Abstract
Fragile X syndrome is caused by loss of FMR1 protein expression. FMR1 binds RNA and associates with polysomes in the cytoplasm; thus, it has been proposed to function as a regulator of gene expression at the posttranscriptional level. Posttranslational modification of FMR1 had previously been suggested to regulate its activity, but no experimental support for this model has been reported to date. Here we report that FMR1 in Drosophila melanogaster (dFMR1) is phosphorylated in vivo and that the homomer formation and the RNA-binding activities of dFMR1 are modulated by phosphorylation in vitro. Identification of a protein phosphorylating dFMR1 showed it to be Drosophila casein kinase II (dCKII). dCKII directly interacts with and phosphorylates dFMR1 in vitro. The phosphorylation site in dFMR1 was identified as Ser406, which is highly conserved among FMR1 family members from several species. Using mass spectrometry, we established that Ser406 of dFMR1 is indeed phosphorylated in vivo. Furthermore, human FMR1 (hFMR1) is also phosphorylated in vivo, and alteration of the conserved Ser500 in hFMR1 abolishes phosphorylation by CKII in vitro. These studies support the model that the biological functions of FMR1, such as regulation of gene expression, are likely regulated by its phosphorylation.
Fragile X syndrome is the most frequent cause of inheritable mental retardation and is also one of the most common single-gene disorders (14). Cognitive deficits reported for fragile X children range from mild to severe, and behavioral disturbances include social and attention deficits, autistic-like behaviors, unusual responses to sensory stimuli, hyperactivity, and sleep problems (12, 14). The gene directly responsible for fragile X syndrome, FMR1, was identified in 1991 (47). In most cases, the syndrome is caused by a trinucleotide repeat expansion in the 5′ untranslated region of the FMR1 gene (49). The expansion of the CGG repeat results in an absence of the encoded protein. It is therefore clear that the pathophysiological mechanisms leading to symptoms in fragile X syndrome can be elucidated by studying the functions of the FMR1 gene.
The fact that the FMR1 protein is a cytoplasmic protein with RNA-binding motifs (two KH domains and an RGG box) (2, 38, 39) and the fact that it associates with ribosomes (6, 9, 21, 41) suggest that this protein functions in the posttranscriptional regulation of some specific mRNA expression (16). Indeed, FMR1 has recently been proposed to be a translational repressor in a cell-free system and in frog oocytes (26, 27). Interestingly, although the inhibitory effect was demonstrated not to be influenced by the features of the mRNAs used in those studies, FMR1 seemed to specifically inhibit its own translation (36). It has been known that FMR1 is found at postsynaptic sites enriched in ribosomes, where some specific mRNAs, including FMR1, are translated in response to synaptic activation (13). It has also been observed that in fragile X patients the numbers of dendritic spines are different and the morphology is also changed from that of those cells in normal individuals (18). These data suggest that FMR1 takes part in normal synaptic formation during development by regulating the expression of specific mRNAs at the postsynaptic sites.
What kind of mRNAs does FMR1 specifically bind to in vivo? It has been reported that FMR1 in vitro binds specifically to poly(G) and poly(U), but not to poly(A) and poly(C) (38), and it was estimated that FMR1 interacts with about 4% of human fetal brain mRNAs (2). These results indicate that FMR1 has an intrinsic ability to distinguish its target molecules from among all the native RNA molecules and to bind specific sequences. Recently, mRNAs to which FMR1 preferentially binds have been identified by several groups (5, 7, 36, 53). In the mRNAs identified, the nucleotide sequence through which FMR1 prefers to bind can form unusual secondary structures known as G quartets (7, 36), which were originally reported as DNA structures at the ends of chromosomes for stabilization (43, 51). The G quartet structure in RNA is known to be stabilized by coordination with particular cations, such as K+. Indeed, FMR1 showed much higher affinity to a G quartet in a buffer containing K+. Interestingly, Darnell et al. reported that only the RGG box in FMR1 is required, and that the two KH domains are dispensable, for the interaction between FMR1 and a G quartet (7). However, the in vivo relevance of these discoveries has not been elucidated.
The discovery of the existence of two FMR1-related genes, FXR1 and FXR2, has revealed an additional level of complexity in the study of FMR1 functions in vertebrates (40, 52). For many human disorders, redundancy of gene function due to gene duplication in the same species makes it difficult to interpret clearly the effects of loss-of-function mutations on cellular physiology and biochemistry. Recently, a Drosophila homolog of FMR1 (dFMR1) has been identified (48). In agreement with the statement of Wan et al. (48), the recently completed Drosophila melanogaster genome sequence (1) revealed that dFMR1 is a unique, single gene with no homologs in the fly genome. The Drosophila and vertebrate FMR1 proteins share a number of topographical landmarks, including RNA-binding motifs (two KH domains and an RGG box), and show similar biological properties such as a binding preference in vitro for certain RNA sequences and ribosomal association in vivo (19, 48; M. C. Siomi and H. Siomi, unpublished data). Moreover, a number of genes related to complex behaviors such as learning and memory are conserved between flies and humans (30, 34). Therefore, Drosophila could be used to define the molecular pathways leading to human neurological diseases such as fragile X syndrome and to identify the genes involved. In fact, several groups have individually produced fly models of fragile X syndrome and have shown that dFMR1 is necessary for normal synaptic activities (53) and for rhythmic circadian output regulating locomotor activities (8, 17, 31).
For a number of proteins, posttranslational modifications have been well studied and are known to be critical for regulating physiological and biochemical functions. So far, however, very little is known about the posttranslational modifications of FMR1 protein and their effects at the functional level. Here we provide the first evidence that FMR1 protein, both in flies and in humans, is phosphorylated in vivo. By purification from the cytoplasmic lysate of a Drosophila culture cell line, Schneider 2 (S2), and by mass spectrometric analysis, we identified Drosophila casein kinase II (dCKII) (35) as a protein phosphorylating dFMR1. Indeed, recombinant dCKII was able to interact with and phosphorylate dFMR1 in vitro. Systematic mutation analyses revealed that the phosphorylation site in dFMR1 is Ser406, located just upstream of the RGG box. This amino acid residue is conserved among FMR1/FXR family members across many species, and mutation to Ala of the Ser residues in dFMR1 and hFMR1 proteins abolishes the phosphorylation by CKII in vitro. Finally, we demonstrate that the efficiency of dFMR1 in forming homomers and binding RNA is profoundly altered by phosphorylation in vitro. Taken together, our data strongly support the hypothesis that phosphorylation by CKII could play an important role in regulating physiological functions of FMR1, such as regulation of gene expression for the normal formation of synapses during development.
MATERIALS AND METHODS
Cell culture.
S2 cells were grown in Schneider's Drosophila medium (Gibco BRL) supplemented with 10% fetal bovine serum (Sigma), 1% l-glutamine 50 U of penicillin/ml, and 50 μg of streptomycin/ml (Gibco BRL) at 26°C. HeLa cells were grown in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum, 1% l-glutamine, 50 U of penicillin/ml, and 50 μg of streptomycin/ml (Gibco BRL) at 37°C.
Subcellular fractionation, CIP treatment, and Western blot analysis.
A cytoplasmic lysate of S2 cells was prepared in a lysis buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 50 μg of digitonin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, and 0.5% aprotinin. To a half-portion of the lysate, which was not to be treated with calf intestine alkaline phosphatase (CIP), NaF was added to 2% immediately after preparation of the lysate. A cytoplasmic lysate from fly ovaries was prepared in a similar lysis buffer, but 50 μg of digitonin/ml was replaced by 0.5% Triton X-100. Treatment of the cytoplasmic lysates with CIP (NEB) was carried out at 30°C for 20 min. The lysates with and without CIP treatment were then mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and proteins were separated on an SDS-7.5% polyacrylamide gel after heat denaturation. Western blotting was carried out essentially as described previously (41) by using an anti-dFMR1 monoclonal antibody (48). Protein bands were visualized with an ECL Western blotting detection kit (Amersham Bioscience).
In vitro phosphorylation assay.
