Abstract
Developmentally regulated initiation of DNA synthesis was studied in the fly Sciara at locus II/9A. PCR analysis of nascent strands revealed an initiation zone that spans ∼8 kb in mitotic embryonic cells and endoreplicating salivary glands but contracts to 1.2 to 2.0 kb during DNA amplification of DNA puff II/9A. Thus, the amplification origin occurs within the initiation zone used for normal replication. The initiation zone left-hand border is constant, but the right-hand border changes during development. Also, there is a shift in the preferred site for initiation of DNA synthesis during DNA amplification compared to that in preamplification stages. This is the first demonstration that once an initiation zone is defined in embryos, its borders and preferred replication start sites can change during development. Chromatin immunoprecipitation showed that the RNA polymerase II 140-kDa subunit occupies the promoter of gene II/9-1 during DNA amplification, even though intense transcription will not start until the next developmental stage. RNA polymerase II is adjacent to the right-hand border of the initiation zone at DNA amplification but not at preamplification, suggesting that it may influence the position of this border. These findings support a relationship between the transcriptional machinery and establishment of the replication initiation zone.
Replication of the genome is a highly regulated process to ensure complete passage of all of the hereditary material from parent to daughter cells. Origins of replication are well defined in the yeast Saccharomyces cerevisiae where extensive studies have identified autonomously replicating sequences (ARS; reviewed in reference 70) that direct the onset of DNA synthesis in plasmids (14, 49). However, when yeast chromosomes are examined, not all ARS elements are active (31, 42), suggesting that chromosomal context is important. In yeast ARS1, element A (66) is essential for sequence recognition by the six-polypeptide origin recognition complex (ORC) (6, 28). ORC serves as the landing pad for other components of the prereplication complex (reviewed in references 9, 32, and 81). We have shown that the nucleotide position in yeast ARS1 where continuous-strand DNA synthesis starts is directly adjacent to the DNA binding site for ORC (7, 8).
The specification of origins of replication in multicellular eukaryotes is less clear. There are only a few cases in which metazoan ARS elements have been identified on episomes (Table 3 in reference 24). Generally, plasmid replication in metazoan cells is sequence independent and just depends on the size of the plasmid (44, 48, 68). Similarly, there is no sequence specificity for replication origins in early embryos of flies and frogs (50, 51, 82), where there is rapid oscillation between S and M phases of the cell cycle.
An additional complexity in metazoan chromosomes is that the size of replicons changes during development. Classic studies with DNA fiber autoradiography revealed that there are many more replicons in early embryos than in differentiated tissues or in germ cells (11, 19, 20). It appears that the number of sites for initiation of DNA synthesis becomes more restricted as development proceeds. Indeed, two-dimensional (2D) gel analysis demonstrated that replication, which had initiated throughout the genome in the early cleavage divisions of embryos, becomes confined to specific initiation zones after zygotic transcription begins at the mid-blastula transition in Xenopus (51) or after cellularization in Drosophila (79). Thus, after these embryonic stages, each replicon has an initiation zone where DNA synthesis starts. Molecular dissection is under way to uncover the cis-acting elements necessary for origin specification in a chromosomal context in several mammalian origins (3, 4, 52, 65).
We have employed locus II/9A of the fungus fly, Sciara coprophila (reviewed in reference 36), as a model system to study initiation zone specification during development. As in other diptera, polytene chromosomes are found in larval salivary glands as a result of cycles of endoreplication that bypass mitosis (85). A unique feature of Sciara salivary gland polytene chromosomes is that certain loci form large “DNA puffs” in late larval life, which are sites not only of intense transcription but also of DNA amplification (reviewed in reference 35). The largest of these is DNA puff 9A on chromosome II (II/9A) with 17-fold amplification of its DNA, which precedes the burst of transcription from genes II/9-1 and II/9-2 (99). Initiation of amplification is confined to a 5.5-kb zone, with the majority of the initiation events occurring in a 1-kb region, as mapped by 2- and 3D gels (59, 60).
Recently, the method of replication initiation point mapping (34, 37) has allowed us and others to identify the sites for initiation of DNA synthesis at the nucleotide level (1, 7, 8, 10, 41). Sciara II/9A is the only metazoan locus where both the start site for continuous strand synthesis and the ORC binding site have been mapped (10) and extends to higher organisms the yeast ARS1 paradigm of ORC binding adjacent to the initiation site (7). However, it is likely that other factors besides ORC play important roles in definition of the initiation zone in metazoa because (i) specification of the initiation zone in frog eggs is not due to a limiting amount of ORC that is present in sufficient amounts (97) and (ii) specification of the initiation zone at the origin decision point in G1 of mammalian cells seems to occur a few hours after formation of the prereplication complex and thus does not seem to be directly regulated by ORC (72).
In this report, we mapped the initiation zone for replication of Sciara locus II/9A in several developmental stages: (i) 9-h embryos undergoing mitosis, (ii) preamplification stage larval salivary gland polytene chromosomes, and (iii) amplification stage larval salivary glands. The data show that the amplification origin resides within the initiation zone for normal replication. Moreover, the initiation zone has the same boundary at its left side throughout development, but the right boundary changes so that a ∼8-kb initiation zone in embryos and preamplification stage larvae contracts to a 1.2- to 2.0-kb initiation zone at DNA amplification stage. Quantitative PCR revealed that the preferred site for initiation within this zone also changes when the zone contracts. We demonstrate that specification of an initiation zone can be independent of active RNA synthesis, since there is no evidence for transcriptional activity at locus II/9A in 9-h embryos, yet an initiation zone with discrete boundaries is found at this stage. Similarly, the burst of transcription at locus II/9A has not yet started when DNA amplification is occurring (99). Nonetheless, the contraction of the initiation zone at DNA amplification correlates with the in vivo occupancy of RNA polymerase II at the gene II/9-1 promoter, adjacent to the right boundary of the replication initiation zone. This suggests that the start sites for DNA replication may be influenced by the transcriptional machinery, even though RNA synthesis has not yet commenced.
Although there are examples of changes in replicon size, origin activation or inactivation, and timing of origin activation during development and in various cell types, this is the first report that an initiation zone can change during development.
MATERIALS AND METHODS
Isolation of nascent DNA.
Salivary glands from 50 amplification stage (10 × 5 and 12 × 6 eyespot stage [33, 99]) or 500 preamplification stage (6 × 2 and 8 × 4 eyespot stage) female larvae from the 6980 stock of the fungus fly Sciara coprophila were dissected in Robert's (78) CR medium (87 mM NaCl, 3.2 mM KCl, 1.3 mM CaCl2, 1 mM MgCl2, 10 mM Tris-HCl; pH 7.3) and placed in Cannon's insect medium (21). For analysis of mitotic cells, 500 mg of embryos 9 h after egg deposition was collected in Cannon's medium. Total chromosomal DNA was recovered by using DNAzol (Gibco-BRL/Life Technologies, Rockville, Md.) as described earlier (37), yielding ∼15-μg DNA from 100 salivary glands. The replicative intermediates were eluted from a 1-ml benzolated naphtholated DEAE (BND) cellulose column (Sigma, St. Louis, Mo.) and precipitated as previously described (37).
