One major mechanism for achieving cell-type-specific functions in somatic cells is the differential usage of stage- and tissue-specific genes. This is in large part regulated by transcription factors that interact directly with DNA elements controlling transcription of these genes. Several groups of transcription factors with key roles in cellular differentiation and function have been characterized, and the list of transactivating proteins with crucial functions in vivo is continuously growing. One group of evolutionarily conserved proteins with defined roles in a large variety of species are the basic helix-loop-helix (bHLH) proteins (42, 44). These factors are involved in a large number of differentiation processes, including such diverse events as sex determination in Drosophila melanogaster and B-lymphopoiesis in mice (9, 15, 38, 42, 48). In 1993, two independent reports presented a novel type of HLH protein with a dimerization domain containing two helices with homology to the second helix of the classical bHLH protein dimerization domain, but with a different type of DNA binding domain (DBD). This factor was cloned both in a Saccharomyces cerevisiae one-hybrid experiment designed to identify factors interacting with the olfactory-restricted olfactory marker protein-1 (OMP-1) promoter (69), and by biochemical purification of a factor interacting with the B-lymphocyte-restricted mb-1 promoter (27). It was accordingly named Olf-1, or early B-cell factor (EBF), which in turn led to the designation of the factor as O/E-1. Later reports showed that mice expressed at least three more members of this family, EBF2 (mMot1/O/E-3), EBF3 (O/E-2) (23, 40, 71), and O/E-4 (70), with a high degree of similarity in the DNA-binding and dimerization domains. Isolation of the O/E homologue Collier from D. melanogaster (12) provided a proof of principle for the existence of a new family of evolutionarily conserved proteins, and family members have now been cloned in several species (7, 17, 24, 27, 46, 52, 69) (Table 1). The biological roles of this protein family are beginning to be unraveled, and here we are compiling some of the available information about the roles of Collier/Olf/EBF (COE) proteins to give an overview of the known functions of this protein family in specific model systems and in different model species.
TABLE 1.
COE proteins identified to datea
| Protein | Vertebrate protein(s)
|
Invertebrate proteins
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | X. laevis | Zebra fish | Chicken | Skate | D. melanogaster | C. elegans | |
| COE1 | EBF, O/E-1 | EBF, O/E-1 | Olf-1 | EBF | EBF1 | ||||
| COE2 | EBF2, O/E-3, Mmot1 | xCOE2 | zCOE2 | ||||||
| COE3 | EBF3, O/E-2 | xCOE3, xEBF3 | |||||||
| COE4 | O/E-4 | ||||||||
| COE | Collier, Knot | unc-3, CeO/E | |||||||
Sequences for chicken EBF and skate EBF1 are published in the EBI database (http://www.ebi.ac.uk). All proteins are referred to in the text and figures with the prefixes h (human), m (mouse), r (rat), z (zebra fish), g (chicken), s (skate), d (Drosophila), or c (C. elegans) COE, COE1, COE2, COE3, and COE4.
BIOCHEMICAL FEATURES AND GENE ORGANIZATION OF COE PROTEINS
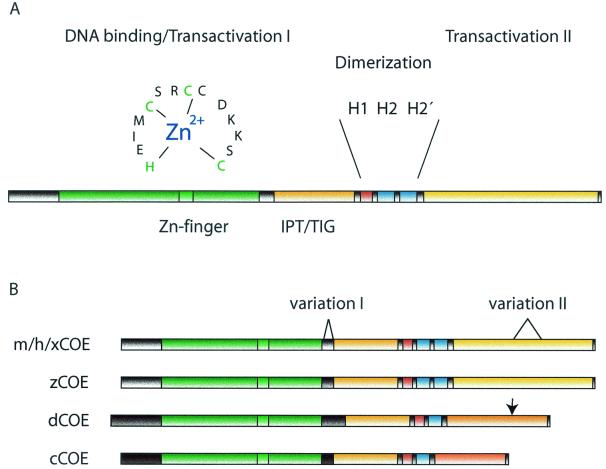
The COE protein family contains several conserved regions and the homology in the DBD between the evolutionarily most distally related proteins still remains above 80%. This is a strong indication that the specific structure is of crucial importance for the function of the protein and that the COE proteins are presenting a novel and unique DBD. COE factors bind DNA through a cysteine- and histidine-rich domain of about 250 amino acids located in the amino-terminal part of the proteins (Fig. 1A). Binding of EBF-1 requires Zn2+ and, even though no sequence homology to known Zn finger domains can be found, three of the cysteines together with one of the histidine residues can be represented as a small Zn2+-stabilized loop with the sequence H-X3-C-X2-C-X5-C (28). The finding that mutations of either of these cysteines or the histidine eliminate DNA binding and the requirement of Zn ions for binding (28) strongly suggests that the COE DBD contains an atypical Zn finger motif obligatory for DNA binding. The Zn coordinating motif is highly conserved in all O/E proteins and has for that reason been referred to as the COE motif. Downstream of the DBD is a second conserved region with apparent similarity to an IPT/TIG (immunoglobulin-like, plexins, transcription factors/transcription factor immunoglobulin) domain (4, 8) (Fig. 1B), present in the DNA binding region of transcription factors such as NFAT and NF-κB (4). Crystallizations of these proteins show that the IPT domain forms an immunoglobulin-like fold of β-sheets connected by loops that both contact DNA and participate in protein-protein interactions (4, 8). The role of this region in COE proteins is unclear, although it has been suggested to be involved in the interaction with Rat O/E-1-associated Zn finger (ROAZ) (66) (see below). Another interesting possibility comes from the findings that the IPT domain-containing plexin proteins are involved in axonal guidance (5, 60), a process suggested to involve also COE proteins (52). The IPT domain could then be a part of a mechanism that integrates external signals like those from semaphorins (60, 73) and activation of genes in the migrating neuron.
FIG. 1.
