by ionizing radiation.
The cellular response to DNA damage is a complex interacting network of pathways that mediate cell cycle checkpoints, DNA repair, and apoptosis. A model lesion for the investigation of these pathways has been DNA double-strand breaks, which rapidly induce cell cycle checkpoints and are repaired by a number of different pathways. In Saccharomyces cerevisiae, these lesions are preferentially repaired by a homologous recombination pathway, which requires proteins from the RAD52 epistasis group. In mammalian cells, both homologous recombination and nonhomologous recombination pathways are utilized. Extensive studies in mammalian cells have shown that complexes of DNA repair and cell cycle checkpoint proteins rapidly localize to sites of double-strand breaks induced by ionizing radiation and create foci that can be detected by immunofluorescent analyses.
Two types of mutually exclusive or nonoverlapping ionizing radiation-induced foci have been observed, those containing the Rad51 protein and those containing the Mre11-Rad50-NBS1 complex (13, 25). Rad51 foci, which contain the tumor suppressor proteins BRCA1 and BRCA2, also appear during S phase in the absence of exogenous induction of DNA damage (5, 29, 43-45). Mre11-Rad50-NBS1 foci can be detected as early as 10 min after irradiation and are clearly present at sites of DNA breaks while DNA repair is ongoing (25, 28, 33). These foci also colocalize with the BRCA1 protein, which has been shown to be required for their formation, possibly through its physical interaction with human Rad50 (hRad50) (53). In addition, coimmunoprecipitation experiments performed with BRCA1 have indicated the presence of a large number of additional proteins in this complex (referred to as the BRCA1-associated surveillance complex) (47). These include the mismatch repair proteins Msh2, Msh6, and Mlh1, the checkpoint kinase ATM, the product of the Bloom's syndrome gene BLM, and replication factor C. The product of the Fanconi's anemia group D2 gene (FANCD2) has also been shown to colocalize with BRCA1 ionizing radiation-induced foci (11). BRCA1, NBS1, and hMre11 have all been shown to be substrates of the ATM kinase and to become phosphorylated in response to the presence of DNA breaks (6, 7, 12, 13, 21, 49, 52).
More recently, two additional proteins have been shown to be involved in the very early stages of ionizing radiation-induced focus formation. H2AX, a variant histone H2A molecule, is phosphorylated within 1 to 3 min after exposure to ionizing radiation, and this modified form, known as γ-H2AX, is associated with the sites of double-strand breaks (35, 39). Also, p53-binding protein 1 (53BP1) is rapidly phosphorylated and relocalized to foci that overlap those containing γ-H2AX, and the Mre11-Rad50-NBS1 complex is subsequently recruited to the same sites (2, 37, 42, 50). The rapid localization of γ-H2AX and 53BP1 to sites of double-strand breaks suggests that they are involved in the initial stages of DNA damage detection and/or processing.
The snm1-1 (for sensitivity to nitrogen mustard) mutant allele of S. cerevisiae was discovered in a screen for mutants lacking resistance to the bifunctional alkylating agent nitrogen mustard (40). Mutations in SNM1 were later found to be allelic to mutations in the PSO2 gene, which had previously been identified by selecting for mutants sensitive to psoralen plus UVA (15). Further characterization of these mutants has shown that they are specifically sensitive to bi- or polyfunctional alkylating agents while exhibiting nearly wild-type levels of sensitivity to UV or ionizing radiation and to monofunctional alkylating agents.
Molecular analysis of snm1 mutants treated with either nitrogen mustard or psoralen plus UVA showed that incision near the sites of interstrand crosslinks proceeded normally; however, a late step in interstrand crosslink repair that is required for restoration of high-molecular-weight DNA appeared to be defective (24, 27). Molecular cloning of the yeast SNM1 gene showed that it encoded a novel 76-kDa protein with a nuclear localization signal and one putative zinc finger motif (14, 38). The SNM1 gene is, under normal conditions, poorly transcribed (about 0.3 transcripts per cell), but a fourfold induction can be seen within 4 h after treatment with agents that induce interstrand crosslinks, but not by monofunctional alkylating agents or the UV-mimetic agent 4-nitroquinoline-1-oxide (48). Interestingly, induction of SNM1 is dependent upon the DUN1 gene, which is known to be required for transcription of genes induced by yeast cell cycle checkpoint pathways activated by DNA damage or replication blocks (10). The biochemical function of the Snm1 protein remains undetermined.
Dronkert et al. (9) reported the identification of three human cDNAs that are homologous to the yeast SNM1 sequence, referred to as hSNM1, hSNM1B, and hSNM1C (now referred to as Artemis). The three proteins encoded by these genes share a region of approximately 300 amino acids that are similar to the yeast protein but are otherwise unique. This homologous region encodes a metallo-β-lactamase domain (31) and has been found in two other mammalian proteins, CPSF73, an mRNA cleavage and polyadenylation factor (16), and ELAC2, a putative prostate cancer protein (46). Dronkert et al. (9) characterized the phenotype of mouse embryonic stem cells that were homozygous for a disrupted SNM1 allele. These cells exhibited a slight increase (twofold) in sensitivity to mitomycin C compared to wild-type cells but not to other crosslinking agents or to ionizing radiation. This low level of sensitivity may be due to functional redundancy between the mammalian homologs or to the fact that this allele did not result in a null phenotype and may have retained partial function.
Mutations in Artemis have recently been associated with a form of radiosensitive severe combined immunodeficiency syndrome that exhibits a deficiency in V(D)J recombination (31). In addition, Artemis has been shown to contain a nuclease in its conserved metallo-β-lactamase domain that cleaves the hairpin at the end of coding joints during V(D)J recombination (23).
Here we report on the cellular dynamics of human Snm1 protein upon treatment with DNA-damaging agents. We show that hSnm1 forms damage-inducible foci that are largely coincident with those formed by hMre11. In addition, hSnm1 colocalizes with the 53BP1 protein before and after exposure to DNA damage and physically associates with this protein. Lastly, we mapped the domain of hSnm1 that is responsible for focus formation to a region of 220 amino acids. These findings suggest that hSnm1 likely plays a role in the cellular response to genotoxic agents.
MATERIALS AND METHODS
Cell culture and DNA transfections.
