Abstract
Expression of the human cytomegalovirus UL4 gene is inhibited by translation of a 22-codon-upstream open reading frame (uORF2). The peptide product of uORF2 acts in a sequence-dependent manner to inhibit its own translation termination, resulting in persistence of the uORF2 peptidyl-tRNA linkage. Consequently, ribosomes stall at the uORF2 termination codon and obstruct downstream translation. Since termination appears to be the critical step affected by translation of uORF2, we examined the role of eukaryotic release factors 1 and 3 (eRF1 and eRF3) in the inhibitory mechanism. In support of the hypothesis that an interaction between eRF1 and uORF2 contributes to uORF2 inhibitory activity, specific residues in each protein, glycines 183 and 184 of the eRF1 GGQ motif and prolines 21 and 22 of the uORF2 peptide, were found to be necessary for full inhibition of downstream translation. Immunoblot analyses revealed that eRF1, but not eRF3, accumulated in the uORF2-stalled ribosome complex. Finally, increased puromycin sensitivity was observed after depletion of eRF1 from the stalled ribosome complex, consistent with inhibition of peptidyl-tRNA hydrolysis resulting from an eRF1-uORF2 peptidyl-tRNA interaction. These results reveal the paradoxical potential for interactions between a nascent peptide and eRF1 to obstruct the translation termination cascade.
Expression of the human cytomegalovirus UL4 gene is controlled by an unusual translational mechanism in which termination is the critical regulatory step (1, 21, 28, 36). The UL4 coding region is present on a transcript that contains three upstream open reading frames (uORFs), the second of which (uORF2) encodes a 22-amino-acid peptide that inhibits downstream translation in cis. By a mechanism that depends on the sequence of the encoded peptide, uORF2 inhibits termination at its own stop codon, resulting in a persistent covalent linkage between the nascent uORF2 peptide and the terminal cognate tRNAPro. The termination arrest causes translating ribosomes to stall over the uORF2 stop codon both in cell-free translation assays and in cells, setting up a blockade that prevents scanning ribosomes from leaking past uORF2 to gain access to the UL4 start codon.
Translation termination is a complex process that involves stop codon recognition, peptidyl-tRNA hydrolysis, and release of ribosome from the message (29). While the termination process is incompletely characterized mechanistically, the participation of protein factors has been elucidated. The class I eukaryotic release factor (eRF1) is responsible for stop codon recognition, ribosome binding, and triggering hydrolysis of the peptidyl-tRNA bond (14). Meanwhile, eRF3 stimulates eRF1 activity in a GTP-dependent fashion (50). In the uORF2 system, the persistent linkage of the nascent uORF2 peptide to tRNAPro indicates that translation of uORF2 interferes with or prior to the peptidyl-tRNA hydrolysis step in the termination cascade. Since translational inhibition depends on the presence of a stop codon at the end of uORF2 (9) and since eRF1 confers stop codon responsiveness on the termination process, we hypothesized that eRF1 might be involved in the uORF2 inhibitory mechanism.
The crystal structure of eRF1 reveals that the protein is composed of three domains; an N-terminal domain implicated in stop codon binding (3, 8, 16), a C-terminal domain responsible for interaction with eRF3 (12, 17, 34), and a middle domain resembling the tRNA acceptor stem and believed to interact with the tRNA linked to the nascent peptide in the ribosomal P site (42). The GGQ minidomain, located in an exposed area at the extreme tip of the middle domain, is invariant in all known class I release factors and is hypothesized to enable hydrolysis of the peptidyl-tRNA bond by the peptidyl transferase center of the ribosome (19, 42). Mutation of GGQ motif residues results in release factors that are defective in their ability to trigger peptidyl-tRNA hydrolysis but that retain the normal eRF1 functions of stop codon recognition, ribosome binding, and eRF3 interaction (19, 41).
In cell-free translation reactions, addition of a loss-of-function eRF1 mutant in which both glycines of the GGQ motif were replaced with alanines (AA-eRF1) resulted in increased translation downstream of uORF2 compared to that realized by addition of wild-type eRF1 (28). This relative enhancement of downstream translation by AA-eRF1 was specific to transcripts containing the wild-type uORF2 sequence. Consistent with the view that ribosomal stalling at the end of uORF2 is responsible for the inhibition of downstream translation, addition of AA-eRF1 also reduced the half-life of the ribosomal stalling event at the uORF2 termination codon in cell-free translation reactions. These results suggested that wild-type eRF1 and the uORF2 peptidyl-tRNA may interact. The present studies were designed to elucidate the role of eRF1 in termination inhibition by uORF2. We found that the interplay between eRF1 and uORF2 required specific residues from each moiety that are predicted to be in close proximity within the ribosome. The findings that eRF1 but not eRF3 accumulates in the stalled uORF2-ribosome complex and that the puromycin sensitivity of uORF2 peptidyl-tRNA is enhanced following eRF1 removal from the complex further implicate eRF1 as a key participant in this unusual regulatory mechanism.
MATERIALS AND METHODS
Plasmids, sequencing, and synthetic RNAs.
The coding sequence for eRF1 was PCR amplified from a clone containing the human TB3-1 cDNA (25) using the forward primer eRF1F 5′-GATCCTGCAGCTGGTGCCACGCGGTTCTATGGCGGACGACCCCAGTGCT-3′ and the reverse primer eRF1R 5′-GATCAAGCTTCTAGTAGTCATCAAGGTCAAA-3′. The PCR product was cloned into the PstI and HindIII sites of pQE9 (Qiagen), which provides an N-terminal six-histidine tag, to yield pEQ814.
PCR mutagenesis was used to construct the AA-eRF1 mutant. The N-terminal portion of eRF1 was amplified using the primers eRF1F and 5′-CTGAGCAGCGCGCCCGTGTTTCTT-3′, while the C-terminal part of eRF1 was amplified with the primers 5′-CGGGCGCGCTGCTCAGTCAGCCTT-3′ and eRF1R. The internal primers alter codons 183 and 184 from glycines to alanines and introduce a BssHI site into codons 181 and 182 through silent mutations. The N-terminal fragment digested with PstI and BssHI, the C-terminal fragment digested with BssHI and HindIII, and pQE9 digested with PstI and HindIII were combined in a three-way ligation to yield pEQ832. Single mutants in the eRF1 GGQ motif were constructed by replacing the 1.2-kb BglII-KpnI fragment in pEQ814 with the corresponding fragment from plasmids containing the complete coding sequence of mutated eRF1 (19). The resulting plasmids pEQ830, pEQ831, and pEQ833 contain G183R, G184S, and G181S mutations of eRF1, respectively.
