Abstract
The SAGA complex is a conserved histone acetyltransferase-coactivator that regulates gene expression in Saccharomyces cerevisiae. SAGA contains a number of subunits known to function in transcription including Spt and Ada proteins, the Gcn5 acetyltransferase, a subset of TATA-binding-protein-associated factors (TAFIIs), and Tra1. Here we report the identification of SLIK (SAGA-like), a complex related in composition to SAGA. Notably SLIK uniquely contains the protein Rtg2, linking the function of SLIK to the retrograde response pathway. Yeast harboring mutations in both SAGA and SLIK complexes displays synthetic phenotypes more severe than those of yeast with mutation of either complex alone. We present data indicating that distinct forms of the SAGA complex may regulate specific subsets of genes and that SAGA and SLIK have multiple partly overlapping activities, which play a critical role in transcription by RNA polymerase II.
The histone components of chromatin are subjected to several posttranslational modifications including acetylation, phosphorylation, ubiquitination, methylation, and ADP-ribosylation that regulate their function and that of the corresponding DNA sequences (16, 56). Of these modifications the acetylation and deacetylation of the ɛ-amino groups of the conserved lysine residues present in the amino terminal tails of all four core histones have long been linked to transcriptional activity and have been the most intensively studied histone modification. The acetylation of histones is catalyzed by histone acetyltransferases (HATs), which are often found to be associated with large multisubunit protein complexes that contain components with identity or homology to known regulators of transcription (17, 47).
The first transcription-related type A HAT to be identified was the Tetrahymena p55 protein, a homologue of Saccharomyces cerevisiae Gcn5 (8). Gcn5 was originally identified through genetic screens as a transcriptional coactivator in yeast (23) and thus provided a strong foundation for the direct coupling of histone acetylation and transcriptional stimulation. Gcn5 has subsequently been identified as a component of high-molecular-weight acetyltransferase/transcriptional adaptor complexes from both yeast and human cells (19, 25, 37, 40, 43, 45, 49, 50). The association of Gcn5 in multiprotein complexes potentiates its nucleosomal HAT activity (3, 19, 43, 49, 58, 59), important for the transcriptional stimulatory activity of these complexes from chromatin templates in vitro and in vivo (31, 54, 61). In addition this association bestows upon Gcn5 an expanded lysine specificity (3, 20, 52, 65). Thus, a common feature of HAT proteins with proposed coactivator function is their frequent association with other subunits that potentiate catalytic function and/or mediate interactions with activators or basal factors (7).
We have previously identified Gcn5 as a component of the 1.8-MDa transcriptional regulatory complex SAGA (Spt-Ada-Gcn5-acetyltransferase). In this complex Gcn5 is associated with the Ada1 to Ada3 members of the Ada (alteration/deficiency in activation) family of proteins. These proteins were originally identified in genetic screens as being related to Gcn5 (Ada4) in that they functionally interact with the transcription factor Gcn4 and the activation domain derived from the herpes simplex virus activator VP16 (23). SAGA also contains all members of the TATA-binding protein (TBP)-related set of Spt (suppressor of Ty) proteins, Spt3, Spt7, Spt8, and Spt20 (Ada5), except TBP (Spt15) itself (19, 21, 25, 36, 46, 55). SPT gene products were originally isolated as suppressors of transcription initiation defects caused by promoter insertions of the transposable element Ty (63). The association of Ada and Spt proteins in a native complex fulfilled a number of genetic predictions, which had indicated that these proteins function in a common pathway (36, 46). In addition to the known Ada and Spt components, mass spectrometry and Western blotting identified a subset of TAFIIs (TBP-associated factors) as integral components of SAGA (21). TAFII proteins were originally discovered and described as factors associated with TBP as components of the conserved general transcription factor TFIID, required for transcriptional regulation at certain promoters (57). Finally, an additional novel subunit of the SAGA complex, named Tra1 (22), has recently been found to serve as a target for activators during recruitment of HAT complexes (7). This protein is a homologue of the human transformation-transcription domain protein (TRRAP), which is a member of the ataxia telangiectasia mutated (ATM) family of phosphatidylinositol kinases. The TRRAP cofactor is essential for c-Myc- and E2F-mediated oncogenic transformation (39, 41).
Interestingly, two human acetyltransferases, the PCAF and hGcn5/STAGA complexes, are remarkably similar to SAGA and contain homologues of many of the Ada, Spt, Tra1, and TAFII proteins found in SAGA (37, 40). Thus, it is very likely that SAGA, PCAF, and hGcn5 complexes are structurally and functionally homologous. A third human HAT complex related to the PCAF, hGcn5, and SAGA complexes in composition is TFTC (TBP-free TAFII-containing complex) (6). The identification of SAGA complexes from yeast and human extracts underscores a mechanism of transcriptional activation from chromatin via a conserved partnership between this group of cofactors and acetyltransferases.
Here we report the characterization of a novel yeast HAT complex, related in composition and substrate specificity to the SAGA complex. We named this complex SLIK, for SAGA-like (22). SLIK notably lacks the Spt8 protein found in SAGA and contains an altered form of Spt7 but potentially includes a number of additional components not found in SAGA. A unique SLIK component has been identified by mass spectrometry as Rtg2, one of three nonessential proteins previously identified as central components of the retrograde response pathway in yeasts. This pathway involves interorganelle communication, whereby the expression of certain nuclear genes is altered in cells with dysfunctional mitochondria. We show that yeast lacking Rtg2 is disrupted in the SLIK complex and that spt7Δ mutant yeast displays a decrease in expression of the classical RTG-regulated CIT2 gene. We present genetic data revealing that SAGA and SLIK double mutants display synthetic phenotypes, similar to those found upon mutation of the shared component Spt7. Finally, we demonstrate that Rtg2 is bound to chromatin at the promoters of the genes that it regulates. Collectively, our data suggest that the SLIK and SAGA complexes perform functions which are partially overlapping but also link the SLIK HAT complex to the retrograde pathway. The identification of two highly related HAT complexes also complicates the earlier interpretation of Spt, Ada, TAFII, and Tra1 function, which had previously been attributed to the SAGA complex.