Nascent DNA was further purified by λ-exonuclease, which degrades DNA with a free 5′ end (parental DNA and broken DNA) but leaves nascent DNA containing an RNA primer at its 5′ end intact (7, 34, 37). This purification is useful as the starting material for PCR analysis of nascent DNA (37, 54, 58). λ-Exonuclease digestion was carried out on replicative intermediate DNA that had been phosphorylated and precipitated (34, 37) and dissolved in 10 μl of 10 mM Tris (pH 8.0) for λ-exonuclease digestion as previously described (37) except that after incubation for 12 h at 37°C, the same amount of λ-exonuclease was added to the sample as a booster and digestion was continued for another 12 h at 37°C.
Size fractionation of nascent DNA.
The nascent DNA was size fractionated on an alkaline low-melting-point agarose gel (AMRESCO, Solon, Ohio) as described earlier (37). The DNA marker lanes (100-bp and 1-kb DNA ladders; Gibco-BRL Life Technologies) were cut out, stained with ethidium bromide, and photographed next to a ruler. The unstained gel lanes with the sample were sliced at 5-mm intervals, and the DNA from each slice was recovered with GELase (Epicentre Technologies, Madison, Wis.) as recommended by the manufacturer. The size of the nascent DNA in each fraction was determined based on the mobility of the DNA markers. The nascent DNA from each fraction was precipitated with ethanol and Pink Pellet carrier (Novagen, Madison, Wis.) and redissolved in 10 μl of TE buffer (10 mM Tris, pH 7.5; 1 mM EDTA).
PCR analysis of nascent strand length.
Equal amounts of nascent DNA from each size fraction were used as the template for PCR by using the primer sets as follows: set A (nucleotide 1 is at the left-hand end of the 5.5-kb EcoRI fragment in locus II/9A), 5′ 978CCT AAT TTC GAA AAT ATG TGT CAT CC953 3′ and 5′ 669GTT ATC TGG CTG CGG CTC TG688 3′; set B (nucleotide 1 is at the left-hand end of the 5.5-kb EcoRI fragment in locus II/9A), 5′ 2020ACT TCA GGT GAA ATC GTG CC2001 3′ and 5′ 1821ACC AAC ACC AAC GAA CCA AC1840 3′; set C (nucleotide −1 is immediately before the transcription start site of gene II/9-1), 5′ 144CGC TGT GTA ATG AGC TG160 3′ and 5′ 374AGA CAA TAA AGC CGT CC358 3′; set D (nucleotide 1 is immediately after the end of transcription of gene II/9-1), 5′ 434CGG TTG AGC CAT ATA AAG CG415 3′ and 5′ 59GCA CCA TGT GGA GTT CC75 3′; and set E (numbering is upstream of the HindIII site at the 3′ end of the HindIII 2.2-kb fragment from locus II/9A), 5′ 6CGA GAG AAG GAA GCT AGG C24 3′ and 5′ 276CCA ATA GAC GTT AAA GAT CCC256 3′.
PCR was performed as described earlier (37) for 30 cycles with 1 min of denaturation at 94°C; 30 s of annealing at 58°C (set A), 55.3°C (set B), 52.5°C (set C), or 56.0°C (sets D and E); and 1 min of elongation at 72°C. The PCR products were purified by using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and electrophoresed in a 2% neutral agarose gel.
Quantitative PCR analysis of nascent strand abundance.
Nascent DNA was obtained by BND cellulose column chromatography and λ-exonuclease digestion as described above, but salivary glands from 1,000 larvae were used as the starting material. After size fractionation by alkaline gel electrophoresis through 1.2% (wt/vol) low-melting-point agarose, nascent DNA from two fractions (860 to 900 bp and 1,500 to 1,570 bp) was recovered by using GELase as described above, ethanol precipitated, redissolved in 40 μl of TE buffer, and stored at −80°C in 1-μl aliquots. Primer sets A to E (see above) were used for quantitative PCR. For each primer set, a competitor DNA fragment was constructed (30), which was exactly the same sequence as the target nascent DNA except for a 20-bp insertion (5′ ACC TGC AGG GAT CCG TCG AC 3′) in the middle.
A fixed amount of nascent DNA (1 μl in 300 μl of PCR Supermix from Gibco-BRL/Life Technologies, enough for six PCRs) was mixed with increasing amounts of competitor for each primer set and amplified with 45 to 55 cycles of PCR by using denaturation at 94°C for 1 min, annealing for 30 s (at 58.5°C for primer sets A and E, 56.0°C for sets B and D, and 55.5°C for set C), and extension at 72°C for 1 min. The PCR products were recovered by using QIAquick as described above, resolved on a 4% MetaPhor agarose gel (FMC BioProducts, Rockland, Maine) in TAE buffer (40 mM Tris-acetate, 1 mM EDTA), stained with ethidium bromide, and photographed. The intensity of each band was determined by using NIH Image 1.61.1 software.
Chromatin immunoprecipitation (ChIP).
A total of 25 to 50 pairs of amplification or preamplification stage salivary glands were dissected in batches of 10 to 15 in Robert's CR medium (78) (where 10 mM HEPES [pH 7.6] replaced the Tris buffer), placed in 0.5 ml of the dissection buffer containing 2% formaldehyde, and incubated 15 min at room temperature on a rotator, followed by 45 min at 12°C. The sample was centrifuged and resuspended in the dissection buffer containing 0.125 mM glycine to stop the reaction. The cross-linked salivary glands were rinsed twice with cold Tris-buffered saline (150 mM NaCl, 20 mM Tris-HCl; pH 7.6) and resuspended in 400 μl of lysis buffer (90) for sonication to an average size of 500 bp.
Subsequently, 3 μl of primary antibody was added to the sample and incubated at 12°C overnight on a rotator. Antibodies used were gAPα-D1 against the 140-kDa subunit of RNA polymerase II (84) (a generous gift from Arno Greenleaf), antibody against calf thymus histones (Chemicon, Temecula, Calif.), or anti-actin (Santa Cruz Biochemicals, Santa Cruz, Calif.). Then 20 μl of protein G-Sepharose beads (Pharmacia, Peapack, N.J.) and 1% Empigen BB (Calbiochem, La Jolla, Calif.) were added for a further incubation on a rotator for 4 h at 12°C. The bead complexes were washed as described before (87), resuspended in 200 μl of elution buffer (50 mM Tris-HCl, pH 8.0; 10 mM EDTA; 1% sodium dodecyl sulfate), and incubated at 65°C for 30 min. After centrifugation at 14,000 rpm for 2 min to pellet the beads, the cross-links in the supernatant material were reversed at 65°C for 12 h. Subsequently, 100 μg of proteinase K (Boehringer Mannheim, Ingelheim, Germany) was added for a 2-h incubation at 37°C, followed by two phenol-chloroform extractions, one chloroform extraction, and ethanol precipitation with Pink Pellet carrier. The DNA was resuspended for PCR analysis as described above, with 37 to 39 cycles after an initial step of 95°C denaturation. The products were analyzed by 2.5% agarose gel electrophoresis.
RESULTS
Developmental analysis of the boundaries for replication initiation.
We utilized PCR analysis of nascent strands (96) to map the initiation zone where DNA replication starts at locus II/9A in Sciara during developmental stages that precede DNA amplification. Like 2D gel mapping, PCR analysis of nascent strands utilizes material from an asynchronous population of cells, thus avoiding potential artifacts from cell cycle synchronization. The PCR approach also allows examination of preamplification stages since it requires much less material than 2D gels. For the current studies, nascent DNA was isolated by BND column chromatography, followed by λ-exonuclease, and then size fractionated by alkaline gel electrophoresis (37).
