Abstract
The transcription factor C/EBPα is crucial for the differentiation of granulocytes. Conditional expression of C/EBPα triggers neutrophilic differentiation, and C/EBPα can block 12-O-tetradecanoylphorbol-13-acetate-induced monocytic differentiation of bipotential myeloid cells. In C/EBPα knockout mice, no mature granulocytes are present. A dramatic increase of c-Jun mRNA in C/EBPα knockout mouse fetal liver was observed. c-Jun, a component of the AP-1 transcription factor complex and a coactivator of the transcription factor PU.1, is important for monocytic differentiation. Here we report that C/EBPα downregulates c-Jun expression to drive granulocytic differentiation. An ectopic increase of C/EBPα expression decreases the c-Jun mRNA level, and the human c-Jun promoter activity is downregulated eightfold in the presence of C/EBPα. C/EBPα and c-Jun interact through their leucine zipper domains, and this interaction prevents c-Jun from binding to DNA. This results in downregulation of c-Jun's capacity to autoregulate its own promoter through the proximal AP-1 site. Overexpression of c-Jun prevents C/EBPα-induced granulocytic differentiation. Thus, we propose a model in which C/EBPα needs to downregulate c-Jun expression and transactivation capacity for promoting granulocytic differentiation.
CCAAT enhancer binding protein α (C/EBPα) is a tissue-specific transcription factor expressed in liver, differentiating adipocytes, and myelo-monocytic cells. Gene targeting experiments revealed a specific defect in the hematopoietic system of C/EBPα knockout mice (56). The C/EBPα null mice lack mature granulocytes, while all of the other blood cell types are present, including monocytes and peritoneal macrophages (56). Increased levels of C/EBPα expressed from an inducible promoter construct directed differentiation along the granulocytic pathway (35, 51). These results demonstrate the indispensability of C/EBPα for the granulocytic differentiation pathway. In addition, ectopic expression of C/EBPα could prevent 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced monocytic differentiation of bipotential myeloid progenitor cells (35). C/EBPα induces proliferation arrest and differentiation in many cell types, suggesting that the two activities are linked (28, 48, 51).
C/EBP factors comprise a family of related basic-region leucine zipper (bZIP) DNA binding proteins that form homodimers or hetrodimers with other C/EBP proteins to regulate transcription of target genes (24, 25, 38). The bZIP domain contains a region rich in basic amino acids linked to a dimer-forming region called the leucine zipper. Several studies show that C/EBP leucine zippers can mediate heteromeric interactions with negative regulators, with other bZIP proteins, and with non-bZIP transcription factors and can directly regulate transcriptional activation (25).
It has been proposed that certain DNA binding proteins (including C/EBP, Jun, and Myc oncogene proteins) share a common structural motif based on helix-promoting regions containing heptad repeat sequences of leucine (8, 26). This structure is critical to the biological activity of these proteins, since it facilitates the formation of functional dimers in a configuration termed the leucine zipper. Dimerization of bZIP factors of one leucine zipper subfamily with those of another subfamily is much more selective and is somewhat unusual (25, 41).
The AP-1 transcription factors are considered immediate-early response genes and are thought to be involved in a wide range of transcriptional regulatory processes linked to cellular proliferation and differentiation (5, 14, 16, 18-20, 27, 39). A combination of in vitro and in vivo molecular genetic approaches has provided evidence to suggest that AP-1 transcription factors play multiple roles in functional development of hematopoietic precursor cells into mature blood cells along most, if not all, of the hematopoietic cell lineages. This includes the monocyte/macrophage, granulocyte, megakaryocyte, mastocyte, and erythroid lineages (7, 14, 39).
The c-jun proto-oncogene encodes the transcriptional activator protein AP-1. c-jun is a member of the early-response gene family genes that are rapidly and transiently activated in response to proliferative stimuli (22). Among the AP-1 members, c-Jun is unique in its ability to positively regulate cell proliferation and to induce partial macrophage-like morphology in U937 cells (44). TPA treatment of HL60 and other human myeloid leukemia cell lines such as U937 is associated with the appearance of c-Jun transcript (14, 19, 23, 24). The level of c-Jun expression is regulated by both transcriptional and posttranscriptional mechanisms. Among important regulatory elements previously identified in the c-Jun promoter, there are two AP-1 sites, a proximal AP-1 site (pAP-1) located between bp −71 and −64 and a distal AP-1 site (dAP-1) located between bp −190 and −183 in the c-Jun promoter (1, 46, 49). Importantly, c-Jun is autoregulated by its product Jun/AP-1 (1).
The bZIP region of NF-IL6 (C/EBPβ) isoforms mediates a direct association with the bZIP regions of Jun and Fos (to a lesser extent) in vitro (20). It was shown that NF-IL6-2 transactivation capacity is reduced in presence of c-Jun. The N-terminal transactivation domain of NF-IL6-1 seems to be important in regulating the repressional effect by c-Jun. However, the effect of this NF-IL6-c-Jun interaction on c-Jun/AP-1 DNA binding capacity was not addressed (20). CHOP, a member of the C/EBP family (lacking the N-terminal transactivation domain) acts as a dominant inhibitor of C/EBPα. CHOP interacts with c-Jun via the leucine zipper domain of CHOP. CHOP-c-Jun synergizes to activate transcription from an AP-1 site (47). ATF-2, another member of the AP-1 family, also has cross-family dimerization capacity with C/EBP family proteins (43). ATF-2-C/EBPα interaction diminishes the transactivation capacity of C/EBPα through the C/EBP consensus DNA binding sites, whereas this heterodimer complex could transactivate through chimeric DNA binding sites. However, the effect of this interaction on ATF-2 transactivation or DNA binding capacity still needs to be addressed.
Here we propose that the proliferation arrest and granulocytic lineage commitment function of C/EBPα (28, 35) involves inactivation of c-Jun function via attenuation of its DNA binding activity. Inactivation of c-Jun might be important for the multifunction of C/EBPα, i.e., to drive granulocytic differentiation, to block monocytic lineage commitment, and for proliferation arrest.
MATERIALS AND METHODS
Cell culture conditions.
Human myeloid U937 cells stably transfected with a zinc-inducible C/EBPα construct (U937 α#2) or vector alone (U937EV) have been described previously (35). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% l-glutamine (Gibco), 1% PenStrep (Gibco), and 850 μg of G418 (Gibco) per ml. C/EBPα expression from the metallothionine promoter was induced upon adding 100 μM ZnSO4 (Sigma). Human promyelocytic HL60 cells (DSMZ ACC 3) and human chronic myeloid leukemia in blast crisis K562 cells (DSMZ ACC 10) were grown in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1% PenStrep (Gibco), and 1% l-glutamine (Gibco). CV-1 (CCL-70) cells, NIH 3T3 (ACC 59) cells, a 293 clone constitutively expressing the proteins encoded by E1A (293E1A cells), CHO (CCL-61) cells, 293T cells, and Phoenix-A cells were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% FBS, 1% Penstrep, and 1% l-glutamine. HeLa cells were grown in RPMI 1640 medium supplemented with 10% FBS, 1% Penstrep, and 1% l-glutamine.
Plasmids and transient transfections.
A human c-Jun promoter bp −1780/+731 construct was generated by amplifying the c-Jun promoter fragment along with XhoI half sites at each end from human genomic DNA and ligated into the pGL3 basic vector (Promega) XhoI site. A series of 5′ deletions were generated as described previously (47). The bp −79/+170 human c-Jun promoter construct, bp −79/+170 AP-1/CRE mutant, bp −1780/+731 proximal AP-1 mutant, and pGL2 basic vector were gifts from W. V. Vedekis (52). The pcDNA3 C/EBPα construct was generated by releasing a BamHI/EcoRI fragment of rat C/EBPα cDNA from the pUC18 vector and ligating this fragment into pcDNA3 (Invitrogen). The reporter construct p(C/EBP)2TK contains two consensus C/EBPα binding sites linked in tandem and cloned into pTK81 luciferase. The AP-1 ×7 luciferase reporter construct, containing seven repeats of AP-1 DNA binding sites, was purchased from Stratagene. Gal-4(1-147), Gal-4-Tel, and Gal-4 DNA binding domain in eukaryotic expression vector SGS424 were kindly provided by S. Bohlander, Göttingen, Germany. C/EBPαmBR (mutation in the basic region) and C/EBPαΔLZ (C/EBPα leucine zipper domain replaced with GCN4 leucine zipper domain) were kind gifts from A. D. Friedman (23, 24, 44). c-JunΔRK (DNA binding domain deletion) and c-JunΔLZ (leucine zipper domain deletion) have been described previously (34).
CV.1, NIH 3T3, 293E1A, CHO, and HeLa cells (104 cells) were transfected with Lipofectamine Plus (Life Technologies) according to manufacturer's instruction. A 0.05-μg amount of promoterless Renilla luciferase reporter plasmid as an internal control was transfected along with the respective amounts of other DNAs mentioned for each set of transfections. Transfected cells were lysed in 1× passive lysis buffer at 30 h posttransfection. Firefly luciferase activities were normalized to the Renilla luciferase values of pRL-null (3-6, 50). The fold promoter activity was calculated as the ratio between the promoter and promoter plus C/EBPα, assigning a value of 1 for the promoter alone. Firefly luciferase and Renilla luciferase were measured by using the dual luciferase reporter assay system (Promega). Results are given as means and standard errors of the means.
Transfection in U937 and K562 cells was performed with Effectene reagent (Qiagen) per the manufacturer's protocol with a few modifications. Briefly, 1 μg of the total DNA was mixed with 34 μl of EC buffer. Ten microliters of Enhancer reagent was added to the plasmid solution and incubated for 5 min at room temperature. Fifteen microliters of Effectene reagent was then added, and the mixture was further incubated at room temperature for 10 min. The DNA-Effectene complex solution was diluted in 500 μl of RPMI medium and gently added to 106 cells previously aliquoted in six-well plates.
Northern blot analysis.