Plasmids pEQ438, pEQ439, pEQ507, and pEQ542 containing the β-galactosidase (β-Gal) gene with the wild-type, P22A-, P21T-, and S12P-mutated uORF2 leader sequences, respectively, have been previously described (5). The P21T and S12P mutant transcripts contain a K10Q mutation that, like other mutations of K10, preserves the uORF2 translational inhibitory activity (2, 9). Plasmid pEQ508, coding for a β-Gal transcript with K18E/Y19H/I20V mutations in the uORF2 leader, was constructed as described for pEQ542, except that the parent vector pEQ398 was used as a template for PCR amplification (9). Each plasmid also contains nucleotide changes that optimize the uORF2 AUG sequence and result in a Q2E mutation (5).
Plasmid sequences were verified by the Big-Dye terminator cycle sequencing method (ABI). In vitro transcription was performed using plasmid templates digested with BamHI or Asp718I to yield full-length or leader transcript, respectively, with the Ampliscribe T3 transcription kit (Epicenter Technologies).
Expression and purification of eRF1.
Cultures harboring eRF1 expression vectors were grown to an optical density at 600 nm of 0.8, after which isopropyl-β-d-thiogalactopyranoside was added to a concentration of 1 mM. Five hours later bacteria were pelleted and stored at −20°C until use.
All purification steps were carried out at 4°C, using a modified method (18) in combination with affinity chromatography. Briefly, bacteria were suspended in buffer A (20 mM Tris-HCl, pH 8.0, 200 mM KCl, 10 mM imidazole, 1% NP-40, 10% glycerol, and 1 mM dithiothreitol [DTT]) with 1 mM phenylmethylsulfonyl fluoride, lysed by sonication, and cleared by centrifugation at 12,000 × g. The cleared lysate was applied to an Ni2+-nitrilotriacetic acid (Qiagen) column that was equilibrated in buffer A, washed with 10 bed volumes of the same buffer, and then eluted with buffer A containing 60 mM imidazole. The eluent was diluted 10-fold in buffer B (20 mM Tris-HCl, pH 7.4, 1 mM DTT, and 5% glycerol) and was applied to a Mono Q (Sigma) column equilibrated in the same buffer. The column was washed with 10 bed volumes of buffer B, and proteins were eluted with a step gradient of 0 to 500 mM KCl in buffer B at 50 mM intervals. Fractions containing eRF1 were pooled and dialyzed against storage buffer (50 mM Tris-HCl, pH 7.4, 50 mM KCl, 2 mM DTT, and 5% glycerol) and were stored at −70°C. The protein concentration was determined by comparison to bovine serum albumin standards.
β-Gal expression in cell-free translation assays.
Cell-free translation was carried out in micrococcal nuclease-treated rabbit reticulocyte lysate (RRL; Promega) as described earlier (20) with the following modifications. Typical translation reactions were 6.25 μl and contained 70% RRL, 20 μM unlabeled amino acids (each), 0.32 μg of full-length transcripts (synthesized from pEQ438, pEQ439, pEQ507, pEQ508, and pEQ542 templates)/ml, and 0.2 μM release factor or the equivalent volume of release factor storage buffer. After translation for 60 min at 30°C, 8 reaction volumes of MUG solution (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, 1 mg of bovine serum albumin/ml, 1% dimethyl sulfoxide, and 150 μg of methyl-umbelliferyl-β-d-galactoside/ml) was added. After incubation for 60 min at 30°C, a 25-μl aliquot was dequenched by dilution in 175 μl of 0.125 N NaOH and 0.2% H2O2 for 5 min at room temperature, after which fluorescence was measured using a microfluorimeter (Dynatech).
Detection of ribosome association of release factors.
Rabbit polyclonal anti-eRF1 antibodies were raised against bacterially expressed and purified His-tagged eRF1. Rabbit antiserum raised against recombinant GSPT1 (eRF3) was kindly provided by S.-I. Hoshino (University of Tokyo) (27). In vitro translation reactions (25 μl) were performed in RRL programmed with 40 μg of leader transcripts synthesized from pEQ438 or pEQ439 template per ml. Hygromycin B (Sigma) at 16 μg/ml or puromycin (Sigma) at 0.1 mM was used in some reactions. Reactions were translated for 5 min at 30°C, and where indicated, elongation inhibitor was added and translation was continued for an additional 5 min. Reaction mixtures were stored on ice and were then centrifuged at 100,000 × g for 75 min at 4°C in a Beckman TL100 centrifuge using a TLA100.3 rotor to obtain the ribosome pellet fraction. Pellets were rinsed before resuspension in 10 μl of diethyl pyrocarbonate (DEPC)-treated deionized water. With this treatment, rRNA is found in the pellet fraction, while free tRNAs are found in the supernatant, and the uORF2 peptidyl-tRNA is found associated with ribosomes in the pellet fraction (6). For each sample, 4 μl (for eRF1) or 10 μl (for eRF3) of resuspended pellet was fractionated by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and was then transferred to nitrocellulose by electroblotting. Immunoblots were processed using the CDP-Star kit (Tropix). Anti-eRF1 antiserum was used at 1:2,500, anti-eRF3 antiserum was used at 1:1,000, and all other reagents were used to manufacturer specifications.
Association of eRF1 with the ribosomal pellet.
In vitro translation was performed in RRL in a total reaction volume of 175 μl containing 40 μg of pEQ438 leader transcript/ml. Translation was carried out for 5 min at 30°C, at which point a 25-μl aliquot was removed to ice and anisomycin was added to 16 μg/ml to the remainder of the reaction. Additional 25-μl aliquots were removed at 2, 5, 10, 15, 30, and 60 min after drug addition. Ribosome pellets were prepared and the eRF1 content in each sample was analyzed by immunoblotting as described above.
Salt wash and puromycin treatment.
Fifty microliters of cell-free translation reactions containing 40 μg of pEQ438 or pEQ439 leader transcript per ml and 300 μCi of [35S]methionine (Amersham)/ml were translated for 5 min at 30°C. Reactions were split in half and both halves were centrifuged to obtain ribosome pellet fractions. One pellet from each set was washed and resuspended in 10 μl of DEPC-treated deionized water. The other pellet was resuspended in 50 μl of salt wash buffer (30 mM HEPES, pH 7.5, 70 mM KCl, 2 mM magnesium acetate, and 1 mM DTT), and then 4 M KCl was added to 0.5 M. After shaking for 1 h at 4 C, the sample was centrifuged. The resulting ribosome pellet was washed and resuspended in 10 μl of DEPC-treated deionized water. Aliquots of 4 μl from each resuspended pellet were analyzed for eRF1 by immunoblotting as described earlier. One microliter of each sample was fractionated on a 16% SDS-Tris-Tricine gel. Gels were treated with En3Hance (New England Nuclear), dried, and exposed to film to visualize uORF2 peptidyl-tRNA.