Two types of PCR analysis were carried out (Fig. 1). The first is a qualitative approach that maps the boundaries of the initiation zone (96). This method takes advantage of the fact that small nascent DNAs are formed at the origin and grow larger as bidirectional replication proceeds. Each fraction of nascent DNA from gel electrophoresis is used as the template for PCR with primer sets from the area of interest (Fig. 1). The primer set that gives a PCR product from the fraction with the smallest nascent DNA denotes the origin where replication initiates (Fig. 1). The second approach, to be described below, is a quantitative approach (30) that allows determination of the frequency of initiation of DNA synthesis at various positions within the initiation zone. Data from the quantitative method showed that the initiation events dropped off at the boundaries of the initiation zone, thus confirming our results from the qualitative method.
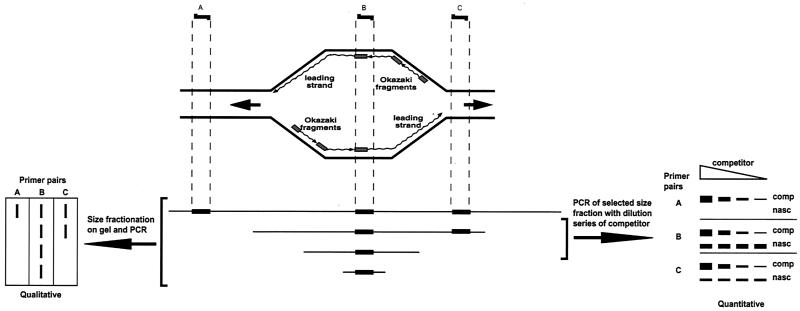
FIG. 1.
Qualitative and quantitative methods to determine regions where DNA synthesis starts. The schematic drawing shows a replication bubble with nascent DNA comprised of the leading strand (continuous synthesis) and the lagging strand (discontinuous Okazaki fragment synthesis and ligation). An asynchronous population of replicating molecules results in replication bubbles of all sizes from each replicon. Nascent DNA is enriched and then separated according to size by alkaline gel electrophoresis. PCR is performed on each size fraction with primer pairs (A to C) from the area of interest. In the qualitative approach (96) (lower left of diagram), all three primer pairs form a product when using the longest nascent DNA as a template, but only primer pair B forms a product from the smallest nascent DNA. In the quantitative approach, a size fraction of nascent DNA is selected for use in competitive PCR (30), using a range of known concentrations of the competitor. The concentration where the product of the competitor is the same as the nascent DNA indicates the concentration of the latter. See the text for further details.
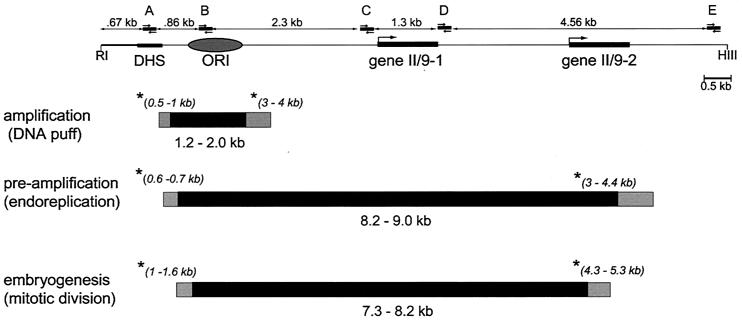
We began our study by use of the qualitative approach (96), by using primer pairs A through E (Fig. 2). As a control for the method, we used PCR analysis of nascent DNA from amplification stage Sciara salivary glands, where the origin of amplification had been mapped before to a 1-kb zone by 2D and 3D gels (59, 60). As can be seen in Fig. 3, primer set B, which is complementary to sequences in the previously mapped origin of amplification, produced PCR products from the smallest nascent DNA analyzed (0.2- to 0.4-kb fraction), confirming that this area is the place where DNA replication begins. Note that DNA of <0.2 kb is not analyzed since it will contain unligated Okazaki fragments that would be detected by all primer pairs. In contrast to primer set B, primer sets A and C, which are to the left and right, respectively, of the amplification origin, did not produce PCR products from small nascent DNA fractions (Fig. 3), indicating that they map beyond the initiation zone. The first observable PCR product utilizing primer set A was produced from 0.5- to 1.0-kb nascent DNA (Fig. 3). Since DNA replication is bidirectional (shown for the Sciara II/9A locus by neutral/alkaline 2D gels [60]) and if we presume that both forks move at the same rate, dividing 0.5 to 1.0 kb by 2 suggests that replication initiated 0.25 to 0.5 kb away from primer set A. Similarly, it can be calculated that replication initiated 1.5 to 2.0 kb away from primer set C, which detected 3- to 4-kb fragments as the smallest nascent DNA (Fig. 3). Therefore, as shown schematically in Fig. 2, it can be deduced from PCR analysis of nascent strand length that the initiation zone for Sciara II/9A DNA amplification is 1.2 to 2.0 kb, coinciding with the area previously mapped by 2D and 3D gels (59, 60).
FIG. 2.
The initiation zone for replication at locus II/9A changes during development. (Top) Map of locus II/9A in Sciara. The DHS occurs concurrently with replication (94), ORI designates the 1-kb amplification origin mapped by 2D and 3D gels (59, 60), and genes II/9-1 and II/9-2 are transcription units from this locus (27). The positions of the five primer pairs (A to E) are shown, as are EcoRI (RI) and HindIII (HIII) restriction sites that flank this 11-kb region. (Bottom) Schematic drawings of the initiation zones calculated from data in Fig. 3. The nascent DNA fractions used to calculate the boundaries of the initiation zones are marked with an asterisk, and the nascent DNA size of that fraction is shown in parentheses. The black area is the initiation zone if the higher value for the smallest detectable nascent DNA is used for the calculation, whereas the gray area delimits the initiation zone calculated from the lower value for the smallest detectable nascent DNA.
FIG. 3.
PCR analysis of nascent strand length from the II/9A locus of Sciara. (A) PCR analysis of nascent strand length at locus II/9A from various developmental stages in Sciara: DNA amplification stage to form DNA puffs in the salivary gland polytene chromosomes (upper panel), preamplification stage salivary glands where endoreplication results in polytenization of the chromosomes (middle panel), and 9-h embryos where mitotic division in cycle 11 occurs (lower panel). Selected size fractions of nascent DNA were pooled and assayed by PCR. Photographs of ethidium bromide-stained agarose gels show the PCR products from nascent strands; the length of nascent DNA in each fraction is indicated. The fractions where a PCR product was obtained from the smallest nascent DNA are marked with an asterisk and were used to estimate the boundaries of the initiation zone (Fig. 2). The pale band seen with primer pair A for the 0.2- to 0.3-kb size fraction of nascent DNA from embryos most likely is due to contamination from unligated Okazaki fragments. The positions of primer sets A to E are shown on Fig. 2. The lanes marked M contain a 100-bp DNA size marker (Gibco-BRL/Life Technologies). (B) Lane 1 of each panel contains a 100-bp size marker (Gibco-BRL/Life Technologies). Lane 2 of each panel shows the efficiency of the indicated primer pair (A to E) in amplifying 20 ng of chromosomal DNA under the same experimental conditions used above in panel A. The size of the PCR product from each primer set is indicated.
