Abstract
Previous studies have suggested that the early-B-cell-specific mb-1(Igα) promoter is regulated by EBF and Pax-5. Here, we used in vivo footprinting assays to detect occupation of binding sites in endogenous mb-1 promoters at various stages of B-cell differentiation. In addition to EBF and Pax-5 binding sites, we detected occupancy of a consensus binding site for E2A proteins (E box) in pre-B cells. EBF and E box sites are crucial for promoter function in transfected pre-B cells, and EBF and E2A proteins synergistically activated the promoter in transfected HeLa cells. Other data suggest that EBF and E box sites are less important for promoter function at later stages of differentiation, whereas binding sites for Pax-5 (and its Ets ternary complex partners) are required for promoter function in all mb-1-expressing cells. Using DNA microarrays, we found that expression of endogenous mb-1 transcripts correlates most closely with EBF expression and negatively with Id1, an inhibitor of E2A protein function, further linking regulation of the mb-1 gene with EBF and E2A. Together, our studies demonstrate the complexity of factors regulating tissue-specific transcription and support the concept that EBF, E2A, and Pax-5 cooperate to activate target genes in early B-cell development.
The regulated expression of stage- and lineage-specific genes is a common theme in the development of differentiated somatic cells. To accomplish this end, tissue-specific and temporally regulated DNA-binding proteins activate a program of gene expression unique to a particular cell type. A major aspect of this regulation is the ability of proteins to work in concert through physical interactions that facilitate DNA binding. Thus, factors that exhibit partially overlapping expression patterns mediate target gene expression through combinatorial association on promoters, enhancers, and other regulatory modules.
B lymphocytes are an important developmental model for understanding how differentiated cells arise from precursor cells with multipotential properties. Although early events in B lymphopoiesis are not yet fully understood, it is accepted that hematopoietic stem cells give rise to common lymphoid progenitor cells with the potential to become T or B lymphocytes. These cells can then differentiate to express early markers of the B-cell lineage. Progression through multiple stages of early differentiation results in rearrangement and expression of immunoglobulin genes, resulting in display of membrane-bound immunoglobulin M (mIgM) as the B-cell receptor for antigen (BCR). Activation of the BCR by antigen results in changes in gene expression, which may involve further maturation in the context of germinal centers and, ultimately, terminal differentiation of B cells to become antibody-secreting plasma cells.
At least three genes encoding sequence-specific DNA-binding proteins are essential for generating early B cells (20). First, E proteins encoded by the E2A gene, E12 and E47, are ubiquitously expressed, yet these alternatively spliced basic helix-loop-helix (bHLH) factors are required for progression of B cells past a very early progenitor stage (4, 54). This is achieved through a combination of cell type-specific multimerization and posttranslational regulation by Id proteins (reviewed in reference 31). Second, mice lacking a tissue-specific regulator, early-B-cell factor (EBF, encoded by the ebf1 gene), fail to rearrange immunoglobulin genes and exhibit a developmental block at a slightly later stage than that observed with E2A deficiency (26). Third, the lack of Pax-5 (also known as B-cell-specific activator protein [BSAP]) in knockout mice results in an accumulation of pre-BI cells that partially rearrange immunoglobulin heavy-chain genes but do not complete rearrangements required for the functional BCR (36, 48). Interestingly, Pax-5-deficient pre-BI cells exhibit a lack of lineage commitment, as evidenced by differentiation into other types of hematopoietic cells (34).
The murine mb-1 promoter is a model system for understanding the control of tissue- and differentiation stage-specific transcription in B-lymphoid development (8, 10, 18, 21, 47, 51). The mb-1 gene encodes Igα, an early B-cell-specific transmembrane protein required for docking of immunoglobulin heavy chains in the B-cell plasma membrane and for transmembrane signaling after stimulation of the BCR by antigen (reviewed in reference 32). In transfection assays, the activity of the TATA-less mb-1 promoter recapitulates the expression pattern of the endogenous mb-1 gene (47). Previous in vitro studies by using DNase I protection and electrophoretic mobility shift assay (EMSA) identified multiple factor binding sites in the promoter, including two sets of sequences recognized by early B-cell-specific factors: the EBF binding site located between positions −178 and −165 (8, 21), and a site that binds Pax-5 in association with Ets family proteins between positions −85 and −66 (10). Between these sites, other investigators identified an octamer-like site similar to that recognized by the POU domain proteins Oct-1 and Oct-2 (28, 29). Two adjacent sequences just upstream of transcription initiation sites (proximal control region) bind Ets proteins (Ets-1, Ets-2, and PU.1) and Sp1, respectively, in vitro (18, 47). Mutation of each of these sites reduced promoter activity in transfected cells, but the question arises as to whether these sites are bona fide regulatory sites in vivo.
To better understand the roles of regulatory sites of the mb-1 gene, we footprinted mb-1 promoters in intact cells (in vivo footprinting) representing different stages of B-cell differentiation. Many of the previously identified factor binding sites, including sequences recognized by EBF, Pax-5:Ets ternary complexes, and Ets proteins (proximal site) in vitro, were selectively protected from chemical modification in intact nuclei. Protection of these sequences was correlated with levels of endogenous mb-1 gene transcripts and developmental stages of the cells. An extended region near the EBF binding site was protected, including a consensus site that binds E2A-encoded proteins in vitro. Transfection studies suggest that sites recognized by EBF and E2A are very important for promoter activity in pre-B cells but are relatively dispensable for activity in pro-B or more mature B cells. Similar to other genes expressed in pre-B cells (e.g., λ5 [44]), our data suggest functional cooperation between EBF and E2A proteins on the mb-1 promoter. Other data suggest that Pax-5:Ets ternary complexes are required for activity in pre-B and mature B cells. We discuss these interactions in the context of B lymphopoiesis and the progression of B-cell differentiation.
MATERIALS AND METHODS
Cell culture conditions.
All cells were grown in RPMI medium supplemented with 7.5% fetal calf serum, 10 mM HEPES, 2 mM pyruvate, 50 μM 2-mercaptoethanol, and 50 μg of gentamicin per ml (Life Technologies AB) at 37°C in 5% CO2. Baf1 and Baf2 subclones of Ba/F3 cells were cultured in the presence of 5% WEHI-3 supernatant containing interleukin-3.
RNA isolation, cDNA synthesis, and quantitative PCR.
Total RNA was prepared by using Trizol (Gibco-BRL). cDNA was prepared by using random hexamers as published previously (24). RNA quantification by “real-time” PCR detection was performed by using the ABI Prism 7700 sequence detection system (Applied Biosystems), and the Stratagene Brilliant core reagent kit (200 nM concentrations of primers and probe, 6 mM MgCl2) with amplification for 40 cycles at 60°C. All reactions were performed in triplicate. Primers and dual-labeled fluorescent probes (Integrated DNA Technologies) for measuring β-actin transcripts were as follows: forward primer, 5′-GACGGCCAAGTCATCACTATTG; reverse primer, 5′-GAAGGAAGGCTGGAAAAGAGC; and FAM-TAMRA fluorogenic probe, 5′-CAACGAGCGGTTCCGATGCCC. For measuring mb-1 transcripts, we used the following: forward primer, 5′-CATCTTGCTGTTCTGTGCAGTG; reverse primer, 5′-TTCTCATTTTGCCACCGTTTC; and FAM-TAMRA fluorogenic probe, 5′-TGCCAGGGACGCTGCTGCTATTC.
All samples were analyzed for control β-actin and mb-1 expression with 2 μl of cDNA (prepared from 200 ng of total RNA). The threshold cycle for expression was determined for each sample and the mean of triplicate tubes calculated. Each sample's mb-1 mean threshold cycle was normalized to its β-actin mean threshold cycle, and expression relative to β-actin was reported as 2 raised to the power of the normalized mean threshold cycle.
DMS protection and/or LM-PCR (in vivo footprinting).
