Abstract
Cholesterol biosynthesis in somatic cells is controlled at the transcriptional level by a homeostatic feedback pathway involving sterol regulatory element binding proteins (SREBPs). These basic helix-loop-helix (bHLH)-Zip proteins are synthesized as membrane-bound precursors, which are cleaved to form a soluble, transcriptionally active mature SREBP that regulates the promoters for genes involved in lipid synthesis. Homeostasis is conferred by sterol feedback inhibition of this maturation process. Previous work has demonstrated the expression of SREBP target genes in the male germ line, several of which are highly up-regulated during specific developmental stages. However, the role of SREBPs in the control of sterol regulatory element-containing promoters during spermatogenesis has been unclear. In particular, expression of several of these genes in male germ cells appears to be insensitive to sterols, contrary to SREBP-dependent gene regulation in somatic cells. Here, we have characterized a novel isoform of the transcription factor SREBP2, which is highly enriched in rat and mouse spermatogenic cells. This protein, SREBP2gc, is expressed in a stage-dependent fashion as a soluble, constitutively active transcription factor that is not subject to feedback control by sterols. These findings likely explain the apparent sterol-insensitive expression of lipid synthesis genes during spermatogenesis. Expression of a sterol-independent, constitutively active SREBP2gc in the male germ line may have arisen as a means to regulate SREBP target genes in specific developmental stages. This may reflect unique roles for cholesterol synthesis and other functional targets of SREBPs during spermatogenesis.
Spermatozoa are highly specialized cells that are formed in the testis through a complex series of developmental stages that can be grouped into three phases: mitotic, meiotic and spermiogenic. Spermatogonial cell types constitute the initial proliferative phase, giving rise to early spermatocytes, which in turn undergo multiple meiotic steps resulting in haploid round spermatids. Finally, spermatids morphologically differentiate into sperm during spermiogenesis. These various events require the elaboration of a well-coordinated developmental program of both cell type- and stage-dependent gene expression (10). Recent studies have begun to identify certain key regulators of these events. For example, the germ cell-specific transcription factor CREMτ is expressed in round spermatids and is required for completion of spermiogenesis (5, 28). However, in general, the mechanisms controlling the cellular specificity and proper timing of gene expression in developing male germ cells remain poorly understood.
In order to explore transcriptional programs controlling spermatogenesis, we have previously examined the cell- and stage-specific regulation of the male germ line proenkephalin promoter in rodents (14, 27). This promoter is expressed exclusively by spermatogenic cells and is highly up-regulated in pachytene spermatocytes and haploid round spermatids (55). We previously identified a regulatory region (GCP1) within the proximal 5′-flanking region of this promoter that is required for its germ cell-specific expression. GCP1 contains two novel direct repeat elements (CTCCAG) that are necessary for promoter activity in vivo. Furthermore, a 50- to 55-kDa protein termed “PACH1” has been identified, which binds specifically to the GCP1 repeats and is unique to spermatogenic cells (26). PACH1 is expressed during meiosis and in early haploid cells with the same developmental pattern during spermatogenesis as the proenkephalin germ line promoter.
Cholesterol is required for numerous cellular processes, including plasma membrane structure and signaling, and as a precursor for steroids and bile acids (42, 44). It also has a central role in fertilization, where its concentrations in the outer sperm membrane appear to regulate capacitation (9). In somatic cells, synthesis of cholesterol and fatty acids is under the control of a family of transcription factors termed “sterol regulatory element binding proteins” (SREBPs). This family consists of three previously identified proteins: SREBP1a and SREBP1c, generated from a common gene by alternative promoter usage and splicing; and SREBP2 (6, 34). These proteins regulate the expression of genes involved in lipid synthesis by binding to sterol regulatory elements (SREs) present within their promoters. SREBP2 plays a predominant role in regulating cholesterol biosynthetic genes, while SREBP1 is more selective for targets involved in fatty acid synthesis (17, 35). Each SREBP is synthesized as a membrane-bound, 125-kDa precursor (SREBPp), which is first transported from the endoplasmic reticulum to the Golgi apparatus via its binding to sterol cleavage-activating protein (SCAP) (6). It is then proteolytically processed within the Golgi apparatus by two distinct proteases (Site 1 and Site 2), to release a soluble, transcriptionally active mature form into the cytoplasm (6). This active form is able to enter the nucleus and regulates SRE-containing promoters. SREBPs are subject to feedback inhibition by sterols, which serves to restrict cholesterol concentrations within a range sufficient to maintain cellular functions without being cytotoxic. Sterols act by blocking the SCAP-dependent transport of SREBPp from the endoplasmic reticulum to the Golgi apparatus, thereby inhibiting formation of the active transcription factor (32).
While the role of SREBPs in the homeostatic control of cholesterol and fatty acid synthesis in somatic cells is clearly established, their function in spermatogenic cells has been less certain. In particular, previous findings have suggested that expression of cholesterol biosynthetic genes during spermatogenesis was determined by SREBP-independent mechanisms (38). In our efforts to clone PACH1, we have isolated the cDNA for a unique isoform of SREBP2. This novel protein, termed “SREBP2gc,” is highly enriched in meiotic spermatocytes and haploid round spermatids. It is translated as a truncated, soluble transcription factor that bypasses the proteolytic processing/sterol feedback pathway and is thus constitutively active. The implications of the expression of this sterol-insensitive SREBP during spermatogenesis and the relationship between SREBP2gc and PACH1 are discussed.
MATERIALS AND METHODS
Cloning of GCP1-binding proteins.
A λgt11 cDNA library from adult rat testis (Clontech, Palo Alto, Calif.) was screened for PACH1 DNA binding activity with a concatenated GCP1 DNA probe. A double-stranded GCP1-32 monomer containing the two repeats (CTCCAG) (27) and BamHI 5′ overhangs was generated by annealing the complementary oligodeoxynucleotides (ODNs) GATCCGAACTCCAGTGTTCGCTCCAGAATCTTGCCC and GATCGGGCAAGATTCTGGAGCGAACACTGGAGTTCG (repeats are underlined). The monomer duplex was end labeled with [γ-32P]ATP and then self-ligated to generate a concatenated probe consisting mainly of approximately six copies of GCP1-32. Southwestern assays confirmed that the concatenated probe bound to the same spermatogenic cell nuclear protein as the monomer (data not shown). A concatenated GCP1-32mut probe with mutations in the two GCP1 repeat elements was generated in the same way with the ODNs GATCCGAATACAGTTGTTCGTACAGTAATCTTGCCC and GATCGGGCAAGATTACTGTACGAACAACTGTATTCG. This mutated probe did not bind to PACH1 on Southwestern blots (data not shown). Protein replica filters were prepared and screened for expression of DNA binding activity by using Y1090 cells. Briefly, plaques were transferred onto Duralose-UV membranes (Stratagene, La Jolla, Calif.) and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C. Proteins were denatured and renatured at 4°C in HEPES binding buffer (HBB: 25 mM HEPES [pH 7.9], 25 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT]) with sequential dilutions of guanidinium-HCl. The transfer membranes were incubated with 5% nonfat dry milk in HBB for 1 h and then with HBB containing 2 × 106 cpm of 32P-labeled probe per ml and freshly sonicated salmon sperm DNA (0.8 μg/ml) overnight at 4oC. Membranes were then washed at 4°C with HBB and submitted to autoradiography. After screening of 5 × 106 plaques with the wild-type GCP1 probe, positive clones were screened with the GCP1mut probe to eliminate nonspecific DNA binding proteins. DNA sequencing of relevant clones was performed by the dideoxy chain-termination method on an Applied Biosystems model 377 DNA sequencer.
5′ and 3′ RACE PCR for rat SREBP2gc cDNA.
Total RNA from enriched rat spermatogenic cells was amplified with a Gene Racer kit (Invitrogen, Carlsbad, Calif.). Sequence-specific primers and the universal primers supplied by the manufacturer were used to extend the 5′ and 3′ ends. Specific 5′ rapid amplification of cDNA ends (RACE) primer 1 (AGCTTTCAAGTCCTGCAGCCTCAAG) and 5′ RACE primer 2 (AGACGGTGATGATCACCCCGACCT) were used for the initial and nested steps, respectively, to generate 5′-end sequences, while 3′ RACE primer 1 (AGCTCCTGAAGGGCATCGACT) and 3′ RACE primer 2 (GTATGAGCTTCGACATTTCTCACACTC) were used for 3′ sequences. The PCR products were subcloned into pCR4-TOPO vector for sequencing with the TOPO TA cloning kit (Invitrogen).
RT-PCR.
Total RNAs from spermatogenic cells and tissues were analyzed by using the Titan One-Step reverse transcription-PCR (RT-PCR) kit (Roche Applied Science, Indianapolis, Ind.). The following primers were used: BP2-1 (ATCATTGAGAAGCGGTACCGGTC), BP2-2 (AAGTCGATGCCCTTCAGGAGCTT), and BP2gc (GTTCAGAGTGTGAGAAATGTCGAAGCTCA).
Preparation of enriched and purified testicular cells.
Adult male germ cells enriched in pachytene spermatocytes and round and condensing spermatids were prepared from rat and mouse testes by enzymatic digestion essentially according to the method of Bellve et al. (4). Purified spermatogenic cell types were prepared from either immature or mature mouse testes by unit gravity sedimentation, as previously described (3, 4). The purities of various germ cell types ranged from 89 to 96%. Sertoli cells were prepared from 19- to 21-day-old rat testes by similar procedures (31), and spermatozoa were isolated from adult mouse epididymis by mincing in the presence of phosphate-buffered saline (PBS) and then filtration through 50-μm-pore-diameter nylon mesh.