COE proteins contain several domains highly conserved in evolution. (A) Schematic structure of mouse COE1. The DBD is colored green and the amino acids and suggested structure of the Zn coordination motif are indicated. The IPT/TIG domain is indicated in orange, putative helix 1 is shown in red, and helix 2 motifs are shown in blue. The carboxy-terminal transactivation domain is colored yellow. (B) Comparison between COE proteins of different species. The color codes are the same as in panel A. All mouse (mCOE), human (hCOE), and Xenopus (xCOE) proteins are homologous throughout the protein. Two sites of variation are indicated where alternative splicing leads to different forms of COE1 and COE3. The (0) forms of murine COE1 and COE3 lack an 8- or 9-amino-acid region (indicated by variation I), which is present in COE1 (8) and COE3 (9). The S form of COE3 lacks the region indicated by variation II, which is present in COE3 L. Mouse COE4 has a more complex splicing pattern and, as COE1 (0) and COE3 (0), lacks the region indicated as variation I. Zebra fish COE (zCOE) is homologous to the other vertebrate proteins in all parts except for an unrelated sequence between the DBD and IPT/TIG domains (shaded in darker grey). Drosophila COE (dCOE) exists in two isoforms of 575 (isoform 1) or 557 (isoform 2) amino acids. They are identical until amino acid 528 (indicated in figure) but differ in their C-terminal sequence.
The mouse EBF/O/E-1, -2, -3, and -4 bind DNA as homo- or heterodimers, and dimerization is primarily dependent on a region downstream of the DBD containing an atypical HLH motif (Fig. 1A). The typical HLH motif, present in, for instance, bHLH proteins (33), contains two dissimilar amphipathic helices, helix 1 and helix 2, connected by a loop. EBF-1 was first reported not to contain any sequence corresponding to helix 1, but rather a duplication of helix 2 resulting in a domain capable of mediating dimerization when coupled to a heterologous protein (28). The helix 2 duplication is only present in vertebrate COEs and not in D. melanogaster Collier (12) or Caenorhabditis elegans unc-3 (52), and dimerization of these proteins is suggested to be mediated by a putative helix 1 present in all the COE proteins (12). However, this region can be deleted without a dramatic effect on EBF-1 DNA binding (CΔ296-367 [27]), and no extensive biochemical analysis of either Collier or unc-3 has been reported, leaving the exact consequences of this discrepancy unclear. Some insight to this issue comes from the identification of a short splice form of O/E-4, which lacks helix 2′ (70). This protein can still bind DNA on its own and cooccupy a perfect COE binding site with a longer O/E-4 splice variant containing helix 2′ (70). The dimeric nature of EBF-1 is also reflected in the palindromic sequence of the DNA binding site (65). O/E-1, -2, and -3 bind variants of the EBF-1 consensus sequence 5′-ATTCCCNNGGGAAT-3′ (65) either as hetero- or homodimers (27, 71). However, EBF-1 with a disrupted HLH domain can still interact with one perfect consensus half-site, although with a dramatically reduced affinity (27, 28).
EBF-1 contains two defined transactivation domains, one context-dependent domain located within the DBD (TS I) and one in the C-terminal part of the protein (TS II) (28). The C-terminal TS II is rich in serine/threonine/proline residues, and this region can activate transcription when coupled to a GAL-4 DBD (28), while TS I was defined based on the ability of an EBF-1 with carboxy-terminal deletions to activate transcription (28). The carboxy-terminal part of the protein is rather poorly conserved among the family members (approximately 50% between mouse O/E-1, -2, and -3), but all these proteins have been shown able to stimulate transcription from a reporter gene controlled by O/E sites (27, 70, 71), although O/E-4 seems to be a poor activator compared to the other proteins (71). Serine-rich carboxy-terminal domains are also present in Collier and unc-3, indicating an evolutionarily conservation of a carboxy-terminal transactivation domain even though the primary amino acid sequence appears to be divergent.
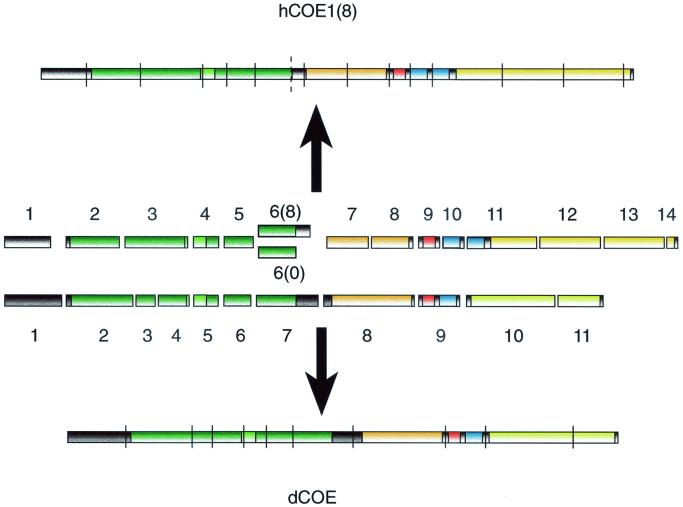
BLAST search of the human genome database for O/E encoding genes suggests the existence of four different genes, situated on chromosomes 5 (ebf-1 related [43]), 8 (ebf-2 related), 10 (ebf-3 related), and 20 (O/E-4 related) (XM038441, XM034639, AL354950, and AL035460, respectively). The structure of the human gene coding for EBF-1 suggests that the protein is encoded by 14 exons which can be compared to the D. melanogaster gene for Collier (13, 14) constructed from 12 exons (Fig. 2). In both genes, exon junctions border the DBD, the IPT domain, and the first two helices of the dimerization region. The DBD is also further subdivided into several exons in both genes. The originally cloned rat Olf-1 and mouse EBF differs in an 8-amino-acid insertion present in EBF (27, 69). This insertion does not seem to be due to species variation but rather to alternative splicing of O/E-1, leading to the inclusion or exclusion of a 24-bp sequence [O/E-1 (8) or O/E-1 (0), respectively]. This is probably explained by a potential splice site in hEBF-1 (dotted line in Fig. 2) that could be alternated with the following site for joining exon 6 to exon 7. In a similar manner, O/E-2 can contain or lack a 9-amino-acid insertion at the same position. O/E-2 has in addition a second alternative splice site 40 amino acids from the C terminus. Alternative splicing gives rise to short (S) or long (L) forms of O/E-2, differing by 36 amino acids. All in all, this gives rise to four different O/E-2 variants, designated O/E-2(0S), -2(0L), -2(9S), and -2(9L) (Fig. 1B), all expressed in embryonic tissue (71). O/E-4 exists in four variants created by alternative splicing at four different sites 3′ of helix 2 (70). Thus, several of the O/E proteins are present in a number of splice forms but the functional consequence of this variation and the expression pattern of the different isoforms remain to be elucidated.