Methylation of DNA in intact cells and ligation-mediated PCR (LM-PCR) was performed by using the protocols of Ausubel et al. and Garrity and Wold, with modifications suggested by Fernandez et al. (3, 9, 12). Briefly, 1 μl of 10% dimethyl sulfate (Aldrich) in ethanol was added to 107 cells in 1 ml of RPMI 1640 with 10% serum and incubated for 90 s at 37°C. Quenching of methylation, isolation of genomic DNA, and cleavage with piperidine was performed as previously described. Control methylation and depurination of genomic DNA (Maxam-Gilbert G or A+G reference ladders) was performed as previously described (17). For LM-PCR of the mb-1 promoter, first-strand synthesis was performed with 2 μg of cleaved genomic DNA and 0.3 μg of primer 1 to detect sense strand (5′-CATTACCCAAACAGGCGTATGACAAG) and to detect antisense strand (5′-GGATCCTTTCTCAGGGATCAGTGGT) as described previously. Reactions were extended one cycle in a thermal cycler (Eppendorf) by denaturing for 5 min at 95°C, annealing for 30 min (64°C for the sense strand and 62°C for the antisense strand), and extending for 10 min at 76°C. Samples were diluted for ligation with annealed linkers and precipitated as described. Amplification of linker-ligated DNA was performed by using LM-PCR.1 (9) and mb-1 primer (sense strand [5′-ACAAGAAGAGGAGGAGAGGCAGGGCTCT] or antisense strand [5′-TTGAACCACCCTCTCCCCGACC]) with 18 cycles of amplification consisting of 1 min at 95°C for 2 min at 68°C for 3 min at 76°C. Amplification reactions were performed with 0.3 pmol of 32P-labeled oligonucleotide (sense strand [5′-GGAGGAGAGGCAGGGCTCTGAGGGCTT] and antisense strand [5′-GAACCACCCTCTCCCCGACCCCA]) with three cycles of 95°C for 2 min and 69°C for 10 min at 76°C. Products were fractionated by electrophoresis on 6% denaturing polyacrylamide gels and detected by using a Molecular Dynamics Storm 960 Phosphorimager (Amersham). Histograms of footprinting data were generated by using ImageQuant software (Amersham).
Plasmid constructs and in vitro mutagenesis.
Plasmids for expression of full-length EBF or E47, or the E47 DBD (amino acids 407 to 649) in transfected cells were described previously (44). The mb-1 promoter reporter plasmids were based on the pGL3-basic reporter plasmid (Promega). The wild-type mb-1 promoter construct was obtained by initially cloning a PCR product spanning the region from −186 to +26 into the SmaI site of pGL3-Basic. Point mutations in the mb-1 promoter were introduced by PCR amplification with mutated sense oligonucleotides to yield either the full-length promoter or the region 3′ of the NdeI site by using the wild-type mb-1 promoter pGL3 construct as a template and 5′-TCTCCCAGTGAGTCGGTTAGTTTG as antisense primer. Wild-type and mutated 5′ oligonucleotides were as follows: wt, 5′-CTAGAGAGAGACTCAAGGGAATTG; M1, 5′-CTAGAGAGAGATTCAAGGGAATTG; M2, 5′-CTAGAGAGAGACTCAATTGAATTGTGGCCAGC; M3, 5′-CTAGAGAGAGACTCAAGGGAATTGTTAAGCCCAGGTGCAGG; M4, 5′-CTAGAGAGAGACTCAAGGGAATTGTGGCCAGCCCTGGTGCAGGGCAG; M5, 5′-CACACATATGGCAAATAAAGGGCCAGGAGTAAGGGCAAATTGAGCCCATCT; M6, 5′-CACACATATGGCAAATAAAGGGCCAGGAGTAAGGGCCACTGGAGCCCATCTAAGGCACGGCTGAAC; and M7, 5′-CACACATATGGCAAATAAAGGGCCAGGAGTAAGGGCCACTGGAGCCCATCTCCGGCACGGCTGAACATCAAGTGAGGCGGAG. The Δdistal mb-1 promoter-reporter includes promoter sequences up to the NdeI site (−112). All mutations were verified by sequencing. The TATA control plasmid, including the TATA box of the murine SP6 Vκ promoter, was described previously (39).
The plasmid for synthesis of recombinant amino-terminal His-tagged EBF(24-429) in Escherichia coli (see below) was constructed by PCR amplification of pEBF17 (17) by using primers 5′-TGAACGCGGTGCGGACGTGGATGCAGGGC and 5′-TCAGACCGAAGTGTTAGCAAGGGCT, followed by ligation into the blunted (Klenow) NdeI site of pET15b (Novagen, Madison, Wis.). The plasmid for synthesis of recombinant hamster E47 DNA-binding domain (amino acids 518 to 649) was produced by amplification of E47FD plasmid DNA (44) with primers 5′-ACCATGGCTCCACGCACGCGCACCA and 5′-TCACAGGTGCCCGGGCGGGTT prior to ligation into the Ecl136II site of pBluescript KS(+).
Production of recombinant proteins.
Recombinant EBF (24-429) was produced in E. coli Rosetta (DE3) (Novagen). Single colonies were picked from plates of freshly transformed bacteria and inoculated into Luria broth supplemented with carbenicillin (500 μg/ml), chloramphenicol (34 μg/ml), and 1 μM zinc acetate. At an optical density at 600 nm of 0.6, cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h with subsequent harvest of bacteria by centrifugation. Bacterial pellets were resuspended in ice-cold binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]), mechanically lysed with a French press, and centrifuged for 60 min in a Sorvall SS34 rotor at 15,000 rpm to remove bacterial debris. Soluble EBF(24-429) protein was purified by using a His-Bind chromatography kit (Novagen) according to the manufacturer's instructions. After dialysis into 20 mM Tris-HCl (pH 8.0), 1 M NaCl, and 5 mM 2-mercaptoethanol, protein was further purified by using Superdex 200 size exclusion chromatography. The protein concentration was determined by the Bradford assay (Bio-Rad). Protein purity was assessed by using 4 to sodium dodecyl sulfate-20% polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining.
The DNA-binding domain of hamster E47 (Pan-1; amino acids 518 to 649) was synthesized by in vitro transcription and translation as described previously (11).
Nuclear protein extracts and EMSAs.
Preparation of nuclear extracts, preparation of DNA probes, and EMSA were performed according to Fitzsimmons et al. (10) with only slight modifications: electrophoretic mobility shift assay (EMSA) reactions did not include detergents, and nondenaturing gels were prepared with 0.5× Tris-borate-EDTA. Pairs of oligonucleotides used for EMSA in Fig. 4 were as follows: murine mb-1 promoter wild-type distal region (5′-TAAAGCCCAGGTGCAGGGCAGTTC and 5′-TAGAACTGCCCTGCACCTGGGCTT); mb-1 promoter mutated distal region (5′-TAAAGCCCTGGTGCAGGGCAGTTC and 5′-TAGAACTGCCCTGCACCAGGGCTT); murine immunoglobulin heavy chain (μ) enhancer μE5 site (5′-TCGAGATCAGAACCGGAACACCTGCAGCA and 5′-TCGATGCTGCAGGTGTTCCGGTTCTGATC); mutated μE5 site (5′-TCGAGATCAGAACCGGAAAACCTTCAGCA and 5′-TCGATGCTGAAGGTTTTCCGGTTCTGATC); and murine μ enhancer μE3 site (5′-TCGAGAAGCAGGTCATGTGGCAGTACT and 5′-TCGAAGTACTGCCACATGACCTGCTTC). Antisera used in EMSA supershift experiments (except for the EBF-specific antiserum produced by M. Sigvardsson) were obtained from Santa Cruz Biotechnology. The probe in Fig. 6 was comprised of the oligonucleotides 5′-CTAGAGAGAGACTCAAGGGAATTGTGGCCAGCCCAGGTGCAGGGCA and 5′-CTAGTGCCCTGCACCTGGGCTGGCCACAATTCCCTTGAGTCTCTCT.
FIG. 4.
Protection of sequences from DMS in footprinting assays correlates with requirements for mb-1 promoter activity in transfected cells. (A) Summary of results from in vivo footprinting and mb-1 promoter mutations tested in panel B. Sequences of the murine mb-1 promoter (−197 to +29, including the translational start codon) are shown with previously mapped sites of transcriptional initiation (47). Approximate binding sites for factors are labeled above and underlined between strands. Guanines and adenines protected in footprinting assays are indicated by open circles for each strand. Guanines showing enhanced cleavage (hypersensitivity) are indicated by closed circles. Mutations tested in panel B (relative to sense strand sequences) are indicated below sequences as lowercase case letters. (B) Functional requirements for mb-1 promoter factor binding sites in transfected cells. Plasmids with wild-type or mutated mb-1 promoter sequences (as in panel A) indicated at left were introduced into Ba/F3 pro-B cells, 18-81 pre-B cells, or A20 lymphoma cells in short-term transfection assays as shown. Luciferase activities were determined relative to the TATA plasmid control (see Materials and Methods) and represent the means of three experiments.
FIG. 6.
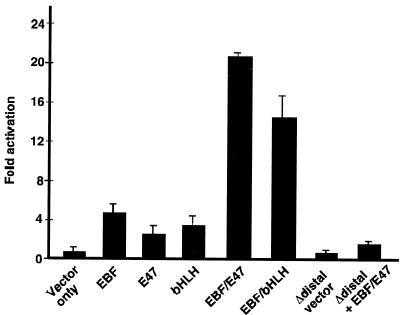
EBF and E47 synergistically activate the mb-1 promoter in transfected HeLa cells. Luciferase reporter plasmids with wild-type or mutated promoters were cotransfected with plasmids for expression of full-length murine EBF, full-length hamster E47 (Pan-1), or E47(407-649) as indicated below. The Δdistal promoter, which lacks mb-1 promoter sequences upstream of the NdeI site (at −112; deleted sequences include the EBF and E2A binding sites) was tested by itself or with coexpressed EBF and E47. All values were normalized to cotransfected Renilla luciferase activity and represent the means of four experiments.