RNA extraction and Northern blots.
Total RNA was extracted from tissues and cells by using Tri-Reagent (Sigma, St. Louis, Mo.). In general, cDNA probes were labeled with [α-32P]dCTP by using the Prime-a-Gene labeling system (Promega, Madison, Wis.). For rat SREBP2gc mRNA mapping studies, probe I was generated from the rat SREBP2gc cDNA by PCR with the primers 5′-UTR (5′ untranslated region) (CGTCTCCCTGAGCTGGACCTCA) and E1 (CTCGTCGATGTCCCCGAGAGT). The PCR product was then used as a template to synthesize a 32P-labeled antisense strand probe with Klenow fragment and primer E1. For probe II, an EcoRI fragment was prepared from SREBP2gc cDNA, while the 3′ RACE PCR product was used for probe III. Twenty micrograms of total RNA was run on formaldehyde-containing agarose gels and transferred to Gene Screen Plus membranes (NEN Life Science, Boston, Mass.). Equivalency of loading and transfer was verified by ethidium bromide staining. Membranes were hybridized with DNA probes (2 × 106 cpm/ml) as described previously (21).
Polysome analysis.
The procedure for polysome fractionation of mouse testicular mRNA was performed as previously described (20). Briefly, a postnuclear fraction was separated on sucrose gradients in buffer (20 mM HEPES [pH 7.6],100 mM KCl, 0.5%Triton X-100, 3 mM β-mercaptoethanol, 300 U of RNAsin per ml) containing either 20 mM MgCl2 (HKM) or 10 mM EDTA (HKE), with the latter used for negative control gradients because EDTA disrupts polysomes. Following RNA extraction, equal percentages of gradient fractions were submitted to Northern analysis.
Protein extraction and Western blotting.
Nuclei were isolated from cell fractions and tissues and extracted as previously described (27). Briefly, samples were homogenized in buffer A (10 mM HEPES [pH 8.0], 1.5 mM MgCl2, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 5 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor cocktail mix [Boehringer Mannheim, Indianapolis, Ind.], 25 μg of N-acetyl-Leu-Leu-Nle-CHO [ALLN] per ml, 0.5% NP-40). Nuclei were centrifuged at 3,000 × g for 3 min at 4°C, and the supernatant was used for cytoplasmic proteins. Nuclei were extracted in NEB (20 mM HEPES [pH 8.0], 25% glycerol, 0.4 M NaCl, 0.2 mM EDTA, 1 mM DTT, 0.1 mM PMSF, protease inhibitor mix, ALLN) on ice for 30 min. Microsomes were prepared by homogenization in buffer A without NP-40 and centrifugation of the postnuclear supernatant at 100,000 × g for 1 h. Following washing, the membrane pellet was solubilized by resuspension in a mixture containing 10 mM Tris-HCl (pH 6.8), 0.1 M NaCl, and 1% (vol/vol) sodium dodecyl sulfate (SDS) and boiled for 5 min prior to electrophoresis.
Protein extracts were separated on 8% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) transfer membranes (Millipore, Bedford, Mass.). The buffer used for dilution of antibodies and the blocking step was 1× PBS buffer with 0.1% Tween 20 and 10% nonfat dry milk. The blocking step was performed at 4°C overnight. Bound antibodies were detected with a Western Lightning Chemiluminescence kit (Perkin-Elmer Life Science, Boston, Mass.) according to the manufacturer's protocol. Rabbit polyclonal antibodies against mouse SREBP2 (residues 1 to 100) and monoclonal antibodies to hamster SREBP2 (immunoglobulin G [IgG]-7D4) (53) were kindly provided by J. Horton; polyclonal antibodies to human SREBP2 (residues 1 to 481) (40) were obtained from Ryuichiro Sato. Rabbit polyclonal antibodies made to human SREBP1 (K-10) that react with both precursor and mature forms of SREBP1 were obtained from Santa Cruz Biotech (Santa Cruz, Calif.).
Southwestern assays.
For Southwestern blotting, nuclear extracts were separated on 8% SDS-polyacrylamide gels and then transferred onto Duralose-UV membranes. Protocols for blocking, binding, and washing steps were as described for cDNA library screening.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), nuclear extracts were assayed under conditions used in earlier studies (27). Double-stranded ODNs containing 5′ overhangs were labeled with [α-32P]dCTP by fill-in reaction with Klenow fragment. DNA probes consisted of GCP1 and GCP1mut sequences (27) and SRE-1 (GATCATCACCCCACTGCA) and SRE-1mut (GATCTGATAAGATATGCA) (core SRE sequence or its mutations are underlined).
Plasmid DNA constructs, bacterial expression, and cell transfection.
The rat SREBP2gc coding region was released from the λ phage DNA by EcoRI digestion and subcloned into pBluescript to generate pBS-RBP2gc for sequencing. The EcoRI fragment was also inserted in frame into pGEX-4T1 expression vector (Phamacia) to create pGEX-RBP2gc for bacterial expression. BL21(DE3) bacteria were transformed with this vector or with empty pGEX-4T1 plasmid and induced with 0.1 mM IPTG. For mammalian expression, the full-length rat SREBP2gc cDNA was first generated by combining the 5′ sequence obtained by 5′ RACE PCR with the pBS-RBP2gc sequence via an internal Blp1 site. The full-length rat SRBP2gc coding sequence was then removed by EcoRI and MscI digestion and ligated into the EcoRI and SmaI sites of pCMV7 to generate pCMV-BP2gc. A truncated, constitutively active human SREBP2 protein (residues 1 to 468) in pCMV7 vector was kindly provided by J. Horton. The following luciferase promoter constructs were created. pLDLR contained the human low-density lipoprotein (LDL) receptor (LDLR) promoter inserted into pGL2 (a gift from J. Horton). pCYP51 contained the human CYP51 promoter inserted into pGL2 (38) (a gift from D. Rozman). Finally, pRPKLuc-0.5 consisted of a 500-bp rat proenkephalin germ line promoter fragment derived from pRPKCAT0.5 (27) inserted into pGL3. DNAs for promoter constructs (0.5 μg), expression vectors (10 ng) and pCMV-β-galactosidase normalization control plasmid (50 ng) were cotransfected into human K293 cells by using Trans-Fast transfection reagent (Promega) and analyzed 40 to 48 h later with commercial kits for luciferase (Promega) and β-galactosidase (Tropix, Bedford, Mass.).
Sterol-depletion studies.
Mouse preadipocyte 3T3-L1 cells were incubated at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS) and penicillin-streptomycin (pen-strep [100 U/ml]). Upon 70 to 80% confluency, the medium was changed to DMEM-pen-strep containing 5% lipoprotein-deficient fetal bovine serum (LPDS), 50 μM compactin, and 50 μM sodium mevalonate, without (sterol depleted) or with (sterol loaded) 10 μg of cholesterol and 1 μg of 25-hydroxycholesterol per ml, and cells were cultured for an additional 9 h. One hour prior to cell harvest, 25 μg of ALLN protease inhibitor per ml was added.
Short-term culturing of spermatogenic cells was performed as previously described (15). Following two differential plating steps, enriched spermatogenic cells from adult mice were cultured in DMEM plus 10% FCS, pen-strep, 1 mM lactate, and 1% nonessential amino acids in 5% CO2 at 33°C. For sterol-depletion studies, cells were incubated for 9 h in the medium described above, in which serum was replaced with either sterol-depleted or sterol-loaded components, as described for 3T3-L1 cells.
RESULTS
Cloning and analysis of SREBP2gc.
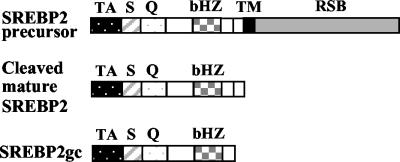
In attempting to clone the cDNA for PACH1, we screened an expression cDNA library from rat testis with wild-type and mutated GCP1 concatemers as positive- and negative-selection probes (see Materials and Methods). One of the clones exhibiting robust and GCP1 repeat-dependent binding was characterized further. DNA sequencing revealed a partial cDNA that was highly related to hamster and human SREBP2 (18, 54). The complete coding sequence as well as 5′- and 3′-UTRs were subsequently generated by RACE RT-PCR (Fig. 1). This cDNA, which we have named SREBP2gc for “germ cell enriched form” (described below), encodes a shortened N-terminal portion of the SREBP2 precursor protein. This includes the conserved transcriptionally active portion consisting of the basic helix-loop-helix (bHLH)-Zip (DNA binding, dimerization, and nuclear localization signal) and N-terminal acidic transactivation domains, but it lacks the membrane-spanning and SCAP binding domains (Fig. 2). SREBP2gc thus is very similar to the mature, proteolytically processed form of SREBP2, which is ubiquitously expressed in somatic cells. One difference relative to the mature form is the absence of 20 amino acid residues from the C terminus of SREBP2gc (Fig. 2 and 3A).
FIG. 1.
Nucleotide and amino acid sequences of rat SREBP2gc cDNA. The numbers to the left refer to the positions of nucleotides and amino acids. The box denotes the conserved bHLH-Zip region, and the acidic N-terminal transactivation domain is underlined. The dashed lines indicate the primers used for 5′ and 3′ RACE-PCR. The dashed-lined boxes indicate potential germ cell-specific polyadenylation signals (52).
FIG. 2.