FIG. 2.
The genomic organization of the human EBF-1 and Drosophila Collier is partially conserved, as shown in this comparison between the exon structure of Drosophila (dCOE) and human (hCOE1) genes. Only the coding parts of the genes are shown. Exons are indicated with numbers, and splice junctions are indicated with lines. The dotted line represents an alternative splice site for joining the human exon 6 to exon 7, creating the 0 form of hCOE1.
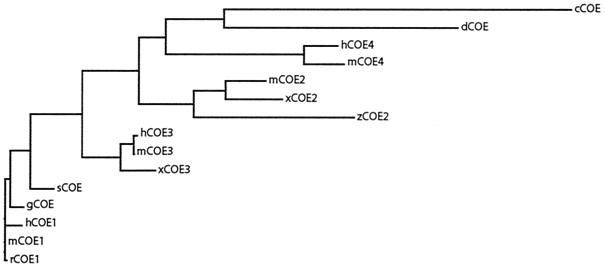
Taken together, the COE family of proteins share several features due to conservation of essential protein structures among the family members. Overall phylogenetic analysis indicates that this family is present in only the animal kingdom, as no related sequence could be found in sequence databases for bacteria, archaebacteria, yeasts, or plants. Furthermore, the O/E-3 proteins and the novel O/E-4 proteins are closest related to the invertebrate Collier and unc-3 (Fig. 3), suggesting that these are the most ancient of the vertebrate family members and that the other proteins have arisen from gene duplications of either of these ancestor genes. This has created a set of COE proteins with restricted expression patterns and in some cases unique roles, which are the focus of the following sections in this review.
FIG. 3.
Phylogenetic tree of the COE family, revealing the genetic relations between the different COE proteins. Sequences were retrieved from the EBI database, and the tree was constructed using the ClustalW method (61). The proteins are referred to in the figure with the prefixes h (human), m (mouse), r (rat), z (zebra fish), g (chicken), s (skate), d (Drosophila), x (Xenopus), or c (C. elegans).
COE PROTEINS IN EMBRYOGENESIS
Some of the most striking examples of differential gene regulation and cellular development occur at the early stages of life in the developing embryo. COE proteins are suggested to play key roles in several different model systems for embryonic development (18). One example of this is the finding that antisense Collier RNA results in abnormal head development in the D. melanogaster embryo (12). This defect, later confirmed by mutant analysis, reflects the Collier requirement for integrating inputs from both the head and trunk segmentation systems at the blastoderm stage and formation of parasegment 0 (13). Collier also appears to be involved in the formation of a specific embryonic somatic muscle (14). The development of muscle-specific properties, such as position and shape, is dependent on a special type of myoblasts, called founder cells, that each fuse with and instruct a defined number of neighboring “naive myoblasts” to form a specific muscle precursor (21). Collier is expressed in one founder myoblast, responsible for formation of the muscle dorsal-achete-3 (DA3). In col mutant embryos, the DA3 founder cell appears unable to recruit neighboring cells, resulting in a lack of DA3 muscle (14). Each founder cell derives from asymmetric division of a progenitor cell, which is itself selected from a cluster of competent cells by the process of lateral inhibition involving Notch/Delta signaling (21). Notch signaling is also required for asymmetric cell division of the progenitors (21). Collier is expressed in the promuscular cluster, giving rise to the progenitor of the DA3 and D05 founder cells (14). Lack of Notch signaling during either specification of the DA3/D05 progenitor or its division results in aberrant expression of Collier and adoption of a DA3 fate by all Col-expressing promuscular cells and specific loss of DO5 muscle development, respectively (14). Collier does not however, appear to be able by itself to induce DA3 fate in the proneural cluster cells, suggesting the necessity of a collaboration with other determining factors in this process (14). COE proteins are also closely linked to Notch signaling events and lateral inhibition in the developing nervous system in Xenopus laevis. Notch signals suppress neuronal differentiation in part by down regulation of the proneural bHLH protein X-neurogenin-1, and Xcoe2 appears to act downstream of this protein in the neural plate of the developing frog embryo, but upstream of the neurogenic bHLH protein XneuroD (17). This positions Xcoe2 as an intermediate factor possibly involved in the maintenance of higher neural potential of the precursor cell at the same time as it has the ability to stimulate commitment into neuronal development by activation of XneuroD (17). Xcoe3 is on the other hand acting downstream of XneuroD, where it appears to stimulate the development of specific neuronal subtypes such as cranial sensory ganglia cells and Rohon Beard cells (51). Involvement of COE proteins in neurogenesis is also apparent in the developing C. elegans embryo, where impaired function of the unc-3 protein results in abnormal development of a specific group of motor neurons formed in the ventral nerve cord (52). In the newly hatched larva, this results in defects in cholinergic DA and DB neurons, which in turn lead to a worm with an abnormal moving behavior. This phenotype correlates with defects in motor neuron axonal path finding and indicates that unc-3 may be involved in fasciculation or growth cone pioneering. A defect is also seen in the axonal projections of D-class motor neurons that normally do not express unc-3, indicating that normal development of these are dependent on signals from adjacent unc-3-expressing cells. The dramatic effect on motor neurons is contrasted by the lack of obvious defects in the outgrowth of ASI chemosensory neurons even though they express unc-3 throughout development (52). The neurons in unc-3-deficient worms also expressed a reporter gene consisting of a fusion of the ASI-specific marker srd-1 and green fluorescent protein, confirming their identity as differentiated ASI neurons (52). The function of these neurons, however, appears to be disturbed, since unc-3-deficient larvae enter the dormant Dauer pathway under inappropriate conditions (1, 52). This process is regulated by pheromones and nutritional and thermal cues and is a biological response to suboptimal growth conditions (1). Defects in Dauer formation connect with the dysfunction of ASI neurons (1, 52), suggesting that even though the chemo sensory neurons are apparently normal, they display a defect in functional performance. This suggests that unc-3 may play roles both in the establishment of neuronal networks and in the physiological functions of neurons in C. elegans.