EMSA in Fig. 5 was performed by using nuclear proteins prepared by the method of Schreiber et al. (41). The oligonucleotides used for EMSAs were as follows: OCT sense (5′-TTCATTGATTTGCATCGCATGAGACGCTAACATCGTACGTTC) and antisense (5′-GAACGTACGATGTTAGCGTCTCATGCGATGCAAATCAATGAA), EBF sense (5′-GAGAGAGACTCAAGGGAATTGTGG) and antisense (5′-CCACAATTCCCTTGAGTCTCTCTC), μE5 (E2A) sense (5′-GGCCAGAACACCTGCAGACG) and antisense (5′-CGTCTGCAGGTGTTCTGGCC) and CD19 (Pax-5) sense (5′-GCAGACACCCATGGTTGAGTGCCCTCCAGG) and antisense (5′-CCTGGAGGGCACTCAACCATGGGTGTCTGC).
FIG. 5.
Expression and DNA-binding activities of E2A, EBF, and Pax-5 proteins in cell lines. (A) Western analysis of protein expression in representative cell lines. Similar numbers of Ba/F3 pro-B, 18-81 pre-B, A20 lymphoma, or S194 plasmacytoma cells were lysed, and proteins were electrophoresed on SDS-PAGE gels. Proteins were Western blotted and probed with antibodies specific for E2A, EBF, or Pax-5. (B) EMSA analysis of Oct-1, Pax-5, E2A, and EBF DNA-binding activities in B-cell line nuclear extracts. Binding of similar amounts of nuclear extract proteins was tested by using 32P-labeled probes.
Transient transfections and luciferase assays.
Lymphoid cells were transiently transfected as previously described (44). For HeLa cell transfections, 500,000 cells were transfected with Lipofectin (Life Technologies) according to the manufacturer's instructions. Cells were harvested after 40 h, and protein extracts were analyzed by using the dual luciferase assay kit (Promega).
Western blot detection of proteins.
Western analysis was performed as previously described (44). Pax-5, E2A, and EBF-specific antibodies were obtained from Santa Cruz Biotechnology. Detection of the secondary antibody was obtained by using the ECL system (Amersham).
DNA microarray analysis.
Affymetrix analysis was performed with biotin-labeled cRNA obtained by in vitro transcription from double-stranded cDNA from the indicated cell lines. Then, 15 μg of cRNA was hybridized to Affymetrix murine genome U74Av2 microarrays (interrogating 12,000 genes and expressed sequence tags) in an Affymetrix GeneChip hybridization oven 320, developed by the addition of fluorescein isothiocyanate-avidin, and washed by using an Affymetrix GeneChip Fluidics Station 400. Detection was performed by using an HP gene array scanner at ×100. All measurements were normalized to zero mean and unity variance. Standard Pearson correlation coefficients were determined for data obtained from 12 different measurements. Calculations were carried out by using the R statistical computing package (ref http://www.r-project.org/).
RESULTS
Expression of the mb-1 gene is modulated during B-cell development.
Prior to identifying sequences that regulate mb-1 promoter function in vivo, we quantitated levels of mb-1 transcripts in murine cell lines representing various stages of B-cell development with a sensitive “real-time” PCR assay. For each cell line, total RNA was converted to cDNA for quantitative PCR analysis by using mb-1 gene-specific primers. All determinations were normalized to levels of β-actin transcripts. The analysis (Table 1) showed that all of the cell lines express significant levels of mb-1 transcripts, with the exceptions of Ba/F3 pro-B cells and terminally differentiated S194 and J558L plasmacytoma/myeloma cells, which had nearly undetectable levels of transcripts. In cell lines representing pre-B cells and intermediate stages of differentiation, levels of transcripts varied over a wide range but were similar between cells at similar stages of development. Abelson virus-transformed pre-B cell lines 40E1 and 18-81, immature (IgM+) B-cell lymphoma (WEHI-231), and a more mature (IgG+) B-cell lymphoma (M12.4.1) expressed high levels of transcripts (ranging from 471.1- to 199.5-fold greater than S194 cells). Treatment of 70Z/3 pre-B cells with bacterial lipopolysaccharide, which activates Igκ transcription in these cells (42), decreased expression by one-half, but levels remained high relative to terminally differentiated cells. In contrast, B lymphoma cells with a germinal center B-cell-like phenotype (A20 and K46 [7]) expressed much lower levels (22.5- to 61.8-fold) of mb-1 transcripts, suggesting changes in the binding of factors to mb-1 gene regulatory elements at this stage of development. Intermediate levels of transcripts were detected in CH12 cells, which decreased somewhat after spontaneous immunoglobulin heavy-chain class switching from IgM- to IgA-expressing cells (156.5 to 113.8-fold). The data suggest that mb-1 transcription is modulated at different stages of B-cell development.
TABLE 1.
Quantitative (real-time) PCR analysis of mb-1 transcripts in murine B-cell lines
| Cell line | Developmental stagea | Immunoglobulin gene statusb | Relative concn of mb-1 transcriptsc |
|---|---|---|---|
| Ba/F3 | Pro-B | All GL | 2.4 ± 0.3 |
| 40E1 | Pre-B | μ, GL | 471.1 ± 0.1 |
| 18-81 | Pre-B | μ, GL | 245.6 ± 0.3 |
| 70Z/3 | Pre-B | μ, κd | 215.3 ± 0.2 |
| 70Z/3 + LPSe | Pre-B | μ, κ | 99.0 ± 0.2 |
| WEHI-231 | Imm-B | μ, κ | 233.9 ± 0.1 |
| M12.4.1 | Mature | γ, κ | 199.5 ± 0.2 |
| A20 | Mature (GC) | γ, κ | 22.5 ± 0.2 |
| K46 | Mature (GC) | γ, κ | 61.8 ± 0.2 |
| CH12-IgM+ | Maturef | μ, κ | 156.5 ± 0.6 |
| CH12-IgA+ | Mature | α, κ | 113.8 ± 0.1 |
| S194 | Plasma cell | α, κ | 1.0 ± 0.3 |
| J558L | Plasma cell | αg, λ | 4.2 ± 0.2 |
ImmB, immature or naive B cell; GC, germinal center.
GL, germ line or unrearranged genes. Expression of heavy-chain (μ, γ, or α) and light-chain (GL, κ, or λ) gene is indicated (1, 7, 24, 25, 38, 42, 50, and 52).
Fold increase versus S194 cells.
κ genes in 70Z/3 cells are only expressed after incubation of cells with bacterial lipopolysaccharide.
LPS, lipopolysaccharide.
CH12 cells undergo heavy-chain class switching in culture to express IgA (α chains). The IgM+ and IgA+ populations were derived by staining and sorted to enrich for the cell surface isotypes.
Heavy-chain genes are deleted in these cells.
In vivo footprinting detects occupancy of mb-1 promoters by multiple factors.
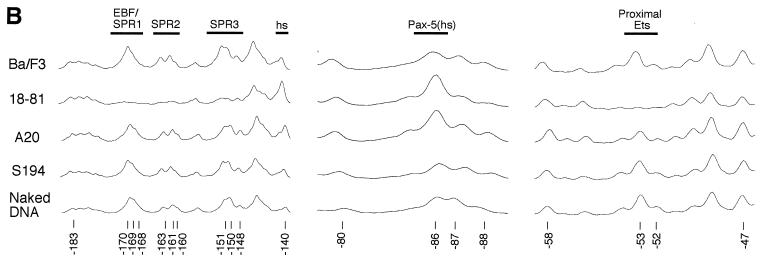
Detection of different levels of mb-1 transcripts in the cell lines suggests that promoter function is modulated by trans-acting factors in a stage-specific fashion during cell differentiation. To identify how occupancy of factor binding sites changes at various stages of differentiation, we performed in vivo footprinting assays on the cell lines shown in Table 1. Figures 1 and 2 show patterns of guanine cleavage after DMS modification and detection by using LM-PCR. In each experiment, patterns obtained from intact viable cells were compared with naked DNA methylated in vitro. A ladder of guanine and adenine cleavage was electrophoresed in parallel to provide reference markers. To quantitate differences observed between cell samples, we prepared histograms of footprinting data from four representative cell lines (Ba/F3, 18-81, A20, and S194) and control DNA by using a Molecular Dynamics Phosphorimager and ImageQuant software.
FIG. 1.
