Abstract
Temperature-sensitive mutants of TFIIB that are defective for essential interactions were isolated. One mutation (G204D) results in disruption of a protein-protein contact between TFIIB and TATA binding protein (TBP), while the other (K272I) disrupts an interaction between TFIIB and DNA. The TBP gene was mutagenized, and alleles that suppress the slow-growth phenotypes of the TFIIB mutants were isolated. TFIIB with the G204D mutation [TFIIB(G204D)] was suppressed by hydrophobic substitutions at lysine 239 of TBP. These changes led to increased affinity between TBP and TFIIB. TFIIB(K272I) was weakly suppressed by TBP mutants in which K239 was changed to hydrophobic residues. However, this mutant TFIIB was strongly suppressed by conservative substitutions in the DNA binding surface of TBP. Biochemical characterization showed that these TBP mutants had increased affinity for a TATA element. The TBPs with increased affinity could not suppress TFIIB(G204D), leading us to propose a two-step model for the interaction between TFIIB and the TBP-DNA complex.
Transcription by RNA polymerase II requires a remarkably large number of accessory factors (33). Basal transcription factors are necessary to correctly position the polymerase at the promoter DNA. The assembly of the transcription complex has been studied predominantly in vitro, but the models derived from these experiments need to be validated by in vivo experiments.
A primary event in transcription is the binding of TATA binding protein (TBP) to the TATA element of the promoter (33). It is generally believed that much transcription regulation occurs at this step. Despite years of study, many questions remain about the TBP binding event. TBP is a saddle-shaped molecule, and DNA binds in a severely distorted manner to the concave face of the protein (27, 32) (Fig. 1). Kinetic and molecular studies indicate that TBP binding occurs in several steps, with bending of the DNA being a critical event (22, 23, 34, 36, 37, 42, 45). TBP may arrive at promoters as part of the larger TFIID complex, although it is not clear how the TBP-associated factors (TAFs) affect binding. In vivo, TBP binding is further complicated by the presence of histones, which can affect DNA accessibility and topology.
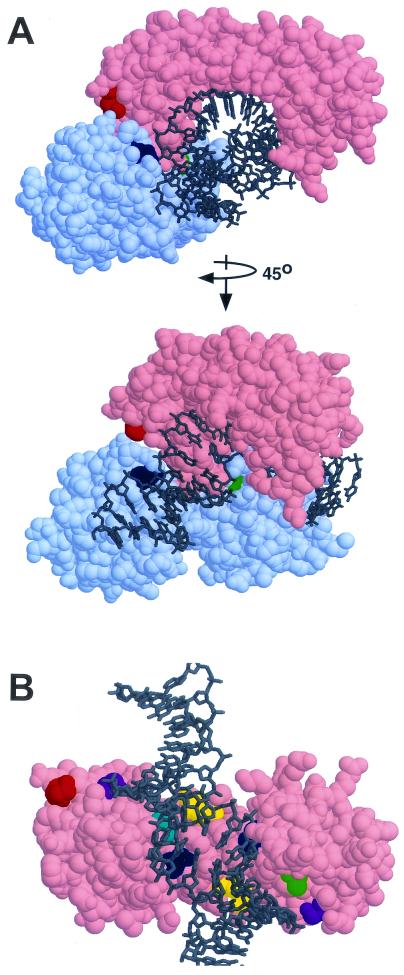
FIG. 1.
Positions of TBP and TFIIB mutants discussed in this paper. (A) Cocrystal structure of TBP, TFIIB, and DNA (31), but note that the amino acid numbers in reference 31 have been changed to those of yeast TBP and TFIIB. TBP is shown in pink, TFIIB is shown in light blue, and DNA is shown in gray. The top view is a perspective from upstream of the transcription complex. The bottom view is the same, except rotated 45° as indicated. TFIIB residue K272 (contactingthe DNA phosphate backbone) is shown in dark blue and G204 (contacting the TBP stirrup) is shown in green. TBP residue K239, which suppresses both TFIIB mutants, is shown in red. (B) TBP suppressor mutations on the DNA binding surface. Starting with the upper view in panel A, TFIIB was removed and the structure was rotated to show a view from the DNA perspective. For orientation, K239 is shown again in dark orange. T124 and T215 are shown in dark blue, V213 is shown in cyan, Q68 and Q158 are shown in yellow, S118 and S209 are shown in purple, and A100 and A101 are shown in green.
The basal transcription factor TFIIB interacts directly with TBP (33). TFIIB has three domains: an N-terminal zinc ribbon and two cyclin-fold domains (4, 6, 8, 19, 21, 46). The zinc ribbon interacts with RNA polymerase II, while the cyclin-fold domains interact with the TBP-DNA complex. A cocrystal structure of TBP, TFIIB, and DNA (31) (Fig. 1) shows that TFIIB makes protein-protein contacts with one of the two TBP “stirrups.” In addition, each cyclin fold of TFIIB contacts the DNA directly. One contacts DNA upstream of TBP, and the other contacts DNA downstream. The TFIIB-DNA contacts are made possible by the TBP-induced bending of DNA.
We generated mutants in TFIIB that were defective in either the TFIIB-TBP or TFIIB-DNA interaction. All of these mutants caused severe growth defects in vivo and were unable to form stable TBP-TFIIB-DNA complexes in vitro. We then selected for altered TBP alleles that could suppress the TFIIB defects. Interestingly, the TFIIB mutants were suppressed by distinct classes of TBP mutations. The most interesting TBP suppressor mutants were found on the DNA binding surface. Surprisingly, these mutants had increased affinity for a TATA element. Our results suggest that the TFIIB-TBP interaction occurs in two steps, one primarily mediated by the protein-protein interaction and the second involving the TFIIB-DNA interactions.
MATERIALS AND METHODS
DNA cloning.