O/E proteins are also expressed in the developing nerve tissues in mice but much of the specific roles for these proteins may be masked in the EBF-1-deficient animals due to complementary expression of other family members. One cell type with specific expression of EBF-1 is the subventricular zone and mantle cells in the lateral ganglionic eminence (22). Postmitotic cells from this region normally leave the subventricular zone to migrate into the mantle zone. In EBF-1-deficient mice, these cells fail to down regulate genes such as ephrin A-4 and Oct6 and to activate expression of mantle-specific genes such as cellular retinoic acid protein 1 and cadherin 8 (22). They also undergo abnormal apoptotic behavior resulting in a dramatic size reduction of the postnatal striatum (22). This also affects the navigation and fasciculation of thalamocortical fibers migrating through the striatum (22), possibly by a mechanism resembling the non-cell autonomous effects observed in neuronal guidance observed in unc-3-deficient C. elegans (52). Defective differentiation is observed among facial branchiomotor neurons that normally undergo a complex migration event in the hindbrain since these cells are unable to down regulate expression of early markers in EBF-1-deficient mice (22).
A role for O/E proteins also in later stages of development has been proposed, based on analysis of D. melanogaster col mutants rescued from embryonic or early larval lethality by transformation with a specifically engineered col transgene. These flies are weak and sterile and display a specific wing patterning phenotype, the lack of central structures (68). A similar phenotype had previously been reported for flies carrying a mutation denoted knot (45), and analysis of col/knot trans-heterozygous flies indicated that Collier and knot correspond to the same gene (68). Antero-posterior patterning of the Drosophila wing involves signaling from the morphogen hedgehog, and experiments using heat-sensitive mutants showed that expression of Collier is dependent on high doses of hedgehog signaling (68). The same signaling systems are involved in the formation of vertebrate limbs and an involvement of O/E proteins in this process might become evident from extended analysis of these signaling pathways.
The role of COE proteins in fetal development of higher animals is largely unknown, but gene bank searches as well as experimental evidence indicate abundant expression of COE proteins at several stages and in several sites of the developing mouse embryo (23, 71). The O/E interacting protein ROAZ (66) (OAZ) is also involved in bone morphogenic protein (BMP) signaling in the developing Xenopus embryo, where it acts together with Smad proteins to stimulate transcription of the gene encoding the homeo-box protein Xvent-2 (31). This functional synergy involves direct protein interactions between ROAZ and Smad1, and even though this interaction involves domains distinct from those essential for the interaction with O/E proteins, it appears as if Smads and O/E protein compete for exiting ROAZ (31). Ectopic expression of O/E protein impaired the ability of ROAZ to stimulate transcription from the Xvent-2 promoter in response to BMP-2 signaling (31), indicating that O/E proteins may be involved in modulation of BMP signaling in the developing embryo. EBF-1-deficient mice were originally reported to develop apparently normally even though they appear to be growth retarded (39). This could indicate that EBF-1 is nonessential for fetal development, but this could also be explained by redundant functions for O/E-2 and -3 (71). Future studies of the COE proteins in fetal development might well provide information about new networks of transcription factors and signaling pathways, increasing the understanding of embryonic development also in humans.
COE PROTEINS IN THE ADULT NERVOUS SYSTEM
In addition to acting in embryonic neural development, COE proteins are also expressed in the adult nervous system in several species. In the mouse, O/E-1 and -3 expression has been detected in the Purkinje cells of the adult cerebellum, and all four O/E proteins are expressed at high levels in olfactory epithelium (71). In situ hybridizations showed that O/E proteins are expressed not only in mature olfactory receptor neurons in the neuronal layer but also in immature cells in the basal layer of olfactory epithelium (71). O/E-1, -2, and -3 but not O/E-4 are also expressed in the vomeronasal organ (70). The expression of O/E proteins in the mature cells correlates with the presence of mRNAs encoding components of the odor detection and signaling cascades in these cells. O/E proteins are also shown to directly interact with promoters regulating the expression of some of these genes, such as the olfactory cyclic nucleotide-gated channel (OcNC), type III adenylyl cyclase (ACIII), and the olfactory enriched G-protein a-subunit (Golf) (36, 69, 71). The lack of expression of these genes in immature cells in the basal layer could be explained by the presence of the O/E interacting protein ROAZ in these but not in the neuronal layer cells (66). This protein contains 29 C2H2 Zn fingers of TFIIA type and appears to have the ability to impair the capacity of O/E proteins to activate target promoters (66), which could thus explain the lack of mature marker gene expression in the immature cells. The role of O/E proteins in the nervous system could be more complex than initially thought since a transgenic mouse carrying a reporter gene controlled by an OMP promoter with a mutated O/E binding site expressed high levels of the transgene marker in olfactory neurons (37). The major phenotype was instead apparently aberrant expression of the reporter in neurons outside of the olfactory system (37). This could indicate a role for the O/E site as a repressor element, ensuring tissue-specific expression of the OMP gene either via an interaction with COE proteins or via other factors. The investigation of the roles of COE proteins in the nervous system is likely to become clearer when mice carrying targeted disruptions of several of the factors to circumvent potential compensatory mechanisms become available.