MATERIALS AND METHODS
Large-scale SAGA and SLIK complex preparations were made from yeast strain CY396 (swi2Δ::HIS3 HO-lacZ SWI2-HA-6HIS::URA3) (42). Yeast strains SFY526 (MATa ade2 lys1 leu2 his3 trp1 gal4Δ gal80Δ Gal1-lacZ::URA3) and SFY526Δrtg2 are as described previously (48). In Fig. 5 and 6, strains used are derivatives of BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (Research Genetics), PGY20 (MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 rtg2Δ::KAN spt8Δ::KAN), PGY21 (MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 rtg2Δ::KAN), PGY22 (MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 spt8Δ::KAN), PGY23 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 spt7Δ::KAN). Glutathione S-transferase (GST)-tagged proteins were expressed in strain EJ758 from Research Genetics.
FIG. 5.
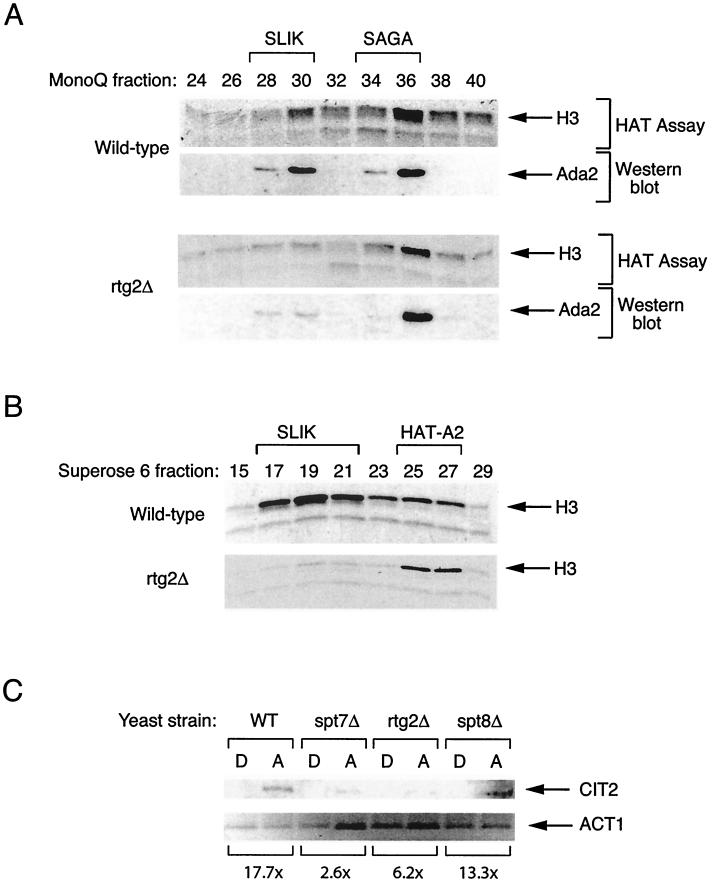
Effect of deletion of RTG2 on the SLIK and SAGA complexes. (A) HAT assay and Western blot analysis of SLIK and SAGA complexes prepared from wild-type (SFY526) or rtg2Δ strains. Mono Q fractions of partially purified SLIK and SAGA were subjected to nucleosomal HAT assay and Western blotting with Ada2 antiserum. (B) SLIK fractions 27 to 31 eluted from the Mono Q column in panel A were separated on a Superose 6 column. Shown is a fluorogram of a nucleosomal HAT assay performed with Superose 6 fractions from wild-type and rtg2Δ strains. (C) Northern blot analysis of CIT2 expression from wild-type (WT, BY4742), spt7Δ, rtg2Δ, and spt8Δ isogenic strains grown in SC medium containing 2% dextrose (D) or 2% acetate (A) as a carbon source. ACT1 (encoding actin) is shown as a loading control. The relative fold increase in CIT2 RNA levels normalized to control RNA for each strain is shown, as estimated by densitometry.
FIG. 6.
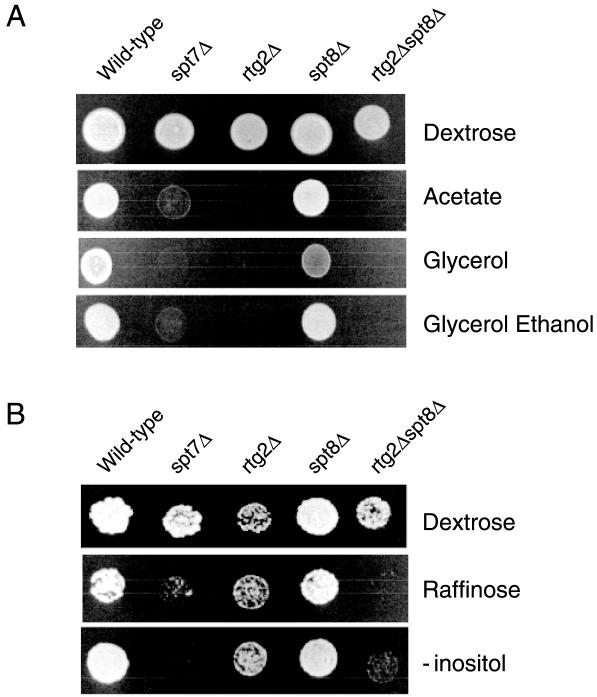
Genetic phenotypes of SLIK and SAGA mutant yeast. (A) Wild-type (BY4742) and spt7Δ, rtg2Δ, spt8Δ, and rtg2Δspt8Δ derivative yeast strains were assayed for their ability to grow on SC medium containing dextrose, acetate, glycerol, or glycerol plus ethanol as carbon sources. (B) Synthetic phenotypes generated in SAGA and SLIK mutant yeast. Wild-type (BY4742), spt7Δ, rtg2Δ, spt8Δ, and rtg2Δ spt8Δ yeast strains were assayed for their ability to grow on SC medium containing 2% dextrose or raffinose carbon sources or in medium lacking inositol.
TAP tagging was performed in yeast strain BY4742, as described previously (44), with the following primers: SPT8 forward, GGGAATTCTACGGACACGACCCTTATTTACGATATAGACTTAGAATCCATGGAAAAGAGAA; SPT8 reverse, CGGAGTAATTATGATTATGATTATGGTTATGATTATTATTACAACTACGACTCACTATAGGG; RTG2 forward, GGAAGTGTAGAGAGGGTTAAAATTGGCGTGCAATTTTATGAAGAATCCATGGAAAAGAGAAG; and RTG2 reverse, TATAAGGATTTCGTATTTATTGTTCAAGTATTTAAAGACTAGATGTACGACTCACTATAGGG.