Either full-length or truncated dFMR1 cDNA (48) was subcloned into the pGEX-5X (Amersham Bioscience) bacterial expression vector to produce glutathione S-transferase (GST) fusion proteins for assay. GST fusion proteins and GST itself were produced and purified as described by the manufacturer. For Fig. 6C, full-length hFMR1 cDNA (38, 47) was used and GST-hFMR1 was produced as well. All the point mutants were constructed by performing PCR using primers specifically designed to alter Ser to Ala. The final products were inserted into the pGEX-5X expression vector, and GST fusion proteins were produced as well. The assay was carried out in P buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 2 mM ATP, and 1 mM dithiothreitol (DTT) in the presence of [γ-32P]ATP (NEN) at 27°C for 1 h. As an enzymatic source, a cytoplasmic lysate of S2 cells after centrifugation at 100,000 × g for 30 min was used for the experiments for which results are shown in Fig. 1B and 2. Purified mammalian CKII (Fig. 6D) was a kind gift from U. Kikkawa (22). After the reaction, proteins were separated on SDS-polyacrylamide gels and autoradiography was carried out to visualize phosphorylated proteins.
FIG. 6.
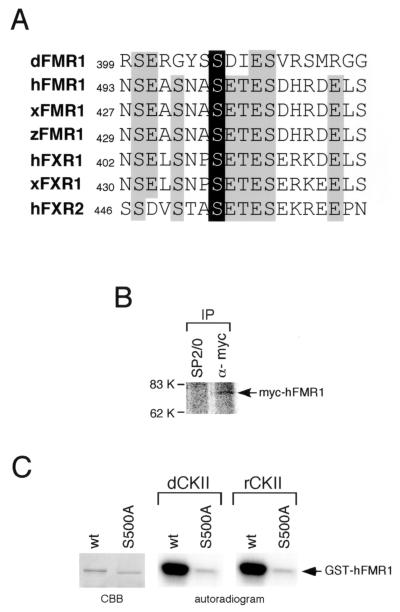
Phosphorylation of FMR1/FXR in humans. (A) The peptide sequence containing S406 of dFMR1 is aligned with the corresponding regions in hFMR1 (47), Xenopus FMR1 (xFMR1) (39), zebra fish FMR1 (zFMR1) (48), human FXR1 (hFXR1) (39), Xenopus FXR1 (xFXR1) (39), and human FXR2 (hFXR2) (52). The phosphorylated residue (S406) in dFMR1 is highly conserved among the sequences and is highlighted on a solid background. Other highly conserved residues are shaded. (B) Phosphorylation of hFMR1 in vivo. HeLa cells 24 h after transfection of myc-hFMR1 were labeled with 32P. Immunoprecipitations were performed with an anti-myc monoclonal antibody (9E10) and a nonimmune control antibody (SP2/0). Autoradiography of the immunoprecipitates separated by SDS-PAGE revealed that one protein band was predominantly labeled with 32P and migrated at the expected size of myc-tagged hFMR1 (∼80 kDa). This band was observed only in the 9E10 immunoprecipitates. (C) The predicted phosphorylation site in hFMR1 (Ser500) was mutagenized to Ala in the context of full-length GST-hFMR1, and an in vitro phosphorylation assay was carried out using the recombinant dCKII shown in Fig. 5A and mammalian CKII (indicated as rCKII). The GST-hFMR1 mutant (S500A) is not phosphorylated, in contrast to wild-type (wt) GST-hFMR1, demonstrating that the conserved Ser residue is the phosphorylation site in hFMR1.
FIG. 1.
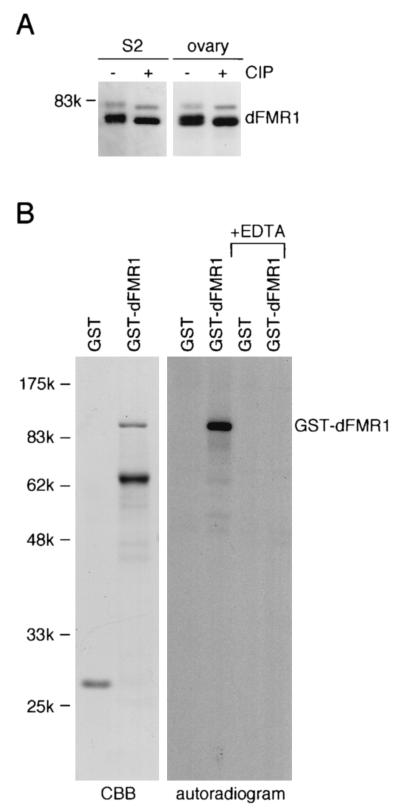
dFMR1 is phosphorylated in vivo and in vitro. (A) dFMR1 proteins in cytoplasmic lysates of S2 cells and of fly ovaries before and after CIP treatment were visualized by Western blotting using an anti-dFMR1 antibody (50). Migration changes of dFMR1 proteins are observed before and after CIP treatment on SDS-7.5% polyacrylamide gels, indicating that dFMR1 proteins both in cultured cells and in animals are phosphorylated in vivo. (B) After production of GST-dFMR1, an in vitro phosphorylation assay was carried out using the cytoplasmic lysate of S2 cells in the presence of [γ-32P]ATP, and the phosphorylated proteins were visualized by autoradiography. The full-length GST fusion protein is clearly phosphorylated, whereas GST alone and the truncated by-product of GST-dFMR1 (∼65 kDa in the CBB panel) (see the text) are not, suggesting that a protein with a dFMR1-phosphorylating activity exists in the cytoplasm of S2 cells and that the activity is sequence specific. It is noted that EDTA abolishes the activity. The left panel shows the protein substrates stained with Commassie brilliant blue (CBB), and the positions of molecular mass markers are indicated on the left of the gel.
FIG. 2.
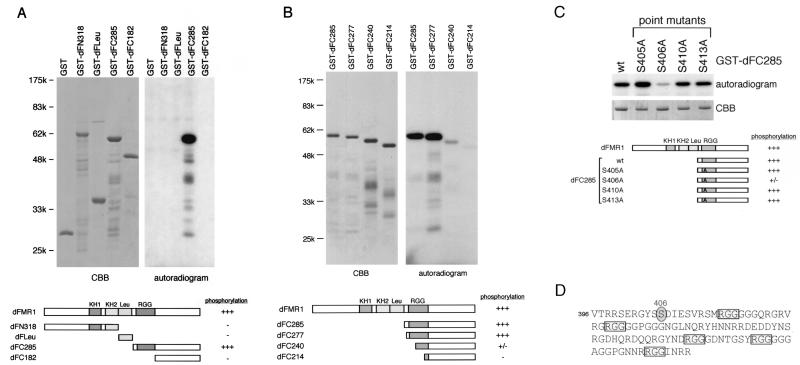
Determination of the phosphorylation site in dFMR1 by systematic mutation analyses. (A) The four parts of dFMR1 (shown in the diagram at the bottom) were fused individually to GST and assayed for in vitro phosphorylation. It may be observed that only GST-dFC285, containing 285 amino acids from the end of dFMR1, becomes phosphorylated, indicating that the phosphorylated region includes the RGG box. In the schematic drawings, the locations of the first and the second KH domains, the Leu-rich region, and the RGG box are indicated as KH1, KH2, Leu, and RGG, respectively. (B) To narrow down the phosphorylation region of dFMR1, the dFC285 fragment was further delineated from its N terminus (shown in the diagram at the bottom), fused to GST, and assayed. GST-dFC277 was phosphorylated efficiently, whereas GST-dFC240 was not, indicating that the region covering amino acid residues 405 to 441 in dFMR1 contains the phosphorylation site. (C) In the region identified in panel B, each of four Ser residues was mutagenized to Ala in the context of GST-dFC285, and mutants were assayed to determine the phosphorylation site in dFMR1. The phosphorylation level of the S406A mutant was detectable but was markedly reduced compared to those of the others, demonstrating that Ser406 is the major phosphorylation site in dFMR1. (D) The amino acid sequence of the region containing the phosphorylation site, Ser406 (circled and shaded), and the RGG box of dFMR1 (48) is shown. Five RGG sequences found in the RGG box are indicated by boxes.