Schematic structures for different forms of SREBP2. Various domains are indicated as follows: acidic N-terminal transactivation domain (TA); serine- and glycine-rich (S) domain, as well as glutamine-rich (Q) domain; conserved bHLH-Zip DNA binding domain (bHZ); transmembrane domain (TM); and regulatory SCAP-binding domain (RSB). A 20-amino-acid sequence missing from the C terminus of SREBP2gc is also indicated by a small open box.
FIG. 3.
Comparison of sequences for rat SREBP2gc and SREBP2 from other species. (A) Protein sequences for SREBP2 from the mutant, sterol-resistant hamster cell line SRD-3 (haSRD-3) (53); rat SREBP2gc (rSREBP2gc); mature human SREBP2 (hSREBP2) (18); and the predicted mature form of mouse SREBP2 (mSREBP2), based on GenBank sequences (accession no. AAK54763) (Note that these sequences are incomplete at the N terminus.) Symbols below the amino acid sequences indicate the following relationships among different species: asterisks, identical amino acids; colons, high similarity; dots or open spaces, poor or no similarity, respectively. (B) Comparison of nucleotide sequences spanning the unique 3′-UTR, splice junction, and coding regions for rat SREBP2gc and SREBP2 from hamster SRD-3 cells. The 5′-splice sequence is underlined, and the stop codon is shown by an asterisk.
The SREBP2gc protein sequence is highly homologous to the corresponding regions of SREBP2 from other species (hamster, 94%; human, 90%; mouse, 94%), especially within the bHLH-Zip and transactivation domains. The greatest variability occurs within the Gly/Ser- and Gln/Pro-rich regions adjacent to the acidic transactivation domain, which also vary in length and/or sequence among mammalian SREBP2s (Fig. 3A). The 5′-UTR of SREBP2gc also shares significant homology with those for previously characterized SREBP2 cDNAs (data not shown), while the 3′-UTR is, in general, divergent (described below). Two potential polyadenylation sequences that are commonly used in spermatogenic, but not somatic, cells (52) were identified upstream of the poly(A) tail (Fig. 1).
Interestingly, another SREBP2 isoform was previously identified in a sterol-resistant, mutant cell line (SRD-3) that is also synthesized as a truncated protein and lacks 20 amino acid residues present at the C terminus of the ubiquitous mature form (53) (Fig. 3A). This mutant cell line was generated from Chinese hamster ovary cells by mutagenesis followed by selection for sterol resistance. The SRD-3 mutant protein apparently arose by selective genomic amplification of only certain exons and introns, generating an mRNA with an unspliced intron sequence starting at the C-terminal Val/stop codon location (Fig. 3B) (53). The first 40 bp of the 3′-UTR of SREBP2gc is 85% identical to the corresponding 3′-unspliced intron sequence of the SRD-3 isoform, including an apparent consensus 5′ splice site (Fig. 3B). It is therefore highly probable that SREBP2gc is generated by alternative splicing in rat testis. SRD-3 mutant cells are sterol resistant due to the unique properties of the truncated SREBP2 mutant protein they express: namely, a soluble, constitutively active transcription factor not subject to feedback inhibition by sterols (53). Based on their essentially identical protein sequences, SREBP2gc is predicted to share these properties, being able to enter the nucleus and constitutively regulate target promoters in expressing cells independent of sterol concentrations.
Analysis of SREBP2 mRNAs in testis and spermatogenic cells.
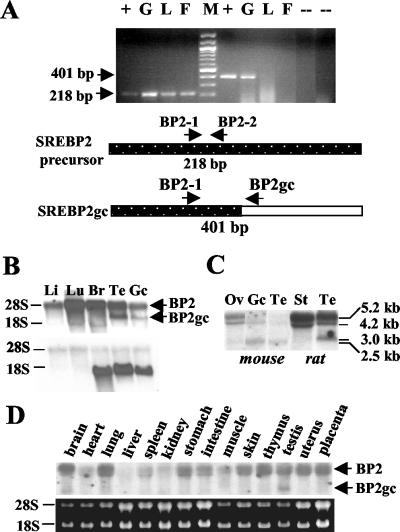
To examine the expression patterns of SREBP2 transcripts in rat and mouse tissues, we first performed RT-PCR studies. Primer pairs were designed that would generate either a 218-bp PCR product from both the SREBP2p and SREBP2gc RNAs or that spanned the 3′-UTR, splice junction, coding region of SREPBP2gc and would generate a unique 401-bp product (Fig. 4A). The common, 218-bp RT-PCR product was detectable in rat somatic tissues, testis, and enriched adult spermatogenic cells (consisting mainly of pachytene spermatocytes, round spermatids, condensing spermatids, and their cytoplasmic fragments) (Fig. 4A). However, the SREBP2gc-specific product was detected only in adult rat germ cells. Sequencing of the latter PCR product showed that it was identical to the predicted coding region of SREBP2gc (data not shown). A similar series of RT-PCR experiments demonstrated the presence of SREBP2gc transcripts in mouse testis and adult male germ cells as well, but not in mouse somatic tissues, such as liver (data not shown). These results thus confirmed that SREBP2gc transcripts are preferentially enriched in adult spermatogenic cells.
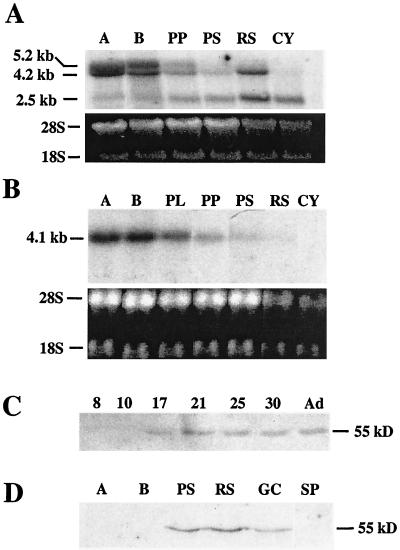
FIG. 4.
SREBP2 transcripts in rat and mouse tissues and male germ cells. (A) RT-PCR analysis. Two micrograms (each) of total RNA from adult rat spermatogenic cells (G), liver (L), and fat tissue (F) was analyzed. pBS-RBP2gc plasmid DNA was used as a positive control (+). No-RNA template and no-reverse transcriptase reactions served as negative controls (−). M, DNA size ladder. Shown are the 218-bp DNA fragment derived from SREBP2 mRNAs by using primers BP2-1 and BP2-2 (lanes to the left of size marker) and the 401-bp SREBP2gc-specific product generated with primers BP2-1 and BP2gc (lanes to the right of the size marker). (B, C, and D) Northern analysis of SREBP2 mRNAs with rat SREBP2gc cDNA as a probe. Twenty micrograms of total RNA was analyzed in each lane. (B) Rat liver (Li), lung (Lu), brain (Br), testis (Te), and enriched spermatogenic cells from adult testis (Gc). The same blot was probed for SREBP2gc (top) and rat proenkephalin (bottom) transcripts. RNA loading in the rat germ cell lane was reduced relative to rat testis, as shown by the lower signal for the germ cell-specific proenkephalin transcript (22) in this lane. (C) Mouse ovary (Ov), enriched adult mouse spermatogenic cells (Gc), mouse testis (Te), rat Sertoli cells (St), and rat testis (Te). (D) Analysis of multiple adult mouse tissues, as indicated. The results of ethidium bromide staining of 28S and 18S RNAs on the blot are shown below.
Northern analysis revealed two major RNA bands in rat testis and enriched adult male germ cells, ∼5 and 3 kb in size (Fig. 4B and C). The ∼5-kb band corresponded to the ubiquitous transcript for the SREBP2 precursor protein and was actually resolvable into two species of 5.2 and 4.2 kb (Fig. 4C), as previously described for human SREBP2 mRNAs (18). The 4.2-kb transcript may arise from the use of an alternative polyadenylation site within the SREBP2 gene (18). These RNAs were detected in rat somatic tissues, including lung, liver, brain, kidney (not shown), and Sertoli cells (Fig. 4B). The 3-kb transcript was not detected in any rat somatic tissues examined, and its size agrees well with that of the rat SREBP2gc cDNA. Analysis of mouse tissues showed a similar expression pattern: both 5.2- and 4.2-kb SREBP2p mRNAs were present in all somatic tissues tested, and a unique, smaller form (∼2.5 kb) was the predominant transcript in mouse testis and adult spermatogenic cells (Fig. 4C and D). Trace amounts of the 2.5-kb RNA also were detected in mouse ovary (Fig. 4C) and brain (Fig. 4D), but not in other tissues. The smaller transcript observed in mouse ovary also may be of germ cell origin. In contrast to the rat, the two larger SREBP2 mRNAs were greatly reduced in mouse testis and adult male germ cells relative to somatic tissues, and the 4.2-kb transcript was more abundant than the 5.2-kb mRNA (Fig. 4C). Thus, both rat and mouse spermatogenic cells contain a novel, shorter SREBP2 transcript that is undetectable or present at very low levels in somatic tissues.