Purification of SLIK and SAGA complexes.
SLIK was purified by a scheme adapted from the work of Grant et al. (21) and illustrated in Fig. 3A. The highly purified SAGA complex used for silver staining has been described previously (21). Elution of SLIK from each column was monitored by HAT assays. In brief, whole-cell extracts were prepared from 40 liters of yeast strain CY396 grown to mid-log phase in yeast extract-peptone-dextrose (YPD) medium and prepared as described elsewhere (18). The extract was bound batchwise with 40 ml of Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen). The resin was then washed in a column with 20 mM imidazole, followed by elution of the bound proteins with 300 mM imidazole. The Ni2+-NTA agarose column flowthrough was directly loaded onto a Mono Q HR 16/10 column (Amersham Pharmacia). Bound proteins were eluted with a 500-ml linear gradient from 100 to 500 mM NaCl. Peak SLIK fractions were pooled, diluted to 100 mM NaCl, and loaded onto a Mono Q HR 5/5 column (Amersham Pharmacia). Bound proteins were eluted with a 25-ml linear gradient from 100 to 500 mM NaCl. Peak fractions from the Mono Q column were pooled and diluted to 100 mM NaCl and loaded onto a Mono S HR 5/5 column (Amersham Pharmacia). Bound proteins were eluted with a 10-ml linear gradient from 100 to 500 mM NaCl. Peak fractions were diluted to 100 mM NaCl and loaded directly onto a 1-ml histone agarose column (Sigma). Bound proteins were eluted with a 10-ml linear gradient from 100 mM to 1 M NaCl. Peak fractions were diluted to 100 mM NaCl and loaded onto a 1-ml native DNA cellulose column (Amersham Pharmacia), and bound proteins were eluted with a 10-ml gradient from 100 mM to 1 M NaCl. Peak fractions were concentrated down to 0.7 ml with a Centriprep-30 concentrator (Millipore). Samples were then loaded on a Superose 6 HR 10/30 column (Amersham Pharmacia) equilibrated with 350 mM NaCl. SLIK eluted from the column with an apparent molecular mass of approximately 1.8 MDa. Peak Superose 6 fractions were diluted to 100 mM NaCl and loaded onto a Mini Q PC 3.2/3 column (Amersham Pharmacia). Bound proteins were eluted with a 4.8-ml gradient from 100 to 500 mM NaCl.
FIG. 3.
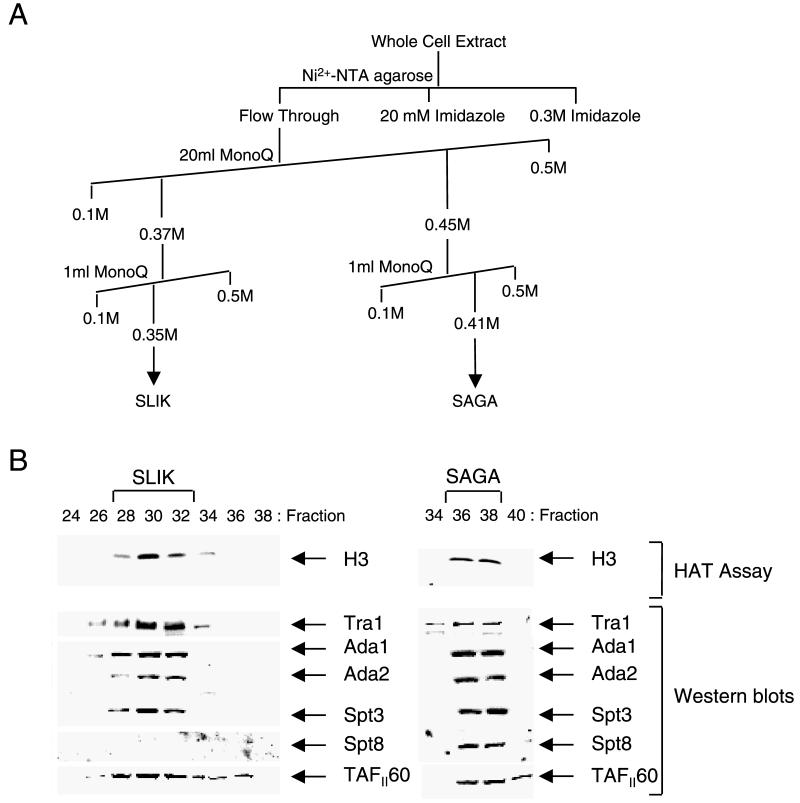
Identification of the components of SLIK. (A) Schematic representation of the chromatographic purification of SLIK, as determined by HAT assays and Western blotting. (B) HAT assay and Western blotting of peak SLIK fractions eluted from the final Mini Q column. Fluorogram and Western blotting analyses indicate that SLIK predominantly elutes in a single protein peak. (C) Silver stain gel of the purified SLIK complex in comparison to SAGA. The peak SLIK fraction from the final Mini Q column was analyzed by silver staining of proteins separated on an SDS-4 to 20% polyacrylamide gel. For comparison a highly purified SAGA complex is shown. The components identified by mass spectrometry are grouped according to their different classes of transcriptional regulators, namely, Spts, Adas, TAFIIs, or others. The position of a protein which likely represents TAFII25 is also shown. Candidate protein bands which appear unique to SLIK are marked with asterisks. Numbers at left of panel C are molecular masses in kilodaltons.
SLIK and SAGA were also partially purified from 4 liters of wild-type (SFY526) or rtg2Δ (SFY526 rtg2Δ) mutant strains, grown to mid-log phase in YPD medium. Whole-cell extracts were prepared by glass bead disruption (18) and bound to 5 ml of nickel-NTA agarose, as described above. SAGA and SLIK were purified from the flowthrough of 5 ml of nickel-NTA, by diluting unbound material to 100 mM NaCl and binding proteins to a Mono Q HR 10/16 column. Bound proteins were eluted with a 500-ml linear gradient from 100 to 500 mM NaCl. Peak SLIK fractions were concentrated and loaded on a Superose 6 column as described above.