Twenty micrograms of total RNA from adult mouse brain, peritoneal macrophages, and day 19 fetal liver from C/EBPα+/+, C/EBPα+/−, and C/EBPα−/− mice was denatured in formamide and fractionated on 1% agarose-2.2 M formaldehyde gel. RNA was transferred to a Biotrans (ICN) membrane in 10 SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the blots were hybridized at 58°C in Church-Gilbert buffer (1 M NaH2PO4, 1 M Na2HPO4, 20% sodium dodecyl sulfate [SDS], 100 mM EDTA). Hybridization probes were prepared with a random priming kit (Roche) with the incorporation of 5′-[α-32P]dATP (3,000 Ci/mmol; Amersham). The blots were washed twice in 1× SSC-0.1% SDS for 15 min at 60°C. A 1.1-kb BamHI fragment from SP6 c-Jun plasmid served as a probe for c-Jun.
Real-time quantitative PCR.
Real-time quantitative PCR was performed using the light cycler technology (Roche Diagnostic) as described previously (12). The primers for c-Jun were 5′ GCA TGA GGA AAC GCA TCG CTG CCT CCA AGT 3′ (forward) and 5′ GCG ACC AAG TCC TTC CCA CTC GTG CAC ACT 3′ (reverse). Glucose-6-phosphate dehydrogenase (G6PD) primers were 5′ CCG GAT CGA CCA CTA CCT GGG CAA C 3′ (forward) and 5′ GTT CCC CAC GTA CTG GCC CAG GAC CA 3′ (reverse). Thirty-five cycles of c-Jun and G6PD amplification were performed as follows: 95°C for 10 min, 95°C for 0.5 s, 64°C for 10 s, and 72°C for 25 s. The samples were run on a 1.2% agarose gel, and the PCR fragment sizes of 400 bp for c-Jun and 340 bp for G6PD were observed.
Electrophoretic mobility shift assays.
In vitro-translated C/EBPα and c-Jun proteins were made using an in vitro translation kit (Promega) according to the company protocol. A bp −82/−53 proximal AP-1 oligonucleotide (5′ AGGCCTTGGGGTGACATCATGGGCTATTTT) and a bp −57/−38 human granulocyte colony-stimulating factor receptor C/EBPα binding site control oligonucleotide (5′ AAGGTGTTGCAATCCCCAGC) were [γ-32P]dATP (Amersham) end labeled by using polynucleotide kinase (New England Biolabs). The binding conditions were as described earlier (5, 6, 50). C/EBPα (SC-61X) and c-Jun (SC-45X) antibodies and normal rabbit immunoglobulin G (IgG) (SC-2027) from Santa Cruz were used for supershift experiments.
Coimmunoprecipitation.
Nuclear extracts from U937, HL60, and 293T cells transfected with various constructs were prepared as follows. Cells were washed in phosphate-buffered saline and lysed in buffer A (20 mM Tris [pH 8.0], 10 mM NaCl, 3 mM MgCl2, 0.1% [vol/vol] NP-40, 10% [vol/vol] glycerol, 0.2 mM EDTA, 1 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride, 2.5 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 5 μg of antipain per ml) for 15 min on ice with occasional mixing. Nuclei were pelleted by centrifugation in an Eppendorf 541SR centrifuge at 2,000 rpm for 5 min at 4°C. Proteins were extracted from nuclei by incubation at 4°C with snap freeze-thawing three times in buffer C (20 mM Tris [pH 8.0], 400 nM NaCl, 0.2 mM EDTA, 20% [vol/vol] glycerol, 1 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride, 2.5 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 5 μg of antipain per ml). Nuclear debris was pelleted by centrifugation at 14,000 × g for 15 min at 4°C, and the supernatant extract was aliquoted, snap frozen, and stored at −70°C. Sixty micrograms of the nuclear extracts was incubated with 40 μl of protein A-agarose beads and 2 μg of C/EBPα-specific antibody (SC-61) for 3 h, followed by extensive washes in coimmunoprecipitation buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.25% NP-40, and proteinase inhibitors). Samples were subjected to SDS-10% polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore). c-Jun was immunodetected with c-Jun-specific antibody (SC-45). For coimmunoprecipitation assays from transfected cells, 5 × 106 cells were plated in 100-mm-diameter dishes and 5 μg of the respective DNA was transfected by using Lipofectamine Plus reagent (Life Technologies). At 48 h posttransfection, cells were trypsinized and nuclear extract was made as described above.
Protein interaction assay.
C/EBPα and c-Jun were in vitro transcribed and translated in the presence of [35S]methionine (Amersham Pharmacia) by using the T7-Sp6 coupled reticulocyte lysate system (Promega) per the manufacturer's instructions. Glutathione agarose pull-down assay was performed as described previously (3) with 60 μg of U937 nuclear extract.
Retroviral transduction.
Phoenix-A cells were transfected with pMV7-neo, pMV7-c-Jun-neo (kindly provided by L. Bakiri), pMSCV-ires-EGFP and pMSCV,-C/EBPα-ires-EGFP vectors by using Lipofectamine Plus reagents (Life Technologies). Viral supernatant was collected as described by Grignani et al. (17). Transduction experiment were performed with the respective provirus infected at the same time. Three rounds of transduction were performed. Pool of stably transfected cells were subjected to fluorescence-activated cell sorter (FACS) and morphological analyses at various time points.
FACS analysis.
Pools of green fluorescent protein (GFP)-positive HL60 and U937 cells were kept in G418 selection medium for 6 to 8 days, followed by FACS analysis and cytospins. For each flow cytometry analysis, 106 cells were washed twice in washing buffer (phosphate-buffered saline, 0.1% [wt/vol] NaN3, 1% FBS) and resuspended in 100 μl of washing buffer with 2 μl of the respective antibody. Incubation was performed at room temperature for 30 min. A minimum of 104 cells were analyzed by flow cytometery. CD15 PE (clone H198), its isotype control IgM κPE (clone G155-228), and CD 11b PE (clone ICRF44) and its isotype control IgG1 κPE (clone MOPC-21) were purchased from BD Biosciences.
RESULTS
Reciprocal C/EBPα and c-Jun expression.
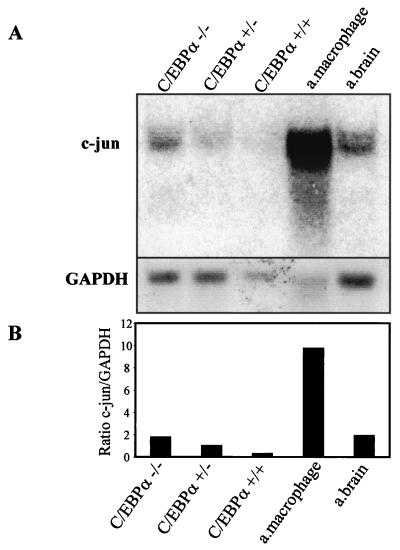
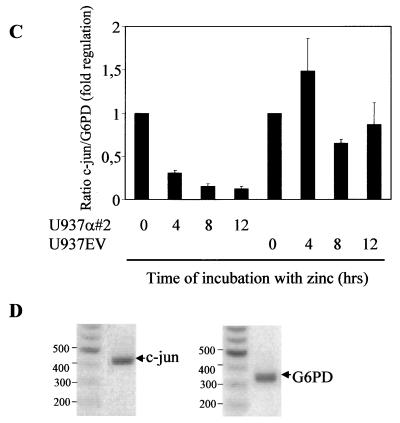
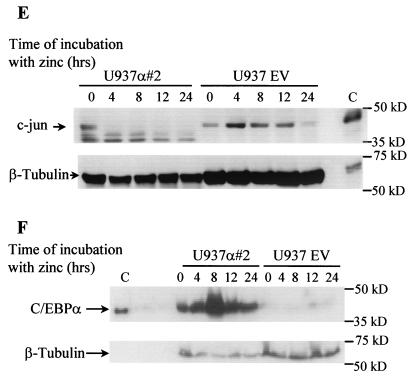
A reciprocal expression pattern of C/EBPα and c-Jun has been observed before in various cell types (13) but has not been not understood clearly. We investigated the level of c-Jun expression in fetal liver of C/EBPα wild-type, heterozygous, and nullizygous mice compared to expression in wild-type adult macrophages (Fig. 1A). About a fivefold decrease in c-Jun mRNA is observed in the wild-type compared to the nullizygous mice (Fig. 1B). Also, a high c-Jun mRNA level was detected in adult macrophages, which is consistent with previous studies. c-Jun expression is regulated by macrophage colony-stimulating factor, which is required for growth and differentiation of mononuclear phagocytes/macrophages (29). Various groups (21, 45) had observed that induction of macrophage differentiation by lipopolysaccharide, tumor necrosis factor alpha (TNF-α), gamma interferon, or interleukin-1 (IL-1) was associated with a decrease in C/EBPα expression. In comparison to wild-type C/EBPα mice, high c-Jun expression was observed in heterozygous mice, whereas the maximum expression was observed in homozygous mice, suggesting that the expression is controlled by both alleles of C/EBPα. The fetal liver samples used for the Northern blot analysis have been shown to have disturbed liver architecture (13). The C/EBPα−/− fetal liver is completely devoid of granulocytes and has a slightly higher percentage of monocytes (56). In the U937 cell line model with inducible C/EBPα expression, the decrease in c-Jun mRNA level is reciprocal to the increase in C/EBPα expression (Fig. 1C). Real-time PCR for c-Jun yielded a specific PCR product of the expected size, indicating that the analysis is specific for c-Jun and G6PD (Fig. 1D). One hundred micrograms of the protein extract was loaded to visualize the c-Jun protein band. At the protein level, a drastic decrease in c-Jun protein is observed from 0 to 4 h of C/EBPα protein induction. The c-Jun protein level is unable to return to basal level even after 24 h of C/EBPα expression induction (Fig. 1E). A faster-migrating band was also detected with the c-Jun-specific antibody. No significant change in the expression of this band was observed upon C/EBPα induction. It is unclear if this band is specific for c-Jun or is an artifact due to the polyclonal nature of the antibody used. The increase in C/EBPα protein expression upon zinc induction is also shown (Fig. 1F) and reference 35. Within 4 h of induction, C/EBPα expression increases; it reaches maximum level by 8 h and then gradually decreases by 24 h. The C/EBPα protein expression level at 24 h was still sufficient to block c-Jun protein expression.