The 3.2-kb EcoRI-ClaI fragment from plasmid pDW5462 (a gift of Mike Hampsey) (38) was cloned into pRS313 (41) to create pRS313-SUA7. The 3.2-kb XbaI-XhoI fragment from pRS313-SUA7 was cloned into pRS413 (41) to create pRS413-SUA7. pRS313-sua7(G204D) was isolated as a temperature-sensitive mutant following hydroxylamine mutagenesis of pRS313-SUA7 and screening for the inability to complement an sua7 deletion in YSB141. pRS313-sua7(K272I) was created by oligonucleotide-directed phagemid mutagenesis of pRS313-SUA7 by using the oligonucleotide SUA7-F (5′GATCAAAGAAACTGCAGGTANATCCCCTATTAC3′) followed by subcloning of the 1.6-kb BstBI fragment into pRS313. Other sua7 mutant alleles were generated by oligonucleotide-directed phagemid mutagenesis of pRS413-SUA7 by using oli-gonucleotides SUA7-B (5′GTGCTGACGTNTCCGGAGTNCAAAGTCTACCC3′) for the C24F and C27Y mutations, SUA7-C2 (5′CTTTGAAGGNGABATCAATGGAG3′) for K166I/T/R (a change from K to I, T, or R at position 166), SUA7-D (5′GATGTTGTATHTGCTCTATNTGGTCTAGTA3′) for C45Y/F, SUA7-E (5′ACAAGGATCCGAMNGGGTGAAACCACGGATA3′) for K98M/R, SUA7-F for K272T/I/R and I269T, and SUA7-G (5′TTTTAAGAGGCABGAGCGAAGATGGTTTC3′) for K217R/M. For protein expression in bacteria, the open reading frames of wild-type, G204D mutant, and K272I mutant Sua7 genes were amplified by PCR and cloned into pET-11d (11).
The plasmid carrying spt15-206(K239E) was a gift of Fred Winston (16). Dominant suppressors of the temperature-sensitive phenotype of sua7(G204D) or of the slow-growth phenotype of sua7(K272I) were isolated as described below from mutant TBP libraries N1 to N6 in vector YCplac22 (a gift of Kevin Struhl) (14) and from hydroxylamine-mutagenized plasmid pUN45-IID (9). The T215 and S209 changes in Spt15 were generated by oligonucleotide-directed phagemid mutagenesis of pUN45-IID by using oligonucleotide TBP-S209 (5′GTTGTTAATTTTTGTTAMCGGAAAGATTGTTC3′) or TBP-T215 (5′AAGATTGTTCTTKTGGGTGCAAAGCAA3′). For protein expression in bacteria, the open reading frames of wild-type and mutant Spt15 genes were amplified by PCR and cloned into pET-11d or pET-24a.
Yeast methods.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Standard methods were used for medium preparation, yeast transformation, and plasmid recovery (18). YDE11, YDE13, and L662 were gifts of Fred Winston (16). YSB141 was used for plasmid shuffling and complementation testing of SUA7 alleles. YDE11 and YDE13 were used for plasmid shuffling and complementation testing of SPT15 alleles. YSB170 and YSB287 were generated by plasmid shuffling of pRS313-sua7(K272I) and pRS313-sua7(G204D), respectively, into YSB141. YSB170 and YSB287 were transformed with pNKY1009 (3) to create the Trp− strains YSB288 and YSB299. Isolation of dominant suppressors of sua7(G204D) and sua7(K272I) was carried out by transformation of YSB289 or YSB288 with mutant TBP libraries or with hydroxylamine-treated pUN45-IID. Transformants were plated at 37°C (YSB289) or 30°C (YSB288), and colonies showing improved growth were selected. Plasmids carrying SPT15 were recovered and sequenced by standard methods.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| YSB141 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pDW5462] |
| YSB170 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS313-sua7(G204D)] |
| YSB287 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS313-sua7(K272I)] |
| YSB288 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 trp1::hisG-URA3-hisG his3Δ200 [pRS313-sua7(K272I)] |
| YSB289 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 trp1::hisG-URA3-hisG his3Δ200 [pRS313-sua7 (G204D)] |
| YSB299 | MATasua7Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS413-sua7(K272T)] |
| YDE11 | MATα spt15Δ::LEU2 ura3-52 leu2-3,112 trp1Δ1 his4-912δ lys2-128δ [pDE38-9] |
| YDE13 | MATa/α spt15Δ::LEU2/spt15Δ::LEU2 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his4-912δ/his4-912δ lys2-128δ/lys2-128δ [pDE38-9] |
| L662 | MATα spt15Δ101::LEU2 ura3-52 leu2Δ1 trp1Δ1 his4-917δ spt3-401 lys2-173r2 [pDE38-9] |
Protein methods.
Recombinant TBP and TFIIB proteins were produced in bacteria as previously described (8, 9, 11). Native gel electrophoresis was also carried out as previously described (7, 8). Gels containing 3 mM MgCl2 were used to assay the TBP-DNA complex, while the TBP-TFIIB-DNA complex was assayed with gels containing no magnesium. Equilibrium dissociation constants were calculated by assaying increasing concentrations of TBP (0.5 to 8 nM) by use of native gel electrophoresis. The fractional occupancy of the probe was measured by a phosphorimager, and data were plotted using the Langmuir isotherm (37). The Kd was derived from the slope of this line. Titrations were done in triplicate to allow determination of error.
RESULTS
Isolation of TFIIB mutant alleles.
In order to explore the functions of TFIIB, mutants with interesting phenotypes were isolated in two ways. First, randomly mutagenized TFIIB plasmids were screened for conditional growth phenotypes. Approximately 50,000 hydroxylamine-mutagenized clones were tested for the ability to grow at 30°C but not at 37 or 15°C. Four clones that were sensitive to cold were isolated. Sequencing of the genes revealed that all of the cold-sensitive alleles corresponded to a change of amino acid 62 from glutamate to lysine. This change is identical to that for the original sua7-1 allele that causes a shift in transcription start sites (38, 39). Biochemical characterization of this mutant has been presented elsewhere (11, 40).
A single temperature-sensitive allele was isolated in the screen which grew slowly even at 30°C (Fig. 2A and data not shown). The protein contained a mutation from glycine to aspartate at residue 204. Based on the cocrystal structure of Arabidopsis TBP, human TFIIB, and DNA, it was determined that TFIIB residue 204 (residue 192 in the human protein) is located near the TBP stirrup. In fact, the crystal structure predicts that leucine 189 of the yeast TBP stirrup makes a van der Waals contact with glycine 204 in TFIIB (31) (Fig. 1A). These amino acids are adjacent to TBP glutamate 188, which makes an essential hydrogen bond between the TBP stirrup and TFIIB (31).