SAGA and SLIK were similarly purified from yeast strains expressing Spt8-TAP and Rtg2-TAP proteins. One liter of each strain was grown to mid-log phase in YPD medium, and whole-cell extracts were prepared by glass bead disruption. Extracts were bound to 200 μl of calmodulin resin (Stratagene), as described previously (44). Fractions eluted from the calmodulin resin were concentrated to 50 μl with an Ultrafree-0.5 centrifugal filter (Millipore) and loaded onto a Superose 6 PC 3.2/30 column (Amersham Pharmacia), equilibrated with 350 mM NaCl. The calibration of the Superose 6 column was as follows: dextran blue (2 kDa), fraction 15; apoferritin (443 kDa), fraction 20/21; alcohol dehydrogenase (150 kDa), fraction 23/24; carbonic acid (29 kDa), fraction 28/29. Peak fractions from the Superose 6 column were diluted to 100 mM NaCl and loaded onto a Mini Q column, as described above.
Mass spectrometry.
The peak SLIK fraction from the final Mini Q column (Fig. 3A) was electrophoresed through a sodium dodecyl sulfate (SDS)-4 to 20% polyacrylamide gradient gel. Coomassie blue-stained protein bands were in-gel trypsin digested, and peptides were identified by microcolumn high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry and database searching as described previously (21).
RNA analysis, HAT assays, and Western blotting.
Total RNA purification and Northern blotting were performed as described previously (2). HAT assays were performed in the presence of HeLa nucleosomes (18). Western blotting was performed as described previously (21).
Microsequencing.
HAT assays were performed in the presence of 5 μg of HeLa nucleosomes, 1 μCi of [3H]acetyl coenzyme A, and equivalent activities of purified SAGA and SLIK complexes. Reaction mixtures were incubated for 40 min at 30°C and were then subjected to SDS-polyacrylamide gel electrophoresis on a 15% polyacrylamide gel and transferred to Immobilon PSQ membrane (Millipore). Histone H3 was excised from the membrane and sequenced as described previously (53).
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed largely as described previously (30). Yeast strains bearing plasmids expressing GST, GST-Rtg2, or GST-Spt8 proteins were grown in synthetic complete (SC) medium containing 2% dextrose to an optical density at 600 nm of 1.0. The yeast was reseeded in SC dextrose-medium, SC medium containing 2% KCl, or SC medium containing 2% galactose and grown for an additional 4 h. Antiserum specific for histone H3 acetylated at lysines 9 and 14 was used for ChIP. In certain experiments glutathione-Sepharose resin (Amersham Pharmacia) was used to precipitate chromatin. Input and precipitated DNA was PCR amplified for regions upstream of the CIT2 and GAL10 genes, with the following primers: GAL1-10, CGCTTAACTGCTCATTGCTATATTG and TTGTTCGGAGCAGTGCGGCGC, and CIT2, CGCACATTCGACGCATTTAT and CAAAAAGTTTTGAGAACCTGTTATG.
RESULTS
Isolation of the novel SLIK complex.
We have previously used nickel-NTA agarose to enrich for HATs and deacetylases from yeast whole-cell extracts (1, 11, 19, 27, 33). In this study, proteins that flowed through the nickel-agarose resin were bound to and eluted from a 20-ml Mono Q ion-exchange column. Analysis of the eluting fractions in HAT assays led to the identification of two distinct nucleosomal HAT complexes. Each activity was pooled and bound to a subsequent 1-ml Mono Q column. Figure 1 illustrates the elution profile of these complexes off the Mono Q columns. One complex was found to be indistinguishable from the SAGA complex by HAT assays and Western blotting (Fig. 1B). A second complex was found to possess the same preference for modification of nucleosomal histones H3 and H2B and to contain a large number of the components found in the SAGA complex. We have named this complex SLIK (SAGA-like) (22). Notably the SLIK complex was found both to be chromatographically distinct from the SAGA complex and to lack the Spt8 protein found in SAGA. Spt8 has been found elsewhere to be required for the normal functioning of SAGA (4, 45, 55).
FIG. 1.
Identification of the SLIK complex. (A) Schematic representation of the isolation of SLIK. Complexes were partially purified from whole-cell extracts prepared from yeast strain CY396. (B) Fluorogram of HAT assays and Western blot analyses performed with SLIK and SAGA complexes. Fractions from the Mono Q columns were incubated with nucleosomes and assayed for HAT activity or assayed by Western blotting with the indicated antisera. The position of histone H3 is indicated, as determined by Coomassie blue staining of the same gel.
Studies of the Gcn5-dependent HAT complexes SAGA and ADA have revealed that the two complexes have overlapping, yet distinct, patterns of acetylation on histone H3 tails. While recombinant Gcn5 acetylates only lysine 14, purified ADA complex acetylates both lysine 14 and lysine 18 and SAGA acetylates to some extent all four commonly acetylated lysines in H3 (20). These results suggested that the association of specific subunits determines the site specificity of Gcn5. We therefore investigated the lysine specificity of purified native SLIK complex in comparison to SAGA. Nucleosomes were in vitro labeled in HAT assays performed with either the SAGA or the SLIK complex, and histone H3 was isolated and subjected to microsequence analysis followed by direct determination of radioactivity at each position in the H3 sequence. The data obtained from the microsequence analysis revealed that SAGA and SLIK generate very similar patterns of nucleosomal H3 acetylation (Fig. 2). Both complexes acetylate Lys-14 > Lys-18 > Lys-9 = Lys-23, a pattern also observed with synthetic H3 peptide substrates with SAGA (20). Collectively these data indicate that the two complexes have similar compositions and substrate specificities.
FIG. 2.
The SLIK and SAGA complexes generate similar patterns of acetylation on histone H3 from nucleosomes. HAT assays were performed with nucleosome substrates incubated together with purified SLIK or SAGA complexes. Radiolabeled histone H3 was subsequently purified and subjected to microsequence analysis followed by direct determination of radioactivity at each position in the H3 sequence. The counts per minute for each cycle are plotted against the amino acid detected for that cycle.
Identification of Rtg2 as a component of SLIK.