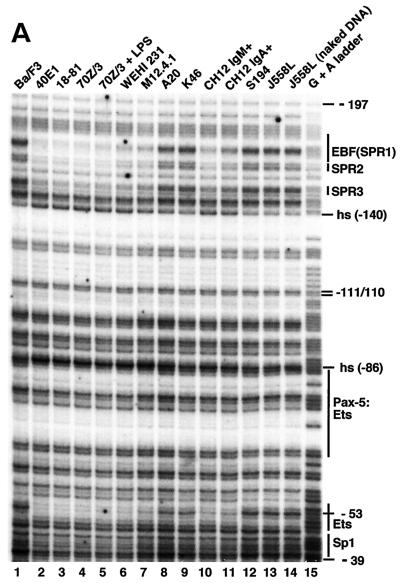
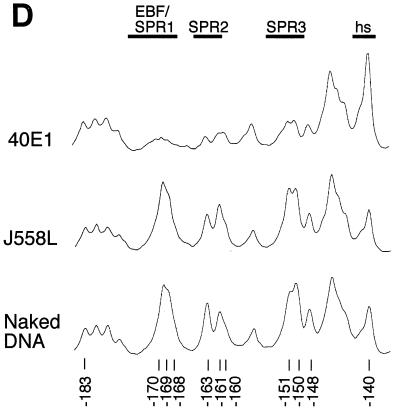
In vivo footprinting analysis of the mb-1 promoter sense strand. Cell lines indicated at top were treated with DMS and cleaved with piperidine prior to amplification of mb-1 promoter sense strand cleavage products by using ligation-mediated PCR. (A) Protection and cleavage of sense strand between −39 and −197. Protected regions and approximate locations of factor binding sites are indicated at right. SPR sense strand protected region. (B) Histograms of cleavage products detected in panel A. Histograms shown for lanes 1 (Ba/F3), 3 (18-81), 8 (A20), 12 (S194), and 14 (J558L naked DNA) were prepared by using ImageQuant software (Molecular Dynamics). All plots are shown to the same scale. Significant differences between peak intensities are highlighted. Factor binding sites and/or sense strand protected regions are indicated above. Note reduced signals at EBF/SPR1, SPR2, and SPR3 sites only in 18-81 cells. Hypersensitivity at −86 is detected in 18-81 and A20 cells. Proximal Ets site (−53/−52) is protected in 18-81, less so in A20 cells. (C) Protection and cleavage products of sense strand between −123 and −197. (D) Histograms of cleavage products detected in panel C. Histograms are shown for lanes 2, 3, and 4.
FIG. 2.
In vivo footprinting analysis of the mb-1 promoter antisense strand. Cell lines indicated at top were treated with DMS and cleaved with piperidine prior to amplification of mb-1 promoter antisense strand cleavage products by using ligation-mediated PCR (see Materials and Methods). (A) Protection and cleavage of antisense strand between −188 and −97. Protected regions and approximate locations of factor binding sites are indicated at right. APR antisense strand protected region. (B) Histograms of cleavage products detected in panel A. See Fig. 1B for details. Histograms are shown for lanes 2, 4, 9, 13, and 15. Note reduced signals in 18-81 cells at EBF/APR1, APR2, and APR3. (C) Protection and cleavage products of antisense strand between −122 and −48. Guanines discussed in the text are indicated at left. The guanine at −74 is hypersensitive to DMS modification in mb-1-expressing cells. (D) Histograms of cleavage products detected in panel C. Histograms are shown for lanes 2, 4, 9, 13, and 15. The bipartite paired domain of Pax-5 makes two sets of contacts.
First, we used the in vivo footprinting assay to detect protected regions on the sense strand of the mb-1 promoter between −197 and −39 (Fig. 1A). Histograms of representative data are shown in Fig. 1B. Patterns of bands detected by using DNA obtained from nonexpressing Ba/F3, S194, and J558L cells (lanes 1, 12, and 13) were indistinguishable from that generated with control DNA (lane 14), suggesting that mb-1 promoters are not occupied by regulatory proteins in these cells, or that occupancy is not stable in a significant fraction of cells. In contrast, evidence of protected sequences was obtained in pre-B, immature B, and more mature cells in three distinct regions of the distal promoter sense strand (SPR1, -2, and -3). Protection of guanines between −170 to −140 was observed in each of the pre-B cell lines (lanes 2 to 5), in WEHI-231 and M12.4.1 cells (lanes 6 and 7), and in CH12 IgM+ cells (lane 10). A slight diminution of protection was observed in CH12 IgA+ cells, which have undergone IgH class switching (lane 11). Weak protection (50% relative to Ba/F3) over this region was observed in cells with germinal center B-cell phenotypes, A20 and K46 (lanes 8 and 9).
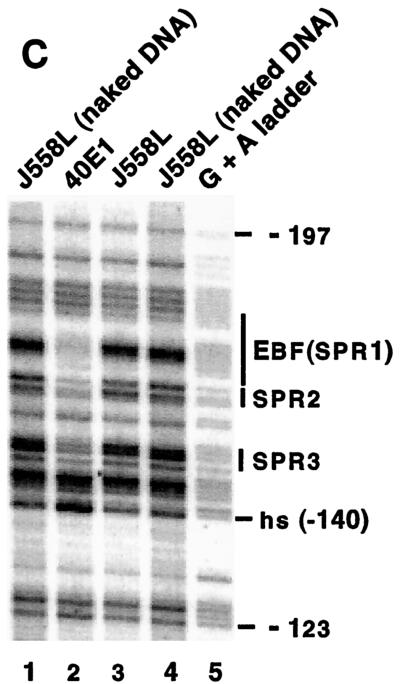
Electrophoresis of samples for an extended period (Fig. 1C and D) resolved details of protection of guanine residues at −170, −169, and −168 in 40E1 pre-B cells (lane 2). Protected bases include predicted contacts within one of the two half sites recognized by EBF (5′-AGACTCAAGGGAAT) in vitro (17, 19). Unexpectedly, additional protected regions, including two protected guanines at −163 and −161 (SPR2), and −151, −150, and (to a lesser degree) −148 (SPR3) were detected that had not been identified previously as factor binding sites. Further downstream, strongly enhanced modification (hypersensitivity) of the guanine at −140 was detected. Overall, the patterns suggest binding of the promoter by EBF and additional factors at downstream sites in early B cells.
Evidence was obtained for factor binding to two other regions of the promoter sense strand in vivo. A single base at −86, which corresponds to a guanine adjacent to the base pair contacted by Ser133 of Pax-5 in the Pax-5:Ets-1 crystal structure (13), is hypersensitive to cleavage in all cell lines expressing high levels of mb-1 transcripts (Fig. 1A and B). Diminished but significant hypersensitivity of this base was detected in A20 (Fig. 1B) and K46 lymphoma cells. In all mb-1-expressing cells, strong protection was observed at −53 (and less at −52) within the Ets protein binding site of the basal control region of the promoter. Protection of these sites was not detected in Ba/F3, S194, or J558L cells. Interestingly, although previously shown to be functionally important in transfection assays (47), protection of the Sp1 binding site identified adjacent the proximal Ets site was not detected. Also, protection of bases −111/−110 within the sequence 5′-ATGGCAAAT, which was reported to be a low affinity binding site for Oct-2 in vitro, was not observed in any of the cell lines (28).
Figure 2A and B show in vivo footprinting and histograms of the antisense strand between −97 and −188. Again, patterns obtained from Ba/F3, S194, and J558L cells are nearly identical with the pattern obtained from methylation of naked DNA in vitro (lanes 2, 13, and 14). In contrast, pre-B cells (lanes 3 to 6), immature B cells (lane 7), and more mature B cells (lanes 8, 11, and 12) showed strong protection of the guanine at −173 (antisense strand protected region 1 [APR1]) within the upstream half site contacted by EBF (5′-GAGTCT [17]). Protection of these regions was lacking in A20 and K46 cells (lanes 9 and 10). We conclude that EBF sites are occupied in pre-B cells, but EBF sites are not occupied in cells representing later stages of development. Protection of the guanine at −153 (APR2) and enhancement of the adjacent guanine at −152 was detected in all cells expressing high levels of mb-1 transcripts, was less pronounced in CH12 cells, and was not detected in A20 and K46 lymphoma cells. Weak protection of the guanine at −147 (APR3) was also detected. Protection of guanines within APR1, APR2, and APR3 was detected coordinately. Thus, similar to data from the sense strand, the data support binding of EBF to the promoter in cells that express high levels of mb-1 transcripts (e.g., 18-81), but not in cells expressing only low (e.g., A20) or absent levels of transcripts (e.g., S194).