Mass spectrometry for identification of in vivo phosphorylation sites.
Coomassie-stained dFMR1 bands were digested with trypsin (37), and the digest solution was desalted with a ZipTip C18 tip (Millipore). The peptide mixture eluted with 70% acetonitrile and 1% formic acid was analyzed by using a Q-Tof2 mass spectrometer (Micromass, Manchester, United Kingdom) equipped with a nanoelectrospray ionization source, and the phosphopeptide sequence was obtained by tandem mass spectrometry (MS/MS). Mass spectra were collected in positive-ion mode. Phosphopeptides were identified by MS data searching against the National Center for Biotechnology Information (NCBI) database by using the Mascot Search program (Matrix Science, London, United Kingdom). MS/MS data were analyzed by the PepSeq program in the BioLynx software package (Micromass) for determination of peptide sequences.
Purification and identification of dCKII.
A cytoplasmic lysate of S2 cells prepared as described above was resolved on a linear sucrose gradient (5 to 30%). The gradient was centrifuged at 4°C in a Beckman MLA-130 rotor at 40,000 rpm for 90 min. Following centrifugation, fractions were collected and assayed for kinase activity toward GST-dFC285. The active fractions from the gradient were pooled, applied to a gel filtration PC-10 column (Amersham Bioscience) to eliminate sucrose, and eluted with buffer A containing 10 mM Tris-HCl (pH 7.5) and 100 mM NaCl prior to being applied to a UnoQ ion-exchange column (Bio-Rad). The column was washed extensively with buffer A, and a 100 mM to 1 M NaCl gradient of buffer A was applied to the column. Fractions (0.5 ml) were collected and assayed for kinase activity. The phosphorylation-active fractions were pooled and diluted with 10 mM Tris-HCl (pH 7.5) to a final 100 mM NaCl concentration prior to being applied to a UnoS ion-exchange column (Bio-Rad). The column was washed extensively with buffer A, and a 100 mM to 1 M NaCl gradient of buffer A was applied to the column. Fractions (0.5 ml) were again collected and assayed for activity. The active fractions were pooled and diluted with 10 mM Tris-HCl (pH 7.5) to a final 100 mM NaCl concentration prior to being applied to a heparin column. The column was washed extensively with buffer A, and proteins were eluted stepwise with 10 mM Tris-HCl (pH 7.5) containing NaCl at the concentrations indicated in Fig. 4C. Each fraction was assayed for kinase activity and for protein content by silver staining after being resolved on an SDS-polyacrylamide gel. Both protein bands (as indicated in Fig. 4C, lane 1 M NaCl) were sent to the Genomic Research Center, Shimadzu Biotech, and mass spectrometric analysis was carried out to identify them.
FIG. 4.
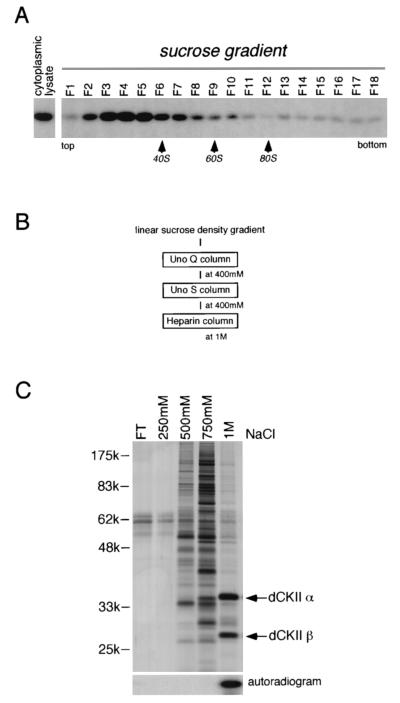
Purification and identification of dCKII as the protein phosphorylating dFMR1. (A) The S2 cytoplasmic lysate with a dFMR1-phosphorylating activity was applied to a linear sucrose density gradient (5 to 30%). After centrifugation, collected fractions were assayed for phosphorylation toward GST-dFC285. Relatively high activities were observed in fractions F3 to F5, which were pooled and employed for further purification steps (see the text). (B) Scheme for purification steps after the sucrose gradient. The NaCl concentrations at which the active fractions were eluted from each column are individually indicated. (C) As the final purification step, a heparin column was used. After stepwise elution with the NaCl concentrations indicated, each fraction was assayed for phosphorylation and for protein contents by silver staining after separation of the proteins on an SDS-polyacrylamide gel. The activity was concentrated in the fraction eluted at 1 M NaCl, in which two proteins (∼35 and ∼28 kDa) were prominent. Both were subjected to MS analysis and were found to be identical to the dCKII alpha and beta subunits (35), respectively.
Production of recombinant alpha and beta subunits of dCKII.
In order to obtain the cDNAs encoding the alpha and beta subunits of dCKII, poly(A)+ RNA was purified from S2 cells and reverse transcription-PCR (RT-PCR) was carried out using primers specifically designed for each clone (35). The cDNAs of the alpha and beta subunits of dCKII were subcloned into pGEX-5X and pET28 (Novagen) to produce GST-dCKII alpha and His-dCKII beta, respectively. Both fusion proteins were produced and purified as described by the manufacturers.
Phosphorylation of hFMR1 in vivo.
HeLa cells were grown as described previously (42). HeLa cells transiently expressing myc-tagged hFMR1 were labeled with [32P]orthophosphate (1 mCi/ml) (Amersham Bioscience) in phosphate-free medium supplemented with 10% fetal calf serum. After incubation for 3 h, the cells were lysed in phosphate-buffered saline buffer containing 1% Empigen, 1 mM EDTA, 0.1 mM DTT, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, and 0.5% aprotinin. After brief sonication, the lysate was clarified by centrifugation at 16,000 × g for 5 min and the supernatant was immunoprecipitated with either an anti-myc antibody (9E10) or a nonimmune control antibody (SP2/0). After extensive washing, the immunoprecipitates were separated on an SDS-polyacrylamide gel and proteins labeled with 32P were visualized by using BAS-2500 (FUJIFILM).
Protein-protein interaction assays.
A fusion construct encoding dFMR1 and a tandem affinity purification (TAP) tag (33) under the control of the Drosophila metallothionein promoter in the pRmHa-3 vector (25) (a kind gift of F. Lafont) was transfected into S2 cells. Expression of the dFMR1-TAP fusion protein was induced by adding copper ions to the medium, and the fusion protein and associated components were recovered from the cytoplasmic lysates of the S2 cells by affinity selection on an immunoglobulin G (IgG) Sepharose 6 Fast Flow column (Amersham Bioscience). After extensive washing with the binding buffer (20 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, 150 mM NaCl, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, and 0.5% aprotinin), the bound materials were separated on an SDS-acrylamide gel and analyzed by Western blotting using an anti-dFMR1 antibody. For the GST pulldown assay, GST-dCKII alpha, GST-dFMR1, and GST alone were bound to 30 μl of glutathione-Sepharose 4B resin (Amersham Bioscience) in 1 ml of binding buffer (20 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, and 150 mM NaCl). Following incubation at 4°C for 1 h, the resins were pelleted and washed with the binding buffer. Either His-dCKII beta or His-dFMR1 was then added to the resins and bound in the absence or in the presence of MgCl2 at 10 mM and ATP at 1 mM at 4°C for 2 h. After extensive washing with the binding buffer, the bound proteins were analyzed by Western blotting to visualize His-dCKII beta and His-dFMR1 by using an anti-His antibody (Santa Cruz). A cytoplasmic lysate of S2 cells prepared as described above was used for the in vivo interaction of dCKII with dFMR1. The antibodies against rat CKII cross-reacting to dCKII (a kind gift from U. Kikkawa) were bound on GammaBind G Sepharose resin (Amersham Bioscience) and incubated with the S2 cytoplasmic lysate at 4°C for 1 h. After extensive washing of the resins, the bound proteins were analyzed by Western blotting using an anti-dFMR1 antibody. For Fig. 7, in vitro-phosphorylated GST-dFMR1 and His-dFMR1 were obtained as described above with a minor modification, namely, that [γ-32P]ATP was replaced with additional 2 mM ATP. In addition, the 1 mM DTT in the buffer was replaced with 0.1 mM tris(2-carboxyethylphosphine)hydrochloride for HIS-dFMR1 phosphorylation. The enzymatic source used in the experiments was the recombinant dCKII shown in Fig. 5A. Phosphorylated GST-dFMR1 was bound to 30 μl of glutathione-Sepharose 4B resin in 1 ml of binding buffer (20 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, and NaCl at the concentration indicated). Following incubation at 4°C for 1 h, the resin was pelleted and washed with the binding buffer. His-dFMR1, either in a phosphorylated state or in an unphosphorylated state, was then added to the resin with 1 ml of the binding buffer and incubated at 4°C for 2 h. After extensive washing with the binding buffer, the bound proteins were detected on Western blots by using an anti-His antibody.