To test puromycin sensitivity, 100-μl cell-free translation reactions containing 300 μCi of [35S]methionine/ml and 40 ng of pEQ438 leader transcript/ml were translated for 5 min at 30°C and were then split in half and centrifuged to collect ribosome pellets. One pellet was washed and resuspended in 20 μl of TMKN buffer (35 mM Tris acetate, pH 8.0, 10 mM magnesium acetate, 60 mM potassium acetate, 30 mM ammonium acetate, and 1 mM DTT) (22) and was then stored on ice until use. The other pellet was salt washed in a total volume of 100 μl as described above and was then centrifuged, washed, and resuspended in 20 μl of TMKN. Each resuspended pellet was split into two 8-μl aliquots. One microliter of each was removed to ice as a 0- min time point. To one sample, puromycin was added to 0.5 mM, while an equal volume of deionized water was added to the other. Samples were incubated at 37°C, and 1.2-μl aliquots (volume adjusted for puromycin addition) were removed to ice after 1, 2.3, 5, 10, and 15 min. The uORF2 peptidyl-tRNA was analyzed as described above and quantified by phosphorimager scan (Storm) and NIH Image software (version 1.62).
RESULTS
Participation of the eRF1 GGQ motif in translational inhibition by uORF2.
Previous results suggested that the termination blockade at the end of uORF2 might result from an interaction of the uORF2 peptidyl-tRNA with the GGQ motif of eRF1 (28). To examine this hypothesis in greater detail, we assessed the effects of a set of eRF1 single mutants on translation downstream from uORF2. Similar to the eRF1 G183A/G184A double mutant (AA-eRF1), two of these mutants, G183R and G184S, are defective for peptidyl-tRNA hydrolysis activity but still retain the ability to recognize stop codons, bind ribosomes, and interact with eRF3 (19, 42). In contrast the G181S mutant of eRF1, which contains a mutation of a conserved glycine just outside the GGQ motif, behaves like wild-type eRF1 and retains all normal eRF1 functions (19).
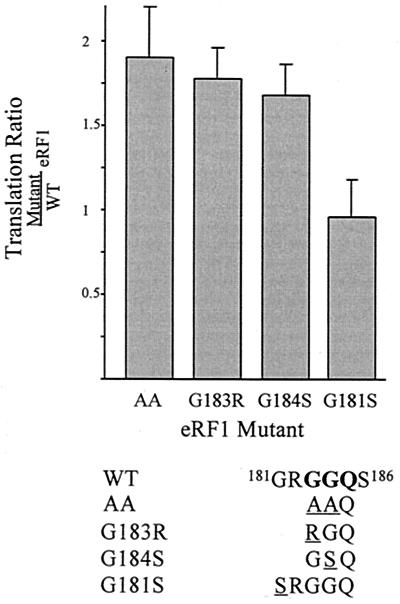
These eRF1 mutants were expressed in and purified from Escherichia coli as His-tagged fusions, and their effects on translation of β-Gal downstream from wild-type uORF2 were assessed in cell-free translation assays (Fig. 1). Mutations in eRF1 that disrupt the putative inhibitory interaction of eRF1 with uORF2 are expected to result in enhanced downstream translation. As previously observed (28), translation downstream from uORF2 was increased approximately twofold after addition of 0.2 μM exogenous AA-eRF1, compared to addition of wild-type eRF1. Similarly, mutation of either glycine within the GGQ motif (G183R or G184S) resulted in an approximately twofold increase in downstream translation relative to that of the wild-type eRF1. In contrast, the G181S mutation did not enhance downstream translation. Therefore, the inhibitory effect of eRF1 appears to depend on glycine residues within the GGQ motif.
FIG. 1.
Mutations in the GGQ motif of eRF1 affect translation downstream of uORF2. Full-length β-Gal transcripts containing the wild-type uORF2 leader were translated in RRL in the presence of exogenous eRF1 as described in Materials and Methods. Downstream translation is expressed as the ratio of β-Gal activity in reactions containing mutated eRF1 to that observed when wild-type eRF1 was added. Each bar displays the mean ratio (plus standard deviation) obtained from an experiment run in triplicate. The relevant portion of the wild-type human eRF1 sequence is shown below, with the GGQ motif indicated in boldface and mutated residues underlined for each variant.
The carboxy-terminal prolines of uORF2 are required for inhibition by wild-type eRF1.
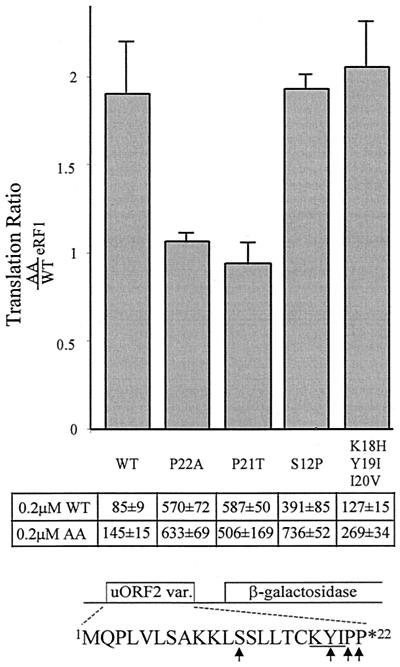
Previous analyses identified specific uORF2 codons that are necessary for inhibition of downstream translation in cells and in cell extracts (2, 5, 9). For example, β-Gal activity resulting from translation of RNA containing wild-type uORF2 was 5- to 10-fold less than from translation of RNAs containing a mutation of the terminal proline to alanine (P22A) or containing no uORF2 (5). Consistent with these previous studies that were performed without addition of eRF1, the uORF2 mutations shown in Fig. 2 augmented downstream translation compared to the wild-type uORF2 leader after the addition of either wild-type eRF1 or AA-eRF1 (Fig. 2).
FIG. 2.
Sensitivity of uORF2 missense mutations to eRF1 effects. Full-length β-Gal transcripts with leaders containing uORF2 mutations were translated in cell-free reactions to which 0.2 μM wild-type eRF1 (WT) or AA-eRF1 (AA) was added. The translation ratio represents β-Gal activity measured after addition of exogenous AA-eRF1 compared to that found after addition of wild-type eRF1 for each RNA. Bars represent the mean ratio (plus standard deviation) obtained from at least two separate experiments in which each RNA was assayed in triplicate. The actual β-Gal activities (means plus or minus standard deviation of triplicate reactions) obtained from representative translations of each RNA are shown below the bars. The wild-type 22 codon uORF2 peptide sequence and amino acids that varied among the transcripts tested are shown below.