A complex pattern of protection and hypersensitivity was detected within the region encompassing binding sites for Pax-5:Ets ternary complexes (Fig. 2C and D). Similar patterns of protection were evident in pre-B cells, WEHI-231, and M12.4.1, and to a lesser degree CH12 cells. As with the hypersensitive base at −86 on the sense strand, A20 (and K46 [data not shown]) exhibited reduced, but significant protection of the Pax-5:Ets binding site on the antisense strand (Fig. 2D). Patterns included protection of the guanine at −85, which is a contact for Ser133 within the Pax-5 carboxy-terminal subdomain (13), and of the guanine at −84, which reflects contacts made on either side by Ser133 and Arg137. Protection of the guanine at −83 was weakly detected. Enhanced methylation of the guanine at −74, which is present within a region of the phosphodiester backbone contacted by the linker connecting the amino- and carboxy-terminal subdomains of Pax-5, correlates with protection of flanking bases. The guanine at −71, although less reactive with DMS overall, was protected in mb-1-expressing cells. This base, which is contacted by His62 in the crystal structure, is a key contact for DNA binding by Pax-5 (51). Very strong protection observed at guanines at −68/-69 corresponds to the GGA core recognized by Ets partners of Pax-5. Protection of these two bases was detected coordinately with protection and/or enhancement at known Pax-5 contacts. Therefore, as predicted by in vitro studies, Pax-5 and Ets binding sites are occupied in concert in vivo. Protection was not observed over the entire Pax-5:Ets ternary complex site in nonexpressing Ba/F3, S194, or J558L cells. In accord with data from the sense strand, protection of the single guanine within the proposed Oct-2 binding site (−109) was not detected in any of the cell lines. Evidence for binding by Pax-5:Ets ternary complexes was obtained in all cells expressing mb-1 transcripts. We could not confirm binding of the promoter by Oct proteins in vivo.
SPR3 and APR2/3 comprise an E box.
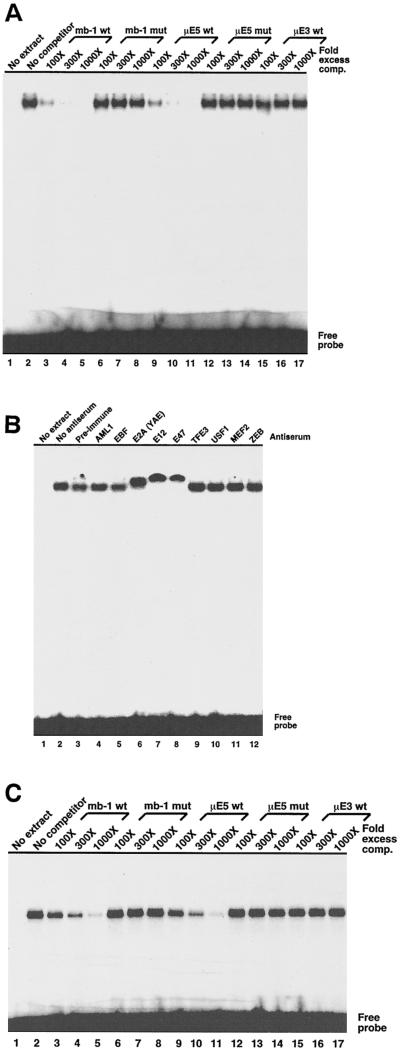
Nucleotide sequences within the newly identified protected regions of the promoter did not suggest the identity(ies) of protein(s) that interact with SPR2, however, the region including SPR3, APR2, and APR3 (5′-CCAGGTGC) includes a consensus binding site (E box) recognized by basic helix-loop-helix proteins (5′-CANNTG). More specifically, the sequence 5′-CAGGTG matches the binding sites identified for the E2A gene-encoded E proteins, which bind this sequence either as E47-E47 homodimers or as E47-E12 heterodimers (6, 49). To identify proteins that bind this site, we performed EMSA by using crude nuclear extract proteins from 40E1 pre-B cells and a 32P-labeled double-stranded oligonucleotide probe comprising −157 to −137 of the wild-type mb-1 promoter. As shown in Fig. 3A, a single low-mobility band was detected. The specificity of complex formation was demonstrated by inhibiting binding with increasing amounts of unlabeled wild-type oligonucleotides (lanes 3 to 5) but not mutated oligonucleotides (lanes 6 to 8 [5′-CAGGTG] is changed to [5′-CTGGTG]). Proteins in the complexes were further identified by competition with oligonucleotides comprising the μE5 site of the murine immunoglobulin heavy-chain enhancer (lanes 9 to 11), which includes an E2A binding site (22, 33). Similar to the mb-1 sequence, mutation of a single base pair of the consensus site blocked competition in vitro (lanes 12 to 14). Assembly of complexes was not inhibited by 1,000-fold excess oligonucleotides comprising the heavy-chain enhancer μE3 E box, which binds related bHLH proteins (TFE3), but not E2A proteins (5).
FIG. 3.
Specific binding of nuclear extract and recombinant E2A proteins to an mb-1 promoter distal region (−136 to −157) probe in vitro. All complexes were detected by using a Molecular Dynamics PhosphorImager. (A) Binding of B-cell nuclear extract proteins to the mb-1 distal region is specifically inhibited by wild type, but not mutated mb-1 promoter sequences. EMSA was performed by incubating 2 μg of crude 40E1 pre-B-cell nuclear protein with the promoter probe in the presence of increasing amounts of unlabeled double-stranded oligonucleotides as shown prior to fractionation on a nondenaturing polyacrylamide gel. Mutated (mut) competitors have a single base change in the E2A consensus site (5′-CAGGTG to 5′-CTGGTG). (B) E2A-specific antisera supershift mb-1 promoter distal region complexes. EMSA was performed as in panel A, except that specific antisera were included (at 1:50 final dilution) during binding reactions.(C) Binding of recombinant E47 to the mb-1 promoter distal region. Hamster E47 (Pan-1) DNA-binding domain (residues 518 to 649) was synthesized by translating synthetic RNA in vitro by using reticulocyte lysates. Binding reactions were performed with 2-μl lysates as shown. “No RNA” indicates unprogrammed lysate. DNA competitors and concentrations are identical to those used in panel A.
The presence of E47 and/or E12 in these complexes was confirmed by the addition of specific antisera to binding reactions prior to EMSA (Fig. 3B). Antisera generated against common epitopes in E12 and E47 (Yae; lane 6), or specific for E12 (lane 7) or E47 (lane 8) further retarded migration of the specific complexes. In contrast, no effect was seen in the presence of preimmune antisera, with specific antibodies raised against AML1 (lane 4), EBF (lane 5), other proteins that bind E boxes including TFE3, USF1, and ZEB (lanes 9, 10, and 12), or MEF2. No reactivity was detected by using antisera raised against cMyc, Pax-5, PU.1, or Ets-1 (data not shown). We conclude that the complexes are comprised of E47 and/or E12, but the significant cross-reactivity of the antisera employed here prevents a more definite identification of these proteins.
E47 and E12 are likely components of mb-1 promoter complexes, but we did not determine whether E2A proteins bind the promoter independently of other proteins. To answer this question, we performed EMSA by using in vitro-translated recombinant E47 (hamster Pan-1) DNA-binding domain (E47-DBD; amino acids 518 to 649) proteins as in Fig. 3A. Specific binding and competition detected was very similar to that detected by using pre-B-cell nuclear extracts (Fig. 3C). Thus, we conclude that the mb-1 promoter includes a target site for E2A proteins.
Functional requirements for factor binding sites.
The in vivo footprinting data (summarized in Fig. 4A) suggest that the mb-1 promoter is differentially occupied at different stages of development. To determine the relative contributions of factor binding sites for mb-1 promoter transcription, we quantitated effects of mutations in a minimal functional promoter segment (−185 to +25) linked to a firefly luciferase reporter gene in short-term transfection experiments. Sequences tested (Fig. 4A) included mutations disrupting either of the EBF half-sites (M1 and M2), Pax-5 (M5), and Ets (M6) ternary complexes, or the proximal Ets site (M7). We also introduced mutations in SPR2 (M3) and APR2/APR3/SPR3 (M4). To obtain an estimate of their relative functional activity, we transfected a reporter plasmid containing only a TATA box upstream of the luciferase gene (TATA). Three different cell lines were transfected representing pro-B (Ba/F3), pre-B (18-81), and more mature B cells (A20), in which different levels of endogenous mb-1 expression (absent, high, or low, respectively) and DMS protection patterns were detected.
We first tested the dependence of promoter activity on factor-binding sites in pro-B cells (Ba/F3). These cells do not express endogenous mb-1 transcripts (Table 1), but activity of the transfected promoter was detected (Fig. 4B). This is somewhat surprising, considering the lack of endogenous mb-1 expression in these cells. However, it is important to note that the transcription detected is exaggerated due to the very high transfection efficiency of Ba/F3 cells. Moreover, these assays test the activities of promoter constructs in the absence of chromatin, which would normally be expected to repress promoter function in the absence of early B-cell-specific activators. Notably, EBF site mutations M1 and M2, E-box mutation M4, and M6, the Ets site involved in cooperative activation with Pax-5, did not affect promoter function; therefore, these sites are not functional in Ba/F3 cells. A small decrease was observed with the M3 mutation of the SPR2 site. Of the mutations tested, only the M7 mutation in the proximal Ets binding site had a large effect on promoter function (decreased to one-eighth of wild-type levels). We conclude that, of the sequences tested, only the proximal region within the basal promoter is fully functional in these cells.