FIG. 7.
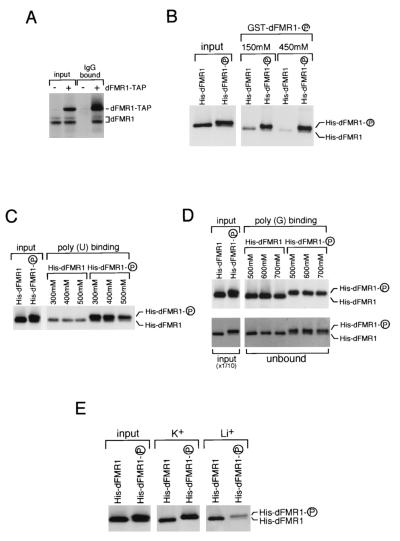
Biochemical properties of dFMR1 are altered by dCKII phosphorylation. (A) An in vivo protein-protein interaction assay was carried out using the TAP tag method. A fusion construct encoding the calmodulin-binding peptide, a TEV protease recognition sequence, two IgG-binding units of protein A of Staphylococcus aureus, and dFMR1 under the control of the Drosophila metallothionein promoter was transfected into S2 cells. The fusion protein and associated components were recovered from cell extracts by affinity selection on an IgG matrix. After extensive washing, the bound material was analyzed on a gel, and Western blotting using anti-dFMR1 antibodies was performed. (B) An in vitro protein-protein interaction assay was carried out. Phosphorylated and unphosphorylated His-dFMR1s (input) were incubated with GST-dFMR1 in a phosphorylated state immobilized on glutathione-Sepharose 4B resin at the NaCl concentrations indicated. After extensive washing, the bound fraction was analyzed by Western blotting using an anti-His antibody. (C) An in vitro RNA-binding assay was carried out using poly(U) beads. Phosphorylated and unphosphorylated His-dFMR1s (input) were incubated with poly(U) at the NaCl concentrations indicated. After extensive washing, the bound fraction was analyzed by Western blotting as for panel B. (D) An in vitro RNA-binding assay was carried out using poly(G) beads. In this experiment, KCl was used instead of NaCl. The unbound fractions are shown below in comparison with the input diluted 1:10, which clearly shows the ratio of the unphosphorylated to the phosphorylated proteins. (E) Poly(G)-binding comparison in the presence of K+ and Li+. Phosphorylated and unphosphorylated His-dFMR1s were incubated with poly(G) in the presence of NaCl and LiCl, both at 150 mM. After extensive washing, the bound fraction was analyzed by Western blotting as for panel D.
FIG. 5.
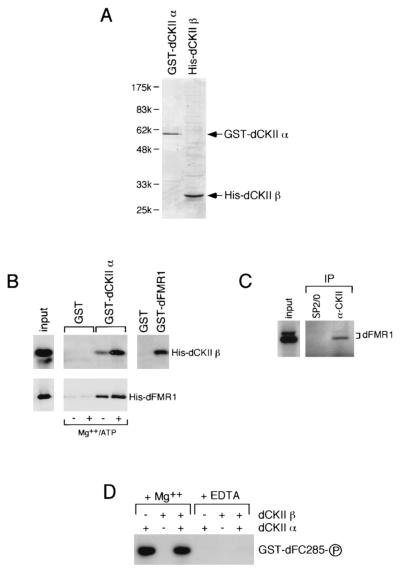
Confirmation of dCKII as the protein with dFMR1-phosphorylating activity. (A) After the cDNAs of dCKII alpha and beta subunits were obtained by RT-PCR, they were subcloned into bacterial expression vectors, and GST-dCKII alpha and His-dCKII beta were produced. Both purified proteins were analyzed by staining with Coomassie brilliant blue on an SDS-polyacrylamide gel. The positions of molecular mass markers are indicated on the left of the gel. (B) Interaction between the dCKII subunits and dFMR1 in vitro. Specific interaction is observed between GST-dCKII alpha and His-dCKII beta or His-dFMR1. In both cases the stronger protein-protein interactions are seen in the presence of MgCl2 and ATP. Not only the alpha but also the beta subunit interacts independently of the other with GST-dFMR1. (C) In vivo interaction of dCKII with dFMR1. Immunoprecipitation was performed from an S2 cytoplasmic lysate using anti-rCKII antibodies and a nonimmune control antibody (SP2/0). Western blotting of the immunoprecipitates using an anti-dFMR1 antibody revealed that dFMR1 specifically coimmunoprecipitated with dCKII. (D) The phosphorylation activities of the recombinant proteins toward GST-dFC285 were assayed. GST-dCKII alpha (shown in panel A) efficiently phosphorylates the substrate, whereas His-dCKII beta has no such activity. It should be noted that the beta subunit added together with the alpha subunit in the reaction mixture did not influence the activity of the alpha subunit itself.
RNA-binding assay.
Binding of His-dFMR1 in phosphorylated and unphosphorylated states was carried out by using 30 μl of either AGPOLY(U) type 6 (Amersham Bioscience) or poly(G) immobilized on polyacrylhydrazido-agarose (Sigma) in 1 ml of binding buffer (20 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, and NaCl [Fig. 7C] or KCl [Fig. 7D] at the concentrations indicated) at 4°C for 2 h. After incubation, the beads were washed with the binding buffer. Bound proteins were then eluted by addition of SDS-PAGE sample buffer to the beads and heat denaturation. Western blotting was performed to visualize His-dFMR1 by using an anti-His antibody.
RESULTS
dFMR1 protein is phosphorylated in vivo and in vitro.
In the course of experiments to elucidate the function of dFMR1, we observed that dFMR1 in S2 cells migrates as several bands when probed with an anti-dFMR1 antibody. This suggested that dFMR1 is posttranslationally modified in vivo and that one such modification is likely phosphorylation. In order to test this hypothesis, the cytoplasmic lysate from S2 cells was treated with CIP, an enzyme known to dephosphorylate both nucleic acids and proteins, and Western blot analysis was performed. In the lysate without CIP, dFMR1 was detected as a strong doublet (∼75 kDa) and a weak upper band (∼80 kDa) on an SDS-7.5% polyacrylamide gel (Fig. 1A, −CIP lane in S2 panel). After CIP treatment, however, the doublet became a discrete single band, apparently comigrating with the lower band of the doublet in the untreated lanes (Fig. 1A, +CIP lane in S2 panel), demonstrating that dFMR1 is phosphorylated in vivo and that the phosphorylation state of dFMR1 is normally heterogeneous in the cells. After CIP treatment, the weak upper band also migrated slightly faster, but did not disappear, indicating that this weak band also represents dFMR1 in a phosphorylated form but that its larger size suggests other modifications and/or alternative splicing for dFMR1. The same phenomenon was observed with fly ovaries (Fig. 1A, right panel), demonstrating that phosphorylation of dFMR1 is not restricted to cultured cells but also occurs in animals. We next investigated if dFMR1 could be phosphorylated in vitro. Recombinant full-length dFMR1 tagged with GST was purified from Escherichia coli and mixed with a cytoplasmic lysate prepared from S2 cells in the presence of Mg2+ and [γ-32P]ATP. As expected, GST-dFMR1 was observed to be phosphorylated under such conditions, whereas GST alone, employed as a negative control, was not (Fig. 1B). When EDTA was added to the reaction mixture to remove Mg2+, the modification was not observed. Interestingly, the truncated peptide of GST-dFMR1 (∼65 kDa; CBB panel in Fig. 1B), which likely lacks the C terminus of the fusion protein, since it was positive by Western blotting using an anti-GST antibody (data not shown), was not phosphorylated under the same circumstances. These results suggest that the phosphorylation is sequence-specific and that the phosphorylation site is likely located towards the C terminus of dFMR1.