FIG. 1.
Reciprocal expression of C/EBPα and c-Jun. (A) Northern blot analysis showing the expression of c-Jun in day 19 fetal livers from C/EBPα+/+, C/EBPα+/−, and C/EBPα−/− mice, adult (a.) mouse brain, and peritoneal macrophages. Total RNA (10 μg) for each sample was electrophoresed, transferred, and hybridized with an α-32P-labeled c-Jun 1.1-kb BamHI-EcoRI cDNA fragment and a G6PD control fragment. (B) Ratio of c-Jun to G6PD from the Northern analysis data in panel A. (C) The U937 α#2 and U937 EV cell lines were induced with 100 μM zinc sulfate, and total RNA was collected at 0, 4, 8, 12, and 16 h. cDNA from 1 μg of total RNA was used for real-time PCR using c-Jun- and G6PD-specific primers. The error bars represent standard errors of the means from three independent experiments. (D) Specific real-time PCR products of c-Jun and G6PD as observed after 1.2% agarose gel electrophoresis. Numbers on the left of each panel are molecular sizes in kilodaltons. (E) Western blot analysis showing the expression of c-Jun protein from whole-cell extracts of the U937 α#2 and U937 EV cell lines after 0, 4, 8, 12, and 24 h of zinc induction. Immunodetection was performed using c-Jun specific antibody. Lane C, in vitro-translated c-Jun positive control. β-Tubulin expression from the same blot is shown as a loading control. (F) Western blot analysis of the whole-cell extracts from panel E for C/EBPα expression with specific antibody. Lane C, in vitro-translated C/EBPα and loading control for the same Western blot.
C/EBPα downregulates c-Jun promoter activity.
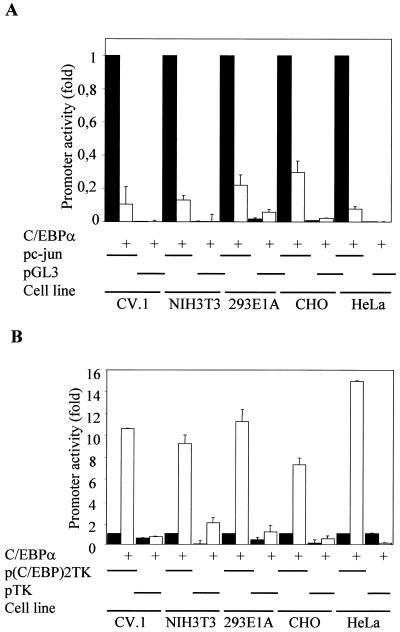
To investigate the ability of C/EBPα as a negative regulator of c-Jun expression, we first asked whether it was through binding to the c-Jun promoter. Human c-Jun promoter (52) bp −1780 to +731 in the pGL3 basic luciferase vector was cotransfected with C/EBPα expression vector in various nonmyeloid cell lines (Fig. 2A). The pGL3 basic luciferase reporter vector into which the c-Jun promoter was cloned served as a vector-alone control. Since we addressed the repression activity of C/EBPα, we also used the p(C/EBP)2TK promoter containing two repeats of the C/EBP consensus DNA binding site as a positive control for C/EBPα transcriptional activity under the same experimental conditions in order to rule out a toxic effect of C/EBPα in transient-transfection experiments (Fig. 2B). These transient-transfection experiments were carried out in various fibroblast cell lines, as shown in Fig. 2, to demonstrate that it was a general phenomenon and not cell line dependent. At least 8-fold downregulation of c-Jun promoter activity in the presence of C/EBPα was observed, whereas the p(C/EBP)2TK promoter was transactivated about 10-fold upon transient expression of C/EBPα.
FIG. 2.
C/EBPα downregulates the c-Jun promoter activity. (A) Transient cotransfection of a c-Jun promoter reporter construct (bp −1780 to +731) and pGL3 with or without C/EBPα in CV.1, NIH 3T3, 293E1A, CHO, and HeLa cells. Solid bars, values for promoter alone; open bars, cotransfection with C/EBPα. The pRL-0 Renilla luciferase construct was cotransfected to normalize for transfection efficiency. Error bars indicate standard errors of the means. (B) Effect of transient cotransfection of C/EBPα on the positive control p(C/EBP)2TK-luciferase reporter construct, indicating the transactivation capacity of C/EBPα in these cell lines. The pTK-luciferase reporter construct served as a negative control. Solid bars, values for promoter alone; open bars, fold promoter activities in the presence of C/EBPα.
C/EBPα does not recruit a TSA-sensitive corepressor complex and blocks TPA-induced c-Jun promoter activity.
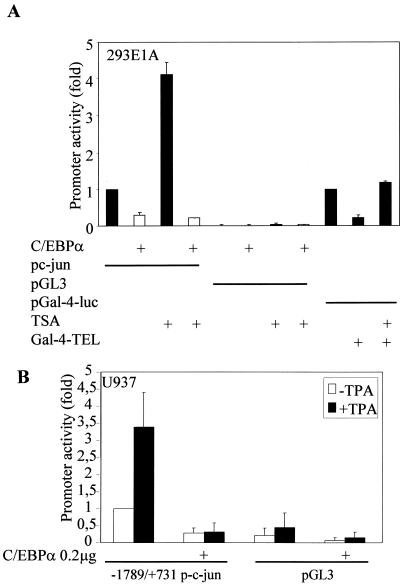
To address the possibility of C/EBPα-mediated c-Jun promoter downregulation by recruiting corepressors, transient-transfection experiments with the c-Jun promoter were carried out with C/EBPα in the presence of trichostatin A (TSA), a potent inhibitor of histone deacetylase-corepressor complex formation on an open transcription promoter machinery. Downregulation of c-Jun promoter activity by C/EBPα is retained even in the presence of 100 nM TSA (Fig. 3A). TSA increases the c-Jun promoter activity by itself. We think that this could be because TSA increases histone H3 acetylation on c-Jun-associated nucleosomes (11). A positive control for TSA showing the loss of recruitment of corepressor complex by transcription factor TEL in the presence of TSA was also included (37).
FIG. 3.
C/EBPα does not recruit a TSA-sensitive corepressor complex and blocks TPA-induced c-Jun promoter activity. (A) Transient-cotransfection experiments in the 293E1A cell line with the c-Jun promoter construct and C/EBPα in the presence or absence of TSA (100 nM). pGal-4-luc with Gal-4-TEL and TSA was used as a positive control for functionally active TSA. (B) U937 cells (106 in six-well plates) were transfected with 0.55 μg of c-Jun promoter construct (bp −1780 to +731) or pGL3, with or without 0.4 μg of C/EBPα expression plasmid or empty vector, and 0.05 μg of pRL-0. The cells were transfected by using the Effectene protocol (Qiagen). At 12 h posttransfection, TPA (100 nM) was added to the respective wells and further incubated at 37°C for 24 to 30 h. The pRL-0 Renilla luciferase construct was cotransfected to normalize for transfection efficiency. The results are the means from three independent experiments, and error bars represent the standard errors of mean values for each set.
TPA, a potent inducer of monocytic differentiation in myeloid bipotential cell lines, has been known to increase c-Jun expression (24). Radomska et al. (35) had earlier demonstrated that C/EBPα can block TPA-induced monocytic differentiation in U937 myeloid cells. We therefore asked whether C/EBPα could inhibit TPA-induced monocytic differentiation capacity by blocking c-Jun expression and activity. As shown in Fig. 3B, human c-Jun promoter activity was downregulated by C/EBPα and, interestingly, the TPA-induced increase in the c-Jun promoter activity was blocked by C/EBPα.
Mapping of the region in the c-Jun promoter that is important for C/EBPα-mediated promoter downregulation.
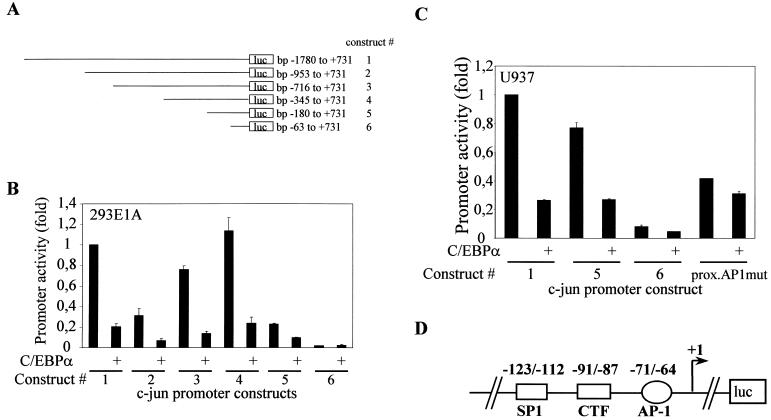
Since C/EBPα does not recruit a TSA-sensitive corepressor complex to the c-Jun promoter, we further asked whether C/EBPα could exert this repressional activity via some specific transcription factor binding sites in the c-Jun promoter. Schematic presentations of various 5′ c-Jun promoter deletion constructs described by Wei et al. (52) are shown in Fig. 4A. These constructs were used for promoter mapping experiments in 293E1A cells (Fig. 4B) and in U937 myeloid cells (Fig. 4C). As observed by Wei et al., each 5′ deletion construct had transcriptional activity different from that of the longest (bp −1780/+731) promoter construct. As seen in Fig. 4B and C, the promoter activity of each c-Jun promoter deletion construct except the bp −63/+731 promoter construct was downregulated by C/EBPα. The bp −63/+731 construct lacks most of the regulatory regions identified so far. Since the bp −180/+731 c-Jun promoter construct was still downregulated by C/EBPα, we concluded that the site important for C/EBPα-mediated transcriptional downregulation would be between bp −180 and −63. Downregulation of the c-Jun promoter activity by C/EBPα was lost when the proximal AP-1 site was mutated (Fig. 4C). This suggested that the proximal AP-1 was important for C/EBPα-mediated downregulation of the c-Jun promoter. A schematic presentation of the c-Jun promoter spanning the bp −180 to −63 region (Fig. 4D) shows the binding sites for various transcription factors. This region includes pAP-1 (proximal AP-1), CTF, and Sp-1 sites.