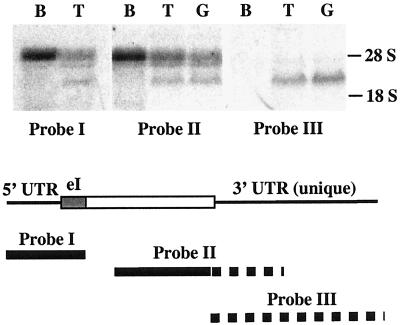
The results presented above suggested that the shorter RNAs enriched in the adult rat and mouse spermatogenic cells contained SREBP2gc sequences. To confirm this, RNA-mapping studies were performed with probes to three different portions of the rat SREBP2gc cDNA: the 5′-UTR and exon I region shared by both the SREBP2p and SREBP2gc RNAs (probe I), the common coding region and unique 3′-UTR (probe II), and the 3′-UTR sequences unique to SREBP2gc transcripts (probe III). While probes I and II hybridized to both the SREBP2p mRNAs as well as the unique 3-kb transcript in rat testis and spermatogenic cells, probe III hybridized only to the germ cell-enriched 3-kb transcript (Fig. 5). We conclude that this shorter novel RNA encodes rat SREBP2gc. The novel 2.5-kb RNA in mouse adult spermatogenic cells also presumably corresponds to the SREBP2gc transcript based on its predominance and expression pattern in these cells (Fig. 4C) (also described below).
FIG. 5.
Identification of SREBP2gc RNA in rat spermatogenic cells. Twenty micrograms (each) of total RNA from rat brain (B), testis (T), and enriched adult rat spermatogenic cells (G) was examined. The positions of probes derived from the SREBP2gc cDNA are shown schematically below the Northern results. Solid lines show sequences common to the SREBP2p and SREBP2gc RNAs, while the dotted lines indicate sequences unique to the SREBP2gc transcript. eI, presumptive exon I region based on sequence homology with the hamster SREBP2 gene.
SREBP2gc RNA is translated.
To address the expression of SREBP2gc protein in male germ cells, we first examined the translational status of its mRNA. Polysomal analysis was performed for SREBP2 transcripts in mouse testis by using sucrose gradient fractionation in the absence or presence of EDTA, which disrupts polysome integrity (Fig. 6A). In general, mRNAs in adult male germ cells are only partially associated with large polysomes (denser gradient fractions) due to a lower overall translation efficiency than is typically found for somatic mRNAs (23). Such a partial polysomal distribution was observed for both the 5.2- or 4.2-kb RNA and the 2.5-kb SREBP2 RNA, the percentage being somewhat higher for the two larger SREBP2p mRNAs (Fig. 6A). This slightly greater polysomal enrichment for the 5.2- and 4.2-kb mRNAs was expected based on the fact that the concentrations of these transcripts are greatly reduced in adult male germ cells of the mouse (Fig. 4C), such that contributions by the more efficiently translated mRNAs in testicular somatic cells (e.g., Sertoli, peritubular, and Leydig cells) are apparent. The distribution of the germ cell-enriched 2.5-kb mRNA in the denser gradient fractions is very similar to that for most other mRNAs translated in adult spermatogenic cells, such as LDH-C and Sp1 (7, 23, 36). Thus, SREBP2gc mRNA is actively translated in mouse adult male germ cells at levels typical of mRNAs expressed during spermatogenesis.
FIG. 6.
Analysis of SREBP2 mRNA translation and protein products in male germ cells. (A) Polysome analysis. Cytoplasmic RNA from mouse testis was fractionated in the presence of Mg2+ (HKM) or EDTA (HKE). Equal percentages of total RNA from fractions within each gradient were examined for SREBP2gc RNA by Northern analysis. RNA in lane 2 of the HKM gradient was underrepresented due to partial loss of sample. Gradient orientations are indicated below the Northern blot results, with larger polysomes localized to the denser (bottom) fractions. (B) Western analysis of SREBP2 protein in mouse and rat tissues and spermatogenic cells. (Left panel) Four micrograms (each) of microsomal protein from enriched adult mouse spermatogenic cells (G), brain (B), liver (Li), and lung (Lu) was examined. (Right panel) Analysis of nuclear extracts (5 μg) from enriched adult mouse spermatogenic cells (G), brain (B), kidney (K), and lung (Lu). (Lower panel) Five micrograms (each) of nuclear extract from enriched adult mouse and rat spermatogenic cells (G), mouse liver (Li), and rat lung (Lu) was examined. (C) Western blot of nuclear extracts (5 μg) from adult mouse spermatogenic cells (G) and sterol-depleted mouse 3T3-L1 cells (3T3). (D) Analysis of SREBP2gc protein in nuclei (Nu) and cytoplasmic (Cyt) fractions of adult mouse spermatogenic cells (5 μg per lane).
Spermatogenic cells express a novel SREBP2 protein.
Immunoblots were performed to determine the nature of SREBP2 proteins in male germ cells of the mouse and rat. As noted earlier, in somatic cells, SREBP2 is synthesized as a 125-kDa precursor, which is then cleaved to release the mature form, which typically runs as a 66-kDa protein on SDS gels (53). However, the levels of the mature protein are generally very low in tissues under normal conditions due to feedback inhibition by sterols. In microsomes, an ∼125-kDa protein was detected in mouse somatic tissues and spermatogenic cells corresponding to the SREBP2 precursor, although the levels were greatly reduced in male germ cells (Fig. 6B, left panel). The latter result is consistent with the very low levels of SREBP2p mRNAs in this cell fraction (Fig. 4C). In contrast, nuclear extracts from adult rat and mouse spermatogenic cells contained abundant amounts of an SREBP2-related protein of ∼55 kDa, while no protein was detected in nuclear extracts from somatic tissues (Fig. 6B, right and lower panels). It should be noted that an identical 55-kDa protein was detected by three different SREBP2 antibodies (see Materials and Methods). As a control, extracts were also analyzed from mouse 3T3 cells cultured under sterol-depleted conditions, which induce SREBP2 precursor processing to form the mature protein (19). Western blotting confirmed that the spermatogenic cell protein runs ∼11 kDa smaller on SDS gels than mouse somatic SREBP2 (∼66 kDa) (Fig. 6C). The apparent size of SREBP2gc from SDS gels is larger than predicted based on its composition (48.6 kDa). The same is true for the 66-kDa, mature somatic form of SREBP2 (24, 53), which in the mouse has a theoretical mass of 51.4 kDa.
Immunoblots also showed that the 55-kDa germ cell protein was present in nuclear but not cytoplasmic fractions of adult mouse spermatogenic cells (Fig. 6D). This finding is consistent with the predicted properties of SREBP2gc as a soluble transcriptional regulator able to directly enter the nucleus.
Spermatogenic cells contain elevated levels of SREBPs.
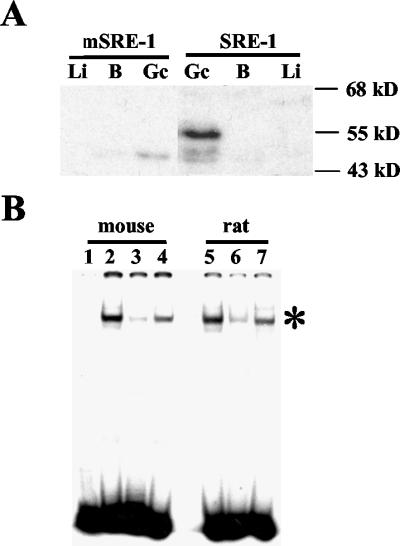
To examine the nature of SREBPs in male germ cells, Southwestern analysis was performed with the SRE from the human LDLR promoter (SRE-1) as a DNA probe. No significant binding was observed with nuclear proteins from mouse somatic tissues (Fig. 7A), consistent with the very low levels of mature SREBPs in these tissues under normal conditions. However, an abundant SREBP was present in nuclear extracts of mouse spermatogenic cells that was identical in size to SREBP2 detected by immunoblots (∼55 kDa). This protein did not bind to DNA in which the SRE-1 core sequence was mutated (Fig. 7A). These findings indicate that the SREBP2-related protein detected on Western blots also possesses SRE binding activity and presumably corresponds to SREBP2gc.
FIG. 7.
Detection of SRE-1 binding proteins in adult mouse spermatogenic cells and tissues. (A) Southwestern analysis of nuclear extracts (5 μg) from enriched spermatogenic cells from adult mouse testis (Gc), brain (B), and liver (Li). Wild-type (SRE-1) and mutant (mSRE-1) probes were used. (B) EMSA of SREBPs in mouse and rat spermatogenic cells. Nuclear extracts (1.5 μg) were examined for spermatogenic cells from adult mouse testis (lanes 2 to 4) and rat testis (lanes 5 to 7) by using the SRE-1 probe. Lanes: 1, probe only; 2 and 5, no competitor; 3 and 6, 10-fold excess of unlabeled SRE-1 wild-type competitor; 4 and 7, 10-fold excess of mutated SRE-1 competitor. The specific SREBP-SRE-1 complex is shown by an asterisk. The residual binding seen with the wild-type SRE-1 competitor was specifically eliminated at larger amounts of competitor (data not shown).
EMSAs detected a single major SRE-1 binding complex in mouse and rat adult spermatogenic cells that was competed well by unlabeled SRE-1 sequences, but not by a mutated SRE-1 competitor (Fig. 7B). Extremely low binding activity was detected with nuclear extracts from somatic tissues such as liver (data not shown). It also should be noted that no SREBP1 (precursor or mature forms) was detectable in adult mouse spermatogenic cells by Western analysis (data not shown), indicating that all SRE binding activity in spermatogenic cells derives exclusively from SREBP2gc (note that none of the available SREBP2 antibodies were able to specifically supershift native SREBP2-DNA complexes from rat or mouse). Based on these various findings, we conclude that rat and mouse adult male germ cells express a unique SREBP corresponding to SREBP2gc. This protein is predicted to function as a sterol-independent, constitutively active transcriptional regulator during spermatogenesis.
Developmental expression of SREBP2gc in male germ cells.