To determine which codons within uORF2 are involved in the interaction with eRF1, we compared the β-Gal activities generated by translation of each RNA after addition of AA-eRF1 versus that of wild-type eRF1. Mutations that destabilize the putative uORF2 peptide interaction with wild-type eRF1 would be expected to express similar levels of β-Gal regardless of whether wild-type or AA-eRF1 is added. The ratio of β-Gal activity after addition of AA-eRF1 to that after addition of wild-type eRF1 for each RNA is shown in the bar graph in Fig. 2. As noted previously (28), translation downstream from wild-type but not from P22A mutant uORF2 was almost twofold greater after addition of AA-eRF1 than after addition of wild-type eRF1 (Fig. 2), suggesting that mutation P22A eliminated the interaction with wild-type eRF1. A mutant in which the penultimate proline of uORF2 was changed to threonine (P21T) also exhibited no enhancement of downstream translation following addition of AA-eRF1 compared to the result of addition of wild-type eRF1, implying that the P21T mutation also disrupted the wild-type eRF1-uORF2 interaction. However, when uORF2 was altered by mutations at serine 12 (S12P) or at codons 18 through 20 (K18H/Y19I/I20V), downstream translation was nearly twofold greater after addition of AA-eRF1 than after addition of wild-type eRF1, indicating that the wild-type eRF1-uORF2 interaction was retained. Thus, even though the S12P and K18H/Y119I/I20V mutations are similar to P21T and P22A in allowing higher levels of downstream translation than does wild-type uORF2, they retain sensitivity to addition of wild-type eRF1, relative to addition of the AA-eRF1 mutant. These results suggest that the inhibitory interaction of the GGQ motif of eRF1 with the uORF2 peptidyl-tRNA depends on the critical C-terminal prolines (P22 and P21) but not on codon S12 or K18/Y19/I20. It is possible that some uORF2 residues like S12 and K18/Y19/I20 contribute to the termination blockade by interacting with factors other than eRF1. Alternatively, such residues may interact with other domains of eRF1 or may interact too weakly with the GGQ motif to be detectable by our assay.
eRF1 but not eRF3 accumulates in the uORF2 peptidyl-tRNA-stalled ribosome complex.
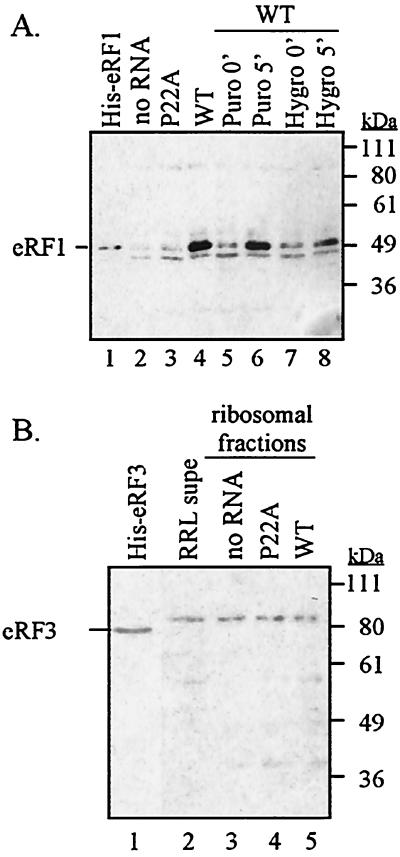
In principle, the uORF2 peptidyl-tRNA could inhibit translation termination at its own stop codon by preventing eRF1 from gaining access to the ribosome. However, the apparent importance of the GGQ motif of eRF1 and the proline residues of uORF2 peptidyl-tRNA suggests that the termination blockade occurs after eRF1 has entered the ribosome but prior to peptidyl-tRNA hydrolysis. To clarify whether the termination blockade occurs before or after eRF1 has entered the ribosome, we investigated whether eRF1 is physically present in the uORF2 peptidyl-tRNA-stalled ribosome complex. We programmed RRL reactions with no RNA or with transcripts containing either the wild-type or P22A mutant uORF2. After incubation, ribosomes and associated factors including uORF2-stalled ribosome complexes were pelleted by centrifugation and were analyzed on immunoblots probed with anti-eRF1 serum. Prior to translation, the vast majority of eRF1 in RRL was present in the supernatant and little was in the ribosomal pellet (data not shown). Similarly, in reactions to which no RNA was added, only a small amount of eRF1 was associated with the ribosome fraction after incubation (Fig. 3A, lane 2). Translation of the P22A mutant uORF2 leader transcripts also resulted in ribosomal pellets with a comparatively low level of eRF1 (Fig. 3A, lane 3). In contrast, the amount of eRF1 in the ribosome pellet was greatly enriched when the wild-type uORF2 leader was used to program the reactions (Fig. 3A, lane 4).
FIG. 3.
Translation of wild-type (WT) uORF2 increases the amount of eRF1 but not of eRF3 associated with the stalled ribosome complex. Pelleted ribosomes and associated proteins from equal volumes of translation reactions were fractionated by SDS-PAGE, transferred to nitrocellulose, and probed using rabbit polyclonal antibody raised against recombinant eRF1 (A) or eRF3 (B) protein. The positions of molecular weight markers are indicated along the right side of the immunoblots, and the position of release factor marked on the left corresponds to migration of purified His-tagged release factor in lane 1. Pellets from a reaction containing no added RNA (no RNA) and reactions containing P22A mutated or wild-type uORF2 leaders are shown. Additional samples in immunoblots (A) are pellets obtained from translation reactions programmed with wild-type uORF2 leader transcripts. The elongation inhibitors puromycin (lanes 5 and 6) and hygromycin (lanes 7 and 8) were added either prior to translation (0′, lanes 5 and 7) or 5 min after the start of translation (5′, lanes 6 and 8), and pellets were harvested after an additional 5 min of translation. A lane containing 2 μl of RRL postribosomal supernatant (RRL supe) is included for comparison in the eRF3 blot.
The recruitment of eRF1 to the ribosome-peptidyl-tRNA complex was dependent on translation of uORF2 since addition of the elongation inhibitors puromycin and hygromycin B prior to translation prevented the accumulation of eRF1 in the ribosomal pellets (Fig. 3A, lanes 5 and 7). However, when the inhibitors were added after a short translation period, eRF1 was enriched in the ribosomal pellets (Fig. 3A, lanes 6 and 8). These results demonstrate that eRF1 accumulates in ribosomes that have translated uORF2 and suggest that inhibition of the termination reaction by uORF2 occurs after entry of eRF1 into the ribosome.
Since eRF1 is enriched in the stalled ribosome complex and since eRF1 and eRF3 are known to associate in vitro (17, 18, 34), we next evaluated whether eRF3 is also a component of the uORF2 peptidyl-tRNA-stalled ribosome complex. We performed immunoblots on the ribosomal pellets from cell-free translation reactions by using a polyclonal antiserum raised against eRF3 (Fig. 3B). Similar to results seen for eRF1, limited amounts of eRF3 were present in the RRL ribosomal pellet fraction compared to those found in the supernatant (Fig. 3B, compare lane 2, corresponding to 2 μl of RRL supernatant, to lane 3, containing the ribosomal pellet from 25 μl of RRL). The eRF3 levels associated with ribosome pellets were similar regardless of whether RNA was added to the reaction (Fig. 3B, compare lane 3 with lanes 4 and 5). Most notably, we did not observe any enrichment of eRF3 in the ribosomal pellet when wild-type uORF2 leaders were translated (Fig. 3B, lane 5). The eRF3 band observed in RRL has a higher apparent molecular weight than does the bacterially expressed marker. This may be due to differences in the amino acid sequence between rabbit eRF3 and human eRF3 or may be caused by a posttranslational modification of eRF3. These results reveal that, unlike eRF1, eRF3 is not enriched in the uORF2 peptidyl-tRNA-stalled ribosome complex.