To identify putative unique components of SLIK, the complex was purified from yeast whole-cell extracts, by a strategy adapted from the purification of the SAGA complex (21). Elution of SLIK from each column, depicted in Fig. 3A, was monitored by a combination of HAT assays and Western blotting utilizing antisera against known components of the complex. SLIK eluting from the final Mini Q column was tested for HAT activity and subjected to Western blotting. SLIK was found to elute in a predominant protein peak, which coeluted with the Tra1, TAFII90, and Ada3 proteins (Fig. 3B). The peak fraction was also analyzed by silver staining, together with purified SAGA complex (21). The two complexes were found to contain remarkably similar patterns of silver staining, consistent with the observation that they share numerous subunits. However, a number of protein bands unique to SLIK were also observed (Fig. 3C).
To identify the components of SLIK, the peak HAT fraction was electrophoresed through an SDS-4 to 20% polyacrylamide gradient gel. Coomassie blue-stained protein bands were excised and in-gel trypsin digested, and proteins were identified by mass spectrometry. Mass spectrometry confirmed the presence of Tra1, Spt3, Spt7, Spt20/Ada5, Ada1, Ada2, Ada3, Gcn5, TAFII17/20, TAFII60, TAFII68/61, and TAFII90. These results were confirmed by Western blotting, which revealed that TAFII25 also copurified with and was present in highly purified SLIK complex. Spt8 was not detected as a component of SLIK by either Western blotting or mass spectrometry. Comparison of the SAGA and SLIK complexes by silver staining revealed that Spt7 showed a different pattern of migration in each complex. A full analysis of Spt7 in SAGA and SLIK has recently been reported (64). In addition to the Ada, Spt, TAFII, and Tra1 proteins we specifically identified Rtg2 as a component of SLIK (Fig. 3C). This protein was found to migrate anomalously at over 100 kDa, perhaps due to incomplete denaturation of Rtg2 in this sample. Mass spectrometry has also revealed other potential components of SLIK (Fig. 3C), which need to be confirmed.
Rtg2 was originally identified as one of three central components of the retrograde response pathway in yeast (35), a stress response mechanism that adjusts various biosynthetic and metabolic activities to alterations in the mitochondrial state. RTG1 and RTG3 encode transcription factors, which activate transcription by binding as a heterodimer to an R box in target promoters (26). Rtg3 has been found to contain an acidic transcriptional domain, which was previously found to be dependent upon Ada2 and Gcn5 for activation (38). How Rtg2 functions in the transcriptional response pathway remained unknown until recently, when Rtg2 was reported elsewhere to regulate the nuclear import of Rtg1 and Rtg3 (28, 51). Our results suggest that Rtg2 may also contribute to transcriptional activation via its association with the SLIK HAT/adaptor complex.
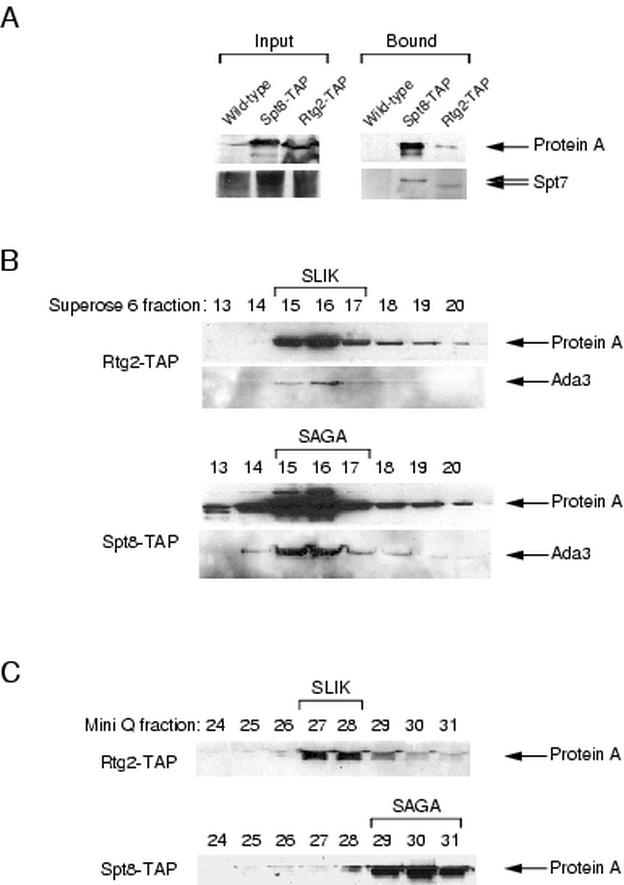
To confirm the stable association of Rtg2 with SLIK, we generated a yeast strain expressing a TAP-tagged Rtg2 protein. The TAP tag is comprised of the calmodulin binding protein and protein A fused to the carboxy terminus of the protein (44). As a control we also generated a TAP-tagged Spt8 protein to analyze the SAGA complex in parallel. Extracts prepared from untagged wild-type, Spt8-TAP, or Rtg2-TAP strains were incubated with calmodulin resin, and fractions eluted from the resin were subjected to Western blotting with protein A antisera. We found that both affinity-purified Rtg2-TAP and affinity-purified Spt8-TAP also precipitated Spt7 protein, while the untagged isogenic strain did not (Fig. 4A). The Spt7 protein coprecipitating with Rtg2-TAP was found to migrate faster by SDS-polyacrylamide gel electrophoresis than Spt7 associated with Spt8-TAP, consistent with the finding that specific forms of Spt7 integrate with SLIK and SAGA (Fig. 3C). To further confirm the association of Rtg2 with SLIK, the eluate from the calmodulin resin was loaded onto a Superose 6 size-exclusion column. We found that both Rtg2 and Spt8 complexes fractionated in high-molecular-mass complexes of approximately 1.8 MDa (Fig. 4B), consistent with their presence in SLIK and SAGA, respectively. Rtg2 was also found to fractionate in a smaller uncharacterized 500-kDa complex from whole-cell extracts (unpublished data). To chromatographically separate and distinguish the Rtg2-TAP and Spt8-TAP complexes, the peak Superose 6 fractions were bound to a Mini Q column. Figure 4C illustrates the elution of these complexes from the Mini Q column. The Rtg2-TAP protein eluted with the SLIK complex, while Spt8-TAP eluted with SAGA.
FIG. 4.