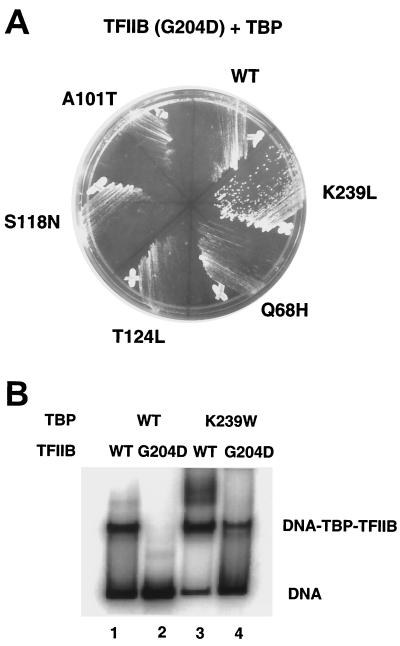
FIG. 2.
Suppression of TFIIB(G204D) by TBP(K239L). (A) Suppression in vivo. Low-copy plasmids carrying the indicated alleles of the TBP gene (SPT15) were transformed into yeast strain YSB289 that carries the TFIIB(G204D) allele. Transformants were streaked for single colonies on selective media and incubated for 3 days at 30°C. (B) Suppression in vitro. Native gel electrophoresis was carried out on binding reactions containing different combinations of TBP [wild type (WT), lanes 1 and 2, or TBP(K239W) mutant, lanes 3 and 4] and TFIIB [wild type (WT), lanes 1 and 3, or TFIIB(G204D) mutant, lanes 2 and 4].
In addition to the random mutagenesis, site-directed mutagenesis of conserved TFIIB residues was performed. We found that changes in the conserved cysteines of the TFIIB zinc finger (C24F, C27Y, C45Y, or C45F) resulted in proteins that could not support viability. Changes in several other conserved residues (G97R, K98 M/R, I269T, or K217R/M) did not result in any mutant phenotypes (data not shown).
Interestingly, each repeat of TFIIB contains a positively charged residue (arginine or lysine) that is absolutely conserved in all TFIIB genes characterized to date. These two residues are predicted to participate in ionic interactions with the phosphate backbone of the DNA (31) (Fig. 1A). Yeast TFIIB mutants with a change at residue K166 or K272 were shuffled into yeast. Interestingly, mutations in the second repeat that removed the positive charge (K272I or K272T) caused a dramatically slow-growth phenotype at 22 and 30°C and resulted in the inability to grow at 37°C (Fig. 3A and data not shown). A conservative substitution mutant (K272R) supported viability similarly to wild-type TFIIB at all temperatures, indicating that a positive charge at this residue is essential for function (data not shown). In contrast to a mutation in the second repeat, that in the first repeat (K166I or K166T) resulted in wild-type phenotypes at all temperatures (data not shown). In the TBP-TFIIB-TATA cocrystal structure (31), the lysine in the first repeat (arginine 154 in human TFIIB) contacts DNA downstream of the TATA box while the residue in the second repeat (arginine 248 in human TFIIB) contacts residues near the 5′ end of the TATA box. Our genetic results agree well with earlier biochemical results from studies using human TFIIB mutants (8) and indicate that these positive residues do not contribute equally to TFIIB function.
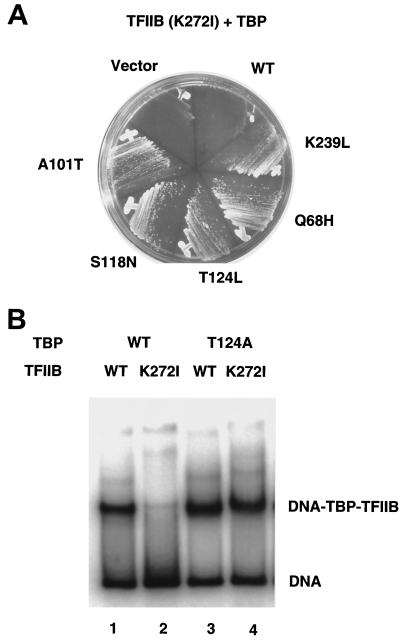
FIG. 3.
Suppression of TFIIB(K272I) by TBP mutant alleles. (A) Suppression in vivo. Low-copy plasmids carrying the indicated alleles of the TBP gene (SPT15) were transformed into yeast strain YSB288 that carries the TFIIB(K272I) allele. Transformants were streaked for single colonies on selective media and incubated for 3 days at 30°C. (B) Suppression in vitro. Native gel electrophoresis was carried out on binding reactions containing different combinations of TBP [wild type (WT), lanes 1 and 2, or TBP(T124A) mutant, lanes 3 and 4] and TFIIB [wild type (WT), lanes 1 and 3, or TFIIB(K272I) mutant, lanes 2 and 4]. Similar results were obtained with the S209N, Q68W, and S118N mutations.
TFIIB mutants are defective for formation of the TFIIB-TBP-DNA complex in vitro.
The TFIIB proteins with the G204D and K272I mutations [TFIIB(G204D) and TFIIB(K272I)] were expressed in bacteria, purified, and tested for the ability to interact with the TBP-DNA complex by use of native gel electrophoresis. Both mutants were severely compromised in their ability to produce a stable complex in a gel shift assay (Fig. 2B and 3B). Under these conditions, the TBP-DNA complex is not seen, but the TBP-TFIIB-DNA complex is stable. Based on the cocrystal structure of the TBP-TFIIB-DNA complex (31) (Fig. 1A), the G204D defect is due to defective protein interaction between TFIIB and TBP. In contrast, the unstable binding of K272I is the result of a loss of interaction between TFIIB and the DNA phosphate backbone. These two mutants with distinct defects were used for further genetic experiments.
TBP suppressors of TFIIB G204D mutation.