HeLa, HEK293, HT-1080, and MCF-7 cells were grown in minimal essential medium (MEM) supplemented with 10% fetal calf serum. Nijmegen breakage syndrome and normal human fibroblast cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Both transient and stable DNA transfections were performed with the FuGENE6 transfection reagent according to the manufacturer's recommendations (Roche Molecular Biochemicals).
Plasmids and constructs.
The original plasmid containing the human SNM1 cDNA (KIAA0086) was obtained from Takahiro Nagase (32). PCR was performed to add the Flag epitope in frame with the 5′ end of the human SNM1 cDNA with the primers 5′-ATCTCGAGCCATGGACTACAAGGACGACGATGACAAGGTATACGTCGACATGTTAGAAGACATTTCCGAA (sense) and CATCATAAACTGGACGTATCT (antisense). The 407-bp PCR product was digested with XhoI and PmlI and inserted into the KIAA0086 plasmid at the same sites. The sequence fidelity of the resulting plasmid (pBS-Flag-hSNM1) was confirmed by DNA sequencing. The Flag-hSnm1 open reading frame was subsequently cloned into the mammalian expression vector pEBS7 (36) and also fused with enhanced green fluorescent protein (EGFP) with the pEGFP-C1 vector from Clontech. The pEGFP-hSNM1 plasmid was cut with combinations of several restriction enzymes and religated, resulting in fusion proteins containing various in-frame deletions or truncations of hSNM1.
The EGFP-hSNM1 open reading frame was subsequently cloned into the pShuttle-CMV plasmid and recombined with the adenovirus genome with the pAd-Easy kit from Stratagene. Recombinant virus was obtained by transfection of the linearized pAd-EGFP-hSNM1 plasmid into HEK293 packaging cells and used according to the manufacturer's protocols.
RNA hybridizations.
Multitissue RNA blots were purchased from Clontech and hybridized with a 32P-labeled probe corresponding to the full-length insert isolated from the hSNM1 clone KIAA0086.
Antibodies and immunoblotting.
A bacterially expressed recombinant protein consisting of a hexahistidine tag fused to the last 268 amino acids of the hSnm1 open reading frame was purified by nickel ion affinity chromatography. Polyclonal antibodies against this polypeptide were raised in rabbits following standard protocols. hSnm1 antiserum was affinity purified against antigen that had been blotted and immobilized on nitrocellulose paper.
Monoclonal antibodies specific for hRad51 were provided by Eva Lee (University of Texas Health Science Center at San Antonio). Monoclonal anti-hMre11 and anti-Rad50 antibodies were provided by John Petrini (Memorial Sloan-Kettering Cancer Center) or purchased from Genetex, and a monoclonal antibody to 53BP1 was provided by Thanos Halozonetis (Wistar Institute) (42). Monoclonal antibodies against BRCA1 were purchased from Oncogene Research. Details on the polyclonal antibodies raised against the first 524 amino acid residues of 53BP1 will be published elsewhere (Morales et al., unpublished data). Monoclonal antibodies against α-tubulin and the Flag epitope (M2) were purchased from Sigma. Antibodies against the GFP protein were purchased from Clontech.
HeLa whole-cell extracts and HEK293 mini-nuclear extracts were prepared as described before (20, 41). Samples were combined with 1 volume of sodium dodecyl sulfate loading buffer and separated by denaturing polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose, and immunoblotting was performed with standard protocols with antibodies diluted in binding solution (1% dry milk protein, 0.5% Tween 20, Tris-buffered saline, pH 7.0), and detection was performed with the enhanced chemiluminescent assay (Amersham Pharmacia Biotech).
In vitro translation and immunoprecipitation.
Flag-hSnm1 protein labeled with [35S]methionine was produced in vitro with the TNT coupled reticulocyte lysate system from Promega. All immunoprecipitations were performed by incubating cell lysates or extracts with affinity-purified primary antibodies or preimmune serum, followed by a second incubation with protein A-agarose beads from Sigma Chemicals. The beads were then washed four times with NET-N buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). The precipitated complexes were resuspended in sodium dodecyl sulfate loading buffer, boiled, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by autoradiography or immunoblotting.
Immunofluorescence.
Cells were grown on glass slides and exposed to the indicated doses of ionizing radiation from a 137Cs source. 4-Hydroperoxycyclophosphamide (4HC) was prepared as a fresh stock at (4 mg/ml) in phosphate-buffered saline. This stock was added directly to the culture medium to a final concentration of 4 μg/ml, and cells were returned to the incubator. After 60 min, the medium was replaced with fresh warm medium. Cells were then allowed to recover for 4 h or 22 h in the incubator. After treatment, cells were washed with Tris-buffered saline (TBS) and fixed with freshly prepared methanol-acetone (1:1) at room temperature for 90 s, and cells were permeabilized by washing with TBS plus 0.1% Tween 20 (TBST) for 15 min at room temperature. Slides were then blocked with 5% goat serum in TBST for 30 min at room temperature in a humidified chamber and incubated with the primary antibodies for 30 min under the same conditions. Samples were washed in TBST and incubated with fluorescently labeled secondary antibodies. Anti-rabbit immunoglobulin G conjugated to fluorescein isothiocyanate and 4′,6′-diamidino-2-phenylindole (DAPI) were obtained from Sigma Chemicals. Anti-mouse immunoglobulin G conjugated to rhodamine was purchased from Jackson Immunologicals. Prepared slides were then analyzed by fluorescent microscopy.
Cell cycle analysis with the laser scanning cytometer.
Human breast cancer MCF-7 cells were seeded onto coverslips and allowed to grow for 48 h before ionizing radiation treatment. Five hours after exposure to 10 Gray, the irradiated cells, together with untreated controls, were fixed and stained with anti-hSnm1 antibodies as described above and counterstained with 10 μg of propidium iodide and 200 μg of RNase A per ml. The stained cells were scanned on a laser scanning cytometer (CompuCyte, Cambridge, Mass.) with the argon ion laser (488 nm) and the standard green and long red filter sets and integration contouring set.
Two-hybrid analysis.