Depletion of eRF1 from the stalled ribosome complex increases the puromycin sensitivity of the uORF2 peptidyl-tRNA.
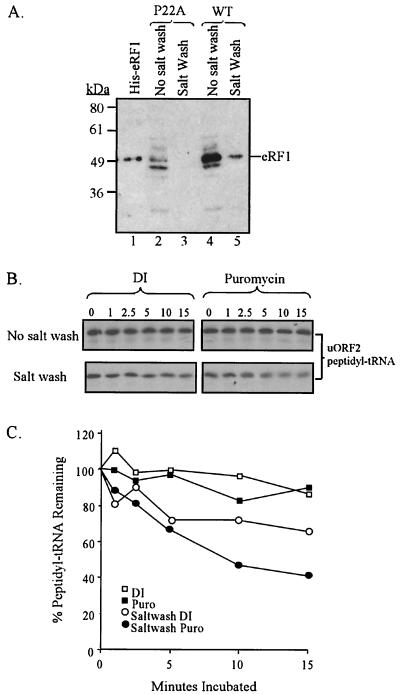
Once formed, the uORF2 peptidyl-tRNA is surprisingly stable to puromycin treatment (7). The interaction of eRF1 with the uORF2 peptidyl-tRNA may stabilize a termination intermediate that prevents puromycin from gaining access to the peptidyl-tRNA ester bond. In such a case, removal of eRF1 from the stalled ribosome complex should convert the uORF2 peptidyl-tRNA to a puromycin-sensitive form. To test this hypothesis, we isolated the uORF2 peptidyl-tRNA-eRF1 ribosome complexes by centrifugation and then salt washed the ribosomal pellet to strip away eRF1 (Fig. 4A). When reactions containing transcripts with the P22A mutant uORF2 were treated in this manner, all detectable eRF1 was removed (Fig. 4A, lanes 2 and 3). The same treatment of reactions programmed with wild-type uORF2 leader transcripts removed most of the accumulated eRF1 (Fig. 4A, lanes 4 and 5). Since the salt wash treatment did not alter the amount of uORF2 peptidyl-tRNA associated with the ribosomal fraction (data not shown), we were able to use this protocol to enrich for stalled ribosome-peptidyl-tRNA complexes depleted of eRF1.
FIG. 4.
Puromycin sensitivity of the uORF2 peptidyl-tRNA in the presence and absence of eRF1. (A) Effect of salt washing on eRF1 association with ribosome pellet fractions. RRL reactions were programmed with P22A-mutated (lanes 2 and 3) or wild-type (WT) uORF2 leader transcripts, and pellets were collected after 5 min of translation. These pellets were either analyzed for eRF1 by immunoblot assay either directly (lanes 2 and 4) or after a salt wash step (lanes 3 and 5). His-tagged eRF1, included as a marker in lane 1, migrates at approximately 50 kDa. (B) Effects of salt washing on ribosomal association of uORF2 peptidyl-tRNA. Ribosome pellets were obtained from RRL reactions programmed with wild-type uORF2 leader transcripts and translated 5 min in the presence of [35S]methionine. Pellets, used directly or after salt washing, were resuspended and treated with 0.5 mM puromycin or deionized water (DI) as a control. Samples collected before puromycin treatment and after 1, 2.5, 5, 10, and 15 min of incubation at 37°C were fractionated on SDS-16% Tris-Tricine gels, and the uORF2 peptidyl-tRNA was band visualized by phosphorimager scan. (C) Graphic representation of the results from panel B. The abundance of ribosome-associated uORF2 peptidyl-tRNA at each time point is expressed as a percentage of the zero-time point signal for each set. Puro, puromycin.
We resuspended ribosome complexes with and without salt washing, treated them with 0.5 mM puromycin, and then examined the amount of [35S]methionine-labeled uORF2 peptidyl-tRNA after various incubation times. An autoradiogram from a representative experiment is shown in Fig. 4B, with the results quantified and shown graphically in Fig. 4C. When we treated stalled ribosome complexes that had not been salt washed with either water or puromycin, the abundance of peptidyl-tRNA remained nearly constant for at least 15 min. In contrast, when eRF1 was removed from the ribosome by salt washing and when puromycin was then added, the uORF2 peptidyl-tRNA signal diminished substantially over time. This decrease was greater than that observed in the control reactions in which deionized water, instead of puromycin, was added to the salt-washed samples. We repeated this experiment three times, and although the absolute values and kinetics of decay of the peptidyl-tRNA were not always the same, in each experiment puromycin accelerated the decay of the peptidyl-tRNA only in the salt-washed samples. Although salt washing may remove additional factors besides eRF1 from the stalled ribosome complexes, the increase in puromycin sensitivity following salt washing is consistent with the hypothesis that an interaction of eRF1 and uORF2 peptidyl-tRNA blocks access of puromycin to its target, the peptidyl-tRNA bond.
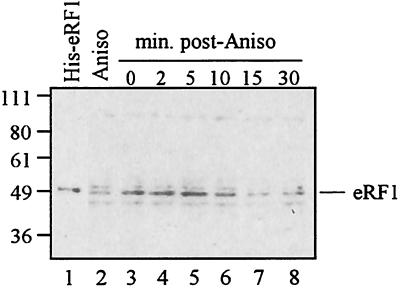
Persistence of eRF1 in the ribosome pellet mirrors the kinetics of the ribosomal stall.
The half-life of the ribosomal association with the mRNA is approximately 7 to 10 min in cell extracts (28). However, previous work demonstrated that the peptidyl-tRNA remains at least partially associated with the ribosome even after the ribosome has departed from the mRNA (7). To investigate whether this persistent association of the uORF2 peptidyl-tRNA with the ribosome might be mediated by eRF1, we examined the kinetics of eRF1 association with the uORF2-stalled ribosome complex. Wild-type uORF2 leaders were translated in RRL for 5 min, and then anisomycin was added to inhibit any further rounds of translation. At various times after drug addition, ribosome fractions were collected and analyzed for the presence of eRF1 by immunoblot assay (Fig. 5). The amount of eRF1 in the peptidyl-tRNA-ribosome complex was fairly constant for 5 min but decreased by 10 min and was near background levels after 15 min. Thus, the kinetics of eRF1 persistence in the ribosome complex resembles the half-life of stalled ribosome association with mRNA. Therefore, the uORF2 interaction with eRF1 does not appear to account for the prolonged association of the uORF2 peptidyl-tRNA with the ribosome; rather an eRF1-independent interaction of the uORF2 peptidyl-tRNA with another ribosomal component is more likely responsible.
FIG. 5.