Because the conditional phenotypes of the TFIIB mutants were due to defects in the ability to form a stable TFIIB-TBP-DNA complex, we decided to screen for mutant TBP alleles that would suppress the temperature sensitivity of TFIIB. We predicted that these would increase the interaction between TBP and TFIIB. Because such TBP alleles would involve a gain of function and should be dominant, the screen was performed by transforming mutant TBP libraries into yeast strains that contained a wild-type copy of the TBP gene. The TBP libraries were mutated on a regional codon basis, and mutations were targeted primarily to non-DNA binding regions of TBP (14).
The strain with the TFIIB(G204D) mutant grows very slowly at 30°C and not at all at 37°C (Fig. 2A). We screened 10,000 to 20,000 TBP mutants from each of six localized mutant libraries for the ability to restore growth at 37°C to the strain containing TFIIB(G204D). Each library represents a highly enriched collection of mutants in a limited region of TBP (14). Strikingly, no suppressing plasmids were obtained from five of the libraries, while over 100 were obtained from a library containing mutations in TBP residues 226 to 240. Forty-eight of the suppressor TBP genes were sequenced. Remarkably, every single suppressing allele involved an amino acid change at lysine 239 (Table 2) (Fig. 2B). Furthermore, all of the changes at K239 were to hydrophobic residues. Most were changes to leucine, isoleucine, or methionine, although phenylalanine and tryptophan substitutions were also isolated. Although some suppressor alleles contained amino acid substitutions in addition to that of lysine 239, these changes followed no pattern and were not required for suppression.
TABLE 2.
TBP suppressors of TFIIB mutants
| TBP allele | Sourcea | Suppression of sua7 (K272I) at 30°C | Suppression of sua7 (G204D) at 37°C | Phenotypeb |
|---|---|---|---|---|
| K239M | Cormack N5 | Very weak | Moderate | Weak TS |
| K239L | Cormack N5 | Very weak | Strong | Weak TS |
| K239F | Cormack N5 | Very weak | Strong | Weak TS |
| K239I | Cormack N5 | None | Strong | Very Weak TS |
| K239W | Cormack N5 | None | Weak | Weak TS |
| Q68H | Cormack N6 | Strong | None | WT |
| Q68W | Cormack N6 | Strong | None | Weak TS, weak Gal− |
| T124V | Cormack N1 | Strong | None | Very weak TS |
| T124L | Cormack N1 | Strong | None | Strong TS, Gal− |
| T124A | Cormack N1 | Strong | Not tested | Strong TS, Gal− |
| T215V | Site directed | Strong | None | Weak TS |
| T215L | Site directed | Weak | None | Weak TS |
| S118T | Cormack N1 | Weak | None | Very weak TS |
| S118N | Cormack N1 | Strong | None | Weak TS |
| S209T | Site directed | Strong | None | Wild type |
| S209N | Site directed | Strong | None | Very weak TS |
| S209K | Site directed | None | None | Lethal |
| A101T | Hydroxylamine | Strong | None | Weak TS |
| A101V | Hydroxylamine | Weak | None | Strong TS, Gal− |
| A100P | Hydroxylamine | Strong | None | Strong TS |
| V2131 | Hydroxylamine | Very weak | None | Weak TS |
| F152W, Q158W | Cormack N3 | Weak | None | TS, weak Gal− |
See reference 14 for details of Cormack libraries. Site directed, site-directed mutagenesis.
TS, temperature sensitive; WT, wild type; Gal−, unable to grow on 2% galactose as a carbon source.
TBP suppressors of TFIIB(K272I).
The TFIIB(K272I) mutant reduces the stability of the TFIIB-TBP-DNA complex due to presumed loss of ionic interactions between the lysine and the DNA phosphate backbone. However, our characterization preceded the development of the crystal structure and the discovery of TFIIB-DNA contacts. Under the assumption that K272 was involved in contacts with TBP, a screen for TBP suppressors of the TFIIB(K272I) mutant was carried out as described above for the TFIIB(G204D) mutant, except that the selection was done at the semipermissive temperature of 30°C. Surprisingly, several dominant suppressing TBP alleles associated with various parts of the protein were isolated (Table 2 and Fig. 3B).
Sequencing of the TBP suppressors of TFIIB(K272I) revealed that they fell into two classes. The first class involved changes at lysine 239 of TBP. As previously observed for suppressors of TFIIB(G204D), changes of the lysine to the hydrophobic residue leucine, methionine, or phenylalanine improved growth in the TFIIB(K272I) strain (Fig. 3B). However, this class of TBP alleles contained the weakest suppressing alleles isolated.
Surprisingly, the second class of suppressors all mapped to the DNA binding face of TBP. Specific changes were isolated at Q68, T124, S118, and F152/Q158 (Table 2). Each of these mutated residues is predicted to make protein-DNA contacts in the cocrystal structure (17, 27, 31, 35, 44) (Fig. 1B). These results were particularly astonishing for two reasons. First, the vast majority of published amino acid substitutions in the DNA binding surface of TBP abolish binding to TATA elements. Second, the mutagenized TBP libraries used in this screen were designed to primarily target the non-DNA binding surfaces of TBP.
Since TBP is a pseudosymmetric protein, we asked whether suppressor amino acid changes in one repeat would also suppress TFIIB(K272I) when constructed in the corresponding residue of the other repeat. In each case, suppression could be mediated by the same amino acid substitution in either repeat (Table 2). For example, both Q68W and Q158W were isolated from the codon-mutagenized libraries. T124 in the first TBP repeat corresponds to T215 in the second repeat. Changing either of these residues to either valine or leucine resulted in suppression of TFIIB(K272I). Mutation of either S118 or S209 to asparagine or threonine had a similar effect. Therefore, the two TBP repeats appear to make roughly equivalent contributions to the suppressor phenotype.
Since the libraries used in the original screen were not mutagenized in large regions of the TBP DNA binding surface, the entire TBP gene was mutagenized using hydroxylamine and the screen for suppressors of TFIIB(K272I) was repeated. Again, suppressors were found in the DNA binding face of TBP. The mutations V213I, A100P, A101T, and A101V were isolated in this screen. These residues were not mutagenized in the libraries of the original screen. Interestingly, these mutations would increase the size of the hydrophobic side chains facing the TATA element minor groove.