Yeast two-hybrid experiments were performed as previously described (19), with the full-length hSNM1 open reading frame fused to the GAL4 activation domain. Plasmids encoding either hMre11, hRad50, or Nbs1 fused to the GAL4 DNA binding domain were generously provided by John Petrini (Memorial Sloan-Kettering Cancer Center).
RESULTS
Characterization of hSNM1 expression.
The human cDNA clone (designated KIAA0086, GenBank accession number D42045) was identified in a database search for genes homologous to the yeast SNM1. Sequence analysis indicated that the largest open reading frame consists of 3,120 bp and that the putative translation initiation codon is at nucleotide residue 919. This long 5′ untranslated region contains an internal ribosome entry site that regulates translation of hSnm1 (51).
Northern blot analysis with the cDNA insert as a probe detected a single transcript of approximately 4.5 kb in all tissues tested, with the highest levels being seen in the pancreas and the brain (Fig. 1A). Similar data by Kikuno et al. were posted in the HUGE database via the World Wide Web (18). Their data include various additional tissues and show elevated hSNM1 message in the testis. In contrast to the significant interstrand crosslink-specific upregulation seen with the yeast SNM1 mRNA (48), no changes in hSNM1 transcript levels or size were observed in HeLa cells within 22 h after treatment with mitomycin C (not shown).
FIG. 1.
(A) Northern blot of mRNAs from various tissues was probed with hSnm1 cDNA. A probe for β-actin was used to control for loading. (B) In vitro-translated Flag-hSnm1 protein labeled with [35S]methionine was precipitated by anti-hSnm1-protein A-agarose beads, protein A beads only, or preimmune-protein A beads. B and SN indicate beads and supernatant, respectively.
We prepared affinity-purified antibodies raised against a recombinant hSnm1 polypeptide encompassing residues 762 through 1040. These antibodies weakly recognized the original recombinant antigen by immunoblotting but did not recognize a band corresponding to the endogenous hSnm1 protein in HeLa whole-cell extracts. Further attempts to detect the full-length protein were similarly unsuccessful, despite the use of several different methods of cell lysate preparation or of extracts from HEK293 cells transiently expressing EGFP-tagged hSnm1 (data not shown). Despite this inability to detect hSnm1 by immunoblotting, these antibodies successfully immunoprecipitated in vitro-translated Flag-hSnm1 protein labeled with [35S]methionine, which migrated at approximately 140 kDa (Fig. 1B) and ectopically expressed EGFP-hSnm1 (see below). This evidence suggests that the failure of these antibodies to detect hSnm1 via immunoblotting is due to an apparent difficulty in recognizing the denatured form of the protein. Dronkert et al. (9) also reported an inability to detect the endogenous protein with antibodies raised against the region between amino acids 644 and 763.
hSnm1 protein localizes to distinct structures in the nucleus.
To determine the subcellular location of the hSnm1 protein, we observed the staining pattern of anti-hSnm1 affinity-purified antibodies by indirect immunofluorescence. Three distinct patterns were observed after staining asynchronous MCF-7 cells. An individual cell showed either diffuse nuclear staining, one or two large nuclear structures that for convenience we refer to as bodies (Fig. 2a), or multiple smaller nuclear foci (Fig. 2b). Similar staining patterns were also observed in the primary human fibroblast line GM08399 (not shown) and the human fibrosarcoma cell line HT-1080. In the latter cell line, we noticed a decrease in the number of cells displaying bodies and a corresponding increase in the population of focus-positive cells (not shown). High-resolution confocal microscopy of the hSnm1 bodies indicated that they had an average diameter of approximately 2 μm and were amorphous or irregular in shape, while the individual foci were usually less than a micrometer in diameter and had a more spherical shape (not shown).
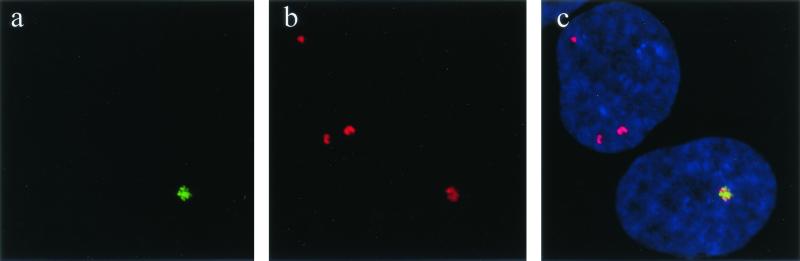
FIG. 2.
hSnm1 is localized to the nucleus. Indirect immunofluorescence of MCF-7 cells probed with hSnm1 affinity-purified polyclonal antibodies displaying (a) diffuse nuclear staining and hSnm1 bodies (as indicated by white arrow) or (b) multiple nuclear foci. Epifluorescence of MCF-7 cells expressing EGFP-hSnm1 fusion protein showing localization to (c) a nuclear body and (d) foci (transfected cell indicated by white arrow). An undamaged HT-1080 cell transiently expressing the Flag-hSnm1 fusion construct was stained with (e) anti-hSnm1 antibodies and (f) anti-Flag M2 monoclonal antibodies. (g) The two images are shown merged after DAPI staining.
To confirm the immunofluorescence results, we transiently transfected HeLa and MCF-7 cells with a plasmid encoding hSnm1 fused to enhanced green fluorescent protein (EGFP-hSnm1). We observed that EGFP-hSnm1 localized to nuclear bodies in both HeLa and MCF-7 cells (Fig. 2c). In certain cells that happened to express higher levels of EGFP-hSnm1, the fusion protein tended to be localized at small foci (Fig. 2d). To further verify these results, we transfected HT-1080 cells with a construct encoding hSNM1 fused to the Flag epitope and doubly stained them with purified anti-hSnm1 antibodies and the monoclonal anti-Flag M2 antibody. In all cells staining positive for Flag foci, anti-hSnm1 also detected foci that completely colocalized with those stained by the Flag antibody (Fig. 2e to g). These observations indicate that the affinity-purified anti-hSnm1 antibodies correctly identified hSnm1 in situ and that ectopically expressed hSnm1 was localized to subnuclear structures, similar to the endogenous protein. Interestingly, we were unable to obtain a cell line stably expressing EGFP-hSnm1 or Flag-hSnm1, suggesting that overexpression of this protein is toxic. A similar finding was reported by Dronkert et al. (9).
hSnm1 nuclear foci are inducible by DNA damage.