Is the origin used for “normal” replication the same as that used for DNA puff amplification? Circumstantial evidence suggests that the mechanism of replication is basically the same for DNA amplification, since proteins required for normal replication are also essential for DNA amplification (reference 80 and references therein). However, the control that regulates once-and-only-once initiation of replication per cell cycle has been overridden. To determine whether the preamplification and amplification origins coincide for Sciara II/9A, we mapped the initiation zone for material from preamplification stages. When nascent DNA from preamplification stage larval salivary glands was used for PCR analysis, products from the smallest DNA fraction (0.2 to 0.4 kb) were detected by using primer set B, as for amplification stage DNA (Fig. 3). Similarly, a PCR product was first detected by primer set A for 0.6- to 0.7-kb nascent DNA (Fig. 3). However, unlike amplification stage DNA, primer sets C and D generated products from all fractions of preamplification DNA, starting from 0.2 to 0.4 kb (Fig. 3). In contrast, PCR products from small nascent DNA were not produced with primer set E, which is much further to the right. The first nascent DNA to be detected by primer set E was 3.0 to 4.4 kb (Fig. 3), indicating that the right-hand boundary for the initiation zone is 1.5 to 2.2 kb away from primer set E. These data show that the left-hand border of the initiation zone is the same at Sciara II/9A for DNA amplification and for polytene endoreplication prior to DNA amplification (Fig. 2). However, the right boundary is further to the right than for amplification, thus resulting in an 8.2- to 9.0-kb initiation zone at II/9A for endoreplication.
The change in the size of the initiation zone noted upon comparing polytene endoreplication with DNA amplification prompted us to examine its size in mitotic cells by using embryos as the source of material. The rapid, syncytial mitotic divisions of Sciara embryos begin to slow down at cycle 9 (25). Cellularization occurs at cycle 11 (9 h after egg deposition, the stage used for the present study) (25). By comparison, the cell cycle in Drosophila embryos begins to slow down at cycle 11, cellularization and an increase in histone H1 occur at cycle 13, and after three more cycles followed by a pause in replication, endoreplication begins, starting first with salivary gland tissue (22, 85). As shown in Fig. 3, nascent DNA from 9 -h Sciara embryos was first detected at a size of 1.0 to 1.6 kb by primer set A and at 4.3 to 5.3 kb and larger by primer set E, and all sizes of nascent DNA were detected by primer sets B, C, and D. From these results it can be calculated that the initiation zone for mitotic embryonic cells is 7.3 to 8.2 kb (Fig. 2). The size and position of the initiation zone in mitotic cells are similar, within the range of experimental error, to those found in preamplification stages of endoreplicating cells, with no significant difference between the initiation zones from these two stages (Fig. 2). In both cases, the left border of the initiation zone is comparable to that for amplifying DNA, but the right border is farther to the right (Fig. 3). The finding that the initiation zone for amplification is located within the initiation zone used prior to amplification is consistent with results from 2D gel analysis of DNA from 3,000 preamplification stage salivary glands. The latter revealed a replication bubble arc in the 5.5-kb EcoRI fragment that contains the amplification origin but not in flanking fragments (94). The number of salivary glands required for 2D gel analysis of preamplification DNA precluded a more extensive study by that method. The PCR analysis presented here not only needs less material but also gives better resolution than 2D gel mapping.
Developmental changes in the preferred initiation sites.
The experiments described above define the boundaries of the initiation zone, but they do not elucidate whether DNA synthesis starts equally throughout this zone, or whether certain sequences within this zone are preferred. To address this question, we utilized quantitative PCR (Fig. 1) to analyze the abundance of nascent DNA throughout the initiation zone (30, 38, 39), by using the same primer sets A to E described above. We created a competitor DNA fragment that was the same as the nascent DNA target except for an insertion of 20 bp of an unrelated sequence. Increasing amounts of the competitor (C) and a fixed amount of the nascent DNA (N) serve as templates for coamplification by PCR with the same primer set. Since the competitor and nascent DNAs compete for the same primers, they are amplified by PCR with the same efficiency. Gel electrophoresis, followed by ethidium bromide staining of the products of the PCR, allowed quantification of the C/N ratio throughout a range of concentrations of the competitor. When C/N = 1, the concentration of nascent DNA can be determined since it is the same as the competitor at that point.
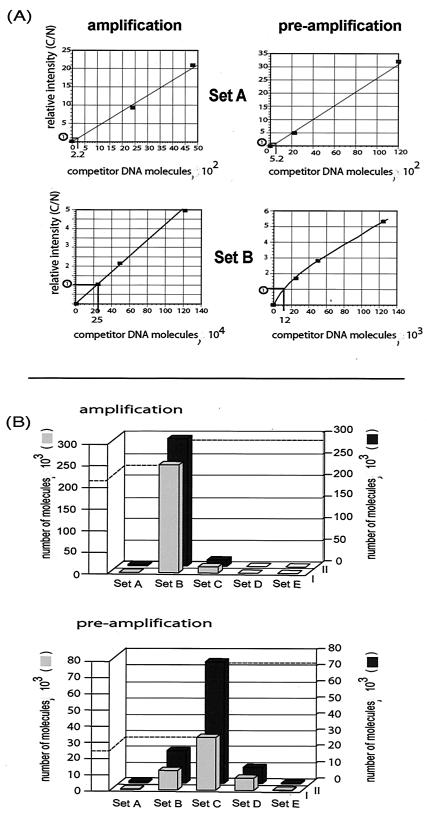
The method of quantitative PCR was used to analyze the abundance of nascent DNA from two size fractions (860 to 900 bp and 1,500 to 1,570 bp), which are sufficiently small to be indicative of initiation of DNA replication but large enough to avoid contamination by Okazaki fragments that are found in all regions of replication. Examples of the data gathered by using amplification stage and preamplification stage salivary gland nascent DNA (860- to 900-bp size fraction) are shown in Fig. 4. As the amount of competitor DNA (C) increases, the amount of PCR product derived from the nascent DNA (N) decreases. Under the PCR conditions used, a linear relationship exists between the ratio of C/N and the increasing concentration of C; the point where N/C = 1 can be interpolated from a graph, as exemplified in Fig. 5A. Such a calculation was done for each primer set for both amplification stage and preamplification stage DNA to determine the number of nascent DNA molecules (Fig. 4). Figure 5B summarizes the number of nascent DNA molecules for the 860- to 900-bp and 1,500- to 1,570-bp fractions (gray and black bars, respectively). As can be seen, at amplification stage the overwhelming majority of small nascent DNA strands are detected by primer set B. In contrast, at preamplification stage, although a significant number of small nascent DNA strands is detected by primer B, the majority are detected by primer set C. These conclusions are the same for independent experiments with the 860- to 900-bp and 1,500- to 1,570-bp fractions. Thus, not only is the initiation zone longer at preamplification than at amplification stage, but also the preferred site for initiation is further to the right within this zone (coinciding with primer set C rather than primer set B).
FIG. 4.
PCR analysis of nascent strand abundance at the Sciara II/9A locus. Nascent DNA from amplification or preamplification stage Sciara salivary glands was fractionated by alkaline agarose gel electrophoresis. Small nascent DNA (N) of 860 to 900 bp (shown here) or 1,500 to 1,570 bp (data not shown) was mixed with increasing amounts of competitor DNA (C) (the number of competitor DNA molecules is indicated) and coamplified by PCR with primer sets A to E. The PCR products were resolved on a 4% MetaPhor agarose gel and stained with ethidium bromide, and the intensity of each band was quantified to allow calculation of the number of small nascent strand DNA molecules (n) per five salivary gland equivalents.