FIG. 4.
c-Jun promoter mapping to identify the region important for C/EBPα-mediated downregulation. (A) Schematic presentation of various c-Jun promoter 5′ deletion constructs used for the transient-transfection experiment. (B) 293E1A cells (104 per well in 24-well plates) were transfected with 0.25 μg of 5′ c-Jun promoter deletion constructs bp −1780/+731, bp −953/+731, bp −716/+731, bp −345/+731, bp −180/+731, and bp −63/+731 with or without 0.2 μg of C/EBPα or empty vector and 0.05 μg of pRL-0. (C) U937 cells (106 per well in six-well plates) were transfected with 0.55 μg of 5′ c-Jun promoter deletion constructs bp −1780/+731, bp −180/+731, bp −63/+731, and bp −1780/+731 proximal AP-1 mutant c-Jun promoter with or without C/EBPα expression plasmid or empty vector and 0.05 μg of pRL-0. The cells were transfected by using the Effectene protocol. The pRL-0 Renilla luciferase construct was cotransfected to normalize for transfection efficiency. The results are the means from three independent experiments, and error bars represent the standard errors of mean values for each set. (D) Schematic presentation of the c-Jun promoter region between bp −180 and −63. This region contains a proximal AP-1 site, CTF site, and SP-1 site.
C/EBPα blocks the autoregulatory capacity of c-Jun by preventing c-Jun from binding to the proximal AP-1 site in the c-Jun promoter.
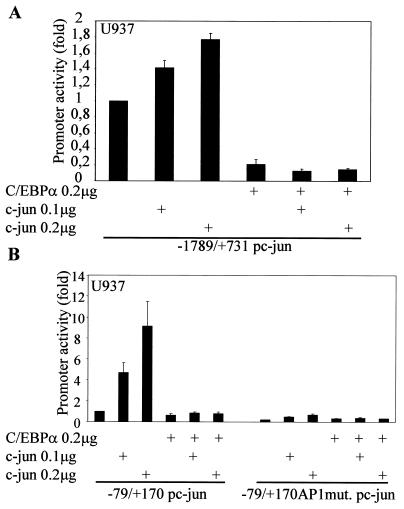
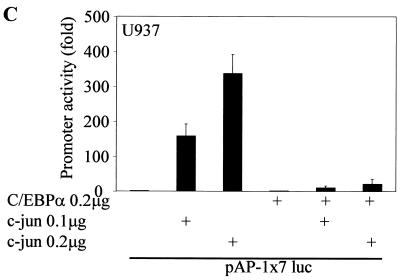
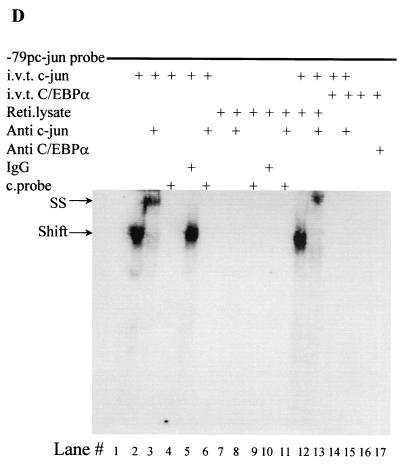
The deletion constructs in Fig. 4B had very low basal transcriptional activity compared to the full-length c-Jun promoter. Hence, we decided to address the importance of the proximal c-Jun promoter in context with the full-length promoter. The bp −1780/+731 c-Jun promoter with a mutated proximal AP-1 site showed higher basal transcriptional activity than the bp −180 and −63 c-Jun promoter deletion constructs (52). Transient-transfection experiments using the bp −1780/+731 c-Jun promoter with mutated proximal AP-1, as shown in Fig. 4C, suggest that in the absence of the proximal AP-1 site, C/EBPα is unable to downregulate the c-Jun promoter. In transient transfections in U937 myeloid cells, c-Jun transactivates its own promoter, an autoregulatory mechanism that was identified by Angel et al. (2). Our data (Fig. 5A) suggest that transactivation of the c-Jun promoter by c-Jun is blocked in the presence of C/EBPα. Based on results from the previous promoter mapping experiments and TPA experiments, we decided to address the importance of the proximal AP-1 site in C/EBPα-mediated c-Jun promoter downregulation. We asked whether C/EBPα could block the transactivation capacity of c-Jun through the proximal AP-1 site in the c-Jun promoter. Using the bp −79/+170 promoter (54) containing only the proximal AP-1 site of the c-Jun promoter (Fig. 5B) and an artificial AP-1 construct containing seven repeats of the consensus AP-1 site (Fig. 5C), we observed that the autoregulatory capacity of c-Jun through the proximal AP-1 binding site was lost in the presence of C/EBPα. Increasing the concentration of c-Jun could not overcome the block by C/EBPα. C/EBPα was transcriptionally active in this set of experiments. To understand how C/EBPα blocks the autoregulatory capacity of c-Jun, we determined the DNA binding capacity of c-Jun to the proximal AP-1 site in the presence of C/EBPα. In a band shift mobility assay using the bp −79 c-Jun promoter oligonucleotide probe, c-Jun binding to the proximal AP-1 site (Fig. 5D, lane 2) was blocked by in vitro-translated C/EBPα (Fig. 5D, lane 14) but not by reticulocyte lysate alone (Fig. 5D, lane 12). Under similar experimental conditions, C/EBPα could bind to the CEBP consensus DNA binding site in the G-CSFR promoter, and the C/EBPα band shift was supershifted in the presence of C/EBPα-specific antibody (data not shown).
FIG. 5.
C/EBPα blocks the autoregulatory capacity of c-Jun by preventing c-Jun from binding to the proximal AP-1 site in the c-Jun promoter. (A) Transient transfections in U937 myeloid cells were performed using a bp −1780/+731 c-Jun promoter construct, 0.2 μg of C/EBPα expression plasmid, and increasing amounts of c-Jun expression plasmid (0.1 and 0.2 μg). Error bars indicate standard errors of the means. (B) bp −79/+190 and bp −79/+190 mutated AP-1 site c-Jun promoter constructs were transiently transfected with 0.2 μg of C/EBPα expression plasmid and increasing concentrations of c-Jun expression plasmid. (C) The AP-1 luciferase construct containing seven repeats of an AP-1 binding site was used for transient transfection with c-Jun and C/EBPα in U937 myeloid cells. (D) Electrophoretic mobility shift assay using [γ-32P]ATP-labeled bp −82/−53 c-Jun promoter oligonucleotide spanning the proximal AP-1 site was performed using in vitro-translated (i.v.t.) c-Jun (lanes 2 to 6 and 12 to 15) and C/EBPα (lanes 12, 13, 16, and 17) proteins, rabbit reticulocyte (Reti.) lysate (lanes 7 to 11, 14, and 15), c-Jun-specific antibody (lanes 3, 6, 8, 11, 13, and 15), normal rabbit IgG (lanes 4 and 9), C/EBPα-specific antibody (lane 17), and self unlabeled competitor probe (lanes 5, 6, 10, and 11). Arrows show the c-Jun shifted band (Shift) and the supershifted higher band with c-Jun-specific antibody (SS).
C/EBPα and c-Jun interact through their leucine zipper domains.
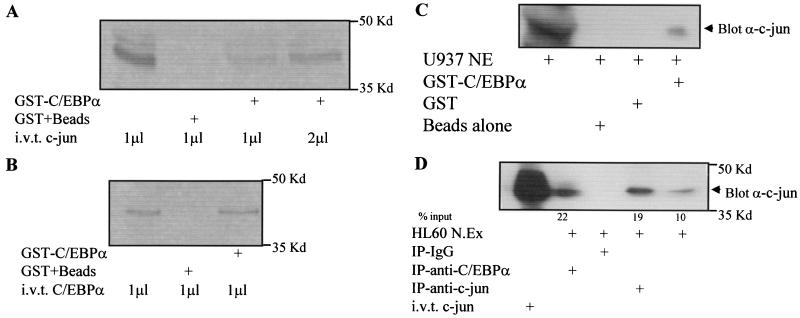
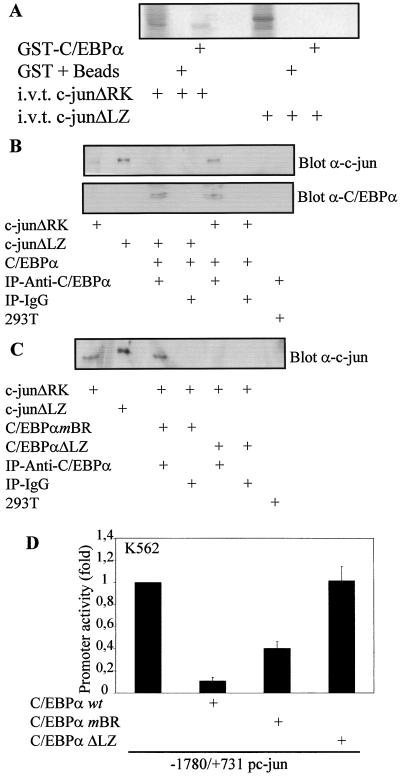
We then asked whether c-Jun could interact with C/EBPα. An in vitro glutathione S-transferase (GST) pull-down assay (Fig. 6A and C) and an in vivo coimmunoprecipitation (Fig. 6D) indicate that C/EBPα and c-Jun interact in vitro and in vivo. An in vivo coimmunoprecipitation experiment with U937 and HL60 cells (Fig. 6C and D) suggested that C/EBPα and c-Jun could interact in myeloid cells. However, a very low basal level of c-Jun is present in U937 cells. Not surprisingly, due to such low levels of c-Jun protein, extremely little C/EBPα complex with c-Jun was observed (Fig. 6C). Figure 6B shows a control for GST-C/EBPα, which can form homodimers with in vitro-translated C/EBPα. We further wanted to investigate which domains of C/EBPα and c-Jun were important for this interaction. c-JunΔRK lacks the DNA binding domain (amino acids 251 to 276), and the c-JunΔLZ mutant construct has the leucine zipper dimerization domain (amino acids 281 to 313) deleted (34). A GST pull-down assay using GST-C/EBPα and in vitro-translated c-JunΔRK and c-JunΔLZ suggested that the leucine zipper dimerization domain of c-Jun was required to interact with C/EBPα (Fig. 7A and B). The C/EBPαmBR construct, carrying a mutation in the basic DNA binding domain, could still bind to c-Jun, but the C/EBPαΔLZ construct (C/EBPα leucine zipper dimerization domain replaced with GCN4 leucine zipper) is unable to bind c-Jun (Fig. 7C). C/EBPα expression in the same experiment was determined to show the presence of the C/EBPα wild-type and mutant protein (Fig. 7B and data not shown). No nonspecific binding was observed with the control IgG coimmunoprecipitation. The C/EBPαmBR construct could downregulate the c-Jun promoter activity, whereas the C/EBPαΔLZ mutant could not (Fig. 7D). These data suggest that the leucine zipper dimerization domains of c-Jun and C/EBPα are important for their interaction.