To determine whether the nuclear localization of hSnm1 was effected by DNA damage, we exposed MCF-7 cells to 10 Gray of ionizing radiation and stained them with anti-hSnm1 at various times postirradiation. Cells were then classified by their anti-hSnm1 staining pattern into three groups: cells with diffuse nuclear staining, cells with hSnm1 bodies, and cells with more than 10 hSnm1 foci. An increase in the fraction of cells containing foci was detected within 30 min, while the fraction of cells containing bodies decreased correspondingly (Fig. 3). Within 5 h, the population of cells containing bodies was virtually abolished. Surprisingly, the population of cells with fewer than 10 foci (including the cells with diffuse nuclear staining) remained relatively unchanged. Upon exposure to the interstrand crosslink-inducing agent 4HC, no significant changes were detected 4 h after drug removal, but a fourfold induction of focus-containing cells was detected 22 h after 4HC exposure.
FIG. 3.
Quantitative analysis of hSnm1 focus induction after DNA damage in MCF-7 cells by indirect immunofluorescence. hSnm1 antibodies were used to probe cells at various times after treatment with either ionizing radiation (IR) or 4HC. The percentages of cells displaying diffuse nuclear staining with fewer than 10 foci (open bars), hSnm1 bodies (hatched bars), and more than 10 foci (black bars) were calculated after scoring at least 100 nuclei for each time point. Reported here are the averages and standard deviations of three data sets.
To determine if the different subnuclear localizations of hSnm1 were related to the cell cycle, we performed laser scanning cytometry on asynchronous MCF-7 cells stained with anti-hSnm1 and propidium iodide. After the x,y coordinates and DNA content (propidium iodide integral) of each cell were recorded, the anti-hSnm1 staining pattern was scored manually by observing at least 150 of the scanned cells through the eyepiece with a standard fluorescein isothiocyanate filter. Due to the characteristic clumpy growth of MCF-7 cells, only cells that could be identified unambiguously as single cells were scored. The correlation between the propidium iodide integral and the anti-hSnm1 staining pattern was then analyzed with the S-Plus statistical software package (Insightful Co.). We observed that in an unirradiated population, the Snm1 bodies occurred most often in G1 cells, high numbers of foci are correlated with G2 cells, and the S-phase population displayed the most cells with diffuse nuclear staining (Fig. 4A). Five hours after exposure to 10 Gray of ionizing radiation, the G1- and S-phase but not G2-phase populations displayed an increase in the number of focus-containing cells (Fig. 4B).
FIG. 4.
Quantitative analysis of hSnm1 nuclear staining during the cell cycle. (A) Histogram displaying the DNA content of asynchronous untreated MCF-7 cells analyzed by laser scanning cytometry and the observed frequencies of the different hSnm1 staining patterns corresponding to the G1, S, and G2 subpopulations. PI, propidium iodide. (B) Similar data obtained from MCF-7 cells taken 5 h after treatment with 10 Gray of ionizing radiation. The percentages of cells displaying diffuse nuclear staining with fewer than 10 foci (white bars), hSnm1 bodies (hatched bars), and more than 10 foci (black bars) were calculated after scoring at least 150 nuclei for each time point.
hSnm1 nuclear foci colocalize with hMre11 after ionizing radiation treatment.
The formation of discrete nuclear foci has been observed for other DNA repair proteins, particularly those involved in recombinational repair pathways. To determine whether the foci containing hSnm1 overlapped either of the previously described foci containing hRad51 or hMre11, we examined HT-1080 and MCF-7 cells by immunofluorescence before and after exposure to ionizing radiation. Staining in unirradiated cells for both hSnm1 and hRad51 indicated that hRad51 was not present at the hSnm1 bodies and that hSnm1 was not localized to hRad51 foci seen in what were presumably cells undergoing S phase (not shown). Furthermore, no colocalization could be seen following irradiation, although both proteins could be seen as nonoverlapping foci forming within the same nuclei (Fig. 5a to c).
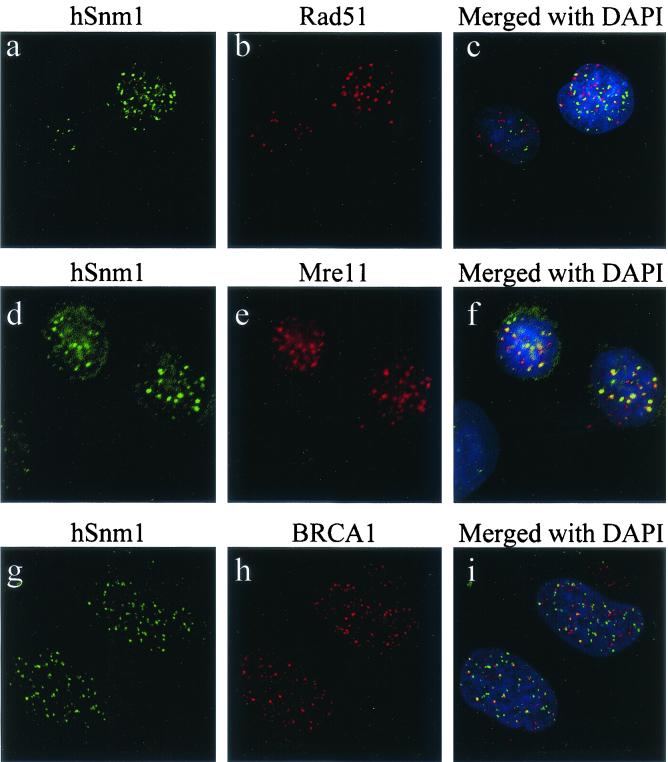
FIG. 5.
Colocalization of hSnm1 foci with hMre11 and BRCA1 but not hRad51, as determined by indirect immunofluorescence. HT-1080 cells were irradiated with 10 Gray and stained 2 h later with (a) anti-hSnm1 and (b) anti-hRad51; (c) merged fields after DAPI staining. HT-1080 cells were irradiated with 15 Gray and stained 9 h later with (d) anti-hSnm1 and (e) anti-hMre11; (f) merged fields after DAPI staining. Partial colocalization with BRCA1 ionizing radiation-induced foci was observed 5 h after 10 Gray, as shown by (g) anti-hSnm1 and (h) anti-BRCA1; (i) both fields merged with DAPI staining.