Although the TBPs with mutations at lysine 239 suppressed both the TFIIB allele that affects protein-protein interactions and the allele that disrupts the TFIIB-DNA contact, this was not true of the TBP binding face mutants. When transformed into the TFIIB(G204D) strain, these TBP alleles did not cause any improvement in cell growth. Therefore, there is allele specificity in the suppression pattern.
Properties of the TBP(K239) suppressors of TFIIB mutants.
In order to characterize the mechanism of the TBP suppressors, the proteins were produced in Escherichia coli and tested by gel shift assay. The TBP(K239) mutants were tested for the ability to bind the adenovirus major late promoter (AdMLP) TATA element. The DNA binding of the mutants was indistinguishable from that of wild-type TBP (data not shown). The ability to form the DNA-TBP-TFIIB complex was also tested by gel shift analysis (Fig. 2B). Whereas wild-type TBP could bind wild-type TFIIB, no stable complex was observed with TFIIB(G204D). The TBP(K239W) protein bound wild-type TFIIB to form a complex with mobility that was identity to that of the wild type. However, the suppressor TBP protein could also stably bind the TFIIB(G204D) protein. Therefore, the in vivo suppression is apparently a direct result of improved interactions between the suppressor TBP and mutant TFIIB.
Suppression of TFIIB mutants by TBP(K239) mutants was predicted to occur by an increase in the affinity of the protein-protein interactions between TBP and TFIIB. Since TBP residue 239 is not predicted to be close to either TFIIB residue 204 or K272 (31) (Fig. 1), it seemed likely that the change of lysine 239 to a hydrophobic residue either created a new favorable contact or removed an unfavorable contact between the proteins. Therefore, the TBP(K239) mutant should also show increased affinity for wild-type TFIIB. In support of this hypothesis, gel shifts using equivalent amounts of TBP (as determined by Coomassie staining and the ability to form a DNA-TBP complex) consistently detected more DNA-TBP-TFIIB complex with the TBP(K239) mutant than with the wild type (Fig. 2B and data not shown).
Properties of the TBP-DNA binding face suppressors of TFIIB mutants.
Although the TBP(K239) mutants were expressed in E. coli at levels comparable to that of wild-type TBP, the DNA binding face mutants were apparently more toxic to the bacteria. When ampicillin was used as the selectable marker for the expression plasmid, the mutant plasmids were lost at high frequency early in log phase growth. The TBP binding face mutants were recloned into an expression vector conferring kanamycin resistance, and this resulted in low but satisfactory levels of protein expression (data not shown).
Because all previously reported TBP mutations in the DNA binding surface have been found to drastically reduce binding to the TATA element, the suppressor TBP proteins were first tested in a gel shift assay. Remarkably, the suppressor proteins did not show any reduction in affinity for the AdMLP TATA element (Fig. 4A). This was particularly surprising because some of the changed amino acids are predicted to directly contact DNA bases (31) (Fig. 1B). For example, T124 and T215 are predicted to make two of the rare hydrogen bonds in the TBP-DNA interface, yet changing either of these residues to alanine does not reduce binding.
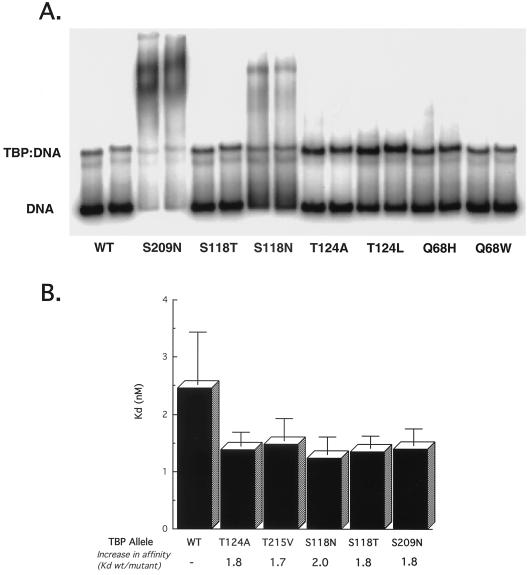
FIG. 4.
TBP suppressors of TFIIB(K272I) have increased DNA binding affinity. (A) Native gel electrophoresis of wild-type and mutant TBP. Thirty nanograms of each protein was incubated with an AdMLP probe. Duplicate lanes for each protein are shown. (B) Kd values for wild-type and mutant TBPs. Titrations of TBP binding to the AdMLP were assayed using native gel electrophoresis, and the Kds were calculated as described in Materials and Methods. Kds for Q68H and Q68W were not significantly different from that for wild-type TBP.
Three possible mechanisms of TFIIB suppression by the TBP binding face mutants were considered when the in vitro tests were designed. First, it was possible that the mutations actually affected TBP dimerization rather than DNA binding. TBP dimers have been demonstrated both in vitro and in vivo, and it has been proposed that the transition to monomer form could be limiting for transcription (12, 13, 24, 25, 43). The ability of the suppressor TBPs to dimerize was tested by glutaraldehyde cross-linking (12, 43). Although this is not a quantitative assay, the TBP mutants all appeared to form dimers in vitro (data not shown). Also, the suppressor mutations on the crystal structure of the yeast TBP dimer are not located at the interface of the TBP molecules (10). Therefore, it is very unlikely that suppression is due to effects on TBP dimerization.
A second possibility was that the suppressor mutations could cause differences in the DNA bending induced by TBP. Since TFIIB recognizes both TBP and the bent DNA, mutations in TBP that change the angle or stability of the bend might increase the affinity of TFIIB for the TBP-DNA complex. To test the bend angle of the TBP-DNA complexes, gel shifts were performed using circularly permuted probes that placed the center of the bend (i.e., the TATA element) at different positions within the DNA fragment (42). TBP-DNA complexes with the mutant proteins migrated in the same way as wild-type complexes, suggesting that the overall shape of the TBP-DNA complex was not affected by the DNA binding face mutations (data not shown).
A third model for suppression postulates that binding energy lost due to the missing TFIIB-DNA contact can be compensated for by stronger contacts between TBP and the DNA (1, 2). The TBP suppressors would therefore be predicted to have increased affinity for TATA elements. To test this, quantitative binding experiments were performed. Equivalent amounts of protein were tested for binding to a consensus TATA element. Binding was plotted using the Langmuir isotherm formula (37), and equilibrium dissociation constants were calculated from the slope of the curves (Fig. 4B). Wild-type TBP had a Kd of approximately 2.5 nM, in good agreement with other published estimates. Most of the mutants did exhibit increased affinity (about twice that of the wild-type protein).