Kinetics of eRF1 presence in the stalled ribosome complex. Cell-free translation reactions programmed with wild-type uORF2 leader transcripts were processed as described in Materials and Methods. The resulting ribosomal pellets were fractionated by SDS-PAGE, transferred onto nitrocellulose, and probed with polyclonal anti-eRF1 antibody. Positions of molecular weight markers and of eRF1 based on the migration of His-tagged eRF1 (lane 1) are indicated in kilodaltons on left. Lane 2 shows the results obtained when anisomycin (Aniso) was added before translation. The remaining reaction was translated for 5 min before the addition of anisomycin. Pellets were collected just before drug addition and at 2, 5, 10, 15, and 30 min after addition.
DISCUSSION
Inhibition of downstream translation by uORF2 occurs via an unusual mechanism (2, 5-7, 9,28). The peptide produced by translation of uORF2 inhibits its own termination, as evidenced by persistence of the uORF2 peptidyl-tRNA linkage (6, 7), and results in ribosomal stalling and inhibition of translation of the downstream cistron. Remarkably, the inhibitory activity is dependent on the uORF2 peptide sequence, with dispersed residues being necessary for full inhibition of downstream translation (2, 9). In this report, we provide evidence that eRF1 is a critical factor in the inhibitory mechanism. We propose a model of the uORF2 mechanism (Fig. 6) in which the two carboxy-terminal prolines of uORF2 interact with glycines of the eRF1 GGQ motif and thereby stabilize an intermediate in the termination cascade, with the uORF2 peptidyl-tRNA in the ribosomal P site and eRF1 in the ribosomal A site. This stabilization inhibits peptidyl-tRNA hydrolysis, resulting in ribosomal pausing at the uORF2 termination codon and, in turn, limited downstream translation. Mutations of eRF1 that destabilize the interaction with uORF2 enable more rapid dissociation of the ribosome-peptidyl-tRNA complex. An alternative interpretation of the relative stimulatory effect of mutant eRF1 on translation downstream from uORF2 is that the mutant eRF1 stimulates nonsense suppression at the end of uORF2, as has been observed in yeast (42). However, the finding that AA-eRF1 reduces the duration of ribosomal stalling at the end of uORF2 in the presence of translation elongation inhibitors argues against this explanation (28).
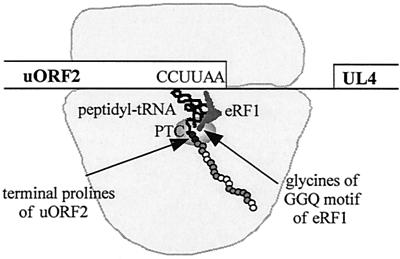
FIG. 6.
Model of translation termination inhibition by uORF2. The ribosome translates through uORF2 until the uORF2 peptide attached to the cognate tRNAPro for the terminal proline codon (CCU) is positioned in the ribosomal P site, while the stop codon (UAA) is in the A site. At this point eRF1 enters the ribosomal A site, recognizes the stop codon, and binds to the ribosome. During the termination reaction, the glycines of the eRF1 GGQ motif and the two terminal prolines of uORF2 (black circles) interact, possibly through the peptidyl-transferase center (PTC, indicated by the shaded oval), to form an intermediate that stabilizes the peptidyl-tRNA bond. This stabilization contributes to prolonged ribosomal stalling, which in turn blocks ribosomes from scanning to the downstream UL4 cistron. Residues of the uORF2 peptidyl-tRNA that are needed for inhibition of downstream translation but that do not interact with eRF1 (shaded circles) may interact with other ribosomal proteins or associated factors.
Peptide sequence-dependent ribosomal stalling, as occurs after translation of uORF2, has been observed in other eukaryotic and bacterial systems (21, 36, 47). Our results raise the question of whether release factors play a role in other uORF-mediated ribosomal stalling events. The S-adenosylmethionine decarboxylase uORF codes for a sequence-dependent polyamine-responsive peptide that, like uORF2, causes ribosomal stalling specifically at the termination codon and results in accumulation of the uORF peptidyl-tRNA (30, 40). While the critical sequences of this uORF do not include the carboxy-terminal residue, the penultimate and antepenultimate residues have been implicated (35). Thus, it is possible that eRF1 acts in concert with the polyamine effector and the nascent peptide to inhibit the termination reaction. Regulation of the bacterial tryptophanase operon gene tnaC also has intriguing similarities to the uORF2 mechanism (22-24). Toeprinting assays show that ribosomes translating tnaC stall over the stop codon. Moreover, the nascent peptide remains covalently attached to a terminal prolyl-tRNA that retains a ribosomal association. The tnaC peptidyl-tRNA when present on washed ribosomes is sensitive to puromycin and is cleaved by release factor only in the absence of tryptophan (22). Thus, it is possible that the tnaC peptide forms a tryptophan-mediated complex with release factor (see Addendum in Proof).
In other examples of uORF peptide-dependent ribosomal stalling, release factors are unlikely to participate in the stalling mechanism. The yeast CPA-1 gene and Neurospora crassa arg2 gene produce sequence-dependent peptides that act in cis to cause ribosomal stalling (13, 48). Although they can affect terminating ribosomes, they also act on elongating ribosomes. Similarly, the cat, cmlA, and ermC uORFs involved in bacterial antibiotic resistance cause ribosomal stalling during elongation, before the stop codon is reached (33, 49). Thus, class I release factors would be unlikely to be involved in these cases. Another bacterial sequence-dependent uORF, secM, causes ribosomes to stall at a proline residue, and the protein product can be detected as a peptidyl-tRNA species (38, 39). However, this stalling event occurs near but not at the carboxy terminus, and thus, release factors are unlikely to participate. In fact, ribosomal protein L22 as well as rRNA has been implicated in this stalling event. Together with our results, analyses in various other systems suggest that nascent peptide interactions with ribosomal or ribosome-associated factors, including but not limited to release factors, can inhibit translation elongation as well as termination.
Intriguingly, several other nascent peptides that alter ribosome function also end with proline codons (24, 26, 32, 38). In E. coli, terminal prolines inhibit termination, as measured by an increase in the competing reaction of nonsense suppression (4). Protein tagging of the YbeL protein mediated by the SsrA RNA in E. coli has a strong requirement for a terminal proline (26), suggesting that this recoding event also is enhanced by inefficient termination after proline addition to the nascent peptide chain. Our results support the possibility that terminal prolines can inhibit termination in eukaryotes. However, in contrast with the observations from study of E. coli, the terminal prolines of uORF2 appear to stabilize eRF1 within the ribosome, presumably at the A site, where it would be predicted to interfere with rather than enhance recoding events. Moreover, terminal prolines do not to stimulate nonsense suppression in yeast (37). If tandem carboxy-terminal prolines have a propensity to inhibit termination, they might be expected to be infrequent. However, a search of 112,657 entries in the SwisProt protein database (release 40.25) identified 191 (∼0.2%) ending with two prolines, not far from the 271 expected if prolines were randomly distributed among these proteins. Finally, the occurrence of prolines at elongation stall sites (38) as well as at termination sites needs to be considered in formulating a model to explain the mechanism(s) by which prolines at the end of nascent peptide chains affect ribosome function.