Double staining of unirradiated cells with antibodies for hSnm1 and hMre11 failed to detect any hMre11 in the hSnm1 bodies, but exposure to ionizing radiation led to a substantial amount of overlap between the foci formed by these two proteins (Fig. 5d to f). We also examined the localization of BRCA1 with respect to hSnm1 ionizing radiation-induced foci and found that colocalization occurred but not to the extent observed with hMre11 (Fig. 5g to i).
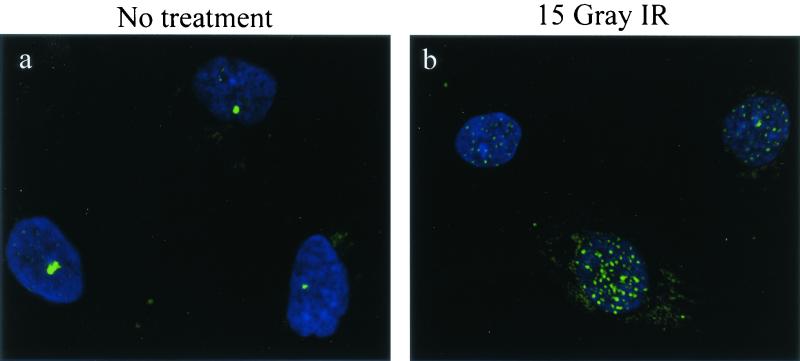
It has been shown previously that cells derived from patients with Nijmegen breakage syndrome do not form hMre11 ionizing radiation-induced foci because they express a mutated form of the NBS1 protein (3). To determine whether hSnm1 foci showed a similar dependence on the NBS1 protein, primary Nijmegen breakage syndrome fibroblast cells were stained with anti-hSnm1 before and after ionizing radiation treatment. Undamaged Nijmegen breakage syndrome cells exhibited hSnm1 bodies and diffuse nuclear staining similar to MCF-7 cells (Fig. 6a). Nine hours after exposure to ionizing radiation, hSnm1 foci could be detected in most cells (Fig. 6b), indicating that the full-length NBS1 protein is not required for the formation of hSnm1 bodies or ionizing radiation-induced foci.
FIG. 6.
hSnm1 focus formation occurs normally in Nijmegen breakage syndrome cells. GM7166 primary fibroblasts were fixed 9 h after mock treatment or after exposure to 15 Gray of ionizing radiation (IR) and subjected to indirect immunofluorescence with anti-hSnm1. (a) Mock-treated cells; (b) after ionizing radiation exposure.
To ascertain whether the colocalization of the hSnm1 and hMre11 ionizing radiation-induced foci was indicative of a physical interaction, we attempted to coimmunoprecipitate hMre11 with anti-hSnm1 antibodies. We failed to detect any hMre11 in hSnm1 immune complexes from lysates prepared from either untreated or irradiated cells (data not shown). Similarly, no interaction was detected between hSnm1 and any of the primary members of the hMre11 complex, including hMre11, hRad50, and NBS1, with the yeast two-hybrid system (data not shown).
Induction of hMre11 ionizing radiation-induced foci has been shown to be normal in cells derived from the cancer-prone syndrome ataxia telangiectasia, which causes defects in the ATM protein (28). Examination of hSnm1 foci in primary ataxia telangiectasia fibroblast cells after exposure to ionizing radiation showed that foci formed normally in these cells (not shown), indicating that ATM is not required for focus formation by hSnm1.
hSnm1 colocalizes and physically associates with h53BP1 protein.
53BP1, a BRCA1 C-terminus (BRCT) domain-containing protein, has recently been reported to localize to nuclear foci of various sizes that also colocalize with phosphorylated histone H2AX after DNA damage (2, 37, 42, 50). After exposure to ionizing radiation, the 53BP1 protein is phosphorylated by ATM and also colocalizes with hMre11 ionizing radiation-induced foci. Given these findings, we examined whether the immunofluorescent staining pattern of hSnm1 overlapped that of 53BP1 before and after exposure to ionizing radiation.
Without treatment, both proteins were highly colocalized to the larger nuclear structures that we refer to as bodies (Fig. 7a to c). Thirty minutes after exposure to ionizing radiation, the 53BP1 foci were well developed, although what appeared to be bodies were still present, whereas the foci formed by hSnm1 were fewer in number and not as robust (Fig. 7d to f). At 90 min after exposure, neither protein was observed in bodies, and both the number of hSnm1 foci per cell and the amount of overlap with 53BP1 foci had increased. At 5 h after exposure, the overlap between the two foci was extensive.
FIG. 7.
Colocalization of hSnm1 and 53BP1 in foci as detected by indirect immunofluorescence. MCF-7 cells were mock treated (a, b, and c) or treated with 10 Gray of ionizing radiation and fixed after 30 min (d, e, and f), after 90 min (g, h, and i), or after 5 h (j, k, and l). (a, d, g, and j) Polyclonal anti-hSnm1 staining with fluorescein isothiocyanate; (b, e, h, and k) monoclonal anti-53BP1 staining with tetramethyl rhodamine isocyanate; (c, f, i, and l) merged fields plus DAPI staining.
Although qualitative in nature, these results suggest that 53BP1 relocalizes to sites of damage more rapidly than hSnm1, but that the ultimate destination of both proteins is the same. This interpretation should be taken with caution because variables such as epitope masking may have affected the strength of the hSnm1 signal at early time points. Nevertheless, these findings indicate an interesting colocalization of 53BP1 and hSnm1 proteins before and after exposure to ionizing radiation. We also examined the localization of 53BP1 in cells expressing EGFP-hSnm1. 53BP1 was colocalized with EGFP-hSnm1 in cells that displayed bodies (Fig. 8), but not in cells containing EGFP-hSnm1 foci. In the latter cells, 53BP1 staining was either very weak compared to neighboring untransfected cells or not colocalized with the EGFP-hSnm1 foci (not shown).
FIG. 8.