Phenotypes of TBP suppressor mutants.
The TBP suppressors of TFIIB mutants were all dominant, since they were isolated in the presence of the wild-type TBP. In order to determine whether these mutants had their own recessive phenotypes, plasmid shuffling was used to introduce them into cells in the absence of any other TBP alleles (Table 2). Several of the mutants with increased affinity (those with the A100P, A101V, A101T, T124A, T124L, and F152W/Q158W mutations) were temperature sensitive and grew poorly on galactose as a carbon source. Many of the others grew more slowly than the wild type at 37°C. This suggests that increased affinity for the TATA element may actually be detrimental to proper gene regulation and growth. Alternatively, these mutations may adversely affect transcription by RNA polymerase I or polymerase III.
DISCUSSION
This study began with the characterization of several TFIIB mutants. Two interesting mutants that caused a severe growth defect in vivo and failed to form a stable TBP-TFIIB-DNA complex in vitro were found. However, the two TFIIB mutants were defective for distinct TFIIB interactions. One disrupts the interaction with TBP, while the other disrupts the TFIIB-DNA interaction.
The TFIIB(G204D) protein is mutated in a residue that makes a van der Waals contact with TBP. The change of glycine to aspartate is predicted to disturb the protein-protein interface between the two factors. When we selected for mutations in TBP that could compensate for this reduction in affinity, we isolated multiple amino acid substitutions at lysine 239 of TBP. Interestingly, all of the changes were to hydrophobic amino acids.
In the TBP-TFIIB-TATA cocrystal structure, the residue corresponding to K239 (K197 in Arabidopsis TBP) hangs down from the C-terminal H1′ helix of TBP and makes a salt bridge with residue D243 of human TFIIB (31) (Fig. 1A). Interestingly, the corresponding TFIIB residue in the yeast protein is lysine 267. This raises the possibility that the TBP suppressors work by removing some charge repulsion between yeast TBP K239 and TFIIB K267, thereby strengthening the TBP-TFIIB interface. One might ask why no suppressors were isolated that change K239 to an acidic residue. It should be noted that TFIIB K267 is surrounded by several other acidic residues that may clash with a negatively charged residue at TBP K239.
Interestingly, a mutation at lysine 239 of TBP has previously been identified in a screen of TBP alleles that can suppress the Spt− phenotype of the spt3-401 allele (15). The TBP(K239E) allele does not have an Spt− phenotype by itself, and the TBP(K239E) mutant could not suppress the temperature-sensitive phenotypes of the TFIIB mutations described in this report (data not shown). Furthermore, the TBP(K239L) and TBP(K239W) alleles, which do suppress the TFIIB mutations, are unable to suppress the Spt− phenotype of an spt3-401 strain. In fact, double-mutant cells [spt3-401, spt15(K239I or K239W)] had a stronger Spt− phenotype (assayed with the his4-912δ and lys2-128δ markers) than cells containing only the spt3 mutation (data not shown). This raises the possibility that the effects of the interacting Spt15 and Spt3 mutations are actually mediated by influencing the TBP-TFIIB interaction. Perhaps TFIIB and Spt3 compete for binding to TBP.
The second interesting TFIIB mutant characterized here was that with the K272I substitution. TFIIB contains two cyclin-fold domains that have significant sequence similarity to each other. Each TFIIB repeat contains a highly conserved pair of residues: a glycine followed by a positively charged residue. The TBP-TFIIB-DNA cocrystal structure shows that the positively charged residues make hydrogen bonds with the phosphate backbone of the DNA (31) (Fig. 1A). The N-terminal repeat contacts DNA downstream of the TATA box, while the C-terminal repeat contacts DNA upstream. Human TFIIB mutants carrying changes in the glycine-arginine pair have been analyzed previously (8). Mutations in the first-repeat pair had little effect in vitro, while mutations in the second-repeat pair led to defects in the formation of the TBP-TFIIB-DNA complex and a reduced ability to support transcription in vitro. In the present study, we provide further evidence that the DNA contact made by the C-terminal TFIIB is important in vivo (leading to the slow growth phenotype of the yeast TFIIB mutants with the K272I and K272T substitutions), while similar changes in the first-repeat residues have little effect.
When TBP suppressors of TFIIB(K272I) were isolated, two classes of mutants were recovered. The first comprised the same TBP mutants that suppressed TFIIB(G204D), i.e., TBPs with lysine 239 changed to a hydrophobic residue. The suppressing phenotype of these TBP alleles can be rationalized by the same mechanism proposed for the suppression of TFIIB(G204D). The TFIIB(K272I) mutant has lost stability because the contact between K272 and DNA is no longer made. Since stability of the interaction between TFIIB and the TBP-DNA complex is dependent upon both protein-protein and protein-DNA interactions, loss of binding energy in one can be compensated for by an increase in the other (1, 2). Therefore, the improved TFIIB contact made by the TBP mutants with changes at K272 compensates for the lost TFIIB-DNA contact in TFIIB(K272I).
The second class of TBP mutants that suppress TFIIB(K272I) mutant phenotypes mapped to the DNA binding surface of TBP (Fig. 1B). The suppression observed with this group was notably stronger than that observed with the TBP(K239) class. It was surprising to isolate mutants on the DNA binding face of TBP, since most previously reported mutations on this highly conserved surface have detrimental effects on TBP binding to DNA. Remarkably, we found that many of these mutant TBP proteins actually had an increased affinity for a consensus TATA element. The proteins with the T124A, T215V, S118N/T, and S209N mutations all bound to the AdMLP with approximately twofold greater affinity than did wild-type TBP. We did not observe increased binding of the proteins with Q68H/W mutations, although it is certainly possible that these mutants would show increased affinity had we tested a variety of TATA elements. We did not assay the proteins with mutations at A100 or A101. However, J. V. Spencer and K. M. Arndt (personal communication) also isolated the A100P mutant in experiments designed to identify TBPs that increased transcription from a “reverse” TATA element (26). They found that this mutant also increased affinity for the AdMLP by two- to threefold.