O/E PROTEINS AND B-LYMPHOCYTE DEVELOPMENT
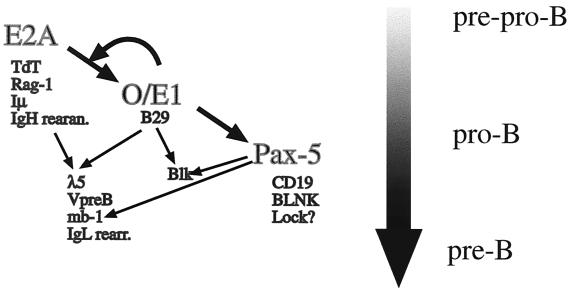
One obvious phenotype of EBF-1-deficient mice is the lack of mature B lymphocytes (39). The pronounced phenotype is likely to be due to the fact that, in contrast to several other cell types, early B cells only express EBF-1. B-cell development in the bone marrow of these mice appears to be arrested at an early stage with the presence of cells expressing B220 and high levels of CD43, defining the pro-B-cell stage (30), but no cells of subsequent differentiation stages (39). The number of cells expressing pro-B-cell markers appeared to be rather normal (39), indicating a differentiation rather then an expansion block of the type observed in, for instance, Sox-4-deficient mice (56). The rather well-defined B-cell phenotype of these mice has allowed for a theoretical positioning of EBF-1 in the functional hierarchy of transcription factors in B-lymphoid development (Fig. 4). The number of cells expressing CD43 is reduced in mice lacking a functional E2A gene (6, 74), indicating that E-box binding proteins precede EBF-1 in the functional hierarchy of the developing B lymphocyte. This idea is further supported by the finding that ectopic expression of the E2A protein, E12, results in activation of the endogenous EBF-1 gene (32) and that these E-proteins also can interact with and activate an EBF-1 promoter region (59). The functional relationship between O/E-1 and E47 is even more extensive because EBF-1 and E2A proteins also appear to act in synergy at the late pro-B-cell stage. This has been shown in mice transheterozygous for mutations in one allele of EBF-1 and one allele of E2A, because they display a more dramatic B-cell phenotype than the single-heterozygote mice do (49). EBF-1 has also been suggested to act in direct synergy with E-proteins in the activation of target promoters controlling the expression of the surrogate light chains VpreB and λ5 (57, 58). The absence of DJ rearrangement and lack of Pax-5 expression in EBF-1-deficient mice (39) suggests that EBF-1 acts upstream of Pax-5 (67). This is further supported by the finding that O/E-1 can interact with the Pax-5 promoter region and has the ability to activate this control element in nonlymphoid cells (49). EBF-1 is also able to bind a functionally important site in its own promoter, suggesting the protein to be involved in an autoregulatory loop (59). This indicates that EBF-1 is a key factor for the specification and progression of the late pro-B-cell stage after entry of the cell into the B-lymphoid pathway but before final commitment of the cell due to expression of Pax-5 (47). The exact explanation for the B-cell phenotype observed in EBF-1-deficient mice is still unclear even though the protein has been suggested to be directly involved in the transcription regulation of several genes encoding components of the pre-B- and B-cell receptor. These include the surrogate light-chain genes (41, 50, 53, 57, 58) and the B-cell receptor signal transducing molecules B29 (2), mb-1 (27, 29) and Blk (3). The ability of EBF-1 to activate promoters controlling expression of the surrogate light chains appears to be conserved also in humans even though the primary DNA sequence of the promoters appears poorly preserved (24, 25). This indicates that the collaboration between EBF-1 and E-proteins is important also in human B-cell development and that the genetic network is conserved between mice and humans. EBF-1 is also involved in induction of germ line transcription from and rearrangement of immunoglobulin genes. This is because ectopic expression of EBF-1 acts in concert with RAG1 and RAG2 to promote (Ig) lambda light-chain and Ig heavy-chain DJ rearrangement in an embryonic kidney cell line (54). EBF-1 is also able to induce germ line transcription believed to precede rearrangement from certain but not all Ig V genes (26). These collected findings suggests that EBF-1 is a pleiotrophic activator of genes defining the pro- and pre-B-cell stage, and the identification of additional target genes for this protein may shed light on the coordinated expression of genes that allows for the progenitor cell to become a committed B-lymphoid precursor.
FIG. 4.
EBF/O/E-1 is a central transcriptional regulator in the developing B lymphocyte. The figure displays the suggested functional relationships between the transcription factors E2A, O/E-1, and Pax-5. Suggested target genes are located under the transcription factors and common target genes are indicated by arrows. The flow and stage of B-cell development are indicated in the right part of the figure by a broad shaded arrow.
COE IN ADIPOCYTE DEVELOPMENT
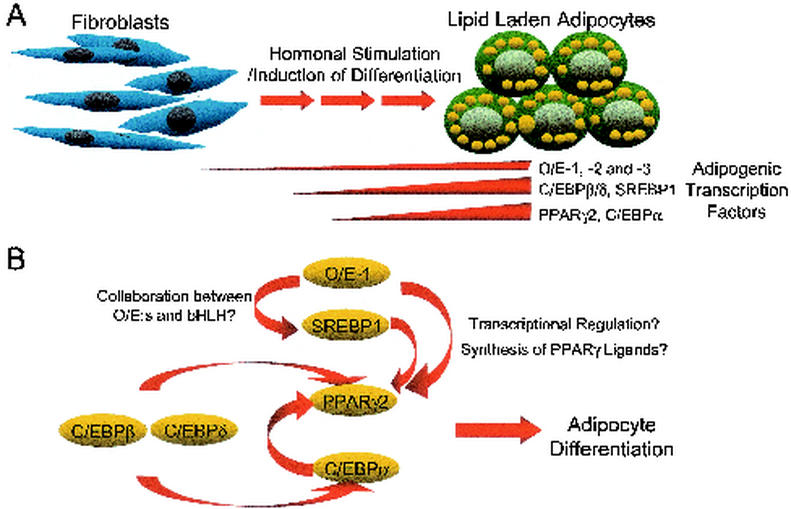
Investigations of the expression pattern of EBF-1 in adult mouse tissue indicated that this protein is expressed at high levels in adipocytes (27). EBF-1 has also been implied to be involved in the regulation of the glucose transporter gene Glut4 (16), suggesting that O/E proteins could participate in the regulation of adipocyte-specific genes. Adipocytes originate from mesodermal stem cells, and it has become increasingly clear that they play important roles not only as a storage for energy but also as an endocrine and paracrine organ secreting a large number of biologically active substances. The molecular mechanisms controlling the development of adipocytes have been studied extensively through the use of immortalized murine fibroblasts and preadipocyte cell lines, and also through various knockout mice (11, 55). By using fibroblasts and preadipocytes as model systems for adipocyte development, it has been possible to dissect this process into distinct stages with coordinated expression of specific genes (11, 55). Several of these temporally expressed genes are transcription factors such as CCAAT/enhancer-binding proteins (C/EBPs) (10, 72), peroxisome proliferator activated receptor γ (PPARγ) (62, 63), and sterol regulatory binding protein 1 (SREBP1/ADD1) (34, 64) (Fig. 5A). C/EBPβ and -δ are considered to function early in adipogenesis to activate the master regulators of adipogenesis, PPARγ and C/ΕβPα (11, 55) (Fig. 5B). These factors then appear to be involved in a regulatory loop where CEBPα and SREBP1 induce further expression and activity of PPARγ (19, 20, 35), thus sustaining and promoting the development of the preadipocyte into a mature cell (Fig. 5B).