The promoter showed relatively higher activity in 18-81 pre-B cells (similar results were obtained in 230-238 pre-B cells [data not shown]) depending on binding sites for early B-cell-specific factors. Either of two (M1 and M2) mutations that disrupt EBF binding (21) reduced promoter function to less than half that of the wild type. A similar effect was observed with mutation of SPR2 (M3). Mutation of the E box (M4) also resulted in reduced promoter activity. The most dramatic decreases in promoter function were observed with mutations of Pax-5 (M5) or the ternary complex Ets sites (M6) and within the proximal Ets binding site (M7).
In contrast with pre-B cells, A20 (and K46) cells express only low levels of endogenous mb-1 transcripts. Footprinting data in these cells suggested a lack of occupancy of EBF and E2A binding sites. In transfected A20 cells, mutations of these sites (M1, M2, or M4) had only small effects on promoter function relative to pre-B cells. In contrast, mutations in the Pax-5 or Ets ternary complex sites (M6 or M7) decreased activity to one-half and one-third of the wild type, respectively, a finding consistent with detection of Pax-5:Ets ternary complexes in these cells (J. Hagman, data not shown). As with other cell lines, the greatest effects were observed with mutation of the proximal region Ets binding site (M7), which decreased promoter function to 7% of that of the wild type. We conclude that EBF and E2A play a lesser role in activating the mb-1 gene in these cells, whereas Pax-5:Ets ternary complexes are important for promoter function.
Functional data from transfection assays suggest that early B-cell-specific factors may be present at different levels in B cells representing different stages of development. Therefore, we performed Western blotting analyses of cell extracts obtained from similar numbers of Ba/F3 pro-B, 18-81 AMuLV-transformed pre-B, K46 lymphoma, and S194 plasmacytoma cells with specific antibodies to quantitate relative concentrations of E2A, EBF, and Pax-5 proteins (Fig. 5A). Concentrations of E2A proteins varied over a two- to threefold range, with higher expression observed in Ba/F3 and 18-81 cells. In contrast, 18-81 cells expressed high levels of EBF, whereas the other cell lines expressed only low levels of this factor. Pax-5 was not detected in Ba/F3 or S194 cells, but similar amounts were detected in 18-81 and K46 cells (and A20 cells, data not shown). Next, we performed EMSA with Pax-5-, EBF-, or E2A-specific probes to assess the levels of functional DNA-binding proteins in nuclear extracts derived from these cells (Fig. 5B). As a control, we detected similar levels of Oct-1 binding to an octamer site with each extract. In support of the Western data, binding of the Pax-5 probe was similar with 18-81 or K46 extracts but was not detected with Ba/F3 or S194 extracts. EBF binding was only significantly detected with 18-81 extracts. In contrast with detection in Western assays, binding of E2A proteins (as BCF-1 [2, 23, 43]) was only detected in 18-81 cells, suggesting that the protein is functional only in the pre-B-cell line (the faster-migrating band detected with S194 cell extracts does not react with anti-E2A antibodies). Overall, patterns of protection observed in footprinting assays (Fig. 1 and 2), detection of proteins in Western blots (Fig. 5A), and detection of functional DNA-binding activities in EMSA (Fig. 5B), are consistent with active E2A proteins in pre-B cells and posttranslational inhibition at other stages of B-cell development (33, 40). In summary, differences in the expression of EBF and Pax-5 were detected that may account for the relative contributions of their binding sites to mb-1 promoter activity in cells at different stages of differentiation.
Synergistic activation and binding of the mb-1 promoter by EBF and E47.
The proximity of E2A and EBF binding sites in the mb-1 promoter suggested that they functionally interact on this control module. To this end, we cotransfected HeLa epithelial cells with vectors for expression of full-length EBF and/or the E2A-encoded E47 protein together with mb-1 promoter/luciferase reporter plasmids. As shown in Fig. 6, EBF activated the promoter fivefold by itself, while full-length E47 activated the promoter 2.5-fold. However, coexpression of these two activators synergistically increased mb-1 transcription by 22-fold. Expression of the bHLH DNA-binding domain of E47 (residues 407 to 649) with EBF also activated the promoter synergistically but to a lesser degree (15-fold) than did full-length E47. This result is explained by the presence of potent activation domains in EBF, which appear to be sufficient for promoter activation in the absence of activation domains of E47. As a control, deletion of the distal region of the promoter (upstream of −112), including EBF and E-box binding sites resulted in the nearly complete loss of transactivation by EBF and E47.
To address whether synergistic activation by EBF and E47 is a function of cooperative binding to the promoter, we performed EMSA to measure binding by recombinant EBF (residues 25 to 429) in the absence or presence of recombinant E47 DNA-binding domain (amino acids 518 to 649). As shown in Fig. 6, increasing amounts of EBF resulted in a single band at lower concentrations (lanes 2 to 4), and a slower-migrating band indicative of higher-order multimers (17) at higher concentrations (EBFx4 in lane 5). Binding of EBF was not affected by the addition of a 1,000-fold excess of E2A binding sites (lane 6). In lanes 7 to 12, the binding of similar concentrations of EBF was tested in the presence of a constant amount of E47 (E47 DBD). At higher concentrations of EBF (lane 11), an additional band was detected that migrated slightly faster than that observed for EBFx4. This band was selectively competed for by excess E2A binding sites, as was the binding of the E47 DBD itself (lane 12). We conclude that, although EBF and E47 each bind the promoter independently, binding by these proteins is at best weakly cooperative. Weak cooperativity between these proteins may explain the synergistic activation of the promoter by these proteins in transfected cells. However, other factors, including proteins that bind the SPR2 site, coactivators, etc., may enhance their functional synergy in vivo.
DNA microarray analysis of regulatory factor and B-cell-specific target gene expression.
Binding of the mb-1 promoter by three cell type-specific factors suggests their combinatorial regulation of its function in early B cells. To establish a functional hierarchy of these genes with regard to their abilities to regulate mb-1 transcription, we performed microarray analysis to simultaneously measure RNA levels from the mb-1 gene and other genes of interest. Data were generated by hybidization of cRNA samples generated from cell lines, including Ba/F3 pro-B cells (three different subclones), pre-B cells 70Z/3, 18-81, 230-238, and 40E1, immature WEHI-231 cells, and later-stage B cells A20, K46, and M12, to Affymetrix chips. The data were then used to investigate how differences in expression levels of transcription factors and other early B-cell-specific genes relate to levels of mb-1 transcripts. The genes were selected based on their suggested ability to direct mb-1 transcription, or potential for coordinated biological functions with the mb-1-encoded protein, Igα. Data were normalized to mean zero and standard deviation one for calculation of the Pearson correlation coefficient (R) between mb-1 and other genes (see Materials and Methods). In this analysis, a value of R = 1 indicates a perfect positive correlation between levels of expression of mb-1 and a second gene (or itself), whereas R = 0 suggests no correlation. A value of R = −1 would indicate a perfect negative correlation.
Table 2 shows correlation coefficients obtained from analysis of microarray data. R values were determined between pairs of determinations, with one always equal to levels of mb-1 expression (therefore, mb-1 is reported on the table as “1”). Notably, the greatest positive correlation was observed between levels of mb-1 and ebf1 transcripts (R = 0.980). Although mb-1 transcripts did not correlate with E2A transcripts (R = −0.120), E2A proteins are primarily regulated posttranslationally. However, the greatest negative correlation was observed between mb-1 and transcripts encoding Id1, a competitive repressor of E2A proteins (R = −0.597). A relatively high correlation, but lower than that of ebf1, was observed between mb-1 and pax-5 transcripts (R = 0.666). Expression of other factors was all significantly less related to mb-1 and ebf1. Of other B-cell-specific target genes, blnk (0.786) and B29 (R = 0.797) showed high positive correlations with mb-1, suggesting that they are partially coregulated. In contrast, only a very low correlation was observed between mb-1 and blk, suggesting that these genes are differentially regulated.
TABLE 2.
DNA microarray analysis of gene expression in B-cell lines
| Gene | Correlation coefficient (R)a |
|---|---|
| mb-1 | 1 |
| ebf1 | 0.980 |
| B29 | 0.797 |
| blnk | 0.786 |
| pax-5 | 0.666 |
| elf-1 | 0.571 |
| ets-1 | 0.514 |
| OCA-B/obf-1/bob-1 | 0.311 |
| elf-3 | 0.254 |
| blk | 0.227 |
| E2A | −0.120 |
| spi-B | −0.149 |
| ld4 | −0.301 |
| elk-4 | −0.333 |
| elk-1 | −0.451 |
| ld1 | −0.597 |
Pearson correlation coefficients (R) were determined by using data obtained from 12 different measurements (see Materials and Methods). Each value was determined for expression of each test gene versus mb-1 for cell lines as shown. Calculations were carried out using the R statistical computing package (http://www.r-project.org/).