As discussed above, the mechanism of TFIIB suppression by the TBP mutants is easily understood by considering that the stability of the TBP-TFIIB-DNA complex derives from the sum of all of its protein-protein and protein-DNA contacts (1, 2). The TFIIB(K272I) mutant loses one TFIIB-DNA interaction, but this can be compensated for by an increase in the affinity of the TBP-TFIIB interaction (via the mutations at K272 of TBP) or the TBP-DNA interaction (via the DNA binding face mutations). It is not clear why the particular amino acid substitutions that were isolated lead to the increased affinity of TBP for DNA. However, it is striking that all of the affected residues make contacts with the DNA.
In some cases (T124L/A, T215V/L, V213I, and Q68H/W), there is an increase in the hydrophobicity of the DNA binding surface. Therefore, it is possible that affinity is increased by making solvation of free TBP more costly energetically, i.e., shifting equilibrium towards bound TBP by making free TBP less favorable. In vivo, it is possible that changing the TBP binding face disrupts a protein-protein interaction that occurs when TBP is not bound to DNA. Both TBP dimerization (12, 13, 24, 25, 43) and an inhibitory TBP-TAF1 interaction (5, 28, 29) occur via the TBP DNA binding face. However, we found that TBP dimerization was not strongly affected in our mutants (data not shown), and our in vitro results showing increased affinity were obtained with purified recombinant TBPs lacking any TAFs.
The S118T/N and S209T/N mutations are not easily explained. These serines make hydrogen bonds to the phosphate backbone. The substituted amino acids can still form hydrogen bonds, and perhaps the difference in side-chain conformation strengthens the bonds. Similarly, it is not clear how the A100P and A101T/V mutations affect DNA binding. The position corresponding to A100 in the second repeat is a proline (P191), and it has been suggested that this asymmetry contributes to the directionality of TBP binding (26). These residues immediately follow the phenylalanines that intercalate between the DNA bases to distort and bend the TATA element. It seems likely that the A100 and A101 positions may influence this intercalation and bending process.
Interestingly, several of the DNA binding face mutants that we isolated occur naturally in the TBP protein of the malaria-causing parasite Plasmodium falciparum. The Q68H, A101T, S118N, and V213I substitutions all appear in the wild-type TBP of P. falciparum (30). Although this TBP is highly diverged from TBPs of other eukaryotes, it is unlikely that these particular substitutions would arise by chance. The P. falciparum genome is extraordinarily A-T rich (90%), suggesting that its TBP must have evolved extremely selective binding for discrimination of true TATA elements from related sequences.
Based on our model of suppression by energetic compensation, one is forced to ask why the TBP mutants with increased affinity for DNA suppress TFIIB(K272I) but not TFIIB(G204D). In contrast, the TBP(K239) mutants suppress both classes of TFIIB mutants. We believe that this allele specificity provides genetic evidence for multiple steps in the formation of the TBP-TFIIB-DNA complex. In one step, a TBP-TFIIB interaction may occur that is not sensitive to the DNA binding state of TBP. The TFIIB(G204D) mutant disrupts this protein-protein interaction and is suppressed only by the TBP(K239) mutants that increase the TBP-TFIIB affinity. Another step may involve DNA contact by both TBP and TFIIB. TFIIB(K272I) decreases the interaction between DNA and the complex formed in the first step, but this can be compensated for by the additional binding affinity provided by the TBP DNA binding face mutants. This step can also be suppressed (at least partially) by the TBP(K239) mutants because the intermediate complex formed in the first step is necessarily a precursor to the second step.
At least three molecular models are consistent with this proposal. In the first, TBP and TFIIB form a complex before TBP completes binding to the DNA. This could mean that TBP and TFIIB bind first as a heterodimer off the DNA. Alternatively, since TBP binding to DNA appears to occur in multiple steps, TFIIB may first recognize an intermediate form of the TBP-DNA complex. Most models of TBP binding to DNA propose one or more intermediates, perhaps with varying degrees of DNA bending. The interaction of K272 of TFIIB with DNA may occur in only one of the later TBP-DNA intermediates.
A third model that might explain our data is one in which TFIIB binding to the preformed TBP-DNA complex occurs in two steps. Some support for this idea comes from comparison of the nuclear magnetic resonance structure of the free TFIIB core domain (4, 20) with TFIIB within the TBP-TFIIB-DNA cocrystal structure (31). The two structures are quite similar when the individual TFIIB cyclin domains are compared, but the relative orientation of the domains is quite different. This suggests that TFIIB might undergo a significant conformation change upon binding to the TBP-DNA complex. TFIIB binding could occur in two steps, with the first mediated primarily by the protein-protein contact with TBP. The second step could then involve a conformation change that allows the TFIIB-DNA contact to form. Future biophysical experiments will help to test these models.
Acknowledgments
We thank K. Dollard and H. Zhou for technical help in the early stages of this study; F. Winston, M. Hampsey, and K. Struhl for strains and plasmids; S. Tan (Pennsylvania State University) for providing a PDB file of the predicted TBP-TFIIA-TFIIB-DNA complex; and K. Arndt (University of Pittsburgh) for communicating unpublished results.
This work was supported by NIH grant GM46498. S.B. is a scholar of the Leukemia and Lymphoma Society.
R.M.B. and J.D. contributed equally to this work.