FIG. 5.
Preferred initiation site for DNA synthesis changes during development. (A) The relative intensity of the competitor and nascent DNA bands (C/N) (see Fig. 4) is plotted against the number of competitor DNA molecules. The point on this linear relationship where C/N = 1 is indicated. (B) The number of nascent DNA molecules (n in Fig. 4) is plotted against primer sets A to E (the distance between primer sets is not drawn to scale; refer to Fig. 2 marking the primer sets on a map of the II/9A locus). The calculated values of small nascent strand abundance are shown for amplification and preamplification stages for nascent DNA fractions of 860 to 900 bp (gray boxes) or 1,500 to 1,570 bp (black boxes). The relatively greater number of nascent DNA molecules detected by primer set C at preamplification stage (black compared to gray boxes) could reflect initiation from an origin not centered at primer set C and present in the 1,500- to 1,570-bp fraction but not in the 860- to 900-bp fraction.
Correlation of the transcriptional machinery and the boundary for initiation of DNA synthesis.
What factors during development might cause contraction of the initiation zone and a change in preference for DNA synthesis initiation sites? Since the left-hand border of the initiation zone remains constant, the question arises: what causes the right-hand border to change during development? After DNA amplification has occurred, there is a burst in transcription of the DNA puff genes (99). Although intense transcription at DNA puff II/9A does not begin until DNA amplification has been completed, some of the transcriptional machinery could already be in place prior to its activation and might affect the right-hand boundary. The promoter for gene II/9-1 is close to the right-hand boundary of the amplification initiation zone, and occupancy by transcriptional components could preclude initiation of DNA synthesis there.
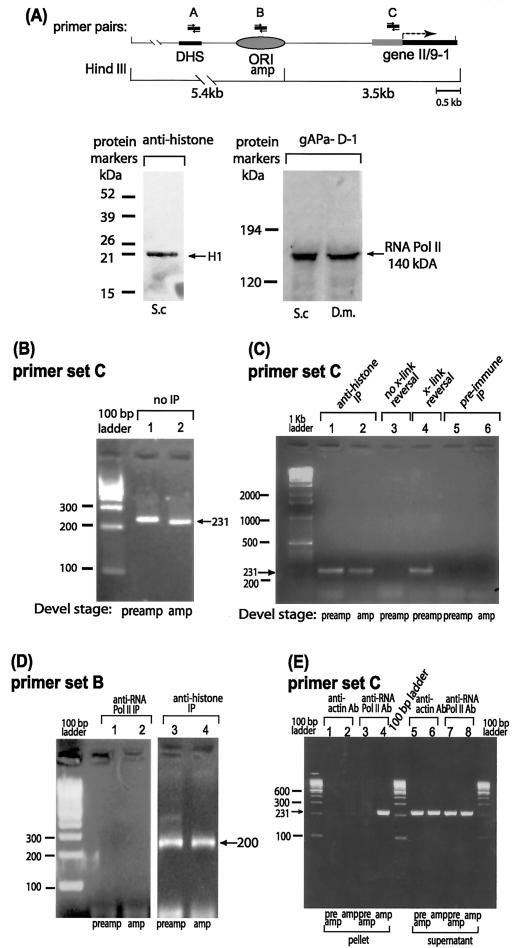
In order to test this idea, ChIP (46, 87, 90) was carried out to explore the developmental profile of gene II/9-1 promoter occupancy by RNA polymerase in vivo. Formaldehyde cross-linking stabilized the in vivo association of proteins with DNA; after immunoprecipitation, the cross-links were reversed and PCR detected the presence of specific DNA sequences in the immunoprecipitate. We utilized an antibody against the RNA polymerase II 140-kDa subunit of Drosophila melanogaster (84). Western blots demonstrated that this antibody reacts specifically with both Drosophila and Sciara (Fig. 6A). Antibody against histones was used as a positive control (Fig. 6C and D), and an anti-actin antibody was used as a negative control (Fig. 6E). The same amount of preamplification or amplification stage DNA was used as the template for PCR, as verified by the observation of comparable amounts of PCR product produced from both developmental stages prior to immunoprecipitation (Fig. 6B) or after immunoprecipitation with the control antibody against histones (Fig. 6C, lanes 1 and 2). Moreover, DNA is not nonspecifically trapped on the beads used for immunoprecipitation, as DNA was not detected on the beads incubated with preimmune serum (Fig. 6C, lanes 5 and 6). As another control, it is apparent that no PCR product is seen when the formaldehyde cross-links are still intact (Fig. 6C, lane 3), but PCR product is formed after cross-link reversal (Fig. 6C, lane 4).
FIG. 6.
ChIP reveals in vivo occupancy by RNA polymerase II near the right-hand border of the initiation zone for DNA amplification. (A) Map of the region of interest in locus II/9A of Sciara, with PCR primer sets A to C indicated. Other details of the map are the same as in Fig. 2. Sites cleaved by HindIII are shown below the map. Western blots are shown for reaction of protein from S. coprophila (S.c.) or D. melanogaster (D.m.) larval homogenate with antibody against the 140-kDa subunit of RNA polymerase II (gAPα-D1) or histones. Only histone H1 is detected in 0.5 M NaCl extracted material. (B to E) Gel electrophoresis of ethidium bromide-stained PCR products with primer set C (panels B, C, and E) or primer set B (panel D) after ChIP with the antibody shown or after the indicated control treatments.
Strikingly, when antibody against RNA polymerase II was used for ChIP, a PCR product was detected only in the amplification stage sample and not in the preamplification sample (Fig. 6E, lanes 4 and 3, respectively). No PCR product was obtained for either developmental stage when using a control antibody against actin (Fig. 6E, lanes 1 and 2). The amount of input DNA was equivalent for samples from both developmental stages, as shown by the similar amounts of PCR product in the supernatant of the immunoprecipitate of the RNA polymerase II antibody (Fig. 6E, lanes 7 and 8) or the actin antibody (Fig. 6E, lanes 5 and 6). These data demonstrate unequivocally that there is in vivo occupancy of the promoter of gene II/9-1 by the 140-kDa subunit of RNA polymerase II at amplification stage but not at preamplification stage. In contrast, there is no in vivo occupancy by RNA polymerase II in the region used as the origin of amplification (detected by primer set B) at either developmental stage (Fig. 6D, lanes 1 and 2) even though similar amounts of input DNA had been used (Fig. 6D, lanes 3 and 4). Thus, there is specific occupancy by the 140-kDa subunit of RNA polymerase II at the promoter of gene II/9-1 but not at sequences further upstream (amplification origin) during DNA amplification stage when the right-hand border of the initiation zone has contracted. Conversely, there is no in vivo occupancy by RNA polymerase II in the promoter at preamplification stage when a larger initiation zone is found. Therefore, the appearance of RNA polymerase II at the gene II/9-1 promoter correlates with contraction of the initiation zone to a position just upstream of the transcriptional machinery.
DISCUSSION
Developmental changes in the DNA synthesis initiation zone.
In this report we have shown for the first time in any system that (i) an origin for DNA amplification resides within the initiation zone for normal replication; (ii) once an initiation zone is defined in early embryogenesis, its size can change during development (e.g., with DNA amplification); and (iii) the preferred start sites for DNA replication within an initiation zone can change during development (cf. DNA amplification to preamplification).