Colocalization of EGFP-hSnm1 protein with 53BP1. MCF-7 cells were transfected with pEGFP-hSnm1 followed by staining with anti-53BP1 and Toto-3. Localization of (a) EGFP-hSnm1 and (b) 53BP1 to a nuclear body in the same cell; (c) the two fields merged with Toto-3.
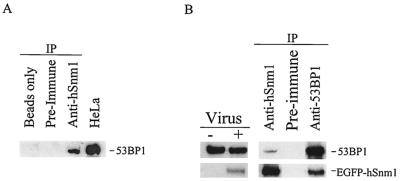
To determine if there is a physical association between 53BP1 and hSnm1, extracts were prepared from untreated HeLa cells, immunoprecipitated with anti-hSnm1 antibodies, and immunoblotted for 53BP1. As shown in Fig. 9A, 53BP1 was immunoprecipitated with the anti-hSnm1 serum. Since we were unable to detect endogenous hSnm1 by immunoblotting, immunoprecipitations with anti-53BP1 were performed with nuclear extracts made from HEK293 cells infected with an adenovirus expressing EGFP-tagged hSnm1. This enabled the use of a monoclonal anti-EGFP antibody to detect the EGFP-hSnm1 fusion protein by immunoblotting. As shown (Fig. 9B), antibodies against 53BP1 were able to coimmunoprecipitate EGFP-hSnm1 protein, confirming that hSnm1 and 53BP1 are members of the same protein complex. When combined with the immunofluorescent results, these data suggest a close and possibly direct association between hSnm1 and 53BP1 before and after DNA damage.
FIG. 9.
Coimmunoprecipitation of 53BP1 and hSnm1. (A) HeLa cell extracts were incubated with beads only, preimmune serum, or affinity-purified anti-hSnm1 antibodies. Polyclonal anti-53BP1 antibodies were used to detect 53BP1 by immunoblotting. (B) HEK293 nuclear extracts were prepared with or without infection with EGFP-hSnm1-expressing adenovirus and immunoblotted with monoclonal antibodies against EGFP or polyclonal antibodies against 53BP1. Immunoprecipitations (IP) were performed with these extracts with anti-hSnm1 and anti-53BP1 antibodies as shown.
Focus-forming region of hSnm1 is localized to a discrete domain.
To define the region of hSnm1 that is required for focus formation, full-length and truncated versions of the EGFP-hSNM1 fusion construct were prepared, transfected into HT-1080 cells, and examined for cellular localization (Fig. 10A). Removal of up to 425 amino acids from the carboxyl terminus of the hSnm1 protein (Δ615-1040) had no effect on focus formation, but removal of 646 amino acids resulted in diffuse nuclear staining and the loss of observable foci (Fig. 10B, panels a and b). These findings suggested that the region between amino acid residues 394 and 615 is involved in focus formation either by binding to DNA directly or by interacting with other proteins present at the foci.
FIG. 10.
Deletion analysis of hSNM1. (A) Various truncations and in-frame deletions were constructed in the hSNM1 segment of the EGFP-hSNM1 fusion gene. The solid black region indicates EGFP, and vertical lines indicate the conserved metallo-β-lactamase domain. Each construct was transfected into HT-1080 cells, and the location of the EGFP signal was determined 24 h later. (B) Expression of EGFP-hSnm1 deletion mutants in HT-1080 cells.
To further examine this issue, we deleted an additional region upstream between residues 109 and 394 in the Δ615-1040 construct, leaving the putative nuclear localization signal between amino acids residues 16 through 20 intact. The resulting EGFP fusion protein was still localized to nuclear foci (Fig. 10B, panel c); however, a minority of cells also displayed diffuse nuclear localization. Deletion of the putative focus-forming region in an otherwise intact hSnm1 protein resulted in unusual localization to a small number of large foci (Fig. 10B, panel d). It is not clear whether these represent novel structures or possibly aberrant versions of the foci or bodies detected previously by immunofluorescence. A slightly larger deletion between residues 311 and 615 resulted in diffuse nuclear staining and loss of the aberrant structures (not shown).
DISCUSSION
We have analyzed the cellular distribution of hSnm1 protein in several cell lines and have shown that it localizes to the nucleus in two apparently distinct structures, which we refer to separately as bodies and foci. The former appear as larger aggregates of protein that typically occur only once or twice per cell, while the latter are smaller and more numerous accumulations of protein. Interestingly, approximately half of the cells usually exhibited neither pattern, but instead showed diffuse nuclear localization.
Upon exposure to ionizing radiation, a time-dependent decrease in the number of bodies and a corresponding increase in the number of foci per cell were observed. These observations suggest that hSnm1 is dispersed from the bodies and migrates to sites of repair, as represented by foci, upon exposure to DNA-damaging agents. In the case of ionizing radiation-induced damage, foci can be seen within 30 min. An equivalently toxic dose of the interstrand crosslinking agent 4HC also induced the formation of foci, but to a lesser extent and only after a relatively long incubation. If double-strand breaks are the focus-inducing lesion in both types of damage, their differences in formation rate may be explained by recent results by Akkari et al. (1). They have shown that in order for an interstrand crosslinking agent to induce a cell cycle checkpoint in human fibroblasts, those cells must pass through a single S phase following exposure. It has also been demonstrated that double-strand breaks occur as intermediates during crosslink repair in mammalian cells (8). Thus, the slower kinetics of focus induction by 4HC may be due to the additional time needed to pass through S phase in order to process these lesions into double-strand breaks.
Cell cycle analysis by laser scanning cytometry of an asynchronous population stained with anti-hSnm1 showed that G1 cells had the highest proportion of cells containing Snm1 bodies and G2 cells had the highest proportion of cells containing foci, while cells in S phase had the highest frequency of diffuse nuclear staining. Furthermore, after exposure to ionizing radiation, the percentages of cells containing foci increased in the G1- and S-phase subpopulations, with the latter group showing the larger relative induction. Interestingly, the G2 subpopulation showed little change in the hSnm1 staining profile before and after ionizing radiation treatment. Since this method allowed us to evaluate the DNA content and hSnm1 localization simultaneously for each individual cell, we are able to conclude that hSnm1 ionizing radiation-induced foci can form in all phases of the cell cycle. The significance of the finding that the bodies largely form in G1 phase is not clear, but this does indicate that the localization of hSnm1 in untreated cells is under cell cycle control.