FIG. 6.
C/EBPα and c-Jun interact in U937 and HL60 myeloid cells. (A) GST-C/EBPα and GST plus beads were incubated with in vitro-translated (i.v.t.) c-Jun as described in Materials and Methods. (B) As a positive control for GST-C/EBPα, GST-C/EBPα was incubated with in vitro-translated C/EBPα. (C) GST-C/EBPα was incubated with 85 μg of U937 nuclear extract (NE). Immunodetection was carried out using c-Jun antibody. GST and glutathione-agarose beads alone were incubated with U937 nuclear extract to determine the specificity of this interaction. (D) Coimmunoprecipitation assays from 60 μg of HL60 nuclear extract (N.Ex) were performed using C/EBPα, c-Jun-specific antibody, or normal rabbit IgG. Immunodetection was carried out using c-Jun-specific antibody. In vitro-translated c-Jun shows the migration of c-Jun protein.
FIG. 7.
C/EBPα and c-Jun interact with their leucine zipper domains. (A) GST-C/EBPα was incubated with 35S in vitro-translated (i.v.t.) c-JunΔRK (lacking the DNA binding domain) and c-JunΔLZ (lacking the dimerization domain). GST plus beads alone incubated with these in vitro-translated proteins served as a negative control. (B) 293T cells were transfected with c-JunΔRK, c-JunΔLZ, or C/EBPα, or mock transfected, and at 24 h posttransfection nuclear extracts from these sets were used for coimmunoprecipitation (IP) assays using either C/EBPα-specific antibody or normal rabbit IgG. The samples were probed with c-Jun- and C/EBPα-specific antibodies. (C) 293T cells were transfected with c-JunΔRK, c-JunΔLZ, C/EBPαmBR (basic region mutated), or C/EBPαΔLZ (dimerization domain replaced with GCN4 leucine zipper) or mock transfected. At 24 h posttransfection, nuclear extracts from these sets were used for coimmunoprecipitation assay with either C/EBPα-specific antibody or normal rabbit IgG. The samples were probed with c-Jun specific antibodies. (C/EBPα blot, data not shown). (D) The bp −1780/+731 c-Jun promoter construct was transiently transfected with and without C/EBPα wild-type, C/EBPαmBR, and C/EBPαΔLZ plasmids in K562 cells. Mean values were normalized to empty vector values. Error bars indicate standard errors of the means from three independent experiments.
Overexpression of c-Jun blocks C/EBPα-induced granulocytic differentiation.
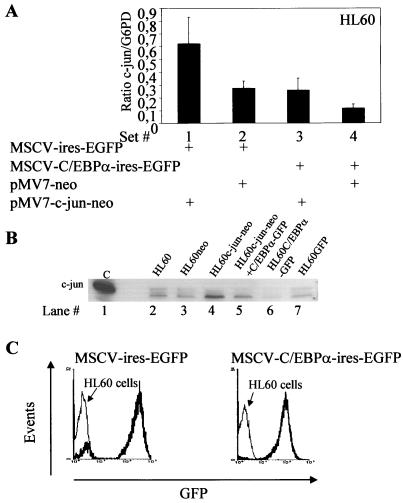
We next wanted to understand the functional implications of C/EBPα-c-Jun interaction. Earlier studies have shown that C/EBPα could block monocytic lineage commitment (35). Here, we address the effect of c-Jun on C/EBPα-induced granulocytic differentiation. Three rounds of retroviral transduction of C/EBPα and c-Jun expression vectors along with their empty vectors were performed in HL60 and U937 cells at the same time. At 24 h after the third transduction, cells were subjected to G418 selection and analyzed consecutively for expression of differentiation markers such as CD11b and CD15. No ongoing differentiation could be detected until day 9 from the start of the experiment. Total RNA from the set of HL60-transduced cells was isolated for real-time PCR for c-Jun and G6PD. As shown in Fig. 1D, specific PCR products for c-Jun and G6PD were observed. The results in Fig. 8A are averages from three independent real-time PCR experiments. HL60 cells transduced with pMV7-c-Jun-neo showed twofold-higher c-Jun mRNA levels than the pMV7-neo vector alone. These two vectors were transduced along with pMSCV-ires-GFP vector (Fig. 8A, sets 1 and 2). When the pMV7-c-Jun-neo and pMV7-neo vectors were transduced along with pMSCV-C/EBPα-ires-EGFP, there was a marked decrease in c-Jun mRNA (Fig. 8A, sets 3 and 4). c-Jun expression from the same set of HL60 samples is shown in Fig. 8B. A decrease in the endogenous c-Jun level is observed upon ectopic expression of C/EBPα. This confirms our previous findings, shown in Fig. 1E. GFP expression of the pMSCV-ires-EGFP vector and pMSCV-C/EBPα-ires-EGFP vector transfected from the same set of HL60 cells was measured by gating the cells in an FL1 window, using FACS program (Fig. 8C).
FIG. 8.
Overexpression of c-Jun blocks C/EBPα-induced granulocytic differentiation. (A) Real-time PCR for c-Jun and G6PD was performed for HL60 cells that were transduced with pMV7-c-Jun-neo (bars 1 and 3), pMV7-neo (bars 2 and 4), pMSCV-C/EBPα-ires-EGFP (bars 3 and 4), and pMSCV-ires-EGFP (bars 1 and 2) to estimate c-Jun expression in each set. Error bars indicate standard errors of the means. (B) Western blotting for c-Jun from the same experimental samples was performed by loading 100 μg of total protein lysates. (C) GFP expression in HL60 cells transduced with MSCV-ires-EGFP and MSCV-C/EBPα-ires-EGFP as analyzed by fluorescence in the FL1 channel. The samples from same HL60 experiment as in panels A, B, and E to G were used for FACS analysis. (D) Western blot analysis of 100 μg of whole-cell extract from the transduced U937 cells. The Western blots were immunoblotted using c-Jun-, C/EBPα-, and β-tubulin specific antibodies. (E) FACS analysis for CD15-PE from the transduced HL60 and U937 cells, along with its isotype control. +ve, positive. (F) FACS analysis for CD11b-PE from transduced U937 cells, along with its isotype control. (G) Wright-Giemsa staining of the transduced cells. HL60 and U937 cells with ectopic expression of C/EBPα showed neutrophils by day 6, which were reduced in the presence of c-Jun expression.
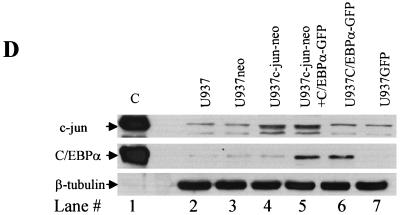
One hundred micrograms of whole-cell extracts from a pool of U937 transduced cells was analyzed for expression of C/EBPα and c-Jun protein (Fig. 8D). High c-Jun protein expression was observed in cells transduced with the c-Jun vector (lanes 4 and 5), whereas the cells transduced with the vector alone showed basal c-Jun expression. Similarly, high C/EBPα expression was observed in cells transduced with the C/EBPα expression vector (lanes 5 and 6). In cells transduced with C/EBPα alone (lane 6), the corresponding c-Jun expression was equivalent to basal levels. We could not observe further downregulation as observed in the U937 inducible cell line model (Fig. 1E). We think that this was due to the low number of GFP-positive cells in this part of the experiment (data not shown).