The phosphorylation site in dFMR1 is Ser406 preceding the RGG box.
To determine the phosphorylation site in dFMR1, we decided to perform systematic mutation analyses. First, dFMR1 was divided into four individual fragments, each of which was fused to GST. After these proteins were purified from E. coli, an in vitro phosphorylation assay was carried out. We found that only one substrate, termed GST-dFC285, which contains 285 amino acid residues at the C terminus of dFMR1 (amino acids 397 to the end), was phosphorylated in this assay (Fig. 2A). Since GST-dFC182, which contains 182 residues at the C terminus of dFMR1 (amino acids 500 to the end), was not phosphorylated, it was determined that a peptide corresponding to amino acids 397 to 499 of dFMR1 (corresponding to the N-terminal region of the dFC285 fragment) contains the phosphorylation site. We next deleted stepwise the dFC285 fragment from its N terminus, fused it to GST, and performed an in vitro phosphorylation assay. GST-dFC277 was phosphorylated as well as GST-dFC285 (Fig. 2B). In contrast, the phosphorylation of GST-dFC240, which lacks 45 residues at the N terminus of dFC285, was markedly reduced (Fig. 2B), demonstrating that a peptide corresponding to amino acids 405 to 441 in dFMR1 contains the phosphorylation site and that a minor site likely exists toward the end of the protein. We then altered each of four Ser residues within the region corresponding to amino acids 405 to 441 to Ala in the context of GST-dFC285. Three point mutants (termed the S405A, S410A, and S413A mutants) were phosphorylated to extents quite similar to that of the wild type(Fig. 2C). Phosphorylation of S406A, on the other hand, was detectable, but the level was markedly reduced compared to the others. Taken together, we determined that Ser406 is the major phosphorylation site in dFMR1.
Ser406 of dFMR1 is phosphorylated in vivo.
We used MS to identify in vivo phosphorylation sites in dFMR1. dFMR1 was immunoprecipitated from a cytoplasmic lysate prepared from S2 cells, immunoprecipitates were resolved on an SDS-PAGE gel, and dFMR1 was analyzed by MS (37). Trypsin digestion of the dFMR1 band identified a phosphopeptide spanning Gly403 to Arg412 with a single phosphate group (data not shown), and MS/MS analysis of the phosphopeptide was used to identify the exact phosphorylation site. The MS/MS spectrum of the phosphopeptide revealed the presence of a phosphate group at Ser406 (Fig. 3).
FIG. 3.
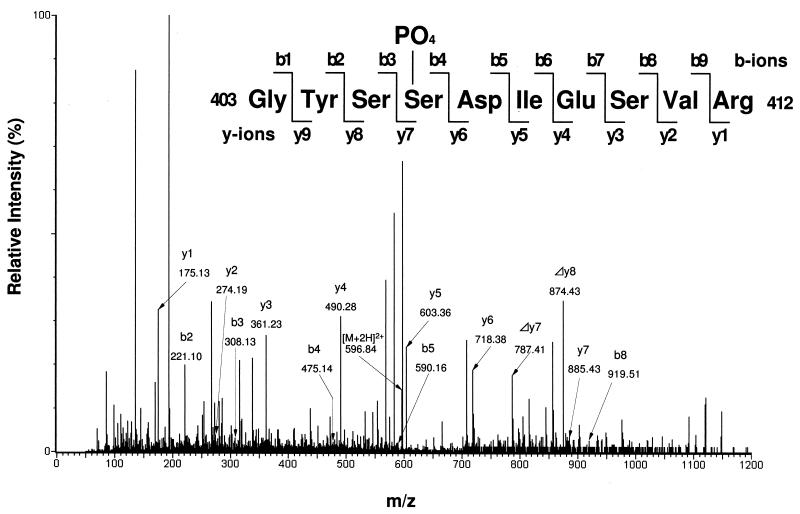
Ser406 of dFMR1 is phosphorylated in vivo. dFMR1 was immunoprecipitated from cytoplasmic lysates of S2 cells and resolved on an SDS-7.5% PAGE gel. The band of dFMR1 was cut out and subjected to analysis of the phosphorylation site. Identification of phosphopeptides was carried out using MS as described in Materials and Methods. The MS/MS spectrum of the tryptic phosphopeptide amino acids 403 to 412 of dFMR1 obtained by collision-induced dissociation of the [M + 2H]2+ precursor ion, m/z 596.84, is shown. Product ions in the spectrum represent mainly single-event preferential cleavage of the peptide bonds, resulting in simultaneous recording of sequence information from the N and C termini (b- and y-type ions, respectively) of the peptide. This spectrum was analyzed by the PepSeq program in the BioLynx software package and was matched to a dFMR1 peptide with additional mass from a phosphate residue (sequence shown on the top). With three potential sites of phosphorylation (three serines), the correct assignment (Ser406) was unambiguously determined based on the presence of ions derived by cleavage at the serine-serine peptide bond. This results in a y7 (m/z 885.43, 805.46 [peptide] + 79.97 [phosphate]) and a Δy7 (m/z 787.41) due to a neutral loss (H2O + phosphate, 97.98) by β-elimination. Δy8 (m/z 874.43), due to a neutral loss (H2O + phosphate) of y8, was detected, and its phosphate should be from Ser406.
The protein phosphorylating dFMR1 is identical to dCKII.
To purify a protein with dFMR1-phosphorylating activity, a cytoplasmic lysate of S2 cells was first fractionated on a linear density sucrose gradient and assayed for phosphorylation activity toward GST-dFC285 (Fig. 4A). Fractions showing the highest activity (F3, F4, and F5 in Fig. 4A) were pooled, and further purification was performed using various chromatography columns (Fig. 4B). Individual fractions eluted from the last column (a heparin column) were assayed and separated on an SDS-polyacrylamide gel, followed by silver staining to visualize the protein pattern (Fig. 4C). The fraction containing kinase activity eluted at 1 M NaCl and contained two predominant proteins with apparent molecular masses of ∼35 and ∼28 kDa (Fig. 4C). Interestingly, these two proteins were observed to exist in an approximately equal molar ratio, suggesting that they might comprise the enzyme that phosphorylates dFMR1. The ∼35- and ∼28-kDa proteins in the fraction at 1 M NaCl were isolated and subjected to MS analysis. We found that they were identical to the alpha and beta subunits of dCKII (35), respectively (Fig. 4C).
Confirmation of dCKII as the protein phosphorylating dFMR1 in vitro.