In the case of uORF2, interactions of the nascent peptide with ribosomal components in addition to eRF1 are likely involved in the inhibitory mechanism. Several residues of uORF2 besides the terminal prolines are required for its ability to inhibit downstream translation. Among the residues tested here (Fig. 2), only the two terminal prolines appear to be involved in the eRF1 GGQ motif-mediated inhibitory effect on downstream translation. For example, the S12P mutation eliminates the inhibitory effect of uORF2 on downstream translation, ribosomal stalling, and accumulation of the uORF2 peptidyl-tRNA (2, 5; unpublished data). Unlike wild-type uORF2 RNA translation (Fig. 3), S12P RNA translation did not result in detectable accumulation of eRF1 in the ribosomal pellet (data not shown). However, the S12P mutant appears to maintain the ability to interact with the GGQ motif of eRF1, presumably through the prolines at codons 21 and 22. In the absence of critical uORF2 codons such as S12, the interactions of the prolines with eRF1 may be too weak and/or transient to be detected by our assays except under conditions of added eRF1 (Fig. 2). Thus, the uORF2 inhibitory mechanism likely depends on interactions between residues of the uORF2 peptidyl-tRNA beside the terminal prolines and cellular factors besides eRF1. Candidate factors include rRNA or ribosomal proteins, especially ones that localize to the nascent peptide exit tunnel of the ribosome, where several critical uORF2 residues such as S12 are expected to be located during termination of uORF2 translation (38).
Ribosomal stalling after translation of uORF2 is not permanent. Rather, the ribosome appears to disengage from the mRNA even when the uORF2 peptide is still linked to tRNAPro (7). The unhydrolyzed uORF2 peptidyl-tRNA appears to remain at least partly associated with the ribosome even after the ribosome has dissociated from the mRNA (7). In our present studies, we found that release of eRF1 from the ribosome follows the same approximate kinetics as disassociation of the mRNA from the ribosome. Thus, the prolonged association of the uORF2 peptidyl-tRNA with the ribosome is not likely due to its interaction with eRF1. Rather, this result supports the hypothesis that the prolonged association of the uORF2 peptidyl-tRNA with the ribosome is due to its interaction with ribosomal or ribosome-associated factors other than eRF1.
Although our results suggest that eRF1 and uORF2 interact, we have not been able to confirm such an interaction or to detect interactions of the uORF2 peptide with other factors using two-hybrid, glutathione transferase pull-down, or far-Western assays (unpublished data). However, each of these assays has limitations. For example, binding sites on the uORF2 peptide may be masked by fusion to a much larger protein, as in the two-hybrid and pull-down assays. The phosphorylation of synthetic uORF2 peptide for use as the probe in our far-Western screening assays may have altered the charge of this small peptide sufficiently to disrupt its normal binding properties. Another possibility is that uORF2 may need to be in its peptidyl-tRNA form and in the context of the translating ribosomes in order to interact with eRF1. The proposed interaction may be weak and thus only occur when the proteins are held in close juxtaposition and in the rotationally constrained conditions provided by the ribosomal A and P site framework. Finally, the interaction of the uORF2 peptide and eRF1 may be indirect. Since peptidyl-tRNA hydrolysis chemistry is carried out by the ribosome, it is possible that the peptidyl-transferase center, or some other ribosomal component, bridges eRF1 and uORF2, resulting in a three-component complex in which eRF1 and uORF2 do not directly contact each other.
eRF1 and eRF3 are known to directly interact both in vitro and in cells, forming a complex through their carboxy termini (12, 18, 34, 45, 50). eRF3 is an essential protein (46) that harbors ribosome- and eRF1-dependent GTPase activity (15). These data suggest that eRF1 and eRF3 act together in the termination reaction. Therefore, we were surprised to detect eRF1 but not eRF3 in ribosomes after translation of wild-type uORF2. There are at least two possible explanations for this discrepancy. eRF1 may not enter the ribosome as an eRF1-eRF3 complex, and in fact, eRF3 might be blocked from entering as a result of the interaction of eRF1 with the uORF2 peptidyl-tRNA. Alternatively, eRF3 might enter with eRF1 but then be released while eRF1 and the uORF2 peptidyl-tRNA continue to interact and stabilize the stalled ribosome complex.
Present experimental evidence suggests that binding of uncomplexed eRF1 to the ribosome is the likelier explanation for our results. While eRF1 and eRF3 are known to interact off the ribosome, there is also evidence indicating that the release factors may bind the ribosome and function separately. In yeast, eRF1 is more tightly associated with the ribosome than is eRF3 (10, 44). Also, in yeast mutants where ribosome binding by eRF1 was diminished, eRF3 association with the ribosome remained mostly unaffected (43). Moreover, the eRF1-eRF3 complex may not be the active form of release factor. In in vitro termination assays, eRF1 is capable of stimulating peptidyl-tRNA hydrolysis in the absence of eRF3 (14, 17), and in RRL and human cell lines, eRF1 alone can result in antisuppression (11, 31).
In this investigation we have provided evidence supporting a role for eRF1 in the autorepression of translation termination by uORF2. Stabilization of an intermediate resulting from an interaction of eRF1 with the nascent uORF2 peptidyl-tRNA appears to delay or prevent peptidyl-tRNA hydrolysis, the core enzymatic step of termination. Compared to what is known of processes such as initiation, relatively little is known about termination and its potential for regulating gene expression. Because uORF2 is one of a small but growing number of sequence-dependent uORFs that exhibit translational control at the level of termination, our work suggests that termination may be a target for translational control in diverse biological systems. As well, these studies reveal that release factors that normally function to facilitate peptidyl-tRNA hydrolysis may, in some situations, participate in inhibition of the termination cascade.
ADDENDUM IN PROOF
A recent report demonstrated that the traC peptide can inhibit elongation when charged tryptophanyl tRNA occupies the A site of the ribosome (G. Gong and C. Yanofsky, Science 297:1864-1867, 2002).
Acknowledgments
We thank Gerald Fuller (University of Alabama) for a plasmid containing the human TB3-1 cDNA for eRF1, Shin-Ichi Hoshino (University of Tokyo) for a plasmid containing the eRF3 cDNA and for rabbit polyclonal anti-eRF3 antiserum, and Lev Kisselev (Engelhardt Institute of Molecular Biology, Moscow, Russia) and Elizabeth Greene and Stephanie Child (Fred Hutchinson Cancer Research Center) for helpful discussions. We also thank the Fred Hutchinson Cancer Research Center Biotechnology, Image Analysis and BioComputing & Informatics Shared Resources for technical assistance.
This work was supported by NIH grants AI26672 and CA91329.