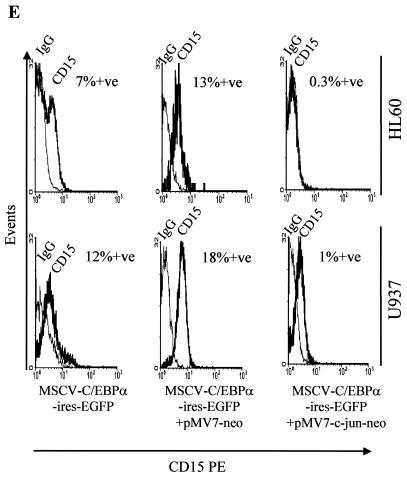
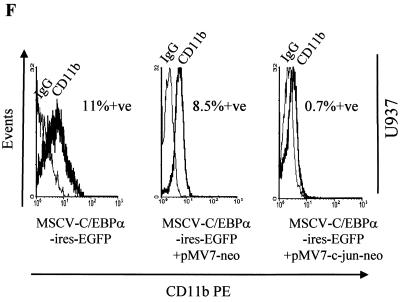
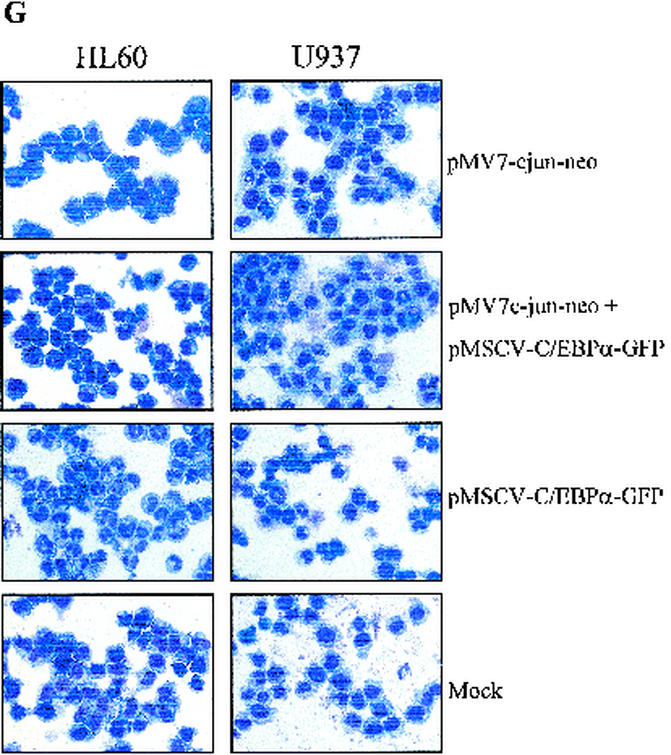
Expression of CD15, a marker for granulocytic differentiation from HL60 and U937 transduced cells, was analyzed. As seen in Fig. 8E, CD15 expression increased in the presence of C/EBPα transduction (left panels), and pMV7-neo vector alone had no effect on C/EBPα-induced granulocytic differentiation (middle panels). The right panels show the CD15 expression in HL60 and U937 cells transduced with MSCV-C/EBPα-ires-EGFP and pMV7-c-Jun-neo. A negative shift of the CD15 peak was observed. This indicated that the increase in CD15 expression by C/EBPα was blocked in the presence of c-Jun (Fig. 8E). Similar results with the CD11b marker were also observed in HL60 cells (Fig. 8F). c-Jun-transduced cells were also investigated for CD11b expression; however, no increase in its expression was observed (data not shown). Morphological changes observed in the presence of C/EBPα and/or c-Jun are shown in Fig. 8G. C/EBPα-induced granulocytic differentiation (about 75 to 80% in both HL60 and U937 cells) was reduced to about 10 and 40% in U937 and HL60, respectively. The mock panels show cells which resembles the blast morphology of untransduced cells. The growth rate of the transduced cells was also investigated. The vector-alone constructs showed growth rates similar to those of the cells alone, whereas the cells expressing c-Jun and/or C/EBPα showed retarded growth (data not shown).
DISCUSSION
C/EBPα downregulates c-Jun expression. In this study, we have investigated the role of C/EBPα as a negative regulator of c-Jun expression and transcriptional activity and its significance in the myeloid lineage commitment. Inducers of monocytic differentiation such as TPA, bryostatin 1, 1,25-dihydroxyvitamin D3, and okadaic acid were shown to increases c-Jun activity by posttranslational events and increased synthesis (1, 7,14, 23, 42). These findings underline the importance of c-Jun as a member of the AP-1 family in the myeloid differentiation program. It has been reported that overexpression of c-Jun in bipotential myeloid cells leads to macrophage-like morphology (44). That report, however, did not address the expression of monocytic/macrophage differentiation markers. In addition, it is not clear why only partial macrophage-like morphology was observed upon c-Jun overexpression. Using a C/EBPα-inducible U937 cell line, we show that an increase in C/EBPα expression results in a significant decrease in the levels of endogenous c-Jun mRNA (Fig. 1C). The c-Jun protein level decreases drastically in the first 4 h of C/EBPα expression (Fig. 1E). At the same time, no change in c-Fos expression was observed upon induction of C/EBPα expression (data not shown). The reciprocal pattern of expression for C/EBPα and c-Jun was also observed in a C/EBPα knockout mouse model and in hepatocytes (Fig. 1A) (13, 36). Although the liver architecture in the C/EBPα newborn is disturbed, the hematopoetic system is not severely affected, except for the granulocytes. The immature hematopoetic cell population is also not affected. This led us to hypothesize that c-Jun downregulation by C/EBPα is important for the granulocytic lineage decision. In addition, C/EBPα is important for hepatocytic and the lung cell differentiation, which is severely disturbed in C/EBPα knockout mice. Figure 1A and B suggest that C/EBPα suppression of c-Jun might also play an important role in hepatocytic differentiation and development. Some reports state that expression of c-Fos, another AP-1 member, also increases on induction of monocytic differentiation. However, the increase in c-Fos mRNA was found to be transient and not myeloid lineage specific (14, 40). c-Fos expression was not sufficient for the process of macrophage differentiation (10, 27, 31). No detectable c-Fos level was observed in the fetal liver samples, whereas adult macrophage and adult brain RNA samples showed minimal c-Fos expression (data not shown). Low levels of C/EBPα mRNA are observed upon treatment of monocytes with inducing agents such as lipopolysaccharide, TNF-α, gamma interferon, and IL-1 (45). Figure 1A also shows a large amount of c-Jun mRNA in adult macrophages. The mechanism of how C/EBPα controls c-Jun expression is observed in the promoter assays. The C/EBPα protein negatively regulates the human c-Jun promoter in transient-transfection assays in fibroblast as well as in myeloid cell lines (Fig. 2), thus indicating that it was a general phenomenon and not cell line specific. As also observed in Fig. 1A, in the fetal livers of the C/EBPα knockout mice, the negative regulation of c-Jun by C/EBPα holds true in various cell types and tissue systems.
Mechanism of c-Jun expression downregulation by C/EBPα through the proximal AP-1 site of the c-Jun promoter.
C/EBPα downregulation of the c-Jun promoter activity was not due to recruitment of a TSA-sensitive corepressor complex (Fig. 3A). The TPA experiment (Fig. 3B) suggested that a transcription factor binding to the c-Jun promoter was important for C/EBPα-mediated c-Jun promoter activity downregulation. Promoter mapping experiments (Fig. 4) suggested that the region between bp −180 and −63 in the c-Jun promoter was important for the c-Jun promoter downregulation by C/EBPα. The proximal AP-1 site (pAP-1) in the promoter lies within the region from bp −180 to −63. Earlier studies with the human c-Jun promoter addressed the importance of the proximal AP-1 site in c-Jun promoter being sufficient for a maximal response to various signals (TPA, serum, UV, E1A, and IL-1) (18, 49). Using the human c-Jun promoter, we show that C/EBPα blocks the autoactivation capacity of c-Jun through the proximal AP-1 site. On mutation of this proximal AP-1 site, C/EBPα was no longer able to downregulate the c-Jun promoter activity (Fig. 4C and 5B). These results led us to hypothesize that C/EBPα and c-Jun might interact. This is the first report showing C/EBPα and c-Jun interaction in myeloid cells (Fig. 6). Furthermore, the leucine zipper domains of both proteins are required for this interaction (Fig. 7), and the leucine zipper domain of C/EBPα was important for downregulating the c-Jun promoter activity (Fig. 7D). DNA binding experiments (Fig. 5D) indicate that C/EBPα binding to c-Jun inhibits latter from binding to its consensus AP-1 site in the c-Jun promoter. This was also confirmed by transient-transfection experiments using the bp −79 c-Jun promoter having only the proximal AP-1 site (Fig. 5B) and the full-length c-Jun promoter with the proximal AP-1 site mutated (Fig. 4C). It still needs to be addressed whether the C/EBPα-c-Jun interaction can block the latter from binding to the AP-1 site in other c-Jun-regulated promoters as well. We think that the presence of other interacting partners of both these proteins (e.g., PU.1, AML-1, p300, and C/EBPβ) might play an important role in such interactions in a promoter-specific context.
Biological implication of C/EBPα-c-Jun interaction in normal myelopoiesis and leukemia.
Previous reports have addressed the indispensability of C/EBPα in driving granulocytic differentiation, as well as its role in acute myeloid leukemia (AML) (9, 15, 32, 33, 35, 53, 56). The results shown in Fig. 8 indicate the importance of c-Jun expression downregulation by C/EBPα in myeloid lineage commitment. If c-Jun expression was high at the time of lineage commitment, it might block C/EBPα from committing these cells to the granulocytic lineage. c-Jun is an important regulator of TPA-mediated or independent macrophage lineage commitment (19, 44). TPA-induced monocytic differentiation was blocked by C/EBPα (35). These reports indicated that a block in c-Jun expression by C/EBPα might be also necessary for preventing macrophage differentiation so as to bias the progenitor cells towards the granulocytic lineage decision. Here we report the functional significance of the c-Jun block by C/EBPα (Fig. 8). The normal granulocytic differentiation capacity of C/EBPα, as observed by an increase in the CD15 and CD11b markers, was abolished upon overexpression of c-Jun (Fig. 8E and F). Morphologically, C/EBPα-induced granulocytic differentiation (75 to 80% in both cell types) was reduced about by 10% (U937) to 40% (HL60), as observed in Fig. 8G. These data indicate that inhibition of c-Jun expression and function is essential for C/EBPα to commit precursor myeloid cells towards the granulocytic lineage.
C/EBPα-c-Jun interaction (Fig. 6 and 7) suggests that C/EBPα might pull c-Jun away from its other interacting partners, thereby inhibiting c-Jun's function, e.g., c-Jun interaction with C/EBPβ in monocytic differentiation by TNF-α (55). Ubeda et al. (47) have reported that CHOP, a dominant negative regulator of C/EBP family members, can interact with c-Jun through its leucine zipper domain. By such interaction, CHOP synergizes with c-Jun to activate transcription through the AP-1/TRE site. In contrast to these findings, we observe that the C/EBPα-c-Jun interaction prevents c-Jun from binding to the AP-1 site in the c-Jun promoter. The C/EBPα-c-Jun complex formation probably pulls c-Jun away from c-Jun-regulated genes.
Transient transfection of the C/EBPα mutant construct with the c-Jun promoter (Fig. 7D) emphasizes the functional importance of the interaction between the leucine zipper domains of C/EBPα and c-Jun (Fig. 6D and 7A to C). From our data and previous studies by other groups, it is clear that the leucine zipper dimerization domain that is conserved in the C/EBP family members is important for interaction with c-Jun. However, the functional outcome of such interactions is diverse. We think that apart from the C-terminal leucine zipper domains, the N-terminal domain of each C/EBP member is critical for the function of such interactions. This hypothesis is well understood, since C/EBP members such as C/EBPα, C/EBPβ, and CHOP interact with c-Jun by their leucine zipper domains. However, C/EBPα antagonizes c-Jun function, whereas the latter two synergize with c-Jun.