REFERENCES
- 1.Ackers, G. K., A. D. Johnson, and M. A. Shea. 1982. Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. USA 79:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackers, G. K., M. A. Shea, and F. R. Smith. 1983. Free energy coupling within macromolecules. The chemical work of ligand binding at the individual sites in co-operative systems. J. Mol. Biol. 170:223-242. [DOI] [PubMed] [Google Scholar]
- 3.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagby, S., S. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. [DOI] [PubMed] [Google Scholar]
- 5.Bai, Y., G. M. Perez, J. M. Beechem, and P. A. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barberis, A., C. W. Muller, S. C. Harrison, and M. Ptashne. 1993. Delineation of two functional regions of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90:5628-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 8.Buratowski, S., and H. Zhou. 1993. Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90:5633-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski, S., and H. Zhou. 1992. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science 255:1130-1132. [DOI] [PubMed] [Google Scholar]
- 10.Chasman, D. I., K. M. Flaherty, P. A. Sharp, and R. D. Kornberg. 1993. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl. Acad. Sci. USA 90:8174-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, E. J., and S. Buratowski. 1999. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 274:25807-25813. [DOI] [PubMed] [Google Scholar]
- 12.Coleman, R. A., and B. F. Pugh. 1997. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc. Natl. Acad. Sci. USA 94:7221-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman, R. A., A. K. Taggart, S. Burma, J. J. Chicca II, and B. F. Pugh. 1999. TFIIA regulates TBP and TFIID dimers. Mol. Cell 4:451-457. [DOI] [PubMed] [Google Scholar]
- 14.Cormack, B. P., and K. Struhl. 1993. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science 262:244-248. [DOI] [PubMed] [Google Scholar]
- 15.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 16.Eisenmann, D. M., C. Dollard, and F. Winston. 1989. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell 58:1183-1191. [DOI] [PubMed] [Google Scholar]
- 17.Geiger, J. H., S. Hahn, S. Lee, and P. B. Sigler. 1996. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272:830-836. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif. [PubMed]
- 19.Ha, I., S. Roberts, E. Maldonado, X. Sun, L. U. Kim, M. Green, and D. Reinberg. 1993. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7:1021-1032. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi, F., R. Ishima, D. Liu, K. I. Tong, S. Kim, D. Reinberg, S. Bagby, and M. Ikura. 1998. Human general transcription factor TFIIB: conformational variability and interaction with VP16 activation domain. Biochemistry 37:7941-7951. [DOI] [PubMed] [Google Scholar]
- 21.Hisatake, K., R. G. Roeder, and M. Horikoshi. 1993. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature 363:744-747. [DOI] [PubMed] [Google Scholar]
- 22.Hoopes, B. C., J. F. LeBlanc, and D. K. Hawley. 1998. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 277:1015-1031. [DOI] [PubMed] [Google Scholar]
- 23.Hoopes, B. C., J. F. LeBlanc, and D. K. Hawley. 1992. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J. Biol. Chem. 267:11539-11547. [PubMed] [Google Scholar]
- 24.Jackson-Fisher, A. J., S. Burma, M. Portnoy, L. A. Schneeweis, R. A. Coleman, M. Mitra, C. Chitikila, and B. F. Pugh. 1999. Dimer dissociation and thermosensitivity kinetics of the Saccharomyces cerevisiae and human TATA binding proteins. Biochemistry 38:11340-11348. [DOI] [PubMed] [Google Scholar]
- 25.Jackson-Fisher, A. J., C. Chitikila, M. Mitra, and B. F. Pugh. 1999. A role for TBP dimerization in preventing unregulated gene expression. Mol. Cell 3:717-727. [DOI] [PubMed] [Google Scholar]
- 26.Juo, Z. S., T. K. Chiu, P. M. Leiberman, I. Baikalov, A. J. Berk, and R. E. Dickerson. 1996. How proteins recognize the TATA box. J. Mol. Biol. 261:239-254. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. L., D. B. Nikolov, and S. K. Burley. 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365:520-527. [DOI] [PubMed] [Google Scholar]
- 28.Kokubo, T., M. J. Swanson, J. I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokubo, T., S. Yamashita, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc. Natl. Acad. Sci. USA 91:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAndrew, M. B., M. Read, P. F. Sims, and J. E. Hyde. 1993. Characterisation of the gene encoding an unusually divergent TATA-binding protein (TBP) from the extremely A+T-rich human malaria parasite Plasmodium falciparum. Gene 124:165-171. [DOI] [PubMed] [Google Scholar]
- 31.Nikolov, D. B., H. Chen, E. D. Halay, A. A. Usheva, K. Hisatake, D. K. Lee, R. G. Roeder, and S. K. Burley. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119-128. [DOI] [PubMed] [Google Scholar]
- 32.Nikolov, D. B., S. H. Hu, J. Lin, A. Gasch, A. Hoffmann, M. Horikoshi, N. H. Chua, R. G. Roeder, and S. K. Burley. 1992. Crystal structure of TFIID TATA-box binding protein. Nature 360:40-46. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 34.Parkhurst, K. M., R. M. Richards, M. Brenowitz, and L. J. Parkhurst. 1999. Intermediate species possessing bent DNA are present along the pathway to formation of a final TBP-TATA complex. J. Mol. Biol. 289:1327-1341. [DOI] [PubMed] [Google Scholar]
- 35.Patikoglou, G. A., J. L. Kim, L. Sun, S. H. Yang, T. Kodadek, and S. K. Burley. 1999. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13:3217-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Howard, G. M., P. A. Weil, and J. M. Beechem. 1995. Yeast TATA binding protein interaction with DNA: fluorescence determination of oligomeric state, equilibrium binding, on-rate, and dissociation kinetics. Biochemistry 34:8005-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petri, V., M. Hsieh, and M. Brenowitz. 1995. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry 34:9977-9984. [DOI] [PubMed] [Google Scholar]
- 38.Pinto, I., D. E. Ware, and M. Hampsey. 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68:977-988. [DOI] [PubMed] [Google Scholar]
- 39.Pinto, I., W. H. Wu, J. G. Na, and M. Hampsey. 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269:30569-30573. [PubMed] [Google Scholar]
- 40.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starr, D. B., B. C. Hoopes, and D. K. Hawley. 1995. DNA bending is an important component of site-specific recognition by the TATA binding protein. J. Mol. Biol. 250:434-446. [DOI] [PubMed] [Google Scholar]
- 43.Taggart, A. K., and B. F. Pugh. 1996. Dimerization of TFIID when not bound to DNA. Science 272:1331-1333. [DOI] [PubMed] [Google Scholar]
- 44.Tan, S., Y. Hunziker, D. F. Sargent, and T. J. Richmond. 1996. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381:127-151. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, X., and W. Herr. 2002. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell 108:615-627. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, W., Q. Zeng, C. M. Colangelo, M. Lewis, M. F. Summers, and R. A. Scott. 1996. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 3:122-124. [DOI] [PubMed] [Google Scholar]