The biological significance of the hSnm1 bodies remains undetermined. Our observations on their occurrence and morphology distinguish them from other subnuclear structures such as Cajal bodies and promyelocytic leukemia bodies, although the occurrence of such objects is also known to be influenced by changes in the cell cycle (22, 26). One possibility is that they function as protein reservoirs to sequester hSnm1 until its function is required. The formation of nuclear foci in cells overexpressing epitope-tagged hSnm1 in the absence of DNA damage could then be a consequence of these reservoirs' filling to capacity. Since we and others have found that even slight overexpression is deleterious to cells, there is reason to believe that hSnm1 activity is tightly regulated (9). Additional posttranslational regulation is controlled at least in part by the presence of an internal ribosome entry site in the 5′ untranslated region of hSNM1 (51).
The apparent colocalization of hSnm1 with Mre11 and, to a lesser extent, with BRCA1 ionizing radiation-induced foci indicates that hSnm1 is relocated to sites of double-strand breaks, but the significance of this relocalization is unclear. The less extensive localization with BRCA1 was expected because this protein also colocalizes with hRad51 ionizing radiation-induced foci (5, 43). There is little evidence to suspect Snm1 of having a direct role in double-strand break repair, since mouse cells carrying an SNM1 allele with an internal deletion displayed wild-type sensitivity to ionizing radiation-induced damage and a slight sensitivity to mitomycin C (9). However, the significance of these findings is uncertain, since this disrupted allele may not have resulted in a null phenotype. In addition, it is not yet known whether there is functional overlap among the mammalian Snm1 homologues.
A recent report by Moshous et al. (31) showing that mutations in Artemis cause sensitivity to ionizing radiation as well as a defect in V(D)J recombination suggests that members of the Snm1 protein family may be involved in the detection and/or repair of double-strand breaks. It was also observed that the conserved domain within the Snm1 homologs represents a metallo-β-lactamase fold, implying that these proteins contain a hydrolase activity that has been reported to encode a nuclease activity that may be involved in processing the ends of double-strand breaks (31). Since studies with yeast snm1 mutants have shown that they are not deficient in the initial incision events that occur at crosslinks but are defective in a later step required for the restoration of high-molecular-weight DNA from fragmented DNA (24, 27), the yeast homolog may be involved in the recognition and/or processing of double-strand breaks that occur as intermediates during interstrand crosslink repair.
The colocalization of hSnm1 and 53BP1 in the nucleus before and after ionizing radiation and their physical association supports the hypothesis that hSnm1 is involved in the DNA damage response. Although the function of 53BP1 in the DNA damage response has yet to be ascertained, somewhat more is known about 53BP1 than about hSnm1. Its observed rapid ionizing radiation-induced focus formation and strong association with γ-H2AX suggest that it may act upstream of the actual repair processes (2, 37, 42, 50), and recent findings by one of us show that 53BP1 null homozygous mice are hypersensitive to ionizing radiation (P. B. Carpenter, unpublished results). A recent report by Celeste et al. (4) has shown that ionizing radiation-induced foci containing BRCA1, 53BP1, and Mre11 do not form in cell lines homozygous (−/−) for H2AX.
We have not as yet determined whether the formation of hSnm1 foci or bodies has a similar dependence on H2AX or on 53BP1. A recent report by Jullien et al. presented evidence for 53BP1's localization to the kinetochores of mitotic cells (17). In addition to its relatively high abundance on kinetochores that failed to capture microtubules, their work discovered that 53BP1 is hyperphosphorylated in response to mitotic spindle poisons. This suggests that 53BP1 may play a role in mitotic checkpoint control in addition to any function it may serve in the response to DNA damage.
Using expression of EGFP-hSNM1 fusions that resulted in truncations or targeted deletions of hSnm1, we mapped a region of 220 amino acids from residues 394 to 615 that is involved in focus formation. This region lies upstream of the conserved metallo-β-lactamase domain and does not contain any known sequence motifs. This ability of a short sequence segment to direct proteins to discrete sites within the nucleus has been observed for other DNA repair-replication proteins. Both DNA ligase I and replication factor C contain a homologous sequence of about 20 amino acids that directs these two proteins to replication factories through an interaction with PCNA (30). Also, a small region has been mapped within the xeroderma pigmentosum group G protein that is responsible for its focus formation after exposure to UV irradiation (34). Presumably, the focus-forming region of hSnm1 either directly interacts with aberrant DNA structures or interacts with one or a small number of proteins at such sites.
Conclusions.
The initial studies of Snm1 and Artemis in mammalian cells are consistent with studies in S. cerevisiae indicating a role for this family of proteins in the cellular response to DNA damage (9, 31). The characterization of a disrupted allele of SNM1 in the mouse showed a mild increase in sensitivity to mitomycin C in embryonic stem cells and an increased rate of mortality in whole animals that had been injected with this drug (9). Our results have demonstrated that before the introduction of DNA damage, hSnm1 protein localizes in the nucleus to a discrete structure that also contains 53BP1. A physical association between hSnm1 and 53BP1 was also shown by immunoprecipitation in extracts from untreated cells. After DNA damage, hSnm1 colocalizes with 53BP1, Mre11-Rad50-NBS1, and to a lesser extent with BRCA1 ionizing radiation-induced foci. While it was previously shown that 53BP1 is phosphorylated and relocalizes to sites of double-strand breaks after ionizing radiation treatment and to kinetochores during mitosis, its role in the response to genotoxic damage remains unknown. Likewise, the role of hSnm1 remains to be elucidated, but taken together, our findings suggest that 53BP1 and hSnm1 may cooperate in these pathways.
Acknowledgments
We thank John Petrini, Eva Lee, and Thanos Halozonetis for generously providing antibodies.
This work was supported by National Institutes of Health grants CA52461, CA75160, CA90270, GM65812-01, and R01 DE 13157 and by National Cancer Institute core grant P30 CA16672. Partial funding for C.T.R.'s stipend came from an American Legion Auxiliary Fellowship and National Institutes of Health predoctoral training grant CA09299. Additional funding was provided by the Welch Foundation (AU-1447) and the Ellison Medical Foundation (NS-0042-99). The DNA sequencing core facility of M. D. Anderson Cancer Center is supported by grant CA16672.