To gain a better understanding of the functional significance of SREBP2gc, we examined its pattern of expression in purified spermatogenic cell types. With respect to SREBP2 mRNAs, the two larger SREBP2p mRNAs were predominant in mitotic spermatogonial stages, with very little of the 2.5-kb (SREBP2gc) mRNA detectable (Fig. 8A). In early (prepubertal) and late (adult) pachytene spermatocytes, the SREBP2p transcripts were reduced in abundance. In contrast, the 2.5-kb mRNA markedly increased in these stages, and was the major SREBP2 mRNA in late pachytene spermatocytes. In haploid round spermatids, the levels of both of the 2.5- and 4.2-kb mRNAs were noticeably increased, with the 2.5-kb mRNA still predominant. Only the 2.5-kb mRNA was observed in cytoplasts (cytoplasmic fragments derived from condensing spermatids).
FIG. 8.
Developmental expression of SREBP2gc in male germ cells. (A) Northern analysis of purified mouse spermatogenic cells with rat SREBP2gc cDNA as a probe. Fifteen micrograms (each) of total RNA from spermatogonia type A (A), spermatogonia type B (B), prepubertal pachytene spermatocytes (PP), pachytene spermatocytes (PS), round spermatids (RS), and cytoplasts (Cy) was examined. Relative RNA loading is shown below by the ethidium bromide staining of the blot prior to hybridization. (B) Northern analysis of purified mouse spermatogenic cells with SREBP1c cDNA as a probe. Fractions are as in panel A, with the addition of preleptotene spermatocytes (PL), and the ethidium bromide-stained blot is shown below. (C) Western analysis of SREBP2gc protein in the developing mouse testis. Five micrograms (each) of testis nuclear extracts was examined from the indicated postnatal day mice and adults (Ad). (D) Western analysis of SREBP2gc protein in purified mouse spermatogenic cells. Five micrograms (each) of nuclear extracts from spermatogonia type A (A), spermatogonia type B (B), pachytene spermatocytes (PS), round spermatids (RS), enriched spermatogenic cells from adult mouse testis (Gc), and mouse sperm (SP) was examined.
The pattern for SREBP1 transcripts during mouse spermatogenesis differed markedly from that for SREBP2. Only the 4.1-kb mRNA for the SREBP1 precursor protein (49) was detected in purified male germ cells, and it declined dramatically in adult pachytene spermatocytes and round spermatids (Fig. 8B). This result is identical to the pattern for the 5.2-kb SREBP2p mRNA (Fig. 8A).
Immunoblotting of nuclear extracts from developing mouse testis was performed to determine the timing of onset for the 55-kDa SREBP2gc protein (Fig. 8C). This protein was first detected on postnatal day 17, when pachytene spermatocytes become predominant in mouse testis (3). SREBP2gc protein increased further by day 21, when round spermatids increase in numbers, and remained relatively constant thereafter. EMSAs revealed a similar time course for SRE-1 binding activity in the developing mouse testis (data not shown). These findings are consistent with SREBP2gc expression mainly in spermatocytes and spermatids, but with no significant expression in testicular somatic cell types, which are more predominant in the immature testis. Analysis of purified spermatogenic stages confirmed this pattern (Fig. 8D). SREBP2gc was not expressed in spermatogonia, but was readily detected in pachytene spermatocytes and round spermatids. Interestingly, SREBP2 was undetectable in epididymal sperm, indicating that it is not retained within spermatozoa during their formation. Thus, the SREBP2gc transcript and protein are expressed in the same stage-dependent manner, suggesting a particularly important role for this protein in transcriptional regulation during meiotic and early haploid spermatogenic stages.
SREBP2gc regulates SRE-containing promoters expressed during late spermatogenesis.
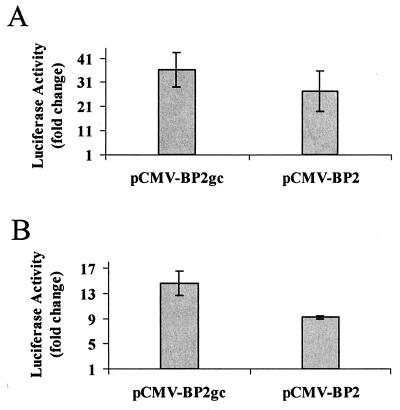
To determine the transcriptional properties of SREBP2gc, we examined its ability to activate promoters which are expressed during the later phases of spermatogenesis. As shown earlier, SREBP2gc recognizes the SRE-1 site within the promoter for the human LDLR gene. Further, expression of LDLR mRNA increases with germ cell maturation during chicken spermatogenesis (25), and in the rat testis, it increases developmentally with the appearance of late spermatogenic cell types (29). We therefore cotransfected an LDLR-luciferase promoter construct together with plasmids expressing either rat SREBP2gc or the mature form of human SREBP2 into K293 cells (Fig. 9A). SREBP2gc and mature SREBP2 stimulated the LDLR promoter 36- and 27-fold, respectively.
FIG. 9.
Transcriptional activity of rat SREBP2gc. K293 cells were cotransfected with luciferase reporter plasmids for the LDLR promoter (A) or CYP51 promoter (B) and expression vectors for rat SREBP2gc (pCMV-BP2gc), constitutively active human SREBP2 (pCMV-BP2), or empty pCMV7 parent vector along with pCMV-β-galactosidase control plasmid. Data are normalized relative to β-galactosidase activity and are expressed as the fold increase in activity relative to the activity of the pCMV7 control plasmid. Shown are the means ± standard errors of three independent experiments.
The CYP51 gene encodes an enzyme required for the synthesis of 4,4-dimethyl-5α-cholesta-8,14,24-triene-3β-ol, an intermediate in the formation of cholesterol from lanosterol, and its promoter is regulated by SREBPs in somatic cells (38). The CYP51 gene is also expressed in a stage-dependent manner during spermatogenesis, where it is up-regulated in round spermatids (45). In cotransfection studies, SREBP2gc stimulated the CYP51 promoter nearly 15-fold, compared with a 9-fold increase induced by mature SREBP2 (Fig. 9B). Thus, SREPB2gc is a highly potent activator of SRE-containing target genes expressed during spermatogenesis and thus may have a critical role in the stage-dependent expression of cholesterol biosynthetic genes and other potential targets in meiotic and early haploid spermatogenic cells.
Cell-type-specific regulation of SREBP2gc expression during spermatogenesis.
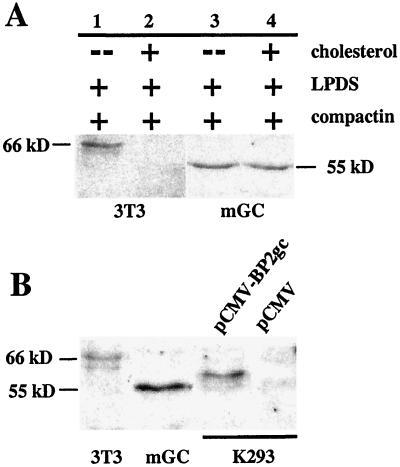
Based on its novel structure, SREBP2gc should be produced in a sterol-independent, constitutive manner in adult male germ cells. To address this question directly, the effects of sterol levels on SREBP2gc protein were examined in primary cultures of adult mouse spermatogenic cells (Fig. 10A). As a control, parallel experiments were also performed with mouse 3T3-L1 cells. The mature, somatic (∼66 kDa) SREBP2 protein was not detected in 3T3-L1 cells cultured with cholesterol, but was highly up-regulated under sterol-depleted conditions. In contrast, levels of the 55-kDa SREBP2gc protein remained unchanged in spermatogenic cells cultured in the presence and absence of sterols. This indicates that SREBP2gc is regulated by distinct cell-specific, cholesterol-insensitive mechanisms during spermatogenesis.
FIG. 10.
Cell-type-specific regulation of SREBP2gc expression during spermatogenesis. (A) Effect of sterols on SREBP2 protein in mouse 3T3-L1 (3T3) and spermatogenic (mGC) cells. Lanes: 1 and 3, sterol-depleted culture conditions with compactin and LPDS alone; 2 and 4, sterol-loaded (plus cholesterol) conditions. (B) Size comparison for SREBP2 proteins from mouse 3T3-L1 cells (3T3), enriched spermatogenic cells from adult mouse testis (mGC), and K293 cells transfected with pCMV-BP2gc or pCMV empty vector. Five micrograms (each) of the nuclear extracts was examined by Western analysis, except for K293 cell samples (1.2 μg).
As noted earlier, SREBP2gc protein in mouse and rat germ cells appears considerably smaller than its 66-kDa somatic counterpart on SDS gels. This mobility difference cannot be accounted for solely by the absence of 20 amino acids from the C terminus of SREPB2gc. Furthermore, mutant SREBP2 from SRD-3 hamster cells, which lacks the same 20 amino acids and has a theoretical mass (49.2 kDa) nearly identical to that of SREBP2gc (48.6 kDa), migrates as a significantly larger protein (∼62 kDa) than SREBP2gc on SDS gels (53). These observations suggested that additional, cell-specific mechanisms may differentially influence the electrophoretic behavior of SREBP2 in spermatogenic cells. To address this, the electrophoretic mobility of rat SREBP2gc was examined after expression in a somatic cell line, human embryonic kidney K293 cells (Fig. 10B). Following transfection with expression vector, K293 cells expressed an SREBP2gc protein that was significantly larger (59 kDa) than the endogenous protein in mouse spermatogenic cells. Thus, significant differences between the levels of synthesis of SREBP2gc protein occur in spermatogenic versus nonspermatogenic cells, apparently at the level of posttranslational modification. The structural basis for this cell-specific difference in mobility remains to be determined, although the N-terminal region was previously implicated in the slower-than-predicted mobility of SREBPs on SDS gels (41). One possibility is that residues within the Ser-rich domain (Fig. 3) undergo cell-type-specific modifications. For example, this region of rat SREBP2gc contains up to 10 potential Ser phosphorylation sites (data not shown). A function for this region of the N terminus has not been defined.