Rtg2 is a unique component of SLIK. (A) Rtg2 associates with the Spt7 protein in vivo. Extracts prepared from strains expressing TAP-tagged Rtg2 or Spt8 or untagged control proteins were incubated with calmodulin resin. Inputs and protein eluted from the resin were subjected to Western blotting with antibodies directed against protein A or Spt7. (B) Size-exclusion chromatography of the Rtg2-TAP and Spt8-TAP complexes isolated from the calmodulin resin. Western blotting of Superose 6 fractions was performed with antibodies against protein A or Ada3. (C) Western blots of Rtg2-TAP and Spt8-TAP fractions eluted from Mini Q columns. Fractions from the Superose 6 column were bound to Mini Q columns, and fractions eluted were assayed by Western blotting with protein A antibodies. The relative positions of the SAGA and SLIK complexes are indicated.
An rtg2Δ mutant specifically perturbs the SLIK complex.
To investigate the contribution of Rtg2 to the integrity of SLIK, we utilized a yeast mutant strain bearing a deletion of the RTG2 gene. Whole-cell extracts were prepared from wild-type and rtg2Δ strains and fractionated over a Mono Q column. The presence of SAGA and SLIK was detected by HAT assay and Western blotting of fractions with antiserum against the common component Ada2. SAGA and SLIK isolated from the wild-type strain were found to show the expected elution profile (Fig. 5A). In comparison, extracts prepared from the rtg2Δ strain were found to be significantly reduced in the amount of HAT activity and Ada2 protein normally coincident with the SLIK complex, while maintaining a similar amount of the SAGA complex. However, some residual HAT activity and Ada2 protein were detected in the rtg2Δ Mono Q fractions 28 to 30 (Fig. 5A). We investigated whether some SLIK complex remained intact in yeast lacking Rtg2 by further purifying peak Mono Q fractions over a Superose 6 size-exclusion column. We found that the HAT activity from the wild-type strain fractionated into two peaks, the predominant SLIK complex and a smaller (250-kDa) H3 HAT activity (Fig. 5B). The latter complex also contains Ada2 protein (unpublished data) and may represent the previously reported HAT-A2 activity (49, 50). Superose fractions prepared from the rtg2Δ strain were found to be specifically depleted in the amount of HAT activity associated with the high-molecular-weight SLIK complex, while maintaining a similar amount of the smaller HAT-A2 complex (Fig. 5B). Collectively these results indicate that Rtg2 is important for the integrity and normal fractionation of the SLIK complex but not for SAGA. Densitometric analysis of the Ada2 Western blots suggests that SAGA is approximately 1.5-fold more abundant than SLIK in our preparation. Extracts prepared from yeast strains having the SPT7 and SPT20 genes deleted were found to be depleted of both SAGA and SLIK, consistent with these proteins being components shared by the two complexes (unpublished data).
The observation that Rtg2 is a component of SLIK suggests that this complex may function in the transcriptional regulation of RTG target genes. A classical Rtg-regulated gene is CIT2, which encodes a peroxisomal isoform of citrate synthase. Both basal and elevated expression of CIT2 are dependent on RTG1, RTG2, and RTG3 (26). Also CIT2 is induced in media containing acetate as a carbon source (60). Wild-type, rtg2Δ, spt7Δ, and spt8Δ yeast strains were grown overnight in SC-dextrose medium and then reseeded in either SC-dextrose or SC-acetate medium and grown for an additional 4 h. RNA was subsequently prepared for Northern blotting with a probe for CIT2. We found that full induction of CIT2 expression was dependent upon functional RTG2 and SPT7 genes but largely independent of SPT8, implying that SLIK rather than SAGA regulates activated CIT2 transcription (Fig. 5C). However, we did not observe a requirement for SPT7 in the basal expression of CIT2, in contrast to RTG2 (unpublished data).
SAGA and SLIK perform partially overlapping functions.
Null alleles in RTG genes result in pleiotropic phenotypes, including loss of CIT2 expression, an inability to use acetate as a carbon source, and glutamate or aspartate auxotrophy (35). Such phenotypes are consistent with cells defective in the tricarboxylic acid and glyoxylate cycles. The observation that proteins such as Spt7 are physically associated with Rtg2 would imply that spt7 mutant yeasts share phenotypes common to those found in an rtg2 mutant. We therefore assayed for the ability of wild-type, spt7Δ (SAGA and SLIK), rtg2Δ (SLIK), and spt8Δ (SAGA) yeast strains to utilize dextrose, acetate, and glycerol or glycerol plus ethanol as a carbon source. Consistent with previous reports, we found that rtg2Δ yeast shows poor growth on plates containing acetate, glycerol, or glycerol plus ethanol in comparison to wild-type yeast (Fig. 6A). However, spt8Δ yeast is able to utilize all carbon sources relatively well. In contrast, spt7Δ yeast is compromised in its ability to utilize carbon sources other than dextrose, a phenotype consistent with a function of the Rtg proteins.
The identification of certain phenotypes unique to either mutation of SPT8 or mutation of RTG2 suggests that SAGA and SLIK perform some distinct functions. However, since the two complexes share at least 13 subunits and have similar substrate specificities in HAT assays, it seems likely that these two activities would also possess certain overlapping functions. That is, loss of function in one complex may be compensated by the activity of the second complex. We therefore sought to identify putative synthetic phenotypes associated with combined SAGA and SLIK mutations by crossing yeasts with mutations in rtg2 and spt8. Figure 6B shows that either rtg2Δ or spt8Δ yeast is able to utilize raffinose as a carbon source and grow normally on medium lacking inositol. However, spt7Δ or rtg2Δ spt8Δ strains display both poor growth on raffinose medium and inositol auxotrophy. These results indicate that certain broad phenotypes associated with mutation of the gene encoding the Spt7, Spt20, or Ada1 protein may be the result of the subsequent disruption of both the SAGA and SLIK complexes. Mutants with combined mutations in unique components of SAGA and SLIK can also synthetically manifest such phenotypes.
Rtg2 is bound to chromatin at the promoter of the derepressed CIT2 gene.