REFERENCES
- 1.Akkari, Y. M., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand crosslinks. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 4.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., D. P. Silver, D. Walpita, S. B. Cantor, A. F. Gazdar, G. Tomlinson, F. J. Couch, B. L. Weber, T. Ashley, D. M. Livingston, and R. Scully. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317-328. [DOI] [PubMed] [Google Scholar]
- 6.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8:137-147. [DOI] [PubMed] [Google Scholar]
- 8.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand crosslinks in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dronkert, M. L., J. de Wit, M. Boeve, M. L. Vasconcelos, H. van Steeg, T. L. Tan, J. H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand crosslinking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 12.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 13.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haase, E., D. Riehl, M. Mack, and M. Brendel. 1989. Mol. cloning of SNM1, a yeast gene responsible for a specific step in the repair of crosslinked DNA. Mol. Gen. Genet. 218:64-71. [DOI] [PubMed] [Google Scholar]
- 15.Henriques, J. A., and E. Moustacchi. 1980. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95:273-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenny, A., L. Minvielle-Sebastia, P. J. Preker, and W. Keller. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274:1514-1517. [DOI] [PubMed] [Google Scholar]
- 17.Jullien, D., P. Vagnarelli, W. C. Earnshaw, and Y. Adachi. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115:71-79. [DOI] [PubMed] [Google Scholar]
- 18.Kikuno, R., T. Nagase, M. Waki, and O. Ohara. 2002. HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 30:166-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, L., S. J. Elledge, C. A. Peterson, E. S. Bales, and R. J. Legerski. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91:5012-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., C. A. Peterson, X. Zhang, and R. J. Legerski. 2000. Requirement for PCNA and RPA in interstrand crosslink-induced DNA synthesis. Nucleic Acids Res. 28:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 22.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 24.Magana-Schwencke, N., J. A. Henriques, R. Chanet, and E. Moustacchi. 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79:1722-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matera, A. G. 1999. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9:302-309. [DOI] [PubMed] [Google Scholar]
- 27.Meniel, V., N. Magana-Schwencke, and D. Averbeck. 1995. Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand crosslinks by 8-methoxypsoralen plus. UVA Mutagenesis 10:543-548. [DOI] [PubMed] [Google Scholar]
- 28.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. (Erratum, 21:1898.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuta, R., J. M. LaSalle, H. L. Cheng, A. Shinohara, H. Ogawa, N. Copeland, N. A. Jenkins, M. Lalande, and F. W. Alt. 1997. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. USA 94:6927-6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 32.Nagase, T., N. Miyajima, A. Tanaka, T. Sazuka, N. Seki, S. Sato, S. Tabata, K. Ishikawa, Y. Kawarabayasi, and H. Kotani. 1995. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 2:37-43. [DOI] [PubMed] [Google Scholar]
- 33.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 34.Park, M. S., J. A. Knauf, S. H. Pendergrass, C. H. Coulon, G. F. Strniste, B. L. Marrone, and M. A. MacInnes. 1996. Ultraviolet-induced movement of the human DNA repair protein, xeroderma pigmentosum type G, in the nucleus. Proc. Natl. Acad. Sci. USA 93:8368-8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C., and R. Legerski. 1991. High-frequency transformation of human repair-deficient cell lines by an Epstein-Barr virus-based cDNA expression vector. Gene 107:279-284. [DOI] [PubMed] [Google Scholar]
- 37.Rappold, I., K. Iwabuchi, T. Date, and J. Chen. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613-620. (Erratum, 154:469, 2001.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter, D., E. Niegemann, and M. Brendel. 1992. Mol. structure of the DNA crosslink repair gene SNM1 (PSO2) of the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 231:194-200. [DOI] [PubMed] [Google Scholar]
- 39.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhland, A., M. Kircher, F. Wilborn, and M. Brendel. 1981. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat. Res. 91:457-462. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber, E. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scully, R., J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D. M. Livingston. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265-275. [DOI] [PubMed] [Google Scholar]
- 44.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 45.Tashiro, S., N. Kotomura, A. Shinohara, K. Tanaka, K. Ueda, and N. Kamada. 1996. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene 12:2165-2170. [PubMed] [Google Scholar]
- 46.Tavtigian, S. V., J. Simard, D. H. Teng, V. Abtin, M. Baumgard, A. Beck, N. J. Camp, A. R. Carillo, Y. Chen, P. Dayananth, M. Desrochers, M. Dumont, J. M. Farnham, D. Frank, C. Frye, S. Ghaffari, J. S. Gupte, R. Hu, D. Iliev, T. Janecki, E. N. Kort, K. E. Laity, A. Leavitt, G. Leblanc, J. McArthur-Morrison, A. Pederson, B. Penn, K. T. Peterson, J. E. Reid, S. Richards, M. Schroeder, R. Smith, S. C. Snyder, B. Swedlund, J. Swensen, A. Thomas, M. Tranchant, A. M. Woodland, F. Labrie, M. H. Skolnick, S. Neuhausen, J. Rommens, and L. A. Cannon-Albright. 2001. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 27:172-180. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 48.Wolter, R., W. Siede, and M. Brendel. 1996. Regulation of SNM1, an inducible Saccharomyces cerevisiae gene required for repair of DNA crosslinks. Mol. Gen. Genet. 250:162-168. [DOI] [PubMed] [Google Scholar]
- 49.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 50.Xia, Z., J. C. Morales, W. G. Dunphy, and P. B. Carpenter. 2001. Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem. 276:2708-2718. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X., C. T. Richie, and R. J. Legerski. 2002. Translation of hSNM1 is mediated by an internal ribosome entry site that upregulates expression during mitosis. DNA Repair 1:379-390. [PubMed] [Google Scholar]
- 52.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, Q., C.-F. Chen, S. Li, Y. Chen, C.-C. Wang, J. Xiao, P.-L. Chen, Z. D. Sharp, and W.-H. Lee. 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285:747-750. [DOI] [PubMed] [Google Scholar]