Relationship between SREBP2gc and PACH1.
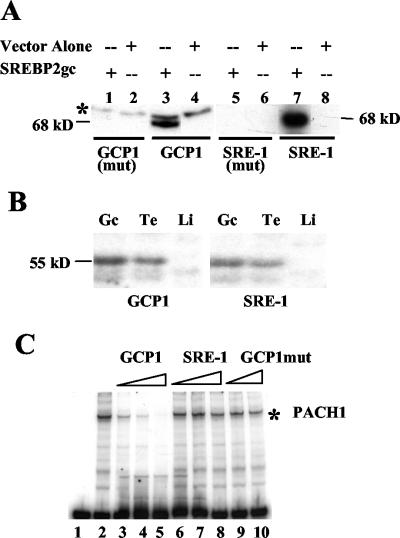
These studies were initiated by the search for the DNA binding protein PACH1, which recognizes the direct repeat sequences within the proenkephalin GCP1 element (26). Like PACH1, SREBP2gc is up-regulated in pachytene spermatocytes and round spermatids. To determine whether SREBP2gc is in fact PACH1, we first expressed rat SREBP2gc in bacteria and examined its DNA binding properties in Southwestern assays (Fig. 11A). The bacterially expressed protein was able to bind to both GCP1 and SRE-1 DNA probes, and this binding was dependent on the relevant core recognition sequences in both cases. Thus, rat SREBP2gc specifically binds to the GCP1 repeat elements, consistent with the isolation of its cDNA based on this property. The GCP1 repeat element (CTCCAG) and the SRE-1 core sequence (ATCACCCCAC) are not highly homologous. However, SREBPs recognize a diverse array of DNA sequences that frequently contain CCA (43), which is present in the GCP1 repeat. In fact, a CCAG sequence forms part of the SREs for squalene synthase and glycerol-3-phosphate acyltransferase (43). These similarities could explain the ability of SREBP2gc to bind this proenkephalin germ line promoter element.
FIG. 11.
Comparative properties of SREBP2gc and PACH1. (A) Southwestern assay of bacterially expressed SREBP2gc protein. Bacterial extracts (7 μg) were assayed by using wild-type or mutant GCP1 or SRE-1 probes (shown below the lanes). The glutathione S-transferase (GST) fusion protein is predicted to be ∼68 kDa in size (29 kDa for GST plus 39 kDa for the SREBP2gc fragment). A nonspecific 70-kDa band (asterisk) was also detected with the GCP1 probe. (B) Southwestern analysis of SRE-1 and GCP1 binding proteins in rat spermatogenic cells and tissues. Five micrograms (each) of nuclear extracts from rat liver (Li), testis (Te), and enriched spermatogenic cells from adult rat testis (Gc) was used. (C) EMSA of GCP1 binding proteins in rat spermatogenic cells. One microgram of nuclear extracts of adult rat spermatogenic cells was examined with a GCP1 DNA probe. Competition experiments were performed with excess unlabeled GCP1, SRE-1, and mutated GCP1 DNAs. The PACH1 complex is shown by an asterisk.
To examine further the possible identity between SREBP2gc and PACH1, comparative Southwestern blots were performed with rat nuclear extracts and the GCP1 and SRE-1 DNA probes (Fig. 11B). For both probes, an ∼55-kDa binding protein was identified in mouse testis and adult male germ cells in equal abundance, but not in liver. No binding was seen in germ cells with mutated versions of these probes (data not shown). This finding suggested that SREBP2gc was the predominant GCP1 binding protein in adult spermatogenic cells, at least as detected by Southwestern assay. However, different results were obtained with EMSAs. The PACH1 complex, as detected with the GCP1 probe (26), was specifically competed by excess amounts of the homologous sequence, but was not by GCP1mut containing mutations in the direct repeat elements (Fig. 11C). Importantly, SRE-1 sequences, which compete well for SREBP2gc binding to the SRE-1 probe in EMSAs (Fig. 7B), were unable to displace PACH1 binding to the GCP1 probe (Fig. 11C). In reciprocal experiments, GCP1 sequences did not compete more strongly than SRE-1 sequences for binding of germ cell extracts to an SRE-1 probe (data not shown). Finally, SREBP2gc was unable to stimulate the rat proenkephalin germ line promoter in cotransfection experiments with K293 cells (data not shown).
These results indicate that, while SREBP2gc is capable of binding to GCP1 sequences, it does not by itself account for the PACH1 DNA binding activity present in adult spermatogenic cells. Based on Southwestern results, PACH1 and SREBP2gc may be distinct proteins having very similar sizes and germ cell abundances. Alternatively, PACH1 may be present in considerably lower concentrations than SREBP2gc in spermatogenic cells or may exist as a multimer of nonidentical subunits, such that SREBP2gc is the major GCP1 binding protein detected on Southwestern blots. The possibility that SREBP2gc is a component of a heterocomplex that binds GCP1 much more avidly than SRE-1 sequences cannot be ruled out at this time.
DISCUSSION
These studies describe the identification of a novel member of the SREBP family in spermatogenic cells that possesses unique properties not found in its somatic counterpart. In particular, SREBP2gc is synthesized as a soluble and constitutively active transcription factor that bypasses the sterol-regulated, proteolysis-dependent mechanisms, which limit its expression in somatic cells (Fig. 12). Thus, novel, sterol-independent mechanisms have evolved to regulate the expression of the SREBP2 transcription factor in male germ cells. SREBP2gc is expressed in a stage-dependent manner during spermatogenesis, with the highest levels occurring during the later transcriptionally active stages, namely pachytene spermatocytes and round spermatids. Apparently this factor does not continue to function in mature sperm (e.g., in DNA packaging or male pronuclear transcription during early embryogenesis). Its stage-dependent expression is controlled at the mRNA level, at least in part, with RNA splicing apparently playing a predominant role based on analogy with the mutant SREBP2 from hamster SRD-3 cells (53). In fact, the SRD-3 mutant protein apparently arose by usurping an alternative splicing mechanism that normally is predominant in male germ cells and is an infrequent event in somatic cells. Alternative splicing is a common mechanism for generating cell-specific mRNAs during spermatogenesis (51), including that for the germ cell-specific isoform of the CREM transcription factor, CREMτ (13). The mechanisms controlling cell- and stage-specific regulation of RNA splicing during spermatogenesis remain unclear. The germ cell protein RBM has been implicated as a potential participant in these processes (12). Since SREBP2 can activate its own promoter, at least in human somatic cells (39), SREBP2gc also may regulate its own expression at the transcriptional level during spermatogenesis. For example, this mechanism could contribute to the increased levels of total SREBP2 transcripts observed in haploid germ cells. However, whether mRNAs for SREBP2gc and SREBP2p arise from a common promoter has not been determined, although they clearly share common 5′-UTR sequences.
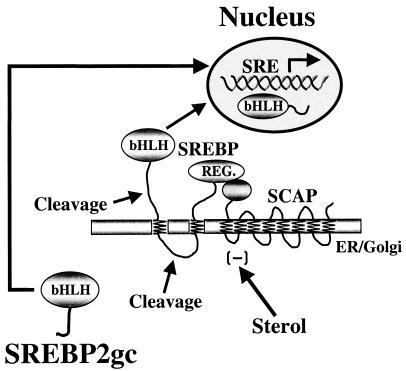
FIG. 12.
Sterol-insensitive regulation of promoters in meiotic and early haploid spermatogenic cells by SREBP2gc. SREBP2gc bypasses the SCAP-dependent, sterol-regulated proteolytic processing pathway and directly enters the nucleus to regulate target genes in a constitutive manner.
The present findings raise important questions about the role of SREBP2 and its target genes during spermatogenesis. In particular, why do spermatogenic cells express a novel, sterol-insensitive form of SREBP2? It is our hypothesis that generation of SREBP2gc provides a mechanism by which spermatogenic cells can regulate SREBP2-dependent genes in selected developmental stages that would otherwise not be possible with the standard sterol-regulated feedback pathway. That is, developmentally regulated synthesis of an alternative transcript encoding a soluble, constitutively active SREBP2 isoform permits its preferential function in specific germ cell stages, which could not be readily achieved via a sterol-dependent inhibitory mechanism. Furthermore, this stage-specific function during later spermatogenesis is limited to SREBP2, since expression of SREBP1 declines dramatically during this process, and no novel isoform is apparent. This suggests that target genes for SREBP2, as opposed to SREBP1, may be particularly important during spermatogenesis. Based on structural similarities, the properties of SREBP2gc are predicted to be identical to those of the mutant SREBP2 protein expressed in SRD-3 cells. In addition to being constitutively active, the protein from SRD-3 cells repressed the cleavage of SREBP precursor proteins, apparently by inhibiting cleavage enzyme expression or activity (53). Thus, SREBP2gc also may repress the processing of the relatively small amounts of SREBP precursors present in adult spermatogenic cells.