REFERENCES
- 1.Alderete, J. P., S. J. Child, and A. P. Geballe. 2001. Abundant early expression of gpUL4 from a human cytomegalovirus mutant lacking a repressive upstream open reading frame. J. Virol. 75:7188-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderete, J. P., S. Jarrahian, and A. P. Geballe. 1999. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol. 73:8330-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram, G., H. A. Bell, D. W. Ritchie, G. Fullerton, and I. Stansfield. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornsson, A., S. Mottagui-Tabar, and L. A. Isaksson. 1996. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 15:1696-1704. [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, J., and A. P. Geballe. 1996. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 16:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, J., and A. P. Geballe. 1996. Inhibition of nascent-peptide release at translation termination. Mol. Cell. Biol. 16:7109-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, J., and A. P. Geballe. 1998. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA 4:181-188. [PMC free article] [PubMed] [Google Scholar]
- 8.Chavatte, L., L. Frolova, L. Kisselev, and A. Favre. 2001. The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 268:2896-2904. [DOI] [PubMed] [Google Scholar]
- 9.Degnin, C. R., M. R. Schleiss, J. Cao, and A. P. Geballe. 1993. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J. Virol. 67:5514-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didichenko, S. A., M. D. Ter-Avanesyan, and V. N. Smirnov. 1991. Ribosome-bound EF-1 alpha-like protein of yeast Saccharomyces cerevisiae. Eur. J. Biochem. 198:705-711. [DOI] [PubMed] [Google Scholar]
- 11.Drugeon, G., O. Jean-Jean, L. Frolova, X. Le Goff, M. Philippe, L. Kisselev, and A. L. Haenni. 1997. Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res. 25:2254-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebihara, K., and Y. Nakamura. 1999. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA 5:739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, P., Z. Wang, and M. S. Sachs. 2000. Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem. 275:26710-26719. [DOI] [PubMed] [Google Scholar]
- 14.Frolova, L., X. Le Goff, H. H. Rasmussen, S. Cheperegin, G. Drugeon, M. Kress, I. Arman, A. L. Haenni, J. E. Celis, M. Philippe, et al. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701-703. [DOI] [PubMed] [Google Scholar]
- 15.Frolova, L., X. Le Goff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334-341. [PMC free article] [PubMed] [Google Scholar]
- 16.Frolova, L., A. Seit-Nebi, and L. Kisselev. 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolova, L. Y., T. I. Merkulova, and L. L. Kisselev. 2000. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolova, L. Y., J. L. Simonsen, T. I. Merkulova, D. Y. Litvinov, P. M. Martensen, V. O. Rechinsky, J. H. Camonis, L. L. Kisselev, and J. Justesen. 1998. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 256:36-44. [DOI] [PubMed] [Google Scholar]
- 19.Frolova, L. Y., R. Y. Tsivkovskii, G. F. Sivolobova, N. Y. Oparina, O. I. Serpinsky, V. M. Blinov, S. I. Tatkov, and L. L. Kisselev. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geballe, A. P., and M. K. Gray. 1992. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 20:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geballe, A. P., and M. S. Sachs. 2000. Translational control by upstream open reading frames, p. 595-614. In W. B. Hershey, M. B. Matthews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc. Natl. Acad. Sci. USA 98:8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong, F., and C. Yanofsky. 2002. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 277:17095-17100. [DOI] [PubMed] [Google Scholar]
- 24.Gong, F., and C. Yanofsky. 2001. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 276:1974-1983. [DOI] [PubMed] [Google Scholar]
- 25.Grenett, H. E., P. Bounelis, and G. M. Fuller. 1992. Identification of a human cDNA with high homology to yeast omnipotent suppressor 45. Gene 110:239-243. [DOI] [PubMed] [Google Scholar]
- 26.Hayes, C. S., B. Bose, and R. T. Sauer. 2002. Proline residues at the C-terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 277:33825-33832. [DOI] [PubMed] [Google Scholar]
- 27.Hoshino, S., M. Imai, M. Mizutani, Y. Kikuchi, F. Hanaoka, M. Ui, and T. Katada. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 273:22254-22259. [DOI] [PubMed] [Google Scholar]
- 28.Janzen, D. M., and A. P. Geballe. 2001. Modulation of translation termination mechanisms by cis- and trans-acting factors. Cold Spring Harbor Symp. Quant. Biol. 66:459-467. [DOI] [PubMed] [Google Scholar]
- 29.Kisselev, L. L., and R. H. Buckingham. 2000. Translational termination comes of age. Trends Biochem. Sci. 25:561-566. [DOI] [PubMed] [Google Scholar]
- 30.Law, G. L., A. Raney, C. Heusner, and D. R. Morris. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276:38036-38043. [DOI] [PubMed] [Google Scholar]
- 31.Le Goff, X., M. Philippe, and O. Jean-Jean. 1997. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol. 17:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomis, W. P., J. T. Koo, T. P. Cheung, and S. L. Moseley. 2001. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 39:693-707. [DOI] [PubMed] [Google Scholar]
- 33.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60:366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkulova, T. I., L. Y. Frolova, M. Lazar, J. Camonis, and L. L. Kisselev. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443:41-47. [DOI] [PubMed] [Google Scholar]
- 35.Mize, G. J., H. Ruan, J. J. Low, and D. R. Morris. 1998. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Biol. Chem. 273:32500-32505. [DOI] [PubMed] [Google Scholar]
- 36.Morris, D. R., and A. P. Geballe. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mottagui-Tabar, S., M. F. Tuite, and L. A. Isaksson. 1998. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 257:249-254. [DOI] [PubMed] [Google Scholar]
- 38.Nakatogawa, H., and K. Ito. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108:629-636. [DOI] [PubMed] [Google Scholar]
- 39.Nakatogawa, H., and K. Ito. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7:185-192. [DOI] [PubMed] [Google Scholar]
- 40.Raney, A., G. L. Law, G. J. Mize, and D. R. Morris. 2002. Regulated translation termination at the upstream open reading frame in s-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 277:5988-5994. [DOI] [PubMed] [Google Scholar]
- 41.Seit-Nebi, A., L. Frolova, J. Justesen, and L. Kisselev. 2001. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29:3982-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song, H., P. Mugnier, A. K. Das, H. M. Webb, D. R. Evans, M. F. Tuite, B. A. Hemmings, and D. Barford. 2000. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100:311-321. [DOI] [PubMed] [Google Scholar]
- 43.Stansfield, I., L. Eurwilaichitr, Akhmaloka, and M. F. Tuite. 1996. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 20:1135-1143. [DOI] [PubMed] [Google Scholar]
- 44.Stansfield, I., G. M. Grant, Akhmaloka, and M. F. Tuite. 1992. Ribosomal association of the yeast SAL4 (SUP45) gene product: implications for its role in translation fidelity and termination. Mol. Microbiol. 6:3469-3478. [DOI] [PubMed] [Google Scholar]
- 45.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stansfield, I., and M. F. Tuite. 1994. Polypeptide chain termination in Saccharomyces cerevisiae. Curr. Genet. 25:385-395. [DOI] [PubMed] [Google Scholar]
- 47.Tenson, T., and M. Ehrenberg. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108:591-594. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Z., A. Gaba, and M. S. Sachs. 1999. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J. Biol. Chem. 274:37565-37574. [DOI] [PubMed] [Google Scholar]
- 49.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]