FIG. 5.
O/E proteins participate in the transcription factor network involved in adipocyte differentiation. (A) Schematic illustration of adipocyte differentiation in which the cells go through several distinct steps of development before becoming mature adipocytes. Temporal expressions of transcription factors important for adipogenesis, including O/E-1, are indicated. (B) Model of the hierarchy of transcription factors controlling adipogenesis, with possible roles of O/E-1 in this network indicated.
All three known mouse EBF/O/E genes are expressed in mouse adipose tissue, and analysis of 3T3 L1 preadipocytes suggests that they are present already in the undifferentiated cells but that the expression levels are increased during terminal adipocyte differentiation (3a). Ectopic expression of EBF-1 is also able to enhance terminal adipocyte differentiation of preadipocytes and to induce adipogenesis in fibroblastic multipotent NIH 3T3 cells. In addition, a dominant negative form of EBF-1 had the ability to suppress differentiation of 3T3-L1 cells, indicating that target genes for EBF proteins are important for terminal adipocyte differentiation (3a). Most of the transcription factors expressed during adipocyte differentiation show a temporal expression pattern which enables them to turn on their target genes at specific time points (55). The finding that the O/E genes are expressed throughout adipogenesis implies that these transcription factors have roles early as well as late in adipocyte development. Temporal activation of target genes may be a result of dosage effects or collaborations with other proteins with more restricted expression patterns (Fig. 5B). The exact mechanisms by which EBF-1 stimulates adipogenesis are under investigation, and even though analysis of mRNA levels of various markers for adipocyte differentiation indicated that preadipocytes infected with retroviruses carrying the EBF-1 gene expressed higher levels of the adipocyte-specific genes PPARγ2, adipocyte fatty acid binding-protein (aFABP), and glycerol phosphate dehydrogenase after hormonal induction, no dramatic differences could be detected between the control and EBF-1 infected cells before the induction of differentiation (3a). This indicates that EBF-1 is not directly controlling any of the examined marker genes and that their increased expression levels rather reflect an enhanced adipogenesis in EBF-1 infected cells. The finding that EBF-1 is capable of initiating adipocyte differentiation of uncommitted NIH 3T3 fibroblasts points towards the notion that EBF-1 can participate in the initiation of the adipogenic program. This effect of EBF-1 was dramatically enhanced by the addition of PPARγ ligands (3a), indicating that at least part of the adipogenic potential of EBF-1 is mediated through PPARγ (Fig. 5B). The role of O/E proteins in adipocyte development in vivo is still unclear, but EBF-1−/− mice are reported to be smaller than wild-type littermates (39), indicating disturbances in the metabolism of these animals. Information from the global gene analysis of EBF-1 infected cells and EBF-deficient mice is likely to identify direct target genes for the O/E proteins that can explain the molecular mechanism behind their role in adipogenesis.
CONCLUDING REMARKS
The available information suggests that the COE proteins belong to a group of ancient proteins that has been evolutionarily conserved from a time before the divergence of insects and mammals, estimated to have occurred about 500 million years ago. We have not been able to find any COE-related proteins in yeasts or plants, indicating that this protein family has arisen in the earliest common animal ancestors and thus is involved in evolutionarily conserved and crucial functions in the animal kingdom. Gene duplications and modifications have then allowed for the creation of a family of genes with restricted expression patterns and functions. In mammals, O/E proteins appear to be involved in the differentiation of cells originating from all three embryonic germ layers. Even though the identification of direct in vivo targets for a transcription factor often is complicated, it appears as if many of the genes controlled by O/E proteins in cell culture systems are lineage and stage specific rather than direct modulators of general features like cell cycle control. A major task lies in revealing the processes allowing these transcription factors to activate target genes selectively within certain cells and tissues. The resolution of this will help us understand complex events such as epigenetic alterations and lineage commitment steps at a molecular level. Much work remains to elucidate the exact roles for COE proteins in normal developmental processes and also the potential role of COE proteins in pathological conditions since disturbance of differentiation is a key mark in human cancer.
Acknowledgments
We thank Alain Vincent for critical reading of the manuscript as well as laboratory members of the Stem Cell Department in Lund and at the Department of Molecular Biology at AstraZeneca in Mölndal for helpful discussions.
The work was funded by The Swedish Cancer Foundation; American Cancer Society Lund Donation; the medical faculty at Lund University; the Magnus Bergwall, Åke Wibergs, and Alfred Österlunds Foundations; The Royal Physiographic Society; Kocks; The Crafoord Foundations; AstraZeneca; and the Swedish Research Council.