Moreover, c-Jun can also function as a coactivator of the PU.1 transcription factor, which is important for the monocytic lineage (5). In the presence of higher C/EBPα expression and TPA, U937 myeloid cells were unable to undergo macrophage differentiation (35). The explanation for this observation could be that C/EBPα inhibits c-Jun from performing its coactivator role for PU.1 or from independent regulation of monocyte-specific genes. This could be either by preventing c-Jun from binding to the AP-1 site in the promoter or by disrupting c-Jun interaction with other transcription factors important for monocytic lineage commitment.
C/EBPα blocking of TPA-induced moncytic differentiation has been addressed before. We wanted to further investigate how c-Jun could affect the granulocytic differentiation capacity of C/EBPα. CD15, an indicator of granulocyte differentiation, is increased upon ectopic expression of C/EBPα. c-Jun alone does not change the expression level of this marker compared to vector alone (data not shown). However, in the presence of c-Jun and C/EBPα expression, a negative shift in the CD15 peak is observed (Fig. 8E). These data suggest that the granulocytic differentiation commitment induced by C/EBPα is blocked by c-Jun. CD11b, a marker for early differentiation, is upregulated irrespective of the lineage specificity. Upon overexpression of C/EBPα, an increase in CD11b expression is observed (Fig. 8F), which is blocked by c-Jun, as also observed with CD15. c-Jun, which has been shown to also induce partial macrophage-like morphology, should also show an increase in CD11b, which was not observed. This negative data was not surprising, because studies (30) have shown that c-Jun is unable to regulate the CD11b promoter on its own. Our data along with that report could explain why c-Jun causes only partial macrophage differentiation. c-Jun probably needs other factors (or needs to act along with other factors, perhaps PU.1) to attain complete monocyte/macrophage differentiation. Morphologically also, c-Jun was observed to inhibit granulocytic differentiation induced by C/EBPα (Fig. 8G).
Recent reports address the important role of C/EBPα in AML (15, 32, 33). Dominant negative mutations in C/EBPα were observed in AML FAB-M2 patient samples in the absence of the AML1-ETO fusion protein. Higher c-Jun mRNA levels in patient samples with C/EBPα mutations than in samples without C/EBPα mutations were observed (data not shown). Recent reports by Gombart et al. (15) have identified C-terminal mutations of C/EBPα in similar AML patient samples. The mutations they characterize show the importance of a functional C-terminal end of the C/EBPα protein. Although these mutants are expressed like the wild-type protein, they have lost their dimerization and/or DNA binding capacity. Although a significant amount of C/EBPα protein is present in AML, due to various modifications, it may be functionally dead and thus unable to rescue leukemic blasts into the normal differentiation program. These findings underline the importance of C/EBPα in controlling c-Jun expression and transcriptional activity in AML. When c-Jun is expressed in a deregulated manner, it might have the potential to act as a proto-oncogene and thus lead to hyperproliferation of the leukemic blasts. The function of each transcription factor is differentiation stage dependent. The concentration of the protein also plays an important role in deciding the cell fate, i.e., lineage commitment versus proliferation state. A few transcription factors, e.g., PU.1, program the cell for a specific lineage depending on their expression level. Similarly, c-Jun could function either as a coactivator for PU.1 leading to differentiation or as a proto-oncogene causing proliferation. As observed in AML blasts, c-Jun might act as a hyperproliferating agent, or, in the case of TPA induced macrophage differentiation, c-Jun might act as a transcription factor driving partial macrophage differentiation. The C/EBPα-c-Jun interaction might disrupt the function of c-Jun, depending on the expression levels of both of these proteins.
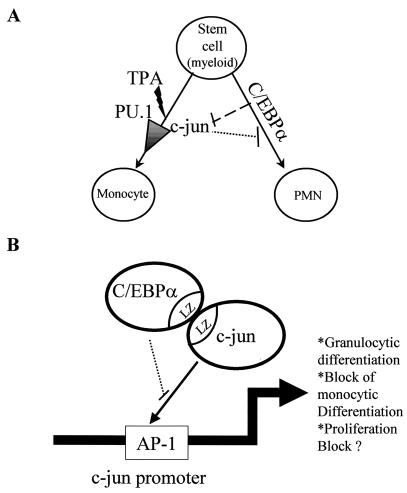
In conclusion, we propose a model for the importance of C/EBPα in blocking c-Jun expression and c-Jun transactivation capacity (Fig. 9). Bipotential myeloid cells differentiate towards the granulocytic lineage upon overexpression of C/EBPα, whereas the same cells have the potential for monocytic lineage commitment in the presence of inducers such as TPA. TPA is known to transactivate c-Jun and increase its expression. One of the important roles of c-Jun is to act as a coactivator of transcription factor PU.1. c-Jun on its own could also drive partial macrophage-like differentiation. However, when C/EBPα and c-Jun interact through their leucine zipper domains, the former prevents c-Jun from functioning as a macrophage differentiation regulator. At the same time, such interaction could also arrest C/EBPα-driven granulocytic lineage commitment (Fig. 9A). The data so far suggest that this interaction blocks c-Jun from binding to the AP-1 site of its own promoter, thereby inhibiting its expression and transcriptional activity (Fig. 9B). The significance of C/EBPα blocking of c-Jun DNA binding capacity for other c-Jun regulated genes still needs to be addressed. Because of such sequestering of c-Jun, C/EBPα might not only commit bipotential myeloid cells to granulocytic lineage but also prevent these cells from becoming monocytes/macrophages.
FIG. 9.
Model for C/EBPα inactivating c-Jun in granulocytic differentiation. (A) Diagrammatic representation of myeloid bipotential stem cells that can differentiate to monocytes/macrophages on induction with TPA or become polymorphonuclear neutrophils on overexpression of C/EBPα. TPA induction for macrophage differentiation requires increases in c-Jun expression and c-Jun transcriptional activity. c-Jun acts as a coactivator of PU.1, leading to monocytic differentiation commitment. C/EBPα blocks the expression and transcriptional activity of c-Jun, thus preventing TPA-induced monocytic lineage commitment. At the same time, c-Jun also blocks C/EBPα-driven granulocytic lineage commitment. (B) Schematic representation showing interaction between C/EBPα and c-Jun via their leucine zipper domains. This interaction prevents c-Jun from binding to the proximal AP-1 site in its own promoter. c-Jun interaction with C/EBPα and the block in binding to its own promoter lead to downregulation of c-Jun expression. This C/EBPα-c-Jun interaction may lead to a block in monocytic lineage differentiation and proliferation.
Acknowledgments
We thank Hanna Radomska for valuable discussions and Diana Dudziak, Falk Nimmerjahn, Joachim Ellwart, Karin Nispel, and Susan King for technical support. Thanks also go to Torsten Haferlach for providing the differentials and imaging of the cytospins.
This work was supported by DFG (German Research Foundation) grant 2042/2-1 to G.B.
REFERENCES
- 1.Angel, P., K. Hattori, T. Smeal, and M. Karin. 1988. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55:875-885. [DOI] [PubMed] [Google Scholar]
- 2.Angel, P., E. A. Allgretto, St. T. Okino, K. Hattori, W. J. Boyle, T. Hunter, and M. Karin. 1988. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature 332:166-171. [DOI] [PubMed] [Google Scholar]
- 3.Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26. [DOI] [PubMed] [Google Scholar]
- 4.Behre, G., P. Zhang, D. E. Zhang, and D. G. Tenen. 1999. Analysis of the modulation of transcriptional activity in myelopoiesis and leukemogenesis. Methods 17:231-237. [DOI] [PubMed] [Google Scholar]
- 5.Behre, G., A. J. Whitmarsh, M. P. Coghlan, T. Hoang, C. L. Carpenter, D. E. Zhang, R. J. Davis, and D. G. Tenen. 1999. c-jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem. 274:4939-4946. [DOI] [PubMed] [Google Scholar]
- 6.Behre, G., S. M. Singh, H. Liu, L. T. Bortolin, M. Christopeit, H. S. Radomska, J. Rangatia, W. Hiddemann, A. D. Friedman, and D. G. Tenen. 2002. Ras signaling enhances the activity of C/EBPα to induce granulocytic differentiation by phosphorylation of serine 248. J. Biol. Chem. 277:26293-26299. [DOI] [PubMed] [Google Scholar]
- 7.Bertani, A., N. Polentarutti, A. Sica, A. Rambaldi, A. Mantovani, and F. Colotta. 1989. Expression of c-jun protooncogene in human myelomonocytic cells. Blood 74:1811-1816. [PubMed] [Google Scholar]
- 8.Brandt-Rauf, P. W., M. R. Pincus, J. M. Chen, and G. Lee. 1989. Conformational energy analysis of the leucine repeat regions of C/EBP, GCN4, and the proteins of the myc, jun, and fos oncogenes. J. Protein Chem. 8:679-688. (Abstract.) [DOI] [PubMed] [Google Scholar]
- 9.Burel, S. A., N. Harakawa, L. Zhou, T. Pabst, D. G. Tenen, and D. E. Zhang. 2001. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol. Cell. Biol. 21:5577-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calabretta, B. 1987. Dissociation of c-Fos induction from macrophage differentiation in human myeloid leukemic cell lines. Mol. Cell. Biol. 7:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos and c-jun-associate nucleosomes upon gene activation. EMBO 19:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emig, M., S. Saussele, H. Wittor, A. Weisser, A. Reiter, A. Willer, U. Berger, R. Hehlmann, N. C. Cross, and A. Hochhaus. 1999. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia 13:1825-1832. [DOI] [PubMed] [Google Scholar]
- 13.Flodby, P., C. Barlow, H. Kylefjord, L. Ährlund-Richter, and K. G. Xanthopoulos. 1996. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J. Biol. Chem. 271:24753-24760. [DOI] [PubMed] [Google Scholar]