To confirm that dCKII is indeed the protein phosphorylating dFMR1, we obtained by RT-PCR the cDNA clones encoding both the alpha and beta subunits of dCKII (35) and inserted them into vectors to produce the GST-dCKII alpha and His-dCKII beta proteins (Fig. 5A). We first examined whether these two recombinant proteins can interact with each other, as might be expected (see Discussion) (11, 35). We found that GST-dCKII alpha interacted with His-dCKII beta and that the affinity of the two proteins was stronger in the presence of Mg2+ and ATP (Fig. 5B). Interaction between dFMR1 and each subunit of dCKII was also examined. Each subunit interacted with dFMR1 independently of the other, and the affinity of the alpha subunit for dFMR1 was again increased by addition of Mg2+ and ATP to the buffer (Fig. 5B). We also examined if dCKII interacts with dFMR1 in vivo. Immunoprecipitation from an S2 cytoplasmic lysate was first performed with anti-rat CKII (rCKII) antibodies cross-reacting with dCKII (data not shown), and then Western blotting was performed using an anti-dFMR1 antibody. A band corresponding to dFMR1 was observed in the lane with anti-rCKII antibodies, but not with the nonimmune control antibody (SP2/0), demonstrating that dFMR1 indeed interacts with dCKII in vivo (Fig. 5C). We next examined the phosphorylation activity of recombinant dCKII toward GST-dFC285. As expected, the alpha subunit efficiently phosphorylated the substrate (see Discussion), whereas the beta subunit did not (Fig. 5D). When GST-dFMR1 was substituted for GST-dFC285, the same data were obtained (data not shown). Interestingly, addition of the beta subunit to the reaction mixture together with the alpha subunit did not appear to alter the kinase activity of the alpha subunit (Fig. 5D). The contribution of the beta subunit to dFMR1 phosphorylation remains unknown.
The Ser406 phosphorylation site in dFMR1 is highly conserved among FMR1/FXR family members across species.
Determining whether FMR1/FXR family members from other species are also phosphorylated by CKII was of great interest. First, we inspected the amino acid sequences of those proteins (40, 47, 48, 52) and found that the region surrounding Ser406 in dFMR1 is highly conserved (Fig. 6A). To analyze whether hFMR1 is phosphorylated in cultured cells, in vivo 32P labeling of HeLa cells expressing myc-tagged hFMR1 was performed, and myc-tagged hFMR1 was immunoprecipitated with anti-myc antibodies. Autoradiography of the immunoprecipitates separated by SDS-PAGE revealed that predominantly one protein band was labeled with 32P and that it migrated at the expected size of myc-tagged hFMR1 (∼80 kDa) (Fig. 6B); this suggests that hFMR1 is a phosphoprotein in vivo. To see if the residue (Ser500) corresponding to Ser406 of dFMR1 (Fig. 6A) is the phosphorylation site in hFMR1, the residue was altered to Ala in the context of full-length hFMR1 and an in vitro phosphorylation assay was carried out using recombinant dCKII. As shown in Fig. 6C, a GST-hFMR1 mutant (S500A) was not phosphorylated, whereas wild-type hFMR1 was. Ser500 in hFMR1 was, therefore, determined to be the phosphorylation site of hFMR1, and there seem to be no other phosphorylation sites in this case. We also tried purified rCKII in the assay and found that the mammalian enzyme phosphorylated wild-type hFMR1, but not the S500A mutant (Fig. 6C). Moreover, we also observed that the mammalian CKII phosphorylated dFMR1 (data not shown).
Phosphorylation of dFMR1 modulates the efficiency of forming homomers and of binding RNA in vitro.
It has been reported that dFMR1 forms homomers in vitro (48), just as mammalian FMR1/FXR proteins do (52). To address the question of whether dFMR1 also forms homomers in vivo, we used the TAP tag method (33). The TAP-tagged dFMR1 protein was expressed in S2 cells, and the fusion protein and associated components were recovered from cell extracts by affinity selection on an IgG matrix and were then analyzed by SDS-PAGE and Western blotting. As shown in Fig. 7A, not only the TAP-tagged dFMR1 but also endogenous dFMR1 was found in the IgG-bound material, indicating that dFMR1 forms homomers in vivo. We then tried to ascertain if the phosphorylation affects the oligomerization of dFMR1. For this experiment, GST- and His-tagged dFMR1s purified from E. coli were phosphorylated by recombinant dCKII. That most of the population of GST- and His-dFMR1 proteins were phosphorylated was confirmed by observing the mobility change on Western blots (see Fig. 7B, input lanes, for His-dFMR1). GST-dFMR1 in a phosphorylated state (data not shown) was first bound to glutathione-Sepharose resins, and after extensive washing, a GST pulldown assay was carried out at 150 and 450 mM NaCl. By performing Western blotting using an anti-His antibody, it was clearly observed that the phosphorylated His-dFMR1 bound much more efficiently than the unphosphorylated His-dFMR1 (Fig. 7B), demonstrating that the oligomerization of dFMR1 is upregulated by dCKII phosphorylation. We next investigated whether the RNA-binding activity of dFMR1 was also influenced by dCKII modification. When the assay was carried out with poly(U) beads, phosphorylated dFMR1 showed a higher affinity for them than did unphosphorylated dFMR1 (Fig. 7C). Considering the result shown in Fig. 7B, the phosphorylated dFMR1 likely forms homomers more efficiently than the unphosphorylated dFMR1 at the NaCl concentrations used in this experiment. Thus, it is speculated that phosphorylation followed by oligomerization confers a higher RNA-binding activity on dFMR1 protein. We also examined dFMR1 binding to poly(G) beads. Recently, it was reported that dFMR1 preferentially binds G quartets in RNA (7, 36). The G quartet is a secondary structure formed in G-rich sequences and is known to be stabilized by certain cations such as K+. Poly(G) is known to form G quartets (23). Thus, this particular experiment was performed in a buffer containing KCl. In this assay, phosphorylated and unphosphorylated dFMR1s showed approximately the same affinity for poly(G) at any given concentration of KCl (Fig. 7D). Interestingly, it was clearly observed that the unphosphorylated form of dFMR1 coexisting with the phosphorylated form in the input (see the right input lane in Fig. 7D) was not detected in the bound fraction but was observed in the unbound fraction (Fig. 7D). When the unphosphorylated form was allowed to bind to the beads first and then the phosphorylated form was added, both forms were detected in the bound fraction (data not shown), suggesting that phosphorylated dFMR1 does not have the ability to displace unphosphorylated dFMR1 from poly(G) beads. It is known that G quartets are stabilized more by K+ than by Li+ and that hFMR1 shows a higher affinity for a G quartet formed in the presence of K+ (36). Thus, the poly(G)-binding activities of phosphorylated and unphosphorylated dFMR1s were compared in the presence of K+ and Li+. Interestingly, the unphosphorylated form of dFMR1 showed no significant difference between K+ and Li+, whereas the phosphorylated form showed much higher affinity for poly(G) in a buffer containing K+ (Fig. 7D), suggesting that in binding to a G quartet, the phosphorylated dFMR1 behaves as does hFMR1.
DISCUSSION
In this study we have shown that dFMR1 protein is phosphorylated in vivo, both in cultured cells and in animals, and that bacterially produced GST-dFMR1 is phosphorylated with a cytoplasmic lysate from S2 cells in vitro. We have also shown that the protein with dFMR1-phosphorylating activity purified from the lysate was dCKII (35) and that recombinant dCKII does indeed phosphorylate dFMR1 in vitro.
CKII is known to phosphorylate both nuclear and nonnuclear proteins (46, 50) and is highly conserved from yeasts to humans. The high degree of conservation suggests that CKII has an essential role in organisms. Indeed, genetic analyses in yeast have demonstrated that the enzyme is essential for viability (32, 44). We found in this study that at least in vitro, both dCKII and mammalian CKII phosphorylate both dFMR1 and hFMR1 at the sites we identified by performing mutation analyses. The cross-species activity implies that the phosphorylation of FMR1 proteins is highly conserved through evolution and is important, perhaps even required, for function of the FMR1/FXR proteins in divergent organisms.
CKII is composed of alpha and beta subunits that are associated as a heterotetramer (α2β2) (22). The alpha subunit contains a protein kinase domain that is homologous to the catalytic domain of other protein kinases (15). The beta subunit has been proposed to play a complex role in regulating the basal activity of the alpha subunit (4). Recently, the physical interaction of the CKII enzyme and its substrates was shown to be mediated by the beta, but not the alpha, subunit (10). In the present study, we observed that both the beta and the alpha subunits are able to interact directly with dFMR1 in vitro and that the affinity of the alpha subunit for dFMR1 is increased in the presence of Mg2+ and ATP. We also found that the alpha subunit is necessary and sufficient for efficient phosphorylation of dFMR1 in vitro; that is, the beta subunit seemed not to influence that activity. The role of the beta subunit of CKII in the phosphorylation of FMR1 remains undefined.