REFERENCES
- 1.Ailion, M., and J. H. Thomas. 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156:1047-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerblad, P., M. Rosberg, T. Leanderson, and M. Sigvardsson. 1999. The B29 (immunoglobulin beta-chain) gene is a genetic target for early B-cell factor. Mol. Cell. Biol. 19:392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerblad, P., and M. Sigvardsson. 1999. Early B cell factor is an activator of the B lymphoid kinase promoter in early B cell development. J. Immunol. 163:5453-5461. [PubMed] [Google Scholar]
- 3a.Akerblad, P., U. Lind, D. Liberg, K. Bamberg, and M. Sigvardsson. 2002. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol. Cell. Biol. 22:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and E. Koonin. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023-1040. [DOI] [PubMed] [Google Scholar]
- 5.Artigiani, S., P. M. Comoglio, and L. Tamagnone. 1999. Plexins, semaphorins, and scatter factor receptors: a common root for cell guidance signals? IUBMB Life 48:477-482. [DOI] [PubMed] [Google Scholar]
- 6.Bain, G., E. C. R. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Kroop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Riel, A. Berns, and C. Murre. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 7.Bally-Cuif, L., L. Dubois, and A. Vincent. 1998. Molecular cloning of Zcoe2, the zebrafish homolog of Xenopus Xcoe2 and mouse EBF-2, and its expression during primary neurogenesis. Mech. Dev. 77:85-90. [DOI] [PubMed] [Google Scholar]
- 8.Bork, P., T. Doerks, T. A. Springer, and B. Snel. 1999. Domains in plexins: links to integrins and transcription factors. Trends Biochem. Sci. 24:261-263. [DOI] [PubMed] [Google Scholar]
- 9.Campos-Ortega, J. A. 1998. The genetics of the Drosophila achaete-scute gene complex: a historical appraisal. Int. J. Dev. Biol. 42:291-297. [PubMed] [Google Scholar]
- 10.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 11.Cowherd, R. M., R. E. Lyle, and R. E. McGehee, Jr. 1999. Molecular regulation of adipocyte differentiation. Semin. Cell Dev. Biol. 10:3-10. [DOI] [PubMed] [Google Scholar]
- 12.Crozatier, M., D. Valle, L. Dubois, S. Ibnsouda, and A. Vincent. 1996. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 6:707-718. [DOI] [PubMed] [Google Scholar]
- 13.Crozatier, M., D. Valle, L. Dubois, S. Ibnsouda, and A. Vincent. 1999. Head versus trunk patterning in the Drosophila embryo; collier requirement for formation of the intercalary segment. Development 126:4385-4394. [DOI] [PubMed] [Google Scholar]
- 14.Crozatier, M., and A. Vincent. 1999. Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: transcriptional response to notch signalling. Development 126:1495-1504. [DOI] [PubMed] [Google Scholar]
- 15.Dambly-Chaudiere, C., and M. Vervoort. 1998. The bHLH genes in neural development. Int. J. Dev. Biol. 42:269-273. [PubMed] [Google Scholar]
- 16.Dowell, P., and D. W. Cooke. 2002. Olf-1/early B cell factor is a regulator of glut4 gene expression in 3T3-L1 adipocytes. J. Biol. Chem. 277:1712-1718. [DOI] [PubMed] [Google Scholar]
- 17.Dubois, L., L. Bally-Cuif, M. Crozatier, J. Moreau, L. Paquereau, and A. Vincent. 1998. XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr. Biol. 8:199-209. [DOI] [PubMed] [Google Scholar]
- 18.Dubois, L., and A. Vincent. 2001. The COE-Collier/Olf1/EBF-transcription factors: structural conservation and diversity of developmental functions. Mech. Dev. 108:3-12. [DOI] [PubMed] [Google Scholar]
- 19.Elberg, G., J. M. Gimble, and S. Y. Tsai. 2000. Modulation of the murine peroxisome proliferator-activated receptor gamma 2 promoter activity by CCAAT/enhancer-binding proteins. J. Biol. Chem. 275:27815-27822. [DOI] [PubMed] [Google Scholar]
- 20.Fajas, L., K. Schoonjans, L. Gelman, J. B. Kim, J. Najib, G. Martin, J. C. Fruchart, M. Briggs, B. M. Spiegelman, and J. Auwerx. 1999. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol. Cell. Biol. 19:5495-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frasch, M. 1999. Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr. Opin. Genet. Dev 9:522-529. [DOI] [PubMed] [Google Scholar]
- 22.Garel, S., F. Marin, R. Grosschedl, and P. Charnay. 1999. Ebf1 controls early cell differentiation in the embryonic striatum. Development 126:5285-5294. [DOI] [PubMed] [Google Scholar]
- 23.Garel, S., F. Marin, M. G. Mattei, C. Vesque, A. Vincent, and P. Charnay. 1997. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev. Dyn. 210:191-205. [DOI] [PubMed] [Google Scholar]
- 24.Gisler, R., S.-E. Jacobsen, and M. Sigvardsson. 2000. Cloning of human early B cell factor and identification of target genes suggest a conserved role for EBF in B cell development. Blood 96:1457-1464. [PubMed] [Google Scholar]
- 25.Gisler, R., and M. Sigvardsson. 2002. The human V-PreB promoter is a target for coordinated activation by early B cell factor and E47. J. Immunol. 168:5130-5138. [DOI] [PubMed] [Google Scholar]
- 26.Goebel, P., N. Janney, J. R. Valenzuela, W. J. Romanow, C. Murre, and A. J. Feeney. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagman, J., C. Belanger, A. Travis, C. W. Turck, and R. Grosschedl. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760-773. [DOI] [PubMed] [Google Scholar]
- 28.Hagman, J., M. J. Gutch, H. Lin, and R. Grosschedl. 1995. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 14:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagman, J., A. Travis, and R. Grosschedl. 1991. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 10:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hata, A., J. Seoane, G. Lagna, E. Montalvo, A. Hemmati-Brivanlou, and J. Massague. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229-240. [DOI] [PubMed] [Google Scholar]
- 32.Kee, B. L., and C. Murre. 1998. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 188:699-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kee, B. L., M. W. Quong, and C. Murre. 2000. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol. Rev. 175:138-149. [PubMed] [Google Scholar]
- 34.Kim, J., and B. Spiegelman. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10:1096-1107. [DOI] [PubMed] [Google Scholar]