Previous studies suggested that SREBPs were not involved in the regulation of cholesterol biosynthetic genes during spermatogenesis (38). This was based in part on the fact that these genes are typically regulated in a coordinate manner by sterols, and this is not the case during spermatogenesis (46). The present findings provide an alternative explanation for the apparent sterol independence of SRE-containing target gene expression in male germ cells: regulation by an SREBP isoform that is not sensitive to sterol concentrations. In fact, a recent study of squalene synthase gene expression in liver and testis provides direct support for this (8). While squalene synthase mRNA was highly regulated by dietary cholesterol levels in liver, no such regulation was observed for its testicular mRNAs. This is consistent with the regulation of the squalene synthase promoter via its SRE sequences in both tissues, in liver via standard sterol inhibition of SREBP2p cleavage to form mature SREBP2, and in testis or spermatogenic cells via stage-specific, sterol-independent activity of SREBP2gc.
Critical questions are the nature of the target genes regulated by SREBP2gc during meiotic and early haploid stages and the functional importance of SREBP2gc transcriptional effects for spermatogenesis. Cholesterol is essential for normal membrane structure and signaling and as a precursor for steroid hormones and bile acids, and it also plays a highly specialized role in sperm function. In order for sperm to undergo the acrosome reaction during fertilization, several cellular modifications must first occur that are collectively referred to as “capacitation.” A major initiating event in this process is the loss of cholesterol from the sperm outer membrane (9). This may involve altered partitioning of lipid rafts necessary for membrane signaling events (50). Thus, incorporation of the correct amounts of cholesterol into the cell membrane of maturing sperm is critical for controlling the fertilization process. Adult spermatogenic cells contain considerable concentrations of free and total cholesterol (greater than those in Sertoli cells) (2), and the expression of several genes involved in the cholesterol biosynthetic pathway increases during male germ cell maturation. These include the mRNAs for CYP51 and farnesyl pyrophosphate synthetase, both of which are up-regulated in round spermatids (38, 47), and squalene synthase mRNA, which is expressed at similar levels in pachytene spermatocytes and round spermatids (38, 46). Other SREBP targets increase in the testis with maturation, which at least suggests their expression in later stages of spermatogenesis. These include the mRNAs for LDLR, HMG coenzyme A (CoA) synthase, HMG CoA reductase, and enzymatic activities for ATP citrate lyase and acetyl-CoA carboxylase (1, 29, 30). Thus, stage-dependent regulation of cholesterol and lipid biosynthetic genes is likely to be an important consequence of SREBP2gc expression during spermatogenesis. A complete analysis of the expression patterns of known SREBP2 target genes in specific germ cell stages is clearly warranted.
The precise impact of the expression of cholesterol biosynthetic genes and other potential SREBP2gc-regulated targets on spermatogenesis remains to be determined. Cholesterol synthesis increases during germ cell maturation in prepubertal pachytene spermatocytes (37). However, no further increase was observed in adult pachytene spermatocytes and round spermatids, when SREBP2gc remains elevated. This may indicate that, in addition to cholesterol, SREBP2gc is important for regulating the synthesis of other lipids in later spermatogenic stages. For example, the mevalonate pathway, which is regulated by SREBP2 and is required for cholesterol synthesis, also provides substrate for the synthesis of farnesyl pyrophosphate used for protein prenylation, dolichol phosphate required for protein glycosylation, and heme A and ubiquinone, which are involved in mitochondrial electron transport (16). It is noteworthy that the synthesis of dolichol phosphate was reported to increase in both mouse pachytene spermatocytes and round spermatids (37), and protein farnesyltransferase transcripts increase during hamster spermatid maturation (33). Farnesyl pyrophosphate synthetase mRNA is also specifically enriched in haploid round spermatids (48). Furthermore, the sterol product of the CYP51 enzyme is converted in the testis to 4,4-dimethyl-5α-cholesta-8,24-diene-3β-ol (also known as “T-MAS,” for “testicular meiosis-activating sterol”), which can induce meiosis in oocytes and has been implicated as an endogenous regulator of spermatocyte maturation (45). The levels of T-MAS increase with testicular development along with the levels of CYP51 mRNA (46). These various observations suggest that SREBP2gc may regulate the synthesis of multiple, key lipid products important for completion of male meiosis and spermiogenesis.
These studies raise an important issue regarding the basis for the differential stage dependence of SREBP target gene expression during spermatogenesis. As noted above, farnesyl pyrophosphate synthetase and CYP51 mRNAs are specifically up-regulated in round spermatids, while squalene synthase transcripts increase to similar levels in pachytene spermatocytes and round spermatids (48). Transcriptional coregulators such as Sp1, CREB/CREM, and NF-Y are required for optimal transactivation by SREBPs (11), and both Sp1 and CREM are expressed in a stage-dependent manner in meiotic and early haploid stages (13, 36). This suggests that stage-specific expression or activity of SREBP2gc coregulators may contribute to the differential regulation of SREBP2gc gene targets during spermatogenesis. For example, the CYP51 gene is activated by CREMτ, and is not expressed in round spermatids of CREMτ-deficient mice (38). Based on this, it was suggested that up-regulation of the CYP51 gene in round spermatids was determined solely by CREMτ. In contrast, the present findings argue that both CREM and SREBP2 are important for CYP51 expression in haploid germ cells. Thus, to fully understand SREBP2gc function during spermatogenesis, it will be important to determine the patterns of expression of various coregulators and target genes for SREBP2gc in purified spermatogenic cell stages.
The highly specialized differentiation and cell-specific transcription occurring during spermatogenesis also raise the possibility that SREBP2gc regulates novel, cell-type-specific genes in male germ cells. This may in part involve interactions with unique spermatogenic cell coregulators yet to be identified. Potentially novel posttranslational modification of SREBP2gc during spermatogenesis could contribute to its germ cell-specific transcriptional properties, although this aspect requires further investigation. Studies in which the function of SREBP2gc is disrupted in vivo should prove extremely informative in determining the roles of this transcription factor and its various target genes and coregulators during spermatogenesis.
Acknowledgments
We thank Jay D. Horton, Department of Internal Medicine, University of Texas Southwestern Medical Center for providing us with numerous reagents as well as helpful advice. We also thank D. Rozman of the University of Ljubljana, Slovenia, and B. Spiegelman of the Dana-Farber Cancer Institute for providing plasmids, as well as R. Sato of the University of Tokyo for antibodies.
This work was supported by PHS grant RO1DK36468 and Center grant DK32520.
REFERENCES
- 1.Bajpai, M., G. Gupta, S. K. Jain, and B. S. Setty. 1998. Lipid metabolising enzymes in isolated rat testicular germ cells and changes associated with meiosis. Andrologia 30:311-315. [DOI] [PubMed] [Google Scholar]
- 2.Beckman, J. K., and J. G. Coniglio. 1979. A comparative study of the lipid composition of isolated rat Sertoli and germinal cells. Lipids 14:262-267. [DOI] [PubMed] [Google Scholar]
- 3.Bellve, A. R., J. C. Cavicchia, C. F. Millette, D. A. O'Brien, Y. M. Bhatnagar, and M. Dym. 1977. Spermatogenic cells of the prepubertal mouse. J. Cell Biol. 74:68-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellve, A. R., C. F. Millette, Y. M. Bhatnagar, and D. A. O'Brien. 1977. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 25:480-494. [DOI] [PubMed] [Google Scholar]
- 5.Blendy, J. A., K. H. Kaestner, G. F. Weinbauer, E. Nieschlag, and G. Schutz. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380:162-165. [DOI] [PubMed] [Google Scholar]
- 6.Brown, M. S., and J. L. Goldstein. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96:11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cataldo, L., K. Baig, R. Oko, M. A. Mastrangelo, and K. C. Kleene. 1996. Developmental expression, intracellular localization, and selenium content of the cysteine-rich protein associated with the mitochondrial capsules of mouse sperm. Mol. Reprod. Dev. 45:320-331. [DOI] [PubMed] [Google Scholar]
- 8.Collins, B. S., T. R. Tansey, and I. Shechter. 2001. Comparative squalene synthase gene expression in mouse liver and testis. Arch. Biochem. Biophys. 395:253-258. [DOI] [PubMed] [Google Scholar]
- 9.Cross, N. L. 1998. Role of cholesterol in sperm capacitation. Biol. Reprod. 59:7-11. [DOI] [PubMed] [Google Scholar]