To determine if SLIK and SAGA are bound to and acetylate the promoters of the genes that we postulate they regulate, we utilized GST-RTG2 and GST-SPT8 strains and performed ChIP experiments. Together with a GST control strain, we first confirmed the expression of our fusion proteins (Fig. 7A). Yeast strains expressing GST, GST-Rtg2, or GST-Spt8 proteins were grown in SC medium with either dextrose or acetate and assayed by Northern blotting for induction of the CIT2 gene. GST pull-downs and ChIPs were also performed with anti-acetyl (Lys-9/14) H3 antiserum from identical samples. Consistent with previous observations, CIT2 transcripts were induced in all strains grown in SC-acetate medium (60), in contrast to the ACT1 control. This induction correlated with the acetylation of histone H3 at the promoter of this gene (Fig. 7B). For the same samples used in ChIP experiments, we substituted glutathione-Sepharose as the affinity resin and performed precipitation experiments. GST pull-down analyses show that GST-Rtg2 is enriched upstream of the CIT2 gene under activating conditions, which correlates with promoter acetylation and transcription activation (Fig. 7B). We did not detect increased binding of GST or GST-Spt8 under the same conditions.
FIG. 7.
Recruitment of SAGA and SLIK complexes to specific promoters is coincident with acetylation and gene activation. (A) Western blot prepared from yeast extracts expressing GST, GST-Spt8, or GST-Rtg2 proteins following binding to glutathione-Sepharose. Anti-GST antiserum was used for immunoblotting. (B) The expression of the CIT2 gene and the control ACT1 gene from the indicated yeast strains grown in SC medium containing dextrose (D) or acetate (A) as carbon sources was monitored by Northern blotting. The presence of histone H3 acetylation was measured through PCR-amplified CIT2 promoter DNA from chromatin immunoprecipitated with anti-acetylated lysine 9/14 H3 antiserum. The binding of GST, GST-Rtg2, or GST-Spt8 on the CIT2 promoter was monitored by PCR amplification of DNA from chromatin precipitated with glutathione-Sepharose affinity resin. PCR amplifications from input and precipitated (bound) DNA are shown. (C) Similar assays were performed to study the induction, acetylation, and promoter binding mediated by SLIK and SAGA at the GAL10 locus with the indicated yeast strains grown in SC medium containing dextrose (D) or galactose (G).
Our genetic analysis of SAGA and SLIK function suggests that these complexes have some overlapping redundant roles (Fig. 6). Such candidate functions include the utilization of galactose and raffinose as a carbon source and regulation of GAL genes (64). Yeast strains expressing GST, GST-Rtg2, or GST-Spt8 proteins were grown in SC medium with either dextrose or galactose and assayed by Northern blotting for induction of the GAL10 gene and by ChIP for acetylation of the GAL1-GAL10 promoter. Consistent with previous observations (5, 10, 29, 32), GAL10 transcription is induced in yeast grown in galactose, which correlates with H3 acetylation of the promoter (Fig. 7C). GST pull-down analysis reveals increased binding of both GST-Rtg2 and GST-Spt8 at the GAL10 promoter under inducing conditions. These observations are consistent with the ability of either SAGA or SLIK to regulate the transcription of certain genes.
DISCUSSION
Identification of the SLIK complex.
We have presented evidence that in yeast two highly related HAT complexes exist, SAGA and SLIK. Consistent with this observation, other studies have previously pointed to the putative existence of two distinct high-molecular-weight Ada-containing complexes (22, 25, 50). A combination of mass spectrometry and Western blotting of highly purified SLIK complex identified a similar set of Ada, Spt, TAFII, and Tra1 proteins as components, which were previously identified in SAGA. Both SAGA and SLIK are high-molecular-weight multiprotein complexes with very similar substrate specificities, preferentially modifying the same lysine residues in the nucleosomal histone H3 tail. However, Spt8 and Rtg2 were identified as unique components of SAGA and SLIK, respectively, suggesting that each complex has a distinct function that is not related to histone substrate specificity.
The RTG1, RTG2, and RTG3 genes are central players in the retrograde response pathway (26, 35). This pathway involves a form of interorganelle communication that signals the functional status of the mitochondrion to the nucleus, resulting in changes in the expression of nuclear genes that encode certain mitochondrial, cytoplasmic, and peroxisomal proteins (14). The CIT2 gene, encoding a peroxisomal citrate synthase, shows a typical retrograde response in that its expression is dramatically increased in cells with dysfunctional mitochondria such as po petite mutants. Both basal and elevated CIT2 expression are dependent on RTG1, RTG2, and RTG3. RTG1 and RTG3 encode basic helix-loop-helix leucine zipper transcription factors that bind as a heterodimer to two sites within the CIT2 promoter (26). Genetic and transactivation studies have indicated that Rtg2 acts upstream of Rtg1 and Rtg3 in the regulation of CIT2 (48). More recently Rtg2 has been found to promote the formation of an active Rtg1-Rtg3 transcription factor by transducing mitochondrial or nitrogen quality signals that affect the phosphorylation state and subcellular localization of Rtg3 (28, 51). Here we present evidence that Rtg2 also functions as an essential component of the SLIK HAT complex, required for its structural integrity. Furthermore, we demonstrate that at least a portion of Rtg2 in yeast is chromatin bound at the promoter of the activated CIT2 gene, which coincides with its acetylation.
It has been previously reported that Rtg3 contains a novel motif within its N-terminal activation domain, termed LDFS, homologous to that of vertebrate class I helix-loop-helix proteins (38). Overexpression of a GAL4-Rtg3 fusion protein in yeast leads to toxicity, which is alleviated in strains lacking Ada2 and Gcn5. Furthermore, the transcriptional activity of the N terminus of Rtg3 is dependent upon Ada2 and Gcn5. These studies implicated the recruitment of the SAGA complex in Rtg3-mediated transcriptional activation, and consistent with this, we observed an in vitro interaction between the Rtg3 N terminus and purified SAGA complex. Our observations that Rtg2, Ada2, and Gcn5 are stable components of SLIK and that Rtg2 is bound to the promoter of the CIT2 gene under inducing conditions also suggest a role for SLIK in Rtg3-mediated transcriptional activation. Consistent with this idea, a number of genes induced in respiration-deficient cells such as ACS1, DIP5, CRC1, TNA1, ADH2, VID24, BAT2, CTF19, AGP2, YNL279W, YER187W, and YDR384C (14) were also found to be regulated by components of the SAGA and SLIK complexes in normal cells (24, 34).