- 14.Gaynor, R., K. Simon, and P. Koeffler. 1991. Expression of c-jun during macrophage differentiation of HL-60 cells. Blood 77:2618-2623. [PubMed] [Google Scholar]
- 15.Gombart, A. F., W. K. Hofmann, S. Kawano, S. Takeuchi, U. Krug, S. H. Kwok, R. J. Larsen, H. Asou, C. W. Miller, D. Hoelzer, and H. P. Koeffl. 2002. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood 99:1332-1340. [DOI] [PubMed] [Google Scholar]
- 16.Gramigni, C., S. Penco, G. Bianchi-Scarra, R. Ravazzolo, and G. Garre. 1998. An upstream negative regulatory element in human granulocyte-macrophage colony-stimulating factor promoter is recognised by AP-1 family members. FEBS Lett. 440:119-124. [DOI] [PubMed] [Google Scholar]
- 17.Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, L. Lanfrancone, C. Peschle, G. P. Nolan, and P. G. Pelicci. 1998. High efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58:14-19. [PubMed] [Google Scholar]
- 18.Hallahan, D. E., E. Dunphy, J. Kuchibhotla, A. Kraft, T. Unlap, and R. Weichselbaum. 1996. Prolonged c-Jun expression in irradiated ataxia telangiectasia fibroblasts. Int. J. Radiat. Oncol. Biol. Phys. 36:355-360. [DOI] [PubMed] [Google Scholar]
- 19.Hass, R., M. Brach, S. Kharbanda, G. Giese, P. Traub, and D. Kufe. 1991. Inhibition of phorbolester-induced monocytic differentiation by dexamethasone is associated with downregulation of c-fos and c-jun (AP-1). J. Cell. Physiol. 149:125-131. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, W., T. K. Kerppola, P. L. Chen, T. Curran, and S. Chen-Kiang. 1994. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol. Cell. Biol. 14:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, H.-M., M. Baer, S. C. Williams, P. F. Johnson, and R. C. Schwartz. 1998. Redundancy of C/EBPalpha, -beta, and delta in supporting the lipopolysaccharide-induced transcription of IL-6 and monocyte chemoattractant protein-1. J. Immunol. 160:2334-2342. [PubMed] [Google Scholar]
- 22.Johnson, R. S., B. van Lingen, V. E. Papaioannou, and B. M. Spiegelman. 1993. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 7:1309-1317. [DOI] [PubMed] [Google Scholar]
- 23.Kharbanda, S., R. Datta, E. Rubin, T. Nakamura, R. Hass, and D. Kufe. 1992. Regulation of c-jun expression during induction of monocytic differentiation by okadaic acid. Cell Growth Diff. 3:391-399. [PubMed] [Google Scholar]
- 24.Lamph, W. W., P. Wamsley, P. Sassone-Corsi, and I. M. Verma. 1988. Induction of protooncogene JUN/AP-1 by serum and TPA. Nature 334:631.. [DOI] [PubMed] [Google Scholar]
- 25.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 26.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1989. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243:1681-1688. [DOI] [PubMed] [Google Scholar]
- 27.Lord, K. A., A. Abdollahi, B. Hoffman-Liebermann, and D. A. Liebermann. 1993. Protooncogenes of the Fos/Jun family of transcription factors are positive regulators of myeloid differentiation. Mol. Cell. Biol. 13:841-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller, C., M. Alunni-Fabbroni, E. Kowenz-Leutz, X. Mo, M. Tommasino, and A. Leutz. 1999. Separation of C/EBPα-mediated proliferation arrest and differentiation pathways. Proc. Natl. Acad. Sci. USA 96:7276-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura, T., R. Datta, S. Kharbanda, and D. Kufe. 1991. Regulation of jun and fos gene expression in human monocytes by the macrophage colony-stimulating factor. Cell Growth Diff. 2:267-272. [PubMed] [Google Scholar]
- 30.Noti, J. D. 1997. Sp3 mediates transcriptional activation of the leukocyte integrin genes CD11c and CD11b and cooperates with c-jun to activate CD11c. J. Biol. Chem. 272:24038-24045. [DOI] [PubMed] [Google Scholar]
- 31.Okada, S., Z. Q. Wang, A. E. Grugoriadis, E. F. Wagner, and T. Ruden. 1994. Mice lacking c-Fos have normal hematopoietic stem cells but exhibit altered B-cell differentiation due to an impaired bone marrow environment. Mol. Cell. Biol. 14:382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabst, T., B. U. Mueller, N. Harakawa, C. Schoch, T. Haferlach, G. Behre, W. Hiddemann, D. E. Zhang, and D. G. Tenen. 2001. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat. Med. 7:444-451. [DOI] [PubMed] [Google Scholar]
- 33.Pabst, T., B. U. Mueller, P. Zhang, H. Radomska, S. Narravula, S. Schnittger, G. Behre, W. Hiddemann, and D. G. Tenen. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 34.Paradis, P., W. R. MacLellan, N. S. Belaguli, R. J. Schwartz, and M. D. Schneider. 1996. Serum response factor mediates AP-1-dependent induction of the skeletal α-actin promoter in ventricular myocytes. J. Biol. Chem. 271:10827-10833. [DOI] [PubMed] [Google Scholar]
- 35.Radomska, H., C. Huettner, P. Zhang, T. Cheng, D. Scadden, and D. G. Tenen. 1998. CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18:4301-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rana, B., D. Mischoulon, Y. Xie, N. Bucher, and S. Farmer. 1994. Cell-extracellular matrix interactions can regulate the switch between growth and differentiation in rat hepatocytes: reciprocal expression of C/EBPα and immediate-early growth response transcription factors. Mol. Cell. Biol. 14:5858-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy, V. A., A. Iwama, G. Iotzova, S. Schulz, A. Elsasser, R. K. Vangala, D. G. Tenen, W. Hiddemann, and G. Behre. 2002. Granulocyte inducer C/EBPα inactivates the myeloid master regulator PU.1: possible role in lineage commitment decisions. Blood 100:483-490. [DOI] [PubMed] [Google Scholar]
- 38.Rorth, P. 1994. Specification of C/EBP function during Drosophila development by the bZIP basic region. Science 266:1878-1881. [DOI] [PubMed] [Google Scholar]
- 39.Rosson, D., and T. G. O'Brien. 1998. AP-1 activity affects the levels of induced erythroid and megakaryocytic differentiation of K562 cells. Arch. Biochem. Biophys. 354:298-305. [DOI] [PubMed] [Google Scholar]
- 40.Sariban, E., T. Mitchell, A. Rambaldi, and D. Kufe. 1998. c-sis but not c-fos gene expression is lineage specific in human myeloid cells. Blood 71:488-493. [PubMed] [Google Scholar]
- 41.Sassone-Corsi, P., L. J. Ransone, W. W. Lamph, and I. M. Verma. 1988. Direct interaction between fos and jun nuclear oncoproteins: role of the ′leucine zipper' domain. Nature 336:692-695. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, M. L., R. M. Stone, D. Rakesh, S. H. Bernstein, and D. W. Kufe. 1990. Transcriptional and posttranscriptional regulation of c-jun expression during monocytic differentiation of human myeloid leukemic cells. J. Biol. Chem. 265:3320-3323. [PubMed] [Google Scholar]
- 43.Shuman, J. D., J. Cheong, and J. E. Coligan. 1997. ATF-2 and C/EBPα can form a heterodimeric DNA binding complex in vitro. Functional implications for transcriptional regulation. J. Biol. Chem. 272:12793-12800. [DOI] [PubMed] [Google Scholar]
- 44.Szabo, E., L. H. Preis, and M. J. Birrer. 1994. Constitutive c-jun expression induces partial macrophage differentiation in U937 cells. Cell Growth Diff. 5:439-446. [PubMed] [Google Scholar]
- 45.Tengku S. T.-M., T. Hughes, H. Ranki, A. Cryer, and D. Ramji. 2000. Differential regulation of macrophage CCAAT-enhancer binding protein isoforms by lipopolysaccharide and cytokines. Cytokine 12:1430-1436. [DOI] [PubMed] [Google Scholar]
- 46.Tornaletti, S., and G. P. Pfeifer. 1995. UV light as a footprinting agent: modulation of UV-induced DNA damage by transcription factors bound at the promoters of three human genes. J. Mol. Biol. 249:714-728. [DOI] [PubMed] [Google Scholar]
- 47.Ubeda, M., M. Vallejo, and J. F. Habener. 1999. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell. Biol. 19:7589-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umek, R., A. D. Friedman, and S. L. McKnight. 1991. CCAAT-enhancer binding protein: a component of a differentiation switch. Science 251:288-292. [DOI] [PubMed] [Google Scholar]
- 49.Unlap, T., C. C. Franklin, F. Wagner, and A. S. Kraft. 1992. Upstream regions of the c-jun promoter regulate phorbol ester-induced transcription in U937 leukemic cells. Nucleic Acids Res. 20:897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vangala, R. K., M. Heiss-Neumann, J. Rangatia, S. M. Singh, C. Schoch, D. G. Tenen, W. Hiddemann, and G. Behre. Master regulator PU.1 is downregulated by fusion protein AML1-ETO in t(8;21) leukemia. Blood, in press. [DOI] [PubMed]
- 51.Wang, X., E. Scott, C. Sawyers, and A. D. Friedman. 1999. C/EBPα bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 52.Wei, P., N. Inamdar, and W. V. Vedeckis. 1998. Transrepression of c-jun gene expression by the glucocorticoid receptor requires both AP-1 sites in the c-jun promoter. Mol. Endocrinol. 12:1322-1333. [DOI] [PubMed] [Google Scholar]
- 53.Westendorf, J. J., C. M. Yamamoto, N. Lenny, J. R. Downing, M. E. Selsted, and S. Hiebert. 1998. The t(8;21) fusion product AML-ETO associates with C/EBPα, inhibits C/EBPα dependent transcription, and blocks granulocytic differentiation. Mol. Cell. Biol. 18:322-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, C., E. M. Rougier-Chappman, J. P. Frederick, M. B. Datto, N. T. Liberati, J.-M. Li, and X. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by TGFβ. Mol. Cell. Biol. 19:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zagariya, A., S. Mungre, R. Lovis, M. Birrer, S. Ness, B. Thimmapaya, and R. Pope. 1998. Tumor necrosis factor α gene regulation: enhancement of C/EBPβ-induced activation by c-Jun. Mol. Cell. Biol. 18:2815-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, D. E., P. Zhang, N.-D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]