DISCUSSION
Multiple factors collaborate to regulate mb-1 transcription.
In this report, we show that mb-1 transcription correlates with occupancy of multiple promoter binding sites by factors in vivo. Using in vivo footprinting, we identified at least six factor binding sites and showed their importance for transcription in early B cells. Each of the sequences protected from DMS modification in intact cells contributed to promoter function in a cell type-specific manner, as evidenced by decreased activity of mutated promoters in short-term transfection assays. These data are supported by Western and EMSA analysis of proteins in representative cell lines, which showed that levels of EBF and Pax-5 proteins in cells correlate with protection of their binding sites in footprinting assays and requirements for these sites in mb-1 promoter function. DNA microarray experiments identified a very close correlation between mb-1 and EBF expression. Thus, the data suggest that mb-1 promoter function is most directly regulated by levels of EBF, but all three factors are important for high level mb-1 transcription in early B cells. At later stages of development, decreased levels of expression are mediated by Pax-5:Ets ternary complexes in the absence of EBF and E2A activity.
Other data detected important roles for other regulatory sites but also suggest a need to reevaluate the roles of sites identified in previous studies. Mutation of the proximal Ets binding site greatly reduced functional promoter activity in all three cell types tested in our transfection studies. In previous studies, mutation of this site greatly decreased promoter activity (47). The sequence 5′-CAGGAAGT comprising the Ets site is perfectly conserved between mice, human, and cattle (16, 47, 53). We conclude that this site is crucial for basal activity of the promoter. Nearby, another factor binding site was identified previously just downstream of the proximal Ets binding site (47). The site binds Sp1 in vitro, and its mutation reduced functional promoter activity in transfected cells. Methylation interference assays detected contacts with the underlined residues in the guanine-rich site 5′-GAGGCGGAG; however, we did not observe protection of these sequences from DMS in intact B cells. The Sp1 site is not well conserved between mb-1 promoters of mice and humans (5′-AGGGTGGGG [16]), which may reflect a lack of functional conservation. Similarly, we did not obtain evidence for occupancy of the octamer-like sequence identified as a potential target site for Oct-1/Oct-2 proteins (28).
Cooperation between EBF and E2A proteins.
Here, we showed that in addition to EBF and Pax-5, transcriptional activity of the mb-1 promoter is critically dependent on binding of bHLH proteins encoded by the E2A gene. The data are supported by studies in the laboratory of Y. Zhuang, who has detected occupancy of the mb-1 promoter by E2A proteins in vivo by chromatin immunoprecipitation (Y. Zhuang, unpublished data). The mb-1 promoter E box (5′-CAGGTGCA) is perfectly conserved between mice, humans, and cattle, suggesting its importance for regulating transcription (16, 47, 53). In vivo footprinting data suggest that EBF, E2A (it is not possible to determine which combination of homo- or heterodimers binds the promoter in vivo), and additional factors bind the promoter as part of higher-order complexes in pre-B cells. EBF and E2A (E47) synergistically activate the promoter in HeLa cells, however, another protein may assist EBF and E2A with assembly of ternary complexes, which is only weakly cooperative in vitro (Fig. 7).
FIG. 7.
DNA binding by EBF and E47. EMSA was performed by using a labeled mb-1 promoter probe (−183 to −140), recombinant EBF(24-429), and recombinant E47(518-649) as indicated above. EBFx4 indicates a higher-order complex of EBF homodimers, likely tetramers (17). EBF:E47 indicates a band that appears only when EBF and E47 are combined. We included 1,000-fold excess oligonucleotides comprising the μE5 E2A binding site in lanes 6 and 12.
The binding of E2A proteins to the promoter contributes additional transcription activation domains and may provide chromatin remodeling functions required for activation of the promoter. Recent studies showed that the conserved LDFS motif in the AD1 domain of these proteins recruits the SAGA histone acetyltransferase (HAT) complex (30). Thus, binding of the promoter by E2A proteins may contribute HAT activity for early events required for activation of mb-1 transcription. EBF also contains multiple activation domains (19) and was shown to alter the higher-order chromatin structure of one target gene (λ5) in a dose-dependent fashion (27). Similar functions of these proteins may be necessary for activation of the mb-1 gene.
Synergy between EBF and E2A controls the expression of multiple genes in early B cells. EBF- and E2A-deficient mice each lack normal B cells and exhibit a developmental block early in B-cell differentiation (4, 26). Synergy between EBF and E2A was inferred by studies in E2A+/−ebf1+/− heterozygous null mice, which exhibited decreased B lymphopoiesis at a later stage than that observed with either homozygous null phenotype (37). E2A+/−ebf1+/− mice show reduced levels of mb-1, λ5, VpreB, RAG1, RAG2, pax-5, CD19, and Lymphoid Enhancer Factor 1 (LEF-1) transcripts, with slightly reduced expression of B29 and E12. Reduced expression appears to be due to the lower frequency of expressing cells, which could reflect decreased cell proliferation or survival but could also reflect the inability of reduced levels of EBF and E2A proteins to efficiently remodel target gene chromatin.
In further support of their functional interactions in vivo, E2A and EBF can activate target genes in nonlymphoid cells. λ5 and VpreB were activated in Ba/F3 pro-B cells by EBF and “forced dimers” of E47 (45), and recent studies suggest that these proteins activate mb-1 transcription in HeLa cells (M. Sigvardsson, unpublished observations). In addition to our studies of the mb-1 promoter, evidence was obtained for cooperative assembly of EBF:E47 ternary complexes on the λ5 (37, 44) and VpreB (15) promoters. The molecular basis for these interactions is not understood, but synergism between the factors on the λ5 promoter involves template-dependent assembly of complexes requiring their respective DNA-binding and dimerization domains (44).
Combinatorial control of mb-1 transcription.
Our experiments suggest that EBF, E2A, and Pax-5:Ets ternary complexes collaborate for efficient transcription of the mb-1 gene in early B cells. Coordinate detection of occupied factor binding sites suggests that these factors assemble higher order structures, or “enhanceosomes” on the promoter similar to those reported on the interferon beta promoter and T-cell receptor α enhancer regions (14, 46). We propose that, together with binding of other factors, coordinated occupancy of the three early B cell-specific factor binding sites results in the stabilization of factor binding and enhanced recruitment of coactivators, etc., that mediate transcriptional activation.
In contrast with pre-B cells, lymphoma cells (A20 and K46) with germinal center B cell phenotypes (IgG+ MHC class II+ CD23+ M17+) showed little if any evidence of binding to the distal region EBF and E2A sites. Reduced but significant protection was detected at Pax-5:Ets ternary complex sites. Detection of Pax-5:Ets site footprints, but not distal region footprints, correlated well with the functional requirements for Pax-5:Ets but not EBF or E2A sites in transfected A20 cells and with levels of EBF (low), E2A (low), and Pax-5 (high) proteins in these cells. Taken together, these data suggest that EBF and E2A may be less important than Pax-5 for regulating low-level mb-1 transcription at later stages of development. In this regard, it will be interesting to determine whether normal germinal center B cells, which undergo immunoglobulin heavy-chain class switching and affinity maturation of antibody genes, use similar mechanisms to downregulate mb-1 transcription. Experiments with normal B-cell populations need to be performed to confirm this hypothesis.
DNA microarray analysis suggests that EBF may be the most crucial for initiating assembly of tissue-specific promoter complexes. mb-1 transcription most closely reflects levels of ebf1 transcripts (R = 0.980), whereas other potential regulators (e.g., pax-5) correlated less well with mb-1 transcripts. It is not possible to accurately assess the level of functional E2A proteins by this method, which is controlled posttranslationally by Id proteins in a developmental stage-specific manner (reviewed in reference 31). However, the greatest negative correlation was observed between mb-1 and Id1, a dominant-negative regulator of E2A protein function. These data further imply that mb-1 transcription is critically dependent on two factors, EBF and E2A.
We propose that mb-1 transcription requires E2A and EBF for its initial activation, but transcription is increased through the binding of Pax-5 and its Ets partners. This hypothesis is supported by studies of Pax-5-deficient mice, which show reduced but significant expression of the mb-1 gene (35). The requirement for coexpression of EBF, E2A, and Pax-5 may prevent transcription of mb-1 in other tissues that express related proteins (i.e., Pax-2 and Pax-8). We currently do not know how these proteins mediate each others binding to target genes in vivo, but the functional contributions of these proteins for chromatin remodeling, assembly of functional higher order complexes, and other requirements for promoter activation will be important areas for future study.