Thus, there is developmental plasticity in origin definition. These findings raise the question of what factors contribute to definition of the initiation zone and determination of where DNA synthesis will start.
Little is known about the developmental specification of replication initiation in metazoans. During the rapid cleavages in early embryos, there is no sequence specificity for DNA synthesis start sites (50, 51, 82). With the onset of zygotic transcription at the loci examined previously (51, 79), initiation of replication becomes more stringent and only begins in certain zones.
We found that the ∼8-kb initiation zone at Sciara locus II/9A is similar for cellular embryos and for salivary gland polytene cells from preamplification stage. Thus, the definition of an initiation zone does not require passage through mitosis, as the initiation zone is the same for cells with a mitotic cycle as for cells undergoing endoreplication where G and S alternate without mitosis. Surprisingly, the situation changes with DNA amplification, and the initiation zone contracts to 1.2 to 2.0 kb. Therefore, sites where DNA synthesis will initiate are not fixed permanently in the organism, but can change, reflecting developmental plasticity. Previously, it was known that some areas where DNA synthesis starts can be silenced as development progresses (11, 19, 20), but the present data are the first example of the boundary of a specified initiation zone changing during development.
One boundary of the initiation zone does not change.
The left-hand boundary of the Sciara II/9A initiation zone remains similar at preamplification and amplification stages and coincides with a strong DNase I-hypersensitive site (DHS) (94). There is no initiation of replication at the DHS (shown here with primer set A), nor to its immediate left as shown previously (59, 60). The DHS might act as a unidirectional barrier, preventing initiation of DNA synthesis to its left. Alternatively, it could play a positive role and stimulate DNA replication to its right. The latter possibility is reminiscent of replication enhancers affecting the complex origins of eukaryotic cells (2, 52, 53). A very interesting example of a replication enhancer is the amplification control element (ACE), which stimulates DNA amplification in the Drosophila chorion loci (23, 73, 74). How ACE3 works is unknown; it binds ORC (5), but replication does not initiate at ACE3 (64). The action of ACE3 to stimulate replication from ori-β located some distance to its right can be blocked by the chromatin boundary element su(hw) (64). It is unknown if ACE flanks the as-yet-unmapped initiation zone for replication at the chorion loci prior to DNA amplification.
Correlation of the developmental change in an initiation zone boundary with the transcriptional machinery.
Although the left boundary of the II/9A initiation zone is fixed during Sciara development, coinciding with DHS, we have shown here that the right boundary changes. The significance of this developmental change will require detailed understanding of the mechanism. At preamplification stages, the initiation zone spans two quiescent transcription units. However, during DNA amplification, initiation events are limited to an area upstream of genes II/9-1 and II/9-2. The right boundary of the amplification initiation zone correlates with occupancy by RNA polymerase II, even though intense transcription will not occur at this locus until the next developmental stage when DNA amplification has ceased (99). Eukaryotic origins are located in intergenic regions (e.g., see Table 4 in reference 24 and Table 1 in reference 93 [40]), suggesting that the machinery for the initiation of replication may not be compatible with transcription, as has been demonstrated in some cases (43, 86, 91, 92, 95). It is probable that evolution has selected for DNA and RNA synthesis to move in the same direction to avoid head-on collisions (13, 63). In the case of bidirectional replication that initiates in the nontranscribed spacer of ribosomal DNA downstream of an active gene (69), a replication fork barrier is located beyond the 3′ end of the ribosomal DNA transcription unit and prevents the replication fork from moving against the direction of rRNA synthesis (15, 61). This fail-safe barrier acts even if transcription is prevented (18), suggesting an epigenetic level of regulation.
Change in the DNA synthesis start site preference.
In addition to a change in the right boundary of the II/9A initiation zone, the data from quantitative PCR analysis of nascent strand abundance presented here show for the first time that the preferred area within the initiation zone where replication starts can change during development. In preamplification stage polytene chromosomes, the greatest preference for initiation of replication exists over the promoter for the quiescent gene II/9-1, but in DNA amplification stage the preferred site where DNA synthesis starts shifts to the left of the II/9-1 promoter. The specified initiation zone seems to comprise preferred sites where DNA synthesis starts (24). For example, application of various mapping methods to the 55-kb initiation zone of the DHFR locus (29) revealed preferred areas where DNA synthesis starts (ori-β and β′, which are 5 kb apart, and ori-γ, which is 22 kb further downstream [references 54 and 76 and references therein]). On rare occasions, synthesis can start at other positions within the zone, which normally are suppressed by the preferred origins. Indeed, it has been shown that DNA synthesis can initiate at these other sites once the preferred site is removed (52). The phenomenon of origin interference (16, 17, 67) makes it unlikely that replication begins at multiple nearby sites in a single molecule. Rather, there is probably population polymorphism, and only a single origin will fire on a given DNA molecule, but which origin that is may vary between molecules. Support for the latter idea comes from 3D gel analysis of the Sciara II/9A amplification origin, suggesting that there is only one active origin per molecule of DNA in this locus (59). As observed in the present study, different molecules of DNA may start DNA synthesis at various sites within the ∼8-kb preamplification initiation zone, but there is preference for initiation to start in the gene II/9-1 promoter region.
A developmental change occurs to switch the preferred start site for replication to the left of the gene II/9-1 promoter during DNA amplification. Previously, we found that DNA synthesis begins adjacent to the ORC binding site in yeast (7, 8) and metazoa (10). At DNA amplification stage, ORC binds to the Sciara II/9A amplification origin but not to the gene II/9-1 promoter region (10). In contrast, our preliminary data (unpublished) show ORC occupancy of the gene II/9-1 promoter only at preamplification stages. Binding by other macromolecules at DNA amplification stage to the gene II/9-1 promoter could prevent ORC from binding there and therefore alter the preferred start site. As a precedent for this, positioning a nucleosome over yeast ARS1 prevented DNA synthesis from starting there (83). Factors other than nucleosomes might also block ORC binding and initiation of DNA synthesis. The ChIP data presented here demonstrate that there is in vivo occupancy of the gene II/9-1 promoter by the 140-kDa subunit of RNA polymerase II at DNA amplification stage, even though intense transcription has not yet started. This suggests that the presence of the transcriptional machinery may block entry of ORC to the promoter region, which had been the preferred start site for DNA synthesis in preamplification polytene chromosomes.
In addition to the occupancy of the gene II/9-1 promoter by RNA polymerase II at amplification stage, other events may also occur to prepare the cell for intense transcription at this locus, which will occur at the next developmental stage, and these could have important ramifications for positioning the start site for DNA replication. Transcription can activate upstream replication, as shown previously in an artificial plasmid system (71). Moreover, the presence of an intact promoter, even without active transcription, can specify the position where replication starts in plasmids injected into Xenopus eggs (E. Danis, S. Menut, C. Girard-Reydet, D. Maiorano, and M. Méchali, unpublished data), indicating that origin activation is not simply due to negative supercoiling in the wake of RNA polymerase that is transcribing DNA. Previous reports link the specification of the replication initiation zone during embryogenesis to the onset of active transcription at that locus (51, 79). We present here the first evidence for specification of a distinct initiation zone (∼8 kb) when no transcription is detected in cellular embryos and preamplification salivary glands at Sciara locus II/9A. Therefore, events other than active transcription may specify the initiation zone. Although one of these events could be the presence of the inactive transcriptional machinery, such as at DNA amplification stage, other events must specify the ∼8-kb initiation zone at earlier stages when RNA polymerase II is absent from this region.