CKII phosphorylates Ser/Thr residues of many protein substrates in vivo, and the consensus sequence is known to be (Ser/Thr)-(Asp/Glu)-X-(Asp/Glu) (24). We determined that the peptide sequence in dFMR1 containing the phosphorylation site (Ser406) was Ser-Asp-Ile-Glu, which perfectly fits the consensus. The phosphorylation site is highly conserved within FMR1/FXR family members from flies to humans. The sequences of these other family members also match the CKII consensus. We have mentioned that there might be minor phosphorylation sites in dFMR1, since the S406A mutation did not result in a total loss of phosphorylation. However, there are no other sequences in dFMR1 that perfectly fit the consensus sequence for CKII phosphorylation. In the case of hFMR1, we could find two other sites that fit the consensus (namely, Thr455 and Thr518). But neither of these is likely to be a site of modification by CKII, at least in vitro, since the GST-hFMR1 mutant (S500A) that contains intact Thr455 and Thr518 residues is not phosphorylated by either dCKII or mammalian CKII. It remains possible, however, that other, unidentified kinases or even CKII itself contributes phosphorylation of the other sites of hFMR1 in vivo. It is known that the negative-charge requirement within the CKII consensus can be supplied by phosphorylated residues (46). If so, once Ser500 in hFMR1 is phosphorylated, Ser497 and consequently Ser494 become appropriate targets for CKII. This sequential phosphorylation may have occurred even in vitro (see Fig. 6C), but at this time we do not have data to support this scenario.
Although it is understood that the phosphorylation activity of CKII appears to be constitutive, there are several studies available showing the regulation of CKII phosphorylation activity in vivo. Recently, it was demonstrated that CKII phosphorylates the human p53 protein in response to UV irradiation and that the chromatin transcriptional elongation factor, which forms a complex with CKII, alters the specificity of CKII to selectively phosphorylate p53 over other substrates including casein (20). In the case of Dishevelled (Dsh), another example of a substrate for dCKII, phosphorylation is regulated in response to Dsh and Dfz2 (a Wingless receptor) expression in the signal transduction pathway of the Wingless protein (50). When we used a linear density sucrose gradient as a first means to purify dCKII, a relatively high activity was obtained in fractions F3 to F5 (see Fig. 4A). This indicated that it was likely that dCKII associates with other molecules, constituting a relatively heavy complex. However, the complex may not be very stable and probably is dissociated through a series of purification procedures, since no other proteins eluted with dCKII from the last heparin column (see Fig. 4C). At this point, we have no data identifying molecules that associate with dCKII. It is possible, as in the case of p53, for example, that the dCKII-interacting proteins and/or nucleic acids regulate the dCKII activity in the phosphorylation of dFMR1. We are currently attempting to isolate the dCKII interactors and understand their involvement in the phosphorylation of dFMR1.
It has been reported that the DNA-binding activity of some nuclear proteins is regulated by CKII phosphorylation (3, 28, 29, 45). In this study we have shown that the RNA-binding activity of dFMR1 to poly(U) is upregulated by phosphorylation by dCKII in vitro. This property may be a result of the favorable homotypic interactions of dFMR1 caused by phosphorylation (see Fig. 7B). We speculate that phosphorylation of dFMR1 induces a conformational change that results in a stronger association with itself and perhaps also with poly(U). Stable oligomerization of dFMR1 simply means that the KH domains and the RGG boxes are also multiplied and are brought close enough together to cooperate, which we think confers the stronger ability to bind poly(U) beads. Surprisingly, different results were obtained when we used poly(G) beads. In such experiments, dFMR1, whether phosphorylated or unphosphorylated, showed approximately the same ability to bind poly(G). On the other hand, when both proteins were added to the beads together, only phosphorylated dFMR1 was detected in the bound fraction. In different experiments, however, where unphosphorylated dFMR1 was allowed to bind poly(G) first, and phosphorylated dFMR1 was added later, both proteins were observed in the bound fraction. One explanation for these observations is that the on-rate and off-rate of phosphorylated or unphosphorylated dFMR1 for poly(G) binding may differ from each other. Data to support this speculation are currently not available, and further detailed studies will be necessary.
These studies constitute the first demonstration that FMR1 is phosphorylated in vivo and that the phosphorylation alters its biochemical properties, such as its interactions with itself and with RNA in vitro. Although the in vivo relevance of these observations is currently unknown, our results should be the starting point for a better understanding of the effects of phosphorylation on the function of FMR1.
Acknowledgments
We thank Gideon Dreyfuss and Lili Wan for generously providing dFMR1 cDNA and the anti-dFMR1 antibody, Ushio Kikkawa for purified mammalian (rat) CKII and the anti-CKII antibody, Bertrand Seraphin for the TAP plasmids, and Frank Lafont for pRmHa-3. We gratefully acknowledge Eiji Majima for help with peptide sequencing by nanoelectrospray MS. We also thank Paul Eder and Tetsuya Taura for critical reading of the manuscript. We are grateful to members of our laboratory, especially Hitomi Ichinose and Yoshinori Kawamura, for technical assistance.
This work was supported by grants from the FRAXA Research Foundation, the Cure Autism Now (CAN) Foundation, the Japan Society for the Promotion of Science (JSPS), the Ministry of Health, Labour and Welfare of Japan, and the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Adam, M. D., S. E. Celniker, R. A. Holt, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Ashley, C. T., K. D. Wilkinson, D. Reines, and S. T. Warren. 1993. FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262:563-566. [DOI] [PubMed] [Google Scholar]
- 3.Bernerich, S. J., and M. Cole. 1991. Casein kinase II inhibits the DNA binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 6:166-176. [DOI] [PubMed] [Google Scholar]
- 4.Bidwai, A. P., D. E. Hanna, and C. V. Glover. 1992. Purification and characterization of casein kinase II (CKII) from delta cka1 delta cka2 Saccaromyces cerevisiae rescued by Drosophila CKII subunits. The free catalytic subunit of casein kinase II is not toxic in vivo. J. Biol. Chem. 267:18790-18796. [PubMed] [Google Scholar]
- 5.Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell, S. A. Tenenbaum, X. Jin, Y. Feng, K. D. Wilkinson, J. D. Keene, R. B. Darnell, and S. T. Warren. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107:477-487. [DOI] [PubMed] [Google Scholar]
- 6.Corbin, F., M. Bouillon, A. Fortin, S. Morin, F. Rousseau, and E. W. Khandjian. 1997. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 6:1465-1472. [DOI] [PubMed] [Google Scholar]
- 7.Darnell, J. C., K. B. Jensen, P. Jin, Y. Brown, S. T. Warren, and R. B. Darnell. 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107:489-499. [DOI] [PubMed] [Google Scholar]
- 8.Dockendorff, T. C., H. S. Su, S. M. J. McBride, Z. Yang, C. H. Choi, K. K. Siwicki, A. Sehgal, and T. A. Jongens. 2002. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34:973-984. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Y., D. Absherm, D. E. Eberhart, Y. Brown, H. E. Malter, and S. T. Warren. 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1:109-118. [DOI] [PubMed] [Google Scholar]
- 10.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 11.Glover, C. V. C., E. R. Shelton, and D. L. Brutlag. 1983. Purification and characterization of a type II casein kinase from Drosophila melanogaster. J. Biol. Chem. 258:3258-3265. [PubMed] [Google Scholar]
- 12.Gould, E. L., D. Z. Loesch, M. J. Martin, R. J. Hagerman, S. M. Armstrong, and R. M. Huggins. 2000. Melatonin profiles and sleep characteristics in boys with fragile X syndrome: a preliminary study. Am. J. Med. Genet. 95:307-315. [PubMed] [Google Scholar]
- 13.Greenough, W. T., A. Y. Klintsova, S. A. Irwin, R. Galvez, K. E. Bates, and I. J. Weiler. 2001. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl. Acad. Sci. USA 98:7101-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagerman, R. J. 1998. Clinical and diagnostic aspects of fragile X syndrome, p. 15-25. In R. D. Wells and S. T. Warren (ed.), Genetic instabilities and hereditary neurological diseases. Academic Press, San Diego, Calif.