- 10.Eddy, E. M. 1998. Regulation of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 9:451-457. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, P. A., D. Tabor, H. R. Kast, and A. Venkateswaran. 2000. Regulation of gene expression by SREBP and SCAP. Biochim. Biophys. Acta 1529:103-113. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, D. J., C. F. Bourgeois, A. Klink, J. Stevenin, and H. J. Cooke. 2000. A mammalian germ cell-specific RNA-binding protein interacts with ubiquitously expressed proteins involved in splice site selection. Proc. Natl. Acad. Sci. USA 97:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foulkes, N. S., B. Mellstrom, E. Benusiglio, and P. Sassone-Corsi. 1992. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature 355:80-84. [DOI] [PubMed] [Google Scholar]
- 14.Galcheva-Gargova, Z., J. P. Tokeson, L. K. Karagyosov, K. M. Ebert, and D. L. Kilpatrick. 1993. The rat proenkephalin germ line promoter contains multiple binding sites for spermatogenic cell nuclear proteins. Mol. Endocrinol. 7:979-991. [DOI] [PubMed] [Google Scholar]
- 15.Gerton, G. L., and C. F. Millette. 1984. Generation of flagella by cultured mouse spermatids. J. Cell Biol. 98:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein, J. L., and M. S. Brown. 1990. Regulation of the mevalonate pathway. Nature 343:425-430. [DOI] [PubMed] [Google Scholar]
- 17.Horton, J. D., I. Shimomura, M. S. Brown, R. E. Hammer, J. L. Goldstein, and H. Shimano. 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 101:2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua, X., C. Yokoyama, J. Wu, M. R. Briggs, M. S. Brown, J. L. Goldstein, and X. Wang. 1993. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA 90:11603-11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue, J., H. Kumagai, T. Terada, M. Maeda, M. Shimizu, and R. Sato. 2001. Proteolytic activation of SREBPs during adipocyte differentiation. Biochem. Biophys. Res. Commun. 283:1157-1161. [DOI] [PubMed] [Google Scholar]
- 20.Kew, D., D. F. Jin, F. Kim, T. Laddis, and D. L. Kilpatrick. 1989. Translational status of proenkephalin mRNA in the rat reproductive system. Mol. Endocrinol. 3:1191-1196. [DOI] [PubMed] [Google Scholar]
- 21.Kew, D., and D. L. Kilpatrick. 1989. Expression and regulation of the proenkephalin gene in rat Sertoli cells. Mol. Endocrinol. 3:179-184. [DOI] [PubMed] [Google Scholar]
- 22.Kilpatrick, D. L., and C. F. Millette. 1986. Expression of proenkephalin messenger by RNA mouse testicular germ cells. Proc. Natl. Acad. Sci. USA 83:5015-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleene, K. C. 1996. Patterns of translational regulation in the mammalian testis. Mol. Reprod. Dev. 43:268-281. [DOI] [PubMed] [Google Scholar]
- 24.Korn, B. S., I. Shimomura, Y. Bashmakov, R. E. Hammer, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1998. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N). J. Clin. Investig. 102:2050-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstedt, K. A., H. Bujo, M. G. Mahon, J. Nimpf, and W. J. Schneider. 1997. Germ cell-somatic cell dichotomy of a low-density lipoprotein receptor gene family member in testis. DNA Cell Biol. 16:35-43. [DOI] [PubMed] [Google Scholar]
- 26.Liu, F., I. Kondova, and D. L. Kilpatrick. 2000. Detection of PACH1, a nuclear factor implicated in the transcriptional regulation of meiotic and early haploid stages of spermatogenesis. Mol. Reprod. Dev. 57:224-231. [DOI] [PubMed] [Google Scholar]
- 27.Liu, F., J. Tokeson, S. P. Persengiev, K. Ebert, and D. L. Kilpatrick. 1997. Novel repeat elements direct rat proenkephalin transcription during spermatogenesis. J. Biol. Chem. 272:5056-5062. [DOI] [PubMed] [Google Scholar]
- 28.Nantel, F., L. Monaco, N. S. Foulkes, D. Masquiller, M. LeMeur, K. Henriksen, A. Dierich, M. Parvinen, and P. Sasson-Corsi. 1996. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380:159-162. [DOI] [PubMed] [Google Scholar]
- 29.Ness, G. C. 1994. Developmental regulation of the expression of genes encoding proteins involved in cholesterol homeostasis. Am. J. Med. Genet. 50:355-357. [DOI] [PubMed] [Google Scholar]
- 30.Ness, G. C., and S. J. Nazian. 1992. Developmental expression of multiple forms of 3-hydroxy-3-methylglutaryl coenzyme A reductase mRNA in rat testes. J. Androl. 13:318-322. [PubMed] [Google Scholar]
- 31.Newton, S. C., and C. F. Millette. 1992. Sertoli cell plasma membrane polypeptides involved in spermatogenic cell-Sertoli cell adhesion. J. Androl. 13:160-171. [PubMed] [Google Scholar]
- 32.Nohturfft, A., D. Yabe, J. L. Goldstein, M. S. Brown, and P. J. Espenshade. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102:315-323. [DOI] [PubMed] [Google Scholar]
- 33.Olson, G. E., S. K. Nagdas, and V. P. Winfrey. 1997. Temporal expression and localization of protein farnesyltransferase during spermiogenesis and posttesticular sperm maturation in the hamster. Mol. Reprod. Dev. 48:71-76. [DOI] [PubMed] [Google Scholar]
- 34.Osborne, T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275:32379-32382. [DOI] [PubMed] [Google Scholar]
- 35.Pai, J. T., O. Guryev, M. S. Brown, and J. L. Goldstein. 1998. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J. Biol. Chem. 273:26138-26148. [DOI] [PubMed] [Google Scholar]
- 36.Persengiev, S. P., P. J. Raval, S. Rabinovitch, C. F. Millette, and D. L. Kilpatrick. 1996. Transcription factor Sp1 is expressed by three different developmentally regulated messenger ribonucleic acids in mouse spermatogenic cells. Endocrinology 137:638-646. [DOI] [PubMed] [Google Scholar]
- 37.Potter, J. E., C. F. Millette, M. J. James, and A. A. Kandutsch. 1981. Elevated cholesterol and dolichol synthesis in mouse pachytene spermatocytes. J. Biol. Chem. 256:7150-7154. [PubMed] [Google Scholar]
- 38.Rozman, D., M. Fink, G. M. Fimia, P. Sassone-Corsi, and M. R. Waterman. 1999. Cyclic adenosine 3′,5′-monophosphate(cAMP)/cAMP-responsive element modulator (CREM)-dependent regulation of cholesterogenic lanosterol 14alpha-demethylase (CYP51) in spermatids. Mol. Endocrinol. 13:1951-1962. [DOI] [PubMed] [Google Scholar]
- 39.Sato, R., J. Inoue, Y. Kawabe, T. Kodama, T. Takano, and M. Maeda. 1996. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271:26461-26464. [DOI] [PubMed] [Google Scholar]
- 40.Sato, R., W. Miyamoto, J. Inoue, T. Terada, T. Imanaka, and M. Maeda. 1999. Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J. Biol. Chem. 274:24714-24720. [DOI] [PubMed] [Google Scholar]
- 41.Sato, R., J. Yang, X. Wang, M. J. Evans, Y. K. Ho, J. L. Goldstein, and M. S. Brown. 1994. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J. Biol. Chem. 269:17267-17273. [PubMed] [Google Scholar]
- 42.Schoonjans, K., C. Brendel, D. Mangelsdorf, and J. Auwerx. 2000. Sterols and gene expression: control of affluence. Biochim. Biophys. Acta 1529:114-125. [DOI] [PubMed] [Google Scholar]
- 43.Shimano, H. 2001. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 40:439-452. [DOI] [PubMed] [Google Scholar]
- 44.Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science 290:1721-1726. [DOI] [PubMed] [Google Scholar]
- 45.Stromstedt, M., M. R. Waterman, T. B. Haugen, K. Tasken, M. Parvinen, and D. Rozman. 1998. Elevated expression of lanosterol 14alpha-demethylase (CYP51) and the synthesis of oocyte meiosis-activating sterols in postmeiotic germ cells of male rats. Endocrinology 139:2314-2321. (Erratum, 139:3771.) [DOI] [PubMed] [Google Scholar]
- 46.Tacer, K. F., T. B. Haugen, M. Baltsen, N. Debeljak, and D. Rozman. 2002. Tissue-specific transcriptional regulation of the cholesterol biosynthetic pathway leads to accumulation of testis meiosis-activating sterol (T-MAS). J. Lipid Res. 43:82-89. [PubMed] [Google Scholar]
- 47.Teruya, J. H., S. Y. Kutsunai, D. H. Spear, P. Edwards, and C. F. Clarke. 1990. Testis-specific transcription initiation sites of rat farnesyl pyrophosphate synthetase mRNA. Mol. Cell. Biol. 10:2315-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teruya, J. H., E. C. Salido, P. A. Edwards, and C. F. Clarke. 1991. Testis-specific transcripts of rat farnesyl pyrophosphate synthetase are developmentally regulated and localized to haploid germ cells. Biol. Reprod. 44:663-671. [DOI] [PubMed] [Google Scholar]
- 49.Tontonoz, P., J. B. Kim, R. A. Graves, and B. M. Spiegelman. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13:4753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travis, A. J., T. Merdiushev, L. A. Vargas, B. H. Jones, M. A. Purdon, R. W. Nipper, J. Galatioto, S. B. Moss, G. R. Hunnicutt, and G. S. Kopf. 2001. Expression and localization of caveolin-1, and the presence of membrane rafts, in mouse and Guinea pig spermatozoa. Dev. Biol. 240:599-610. [DOI] [PubMed] [Google Scholar]
- 51.Walker, W. H., F. J. Delfino, and J. F. Habener. 1999. RNA processing and the control of spermatogenesis. Front. Horm. Res. 25:34-58. [DOI] [PubMed] [Google Scholar]
- 52.Wallace, A. M., B. Dass, S. E. Ravnik, V. Tonk, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, and C. C. MacDonald. 1999. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. USA 96:6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, J., M. S. Brown, Y. K. Ho, and J. L. Goldstein. 1995. Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines. Role of introns in protein evolution. J. Biol. Chem. 270:12152-12161. [DOI] [PubMed] [Google Scholar]
- 54.Yang, J., R. Sato, J. L. Goldstein, and M. S. Brown. 1994. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 8:1910-1919. [DOI] [PubMed] [Google Scholar]
- 55.Zinn, S. A., K. M. Ebert, N. D. Mehta, J. Joshi, and D. L. Kilpatrick. 1991. Selective transcription of rat proenkephalin fusion genes from the spermatogenic cell-specific promoter in testis of transgenic mice. J. Biol. Chem. 266:23850-23855. [PubMed] [Google Scholar]