- 35.Kim, J. B., H. M. Wright, M. Wright, and B. M. Spiegelman. 1998. ADD1/SREBP1 activates PPAR gamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA 95:4333-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudrycki, K., C. Stein-Iszak, C. Behn, M. Grillo, R. Akeson, and F. L. Margolis. 1993. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol. Cell. Biol. 13:3002-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudrycki, K. E., O. Buiakova, G. Tarozzo, M. Grillo, E. Walters, and F. L. Margolis. 1998. Effects of mutation of the Olf-1 motif on transgene expression in olfactory receptor neurons. J. Neurosci. Res. 52:159-172. [DOI] [PubMed] [Google Scholar]
- 38.Lee, J. E. 1997. Basic helix-loop-helix genes in neural development. Curr. Opin. Neurobiol. 7:13-20. [DOI] [PubMed] [Google Scholar]
- 39.Lin, H., and R. Grosschedl. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376:263-267. [DOI] [PubMed] [Google Scholar]
- 40.Malgaretti, N., O. Pozzoli, A. Bosetti, A. Corradi, S. Ciarmatori, M. Panigada, M. E. Bianchi, S. Martinez, and G. G. Consalez. 1997. Mmot1, a new helix-loop-helix transcription factor gene displaying a sharp expression boundary in the embryonic mouse brain. J. Biol. Chem. 272:17632-17639. [DOI] [PubMed] [Google Scholar]
- 41.Martensson, A., and I. L. Martensson. 1997. Early B cell factor binds to a site critical for lambda5 core enhancer activity. Eur. J. Immunol. 27:315-320. [DOI] [PubMed] [Google Scholar]
- 42.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milatovich, A., R. G. Qiu, R. Grosschedl, and U. Francke. 1994. Gene for a tissue-specific transcriptional activator (EBF or Olf-1), expressed in early B lymphocytes, adipocytes, and olfactory neurons, is located on human chromosome 5, band q34, and proximal mouse chromosome 11. Mamm. Genome 5:211-215. [DOI] [PubMed] [Google Scholar]
- 44.Murre, C., G. Bain, M. A. van Dijk, I. Engel, B. A. Furnari, M. E. Massari, J. R. Matthews, M. W. Quong, R. R. Rivera, and M. H. Stuiver. 1994. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta 1218:129-135. [DOI] [PubMed] [Google Scholar]
- 45.Nestoras, K., H. Lee, and J. Mohler. 1997. Role of knot (kn) in wing patterning in Drosophila. Genetics 147:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieminen, P., J. Liippo, and O. Lassila. 2000. Pax-5 and EBF are expressed in committed B-cell progenitors prior to the colonization of the embryonic bursa of fabricius. Scand. J. Immunol. 52:465-469. [DOI] [PubMed] [Google Scholar]
- 47.Nutt, S. L., B. Heavey, A. G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556-562. [DOI] [PubMed] [Google Scholar]
- 48.Olson, E. N. 1993. Regulation of muscle transcription by the MyoD family. The heart of the matter. Circ. Res. 72:1-6. [DOI] [PubMed] [Google Scholar]
- 49.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 50.Persson, C., A. Martensson, and I. L. Martensson. 1998. Identification of a tissue- and differentiation stage-specific enhancer of the VpreB1 gene. Eur. J. Immunol. 28:787-798. [DOI] [PubMed] [Google Scholar]
- 51.Pozzoli, O., A. Bosetti, L. Croci, G. G. Consalez, and M. L. Vetter. 2001. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev. Biol. 233:495-512. [DOI] [PubMed] [Google Scholar]
- 52.Prasad, B. C., B. Ye, R. Zackhary, K. Schrader, G. Seydoux, and R. R. Reed. 1998. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125:1561-1568. [DOI] [PubMed] [Google Scholar]
- 53.Rolink, A., D. Haasner, F. Melchers, and J. Andersson. 1996. The surrogate light chain in mouse B-cell development. Int. Rev. Immunol. 13:341-356. [DOI] [PubMed] [Google Scholar]
- 54.Romanow, W. J., A. W. Langerak, P. Goebel, I. L. Wolvers-Tettero, J. J. van Dongen, A. J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5:343-353. [DOI] [PubMed] [Google Scholar]
- 55.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 56.Schilham, M. W., M. A. Oosterwegel, P. Moerer, J. Ya, P. A. de Boer, M. van de Wetering, S. Verbeek, W. H. Lamers, A. M. Kruisbeek, A. Cumano, and H. Clevers. 1996. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380:711-714. [DOI] [PubMed] [Google Scholar]
- 57.Sigvardsson, M. 2000. Overlapping expression of early B-cell factor and basic helix-loop-helix proteins as a mechanism to dictate B-lineage-specific activity of the lambda5 promoter. Mol. Cell. Biol. 20:3640-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 59.Smith, E. M., R. Gisler, and M. Sigvardsson. 2002. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J. Immunol. 169:261-270. [DOI] [PubMed] [Google Scholar]
- 60.Tamagnone, L., and P. M. Comoglio. 2000. Signalling by semaphorin receptors: cell guidance and beyond. Trends Cell Biol. 10:377-383. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J., D. Higgins, and T. Gibson. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1995. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr. Opin. Genet. Dev. 5:571-576. [DOI] [PubMed] [Google Scholar]
- 63.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 64.Tontonoz, P., J. Kim, R. Graves, and B. Spiegelman. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13:4753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Travis, A., J. Hageman, L. Hwang, and R. Grosschedl. 1993. Purification of early-B-cell factor and characterization of its DNA-binding specificity. Mol. Cell. Biol. 13:3392-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai, R. Y., and R. R. Reed. 1997. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J. Neurosci. 17:4159-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urbánek, P., Z.-Q. Wang, I. Fetka, E. F. Wagner, and M. Busslinger. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901-912. [DOI] [PubMed] [Google Scholar]
- 68.Vervoort, M., M. Crozatier, D. Valle, and A. Vincent. 1999. The COE transcription factor Collier is a mediator of short-range Hedgehog-induced patterning of the Drosophila wing. Curr. Biol. 9:632-639. [DOI] [PubMed] [Google Scholar]
- 69.Wang, M. M., and R. R. Reed. 1993. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364:121-126. [DOI] [PubMed] [Google Scholar]
- 70.Wang, S. S., A. G. Betz, and R. R. Reed. 2002. Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol. Cell. Neurosci. 20:404-414. [DOI] [PubMed] [Google Scholar]
- 71.Wang, S. S., R. Y. L. Tsai, and R. R. Reed. 1997. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J. Neurosci 17:4149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeh, W., Z. Cao, M. Classon, and S. McNight. 1995. Cascade regulation of terminal adipoyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 15:168-181. [DOI] [PubMed] [Google Scholar]
- 73.Yu, H. H., and A. L. Kolodkin. 1999. Semaphorin signaling: a little less per-plexin. Neuron 22:11-14. [DOI] [PubMed] [Google Scholar]
- 74.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]