- 15.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, S. B., M. C. Siomi, and H. Siomi. 2000. Molecular mechanisms of fragile X syndrome. J. Med. Investig. 47:101-107. [PubMed] [Google Scholar]
- 17.Inoue, S. B., M. Shimoda, I. Nishinokubi, M. C. Siomi, M. Okamura, A. Nakamura, S. Kobayashi, N. Ishida, and H. Siomi. 2002. A role for the Drosophila fragile X-related gene in circadian output. Curr. Biol. 12:1331-1335. [DOI] [PubMed] [Google Scholar]
- 18.Irwin, S. A., B. Patel, M. Idupulapati, J. B. Harris, R. A. Crisostomo, B. P. Larsen, F. Kooy, P. J. Willems, P. Cras, P. B. Kozlowski, R. A. Swain, I. J. Weiler, and W. T. Greenough. 2001. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 98:161-167. [DOI] [PubMed] [Google Scholar]
- 19.Ishizuka, A., M. C. Siomi, and H. Siomi. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16:2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller, D. M., X. Zeng, Y. Wang, Q. H. Zhang, M. Kapoor, H. Shu, R. Goodman, G. Lozano, Y. Zhao, and H. Lu. 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7:283-292. [DOI] [PubMed] [Google Scholar]
- 21.Khandjian, E. W., F. Corbin, S. Woerly, and F. Rousseau. 1996. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12:91-93. [DOI] [PubMed] [Google Scholar]
- 22.Kikkawa, U., S. K. O. Mann, R. A. Firtel, and T. Hunter. 1992. Molecular cloning of casein kinase II α subunit from Dictyostelium discoideum and its expression in the life cycle. Mol. Cell. Biol. 12:5711-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., C. Cheong, and P. B. Moore. 1991. Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature 351:331-332. [DOI] [PubMed] [Google Scholar]
- 24.Kuenzel, E. A., J. A. Mulligan, J. Sommercorn, and E. G. Krebs. 1987. Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J. Biol. Chem. 262:9136-9140. [PubMed] [Google Scholar]
- 25.Lafont, F., S. Lecat, P. Verkade, and K. Simons. 1998. Annexin XIIIb associates with lipid microdomains to function in apical delivery. J. Cell Biol. 142:1413-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laggerbauer, B., D. Ostareck, E. M. Keidel, A. Ostareck-Lederer, and U. Fischer. 2000. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10:329-338. [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., Y. Zhang, L. Ku, K. D. Wilkinson, S. T. Warren, and Y. Feng. 2000. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 29:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, A., J. Frost, T. Deng, T. Smeal, N. Al-Alawi, U. Kikkawa, T. Hunter, D. Brenner, and M. Karin. 1992. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell 70:777-789. [DOI] [PubMed] [Google Scholar]
- 29.Marais, R. M., J. J. Hsuan, C. McGuigan, J. Wynne, and R. Treisman. 1992. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. EMBO J. 11:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miklos, G. L., and G. M. Rubin. 1996. The role of the genome project in determining gene function: insights from model organisms. Cell 86:521-529. [DOI] [PubMed] [Google Scholar]
- 31.Morales, J., P. R. Hiesinger, A. J. Schroeder, K. Kume, P. Verstreken, F. R. Jackson, D. L. Nelson, and B. A. Hassan. 2002. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron 34:961-972. [DOI] [PubMed] [Google Scholar]
- 32.Padmanabha, R., J. L. Chen-Wu, D. E. Hanna, and C. V. Glover. 1990. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:4089-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 34.Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. Gabor Miklos, C. R. Nelson, I. K. Hariharan, M. E. Fortini, P. W. Li, R. Apweiler, W. Fleischmann, J. M. Cherry, S. Henikoff, M. P. Skupski, S. Misra, M. Ashburner, E. Birney, M. S. Boguski, T. Brody, P. Brokstein, S. E. Celniker, S. A. Chervitz, D. Coates, A. Cravchik, A. Gabrielian, R. F. Galle, W. M. Gelbart, R. A. George, L. S. Goldstein, F. Gong, P. Guan, N. L. Harris, B. A. Hay, R. A. Hoskins, J. Li, Z. Li, R. O. Hynes, S. J. Jones, P. M. Kuehl, B. Lemaitre, J. T. Littleton, D. K. Morrison, C. Mungall, P. H. O'Farrell, O. K. Pickeral, C. Shue, L. B. Vosshall, J. Zhang, Q. Zhao, X. H. Zheng, and S. Lewis. 2000. Comparative genomics of the eukaryotes. Science 287:2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena, A., R. Padmanabha, and C. V. Glover. 1987. Isolation and sequencing of cDNA clones encoding alpha and beta subunits of Drosophila melanogaster casein kinase II. Mol. Cell. Biol. 7:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaeffer, C., B. Bardoni, J. L. Mandel, B. Ehresmann, C. Ehresmann, and H. Moine. 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 38.Siomi, H., M. C. Siomi, R. L. Nussbaum, and G. Dreyfuss. 1993. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74:291-298. [DOI] [PubMed] [Google Scholar]
- 39.Siomi, H., M. Choi, M. C. Siomi, and G. Dreyfuss. 1994. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77:33-39. [DOI] [PubMed] [Google Scholar]
- 40.Siomi, M. C., H. Siomi, W. H. Sauer, S. Srinivasan, R. L. Nussbaum, and G. Dreyfuss. 1995. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14:2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siomi, M. C., Y. Zhang, H. Siomi, and G. Dreyfuss. 1996. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interaction among them. Mol. Cell. Biol. 16:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siomi, M. C., P. S. Eder, N. Kataoka, L. Wan, Q. Liu, and G. Dreyfuss. 1997. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138:1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, F. W., and J. Feigon. 1992. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature 356:164-168. [DOI] [PubMed] [Google Scholar]
- 44.Snell, V., and P. Nurse. 1994. Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 13:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott, R. L., M. Kalive, Z. Paroush, and A. P. Bidwai. 2001. Drosophila melanogaster casein kinase II interacts with and phosphorylates the basic helix-loop-helix proteins m5, m7, and m8 derived from the enhancer of split complex. J. Biol. Chem. 276:2159-2167. [DOI] [PubMed] [Google Scholar]
- 46.Tuazon, P. T., and J. A. Traugh. 1991. Casein kinase I and II-multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 23:123-164. [PubMed] [Google Scholar]
- 47.Verkerk, A. J., M. Pieretti, J. S. Sutcliffe, Y. H. Fu, D. P. Kurl, A. Pizzuti, O. Reiner, S. Richards, M. F. Victoria, F. P. Zhang., et al. 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905-914. [DOI] [PubMed] [Google Scholar]
- 48.Wan, L., T. Dockendorff, T. A. Jongens, and G. Dreyfuss. 2000. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 20:8536-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren, S. T., and C. T. Ashley. 1995. Triplet repeat expansion mutations: the example of fragile X syndrome. Annu. Rev. Neurosci. 18:77-99. [DOI] [PubMed] [Google Scholar]
- 50.Willert, K., M. Brink, A. Wodarz, H. Varmus, and R. Nusse. 1997. Casein kinase 2 associates with and phosphorylates Dishevelled. EMBO J. 16:3089-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson, J. R., M. K. Raghuraman, and T. R. Cech. 1989. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59:871-880. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., J. P. O'Connor, M. C. Siomi, S. Srinivasan, A. Dutra, R. J. Nussbaum, and G. Dreyfuss. 1995. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14:5358-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y. Q., A. M. Bailey, H. J. Matthies, R. B. Renden, M. A. Smith, S. D. Speese, G. M. Rubin, and K. Broadie. 2001. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107:591-603. [DOI] [PubMed] [Google Scholar]