Certain protein factors stimulate replication, as well as transcription, as first documented for eukaryotic viruses (reviewed in reference 95). Similarly, in yeast the transcription factor Abf1p stimulates replication and can be functionally replaced by some but not other transcription factors (57, 66, 77, 98). Metazoan origins also contain transcription factor binding sites (26, 47). Transcription factors might stimulate replication either by recruitment of RNA polymerase (88) or by contact with the replication machinery. For example, transcriptional activators can interact with replication protein A (45, 55, 56). In addition, it has been shown in Drosophila that the transcription factor E2F is in a complex with ORC and can influence its activity (12). Chromatin remodeling may also occur and/or repositioning of nucleosomes; changes in chromatin can activate a silent telomeric origin in yeast (89). It is known that nucleosomes can facilitate initiation of DNA synthesis, as shown for yeast ARS1 (62). The events above are not mutually exclusive and might all occur together to position the replication start sites in the intergenic region where it will not be compromised by transcription. Our data support a link between the transcriptional machinery and definition of the initiation zone of DNA synthesis and set the stage for further elucidation of the coordination of replication and transcription.
Acknowledgments
We are extremely grateful to Arno L. Greenleaf for Drosophila RNA polymerase II antibodies. We thank John Waggener for help in mapping the primer pairs; Anton Borovjagin for help with the figures; and Zaklina Strezoska, Thilo Sascha Lange, and Rob Burgess for helpful comments on the manuscript.
This work was supported by NIH grant GM35929 to S.A.G.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the α-globin locus control region in initiation of DNA replication. Science 270:815-819. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 4.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin, R. J., T. L. Orr-Weaver, and S. P. Bell. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 7.Bielinsky, A.-K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 8.Bielinsky, A.-K., and S. A. Gerbi. 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3:477-486. [DOI] [PubMed] [Google Scholar]
- 9.Bielinsky, A.-K., and S. A. Gerbi. 2001. Where it all starts: eukaryotic origins of DNA replication. J. Cell Sci. 114:643-651. [DOI] [PubMed] [Google Scholar]
- 10.Bielinsky, A.-K., H. Blitzblau, E. L. Beall, M. Ezrokhi, H. S. Smith, M. R. Botchan, and S. A. Gerbi. 2001. Origin recognition complex binding to a metazoan origin. Curr. Biol. 11:1-20. [DOI] [PubMed] [Google Scholar]
- 11.Blumenthal, A. B., H. J. Kriegstein, and D. S. Hogness. 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp. Quant. Biol. 38:205-223. [DOI] [PubMed] [Google Scholar]
- 12.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 13.Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. [DOI] [PubMed] [Google Scholar]
- 14.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 15.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 16.Brewer, B. J., and W. L. Fangman. 1993. Initiation at closely spaced replication origins in a yeast chromosome. Science 262:1728-1731. [DOI] [PubMed] [Google Scholar]
- 17.Brewer, B. J., and W. L. Fangman. 1994. Initiation preference at a yeast origin of replication. Proc. Natl. Acad. Sci. USA 91:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. [DOI] [PubMed] [Google Scholar]
- 19.Callan, H. G. 1972. Replication of DNA in the chromosomes of eukaryotes. Proc. R. Soc. Lond. Biol. Sci. 181:19-41. [DOI] [PubMed] [Google Scholar]
- 20.Callan, H. G. 1974. DNA replication in the chromosomes of eukaryotes. Cold Spring Harbor Symp. Quant. Biol. 38:195-203. [DOI] [PubMed] [Google Scholar]
- 21.Cannon, G. B. 1964. Culture of insect salivary glands in a chemically defined medium. Science 146:1063.. [DOI] [PubMed] [Google Scholar]
- 22.Carminati, J. L., and T. L. Orr-Weaver. 1996. Changes in DNA replication during animal development, p. 409-434. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Carminati, J. L., C. G. Johnston, and T. L. Orr-Weaver. 1992. The Drosophila ACE3 chorion element autonomously induces amplification. Mol. Cell. Biol. 12:2444-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePamphilis, M. L. 1996. Origins of DNA replication, p. 45-86. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.de Saint Phalle, B., and W. Sullivan. 1996. Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early Sciara coprophila embryogenesis. Development 122:3775-3784. [DOI] [PubMed] [Google Scholar]
- 26.de Stanchina, E., D. Gabellini, P. Norio, M. Giacca, F. A. Peverali, S. Riva, A. Falaschi, and G. Biamonti. 2000. Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol. 299:667-680. [DOI] [PubMed] [Google Scholar]
- 27.DiBartolomeis, S. M., and S. A. Gerbi. 1989. Molecular characterization of DNA puff II/9A genes in Sciara coprophila. J. Mol. Biol. 210:531-540. [DOI] [PubMed] [Google Scholar]
- 28.Diffley, J. F., and J. H. Cocker. 1992. Protein-DNA interactions at a yeast replication origin. Nature 357:169-172. [DOI] [PubMed] [Google Scholar]
- 29.Dijkwel, P. A., and J. L. Hamlin. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 15:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Divacco, S., P. Norio, L. Zentilin, S. Menzo, M. Clementi, G. Biamonti, S. Riva, A. Falaschi, and M. Giacca. 1992. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene 122:313-320. [DOI] [PubMed] [Google Scholar]
- 31.Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. Zhu, J. R. Broach, C. S. Newlon, and J. A. Huberman. 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 11:5346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta, A., and S. P. Bell. 1997. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 13:293-332. [DOI] [PubMed] [Google Scholar]
- 33.Gabrusewycz-Garcia, N. 1964. Cytological and autoradiographic studies in Sciara coprophila salivary gland polytene chromosomes. Chromosoma 15:312-344. [DOI] [PubMed] [Google Scholar]
- 34.Gerbi, S. A., and A.-K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271-280. [DOI] [PubMed] [Google Scholar]
- 35.Gerbi, S. A., and F. D. Urnov. 1996. Differential DNA replication in insects, p. 947-969. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Gerbi, S. A., C. Liang, N. Wu, S. M. DiBartolomeis, B. Bienz-Tadmor, H. S. Smith, and F. D. Urnov. 1993. DNA amplification in DNA puff II/9A of Sciara coprophila. Cold Spring Harbor Symp. Quant. Biol. 58:487-494. [DOI] [PubMed] [Google Scholar]
- 37.Gerbi, S. A., A.-K. Bielinsky, C. Liang, V. V. Lunyak, and F. D. Urnov. 1999. Methods to map origins of replication in eukaryotes, p. 1-42. In S. Cotterill (ed.), Eukaryotic DNA replication. Oxford University Press, Oxford, United Kingdom.