Acknowledgments
We thank Holly Maier and Arthur Gutierrez-Hartmann for critically reading the manuscript and Yuan Zhuang for sharing unpublished data. We gratefully thank Peter Engler, Ursula Storb, Kathryn Calame, Rudolf Grosschedl, and Marian Koshland for providing cell lines. We acknowledge the helpful advice of Umarani Pugazhanthi with primer design for real-time PCR, Philippa Marrack and Brad Swanson for providing instrumentation and advice on the detection of β-actin transcripts, and Scott McNeff and Boyd Jacobson for help with illustrations.
M.S. thanks The Swedish Cancer Foundation and the Swedish Science Council (VR) for their generous support of this work. D.R.C. was the recipient of a Fellowship from the Cancer League of Colorado, Inc., and is supported by National Institutes of Health training grant T32 AI00048. D.F. is supported by funds from NJMRC. J.H. is supported by grants from the National Institutes of Health (R01 AI37574 and P01 AI22295) and also thanks the Monfort Family Foundation, the Cancer League of Colorado, Inc., and the Rocky Mountain Chapter of the Arthritis Foundation for their generous support.
REFERENCES
- 1.Alt, F. W., N. Rosenberg, R. J. Casanova, E. Thomas, and D. Baltimore. 1982. Immunoglobulin heavy chain class expression and class switching in a murine leukemia cell line. Nature 296:325-331. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim, A., R. Shiran, A. Rosen, and M. D. Walker. 1993. Cell-specific expression of helix-loop-helix transcription factors encoded by the E2A gene. Nucleic Acids Res. 21:1601-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bain, G., E. C. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Reile, A. Berns, and C. Murre. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann, H., L. K. Su, and T. Kadesch. 1990. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 4:167-179. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell, T. K., and H. Weintraub. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250:1104-1110. [DOI] [PubMed] [Google Scholar]
- 7.Christoph, T., R. Rickert, and K. Rajewsky. 1994. M17: a novel gene expressed in germinal centers. Int. Immunol. 6:1203-1211. [DOI] [PubMed] [Google Scholar]
- 8.Feldhaus, A., D. Mbangkollo, K. Arvin, C. Klug, and H. Singh. 1992. BlyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol. Cell. Biol. 12:1126-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández, L. A., M. Winkler, and R. Grosschedl. 2001. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21:196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzsimmons, D., W. Hodsdon, W. Wheat, S.-M. Maira, B. Wasylyk, and J. Hagman. 1996. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10:2198-2211. [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimmons, D., R. Lutz, W. Wheat, H. M. Chamberlin, and J. Hagman. 2001. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 29:4154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity, P. A., and B. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 89:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvie, C. W., J. Hagman, and C. Wolberger. 2001. Structural studies of Ets-1/Pax-5 complex formation on DNA. Mol. Cell 8:1267-1276. [DOI] [PubMed] [Google Scholar]
- 14.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 15.Gisler, R., and M. Sigvardsson. 2002. The human V-preB promoter is a target for coordinated activation by early B cell factor and E47. J. Immunol. 168:5130-5138. [DOI] [PubMed] [Google Scholar]
- 16.Ha, H., B. L. Barnoski, L. Sun, B. S. Emanuel, and P. D. Burrows. 1994. Structure, chromosomal localization, and methylation pattern of the human mb-1 gene. J. Immunol. 152:5749-5757. [PubMed] [Google Scholar]
- 17.Hagman, J., C. Belanger, A. Travis, C. W. Turck, and R. Grosschedl. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760-773. [DOI] [PubMed] [Google Scholar]
- 18.Hagman, J., and R. Grosschedl. 1992. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc. Natl. Acad. Sci. USA 89:8889-8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagman, J., M. J. Gutch, H. Lin, and R. Grosschedl. 1995. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 14:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagman, J., and G. Siu. 2001. Transcriptional regulation of early B cell development, p. 137-165. In J. Locker (ed.), Transcription factors. Bios Scientific Publishers, Ltd., Oxford, United Kingdom.
- 21.Hagman, J., A. Travis, and R. Grosschedl. 1991. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 10:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henthorn, P., M. Kiledjian, and T. Kadesch. 1990. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κ2 motif. Science 247:467-470. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, Y., X.-Q. Xin, K. Dorshkind, and C. Nelson. 1994. Pan/E2A expression precedes immunoglobulin heavy-chain expression during B lymphopoiesis in nontransformed cells, and Pan/E2A proteins are not detected in myeloid cells. Mol. Cell. Biol. 14:4087-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller, G., M. Kennedy, T. Papayannopoulou, and M. Wiles. 1993. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 13:473-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, K. J., C. Kanellopoulos-Langevin, R. M. Merwin, D. H. Sachs, and R. Asofsky. 1979. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 122:549-554. [PubMed] [Google Scholar]
- 26.Lin, H., and R. Grosschedl. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376:263-267. [DOI] [PubMed] [Google Scholar]
- 27.Lundgren, M., C.-M. Chow, P. Sabbatini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 28.Malone, C. S., L. Patrone, and R. Wall. 2001. An essential octamer motif in the mb-1 (Igα) promoter. Mol. Immunol. 37:321-328. [DOI] [PubMed] [Google Scholar]
- 29.Malone, C. S., and R. Wall. 2002. Bob1 (OCA-B/OBF-1) differential transactivation of the B cell-specific B29 (Igβ) and mb-1 (Igα) promoters. J. Immunol. 168:3369-3375. [DOI] [PubMed] [Google Scholar]
- 30.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 31.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuuchi, L., and M. R. Gold. 2001. New views of BCR structure and organization. Curr. Opin. Immunol. 13:270-277. [DOI] [PubMed] [Google Scholar]
- 33.Murre, C., A. Voronova, and D. Baltimore. 1991. B-cell and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol. Cell. Biol. 11:1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nutt, S. L., B. Heavy, A. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556-562. [DOI] [PubMed] [Google Scholar]
- 35.Nutt, S. L., A. M. Morrison, P. Dörfler, A. Rolink, and M. Busslinger. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nutt, S. L., P. Urbánek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476-491. [DOI] [PubMed] [Google Scholar]
- 37.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 38.Oi, V. T., S. Morrison, L. A. Herzenberg, and P. Berg. 1983. Immunoglobulin gene expression in transformed lymphoid cells. Proc. Natl. Acad. Sci. USA 80:825-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersson, K., F. Ivars, and M. Sigvardsson. 2002. The pTα promoter and enhancer are direct targets for transactivation by E box-binding proteins. Eur. J. Immunol. 32:911-920. [DOI] [PubMed] [Google Scholar]
- 40.Saisanit, S., and X.-H. Sun. 1997. Regulation of the pro-B-cell-specific enhancer of the Id1 gene involves the C/EBP family of proteins. Mol. Cell. Biol. 17:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sen, R., and D. Baltimore. 1986. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell 47:921-928. [DOI] [PubMed] [Google Scholar]
- 43.Shen, C.-P., and T. Kadesch. 1995. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 15:4518-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sigvardsson, M. 2000. Overlapping expression of early B cell factor and basic helix-loop-helix proteins as a mechanism to dictate B lineage specific activity of the λ5 promoter. Mol. Cell. Biol. 20:3640-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 46.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 47.Travis, A., J. Hagman, and R. Grosschedl. 1991. Heterogeneously initiated transcription from the pre-B- and B-cell-specific mb-1 promoter: analysis of the requirement for upstream factor-binding sites and initiation site sequences. Mol. Cell. Biol. 11:5755-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urbánek, P., Z.-Q. Wang, I. Fetka, E. F. Wagner, and M. Busslinger. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901-912. [DOI] [PubMed] [Google Scholar]
- 49.Voronova, A., and D. Baltimore. 1990. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA 87:4722-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner, N. L., A. W. Harris, and G. A. Gutman. 1975. Membrane immunoglobulin and Fc receptors of murine B- and T-cell lymphomas, p. 203-216. In M. Seligmann, J. L. Preud'homme, and F. M. Kourilsky (ed.), Membrane receptors of lymphocytes. American Elsevier Publishing Co., New York.
- 51.Wheat, W., D. Fitzsimmons, H. Lennox, S. R. Krautkramer, L. N. Gentile, L. P. McIntosh, and J. Hagman. 1999. The highly conserved β-hairpin of the paired DNA-binding domain is required for the assembly of Pax:Ets ternary complexes. Mol. Cell. Biol. 19:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yancopoulos, G. D., S. V. Desiderio, M. Paskind, J. F. Kearney, D. Baltimore, and F. W. Alt. 1984. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature 311:727-733. [DOI] [PubMed] [Google Scholar]
- 53.Youn, H. Y., K. W. Cho, Y. Nishimura, R. Goitsuka, T. Watari, H. Tsujimoto, and A. Hasegawa. 1997. Genomic structure of the bovine mb-1 gene encoding the Ig-alpha subunit of the B cell antigen receptor. Vet. Immunol. Immunopathol. 56:247-257. [DOI] [PubMed] [Google Scholar]
- 54.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]