- 38.Giacca, M., L. Zentilin, P. Norio, S. Divacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, and A. Falaschi. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giacca, G., C. Pelizon, and A. Falaschi. 1997. Mapping replication origins by quantifying the relative abundance of nascent DNA strands using the competitive polymerase chain reaction. Methods 13:301-312. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez, M., and F. Antequera. 1999. Organization of DNA replication origins in the fission yeast genome. EMBO J. 18:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenfeder, S. A., and C. S. Newlon. 1992. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell 3:999-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase, S. B., S. S. Heinzel, and M. P. Calos. 1994. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol. Cell. Biol. 14:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harland, R. M., and R. A. Laskey. 1980. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell 21:761-771. [DOI] [PubMed] [Google Scholar]
- 45.He, Z., T. B. Brinton, J. Greenblatt, J. A. Hassell, and C. J. Ingles. 1993. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 73:1223-1232. [DOI] [PubMed] [Google Scholar]
- 46.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 47.Heintz, N. H. 1996. DNA replication in mammals, p. 983-1004. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Heinzel, S. S., P. J. Krysan, C. T. Tran, and M. P. Calos. 1991. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Cell. Biol. 11:2263-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huberman, J. A., L. D. Spotilla, K. A. Nawotka, S. M. el-Assouli, and L. R. Davis. 1987. The in vivo replication origin of the yeast 2 μm plasmid. Cell 51:473-481. [DOI] [PubMed] [Google Scholar]
- 50.Hyrien, O., and M. Méchali. 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12:4511-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyrien, O., C. Maric, and M. Méchali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 52.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 53.Kim, S.-M., and J. A. Huberman. 1999. Influence of a replication enhancer on a hierarchy of origin efficiencies within a cluster of DNA replication origins. J. Mol. Biol. 288:867-882. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leiter, L. M., J. Chen, T. Marathe, M. Tanaka, and A. Dutta. 1996. Loss of transactivation and transrepression function, and not RPA binding, alters growth suppression by p53. Oncogene 12:2662-2668. [PubMed] [Google Scholar]
- 56.Li, R., and M. R. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p54 bind the cellular replication factor A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 57.Li, R., D. S. Yu, M. Tanaka, L. Zheng, S. L. Berger, and B. Stillman. 1998. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol. Cell. Biol. 18:1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, C., J. A. Bogan, D. A. Natale, and M. L. DePamphilis. 2000. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci. 113:887-898. [DOI] [PubMed] [Google Scholar]
- 59.Liang, C., and S. A. Gerbi. 1994. Analysis of an origin of DNA amplification in Sciara coprophila by a novel three-dimensional gel method. Mol. Cell. Biol. 14:1520-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang, C., J. D. Spitzer, H. S. Smith, and S. A. Gerbi. 1993. Replication initiates at a confined region during DNA amplification in Sciara DNA puff II/9A. Genes Dev. 7:1072-1084. [DOI] [PubMed] [Google Scholar]
- 61.Linskens, M. H., and J. A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7:21-30. [DOI] [PubMed] [Google Scholar]
- 63.Liu, B., and B. M. Alberts. 1995. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267:1131-1137. [DOI] [PubMed] [Google Scholar]
- 64.Lu, L., H. Zhang, and J. Tower. 2001. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malott, M., and M. Leffak. 1999. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-823. [DOI] [PubMed] [Google Scholar]
- 67.Marahrens, Y., and B. Stillman. 1994. Replicator dominance in a eukaryotic chromosome. EMBO J. 13:3395-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Méchali, M., and S. Kearsey. 1984. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 38:55-64. [DOI] [PubMed] [Google Scholar]
- 69.Muller, M., R. Lucchini, and J. M. Sogo. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5:767-777. [DOI] [PubMed] [Google Scholar]
- 70.Newlon, C. S., and J. F. Theis. 1993. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3:752-758. [DOI] [PubMed] [Google Scholar]
- 71.Ohba, R., K. Matsumoto, and Y. Ishimi. 1996. Induction of DNA replication by transcription in the region upstream of the human c-myc gene in a model replication system. Mol. Cell. Biol. 16:5754-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orr-Weaver, T. L., and A. C. Spradling. 1986. Drosophila chorion gene amplification requires an upstream region regulating s18 transcription. Mol. Cell. Biol. 6:4624-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orr-Weaver, T. L., C. G. Johnston, and A. C. Spradling. 1989. The role of ACE3 in chorion gene amplification. EMBO J. 8:4153-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan, W.-J., R. C. Gallagher, and E. H. Blackburn. 1995. Replication of an rDNA gene origin plasmid in the Tetrahymena thermophila macronucleus is prevented by transcription through the origin from an RNA polymerase I promoter. Mol. Cell. Biol. 15:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pelizon, C., S. Divacco, A. Falashi, and M. Giacca. 1996. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16:5358-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rhode, P. R., S. Elsasser, and J. L. Campbell. 1992. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1064-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robert, M. 1971. Einfluss von Ionerstärke und pH auf die differentialle Dekondensation der Nukleoproteide isolieter Speichedrüsen-Zellkerne und Chromosomen von Chironomus thummi. Chromosoma 36:1-33. [DOI] [PubMed] [Google Scholar]
- 79.Sasaki, T., T. Sawado, M. Yamaguchi, and T. Shinomiya. 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwed, G., N. May, Y. Pechersky, and B. R. Calvi. 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seki, T., and J. F. X. Diffley. 2000. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 97:14115-14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shinomiya, T., and S. Ina. 1991. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 19:3935-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simpson, R. T. 1990. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 343:387-389. [DOI] [PubMed] [Google Scholar]
- 84.Skantar, A. M., and A. L. Greenleaf. 1995. Identifying a transcription factor interaction site on RNA polymerase II. Gene Expr. 5:49-69. [PMC free article] [PubMed] [Google Scholar]
- 85.Smith, A. V., and T. L. Orr-Weaver. 1991. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112:997-1008. [DOI] [PubMed] [Google Scholar]
- 86.Snyder, M., R. J. Sapolsky, and R. W. Davis. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Solomon, M. J., P. L. Larsen, and A. Varshavsky. 1988. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53:937-947. [DOI] [PubMed] [Google Scholar]
- 88.Stagljar, I., U. Hübscher, and A. Barberis. 1999. Activation of DNA replication in yeast by recruitment of the RNA polymerase II transcription complex. Biol. Chem. 380:525-530. [DOI] [PubMed] [Google Scholar]
- 89.Stevenson, J. B., and D. E. Gottschling. 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 13:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 91.Stueber, D., and H. Bujard. 1982. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanaka, M., and S. Hiraga. 1985. Negative control of oriC plasmid replication by transcription of the oriC region. Mol. Gen. Genet. 200:21-26. [DOI] [PubMed] [Google Scholar]
- 93.Todorovic, V., A. Falaschi, and M. Giacca. 1999. Replication origins of mammalian chromosomes. Front. Biosci. 4:d859-d868. [DOI] [PubMed] [Google Scholar]
- 94.Urnov, F. D., C. Liang, H. Blitzblau, H. S. Smith, and S. A. Gerbi. A DNase I hypersensitive site flanks an origin of DNA replication and amplification in Sciara. Chromosoma, in press. [DOI] [PubMed]
- 95.van der Vliet, P. C. 1996. Roles of transcription factors in DNA replication, p. 87-118. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 96.Vassilev, L., and E. M. Johnson. 1989. Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res. 17:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walter, J., and J. W. Newport. 1997. Regulation of replicon size in Xenopus egg extracts. Science 275:993-995. [DOI] [PubMed] [Google Scholar]
- 98.Wiltshire, S., S. Raychaudhuri, and S. Eisenberg. 1997. An Abf1p C-terminal region lacking transcriptional activation potential stimulates a yeast origin of replication. Nucleic Acids Res. 25:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu, N., C. Liang, S. M. DiBartolomeis, H. S. Smith, and S. A. Gerbi. 1993. Developmental progression of DNA puffs in Sciara coprophila: amplification and transcription. Dev. Biol. 160:73-84. [DOI] [PubMed] [Google Scholar]