The SPT8 gene was originally identified as a suppressor of retrotransposon insertion mutations in the 5′ regions of the HIS4 and LYS2 genes (62). Mutations in the SPT8 gene were found to give phenotypes similar to those of spt3 mutants or in particular mutations in SPT15, which encodes TBP. SPT genes have been grouped into classes based on distinct mutant phenotypes (63). One such group contains five members that are functionally related to TBP function. Four of the SPT genes (SPT3, SPT7, SPT8, and SPT20) encode components of SAGA, while the fifth gene encodes TBP itself. Subsequently the ADA1 gene product was identified as a component of SAGA and ada1Δ mutants were shown to have phenotypes similar to those of spt7Δ and spt20Δ (25, 55). It is notable that SPT3, SPT7, SPT8, and SPT20 and ADA1 have been identified elsewhere as suppressors of his4-917δ, while RTG2 has not (unpublished data). This may suggest that SAGA rather than SLIK distinctly functions at the HIS4 allele. An alternative explanation is that either complex may function at the HIS4 allele but that individual components of the complexes do not contribute equally to this function. This idea is validated in the observation that deletion of SAGA components results in three major subsets of growth phenotypes in vivo (15, 23, 25, 36, 45, 46). These are the Ada−-related moderate (Ada2/Ada3/Gcn5), Spt−-related moderate (Spt3/Spt8), and the Ada− Spt− severe (Ada1/Spt7/Spt20) subsets. The last group is suggested to be most crucial for SAGA, being required for the integrity of the complex, while the first two are required for distinct subsets of functions (23, 63). However, evidence for a division of labor between the SLIK and SAGA complexes is that synthetic phenotypes are observed in an rtg2 spt8 null strain which are similar to those for mutation of the shared component Spt7 (Fig. 6B). Individual rtg2Δ or spt8Δ mutants show phenotypes partially overlapping but not as severe as that of an spt7Δ mutant. Furthermore, components of both SLIK and SAGA are bound to the GAL1-10 promoter during gene activation, suggestive of the ability of either complex to regulate GAL10 (Fig. 7).
Functional interactions among Spt3, Spt8, and TBP have been reported elsewhere (12, 13). Spt8 and Spt3 are not required for the integrity of SAGA; however, SAGA prepared from an spt8Δ mutant fails to bind TBP in vitro (55). Consistent with this observation, Spt3 is required for the recruitment of TBP and preinitiation complex formation at the GAL1 promoter (5, 10, 32) and for full Gcn4- and Gcr1-mediated activation of the his4-912δ promoter (9). Surprisingly a recent study has shown that mutations in Spt3 and Spt8 cause derepression of HIS3 and TRP3 transcription in uninduced cells (4). This suggested that Spt3 and Spt8 inhibit TBP function at these two genes. In cells induced for HIS3 transcription with the drug 3-aminotriazole (3-AT), a form of SAGA, termed SAGAalt, was identified which lacked Spt8. Exposure of yeast to 3-AT inhibits the HIS3 gene product, causing histidine starvation. SAGAalt was postulated to be an alternate form of SAGA activating the HIS3 and TRP3 genes during starvation conditions. SAGAalt may represent the same complex as SLIK, which we find already to be preformed in rich medium. Consistent with this idea, the SLIK complex can also be found to bind nickel-NTA agarose under less competitive conditions such as in 3-AT-treated starved cells, which have lowered total protein amounts (unpublished data). Unfortunately, we have been unable to perform experiments to test for a putative function for SLIK in the starvation response since this compound is toxic to rtg2Δ mutant cells, irrespective of whether the medium is supplemented with histidine.
Acknowledgments
We thank Fred Winston, David Allis, Ron Butow, Joe Reese, Bertrand Seraphin, and Shelley Berger for yeast strains, antibodies, and plasmids and Fred Winston, Jeff Smith, David Auble, Pei-Yun Wu, and Wang Cheung for technical advice and helpful discussions.
P.A.G. is the recipient of a Burroughs Wellcome Fund Career Award in Biomedical Sciences. This work is supported by grant RR11823-03 awarded to the NCRR Yeast Center, NIDDK grant DK58646 to P.A.G., and NIGMS grant GM47867 to J.L.W.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. Brandl, L. Pillus, J. L. Workman, and J. Côté. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apone, L. M., C. M. Virbasius, J. C. Reese, and M. R. Green. 1996. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10:2368-2380. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 288:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 8.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, A. M., L. J. Gansheroff, and F. Winston. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics 151:1365-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenmann, D. M., K. M. Arnt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 13.Eisenmann, D. M., C. Chapon, S. M. Roberts, C. Dollard, and F. Winston. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein, C. B., J. A. Waddle, W. Hale, V. Dave, J. Thornton, T. L. Macatee, H. R. Garner, and R. A. Butow. 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, P. A. 2001. A tale of histone modifications. Genome Biol. 2:0003.1-0003.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, P. A., and S. L. Berger. 1999. Histone acetyltransferase complexes. Semin. Cell Dev. Biol. 10:169-177. [DOI] [PubMed] [Google Scholar]
- 18.Grant, P. A., S. L. Berger, and J. L. Workman. 1999. Identification and analysis of native nucleosomal histone acetyltransferase complexes. Methods Mol. Biol. 119:311-317. [DOI] [PubMed] [Google Scholar]
- 19.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 20.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 21.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcription stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 22.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 23.Grant, P. A., D. E. Sterner, L. J. Duggan, J. L. Workman, and S. L. Berger. 1998. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8:193-197. [DOI] [PubMed] [Google Scholar]
- 24.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 25.Horiuchi, J., N. Silverman, B. Piña, G. A. Marcus, and L. Guarente. 1997. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol. Cell. Biol. 17:3220-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia, Y., B. Rothermel, J. Thornton, and R. A. Butow. 1997. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol. 17:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 28.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 13:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M.-H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 31.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter, D. Vannier, G. Brosch, D. J. Stillman, D. Shore, and J. L. Workman. 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 275:40961-40966. [DOI] [PubMed] [Google Scholar]
- 34.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 35.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 36.Marcus, G. A., J. Horiuchi, N. Silverman, and L. Guarente. 1996. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol. Cell. Biol. 16:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 38.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 39.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 40.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 41.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, S. M., and F. Winston. 1997. Essential functional interaction of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 48.Rothermel, B. A., J. L. Thornton, and R. A. Butow. 1997. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J. Biol. Chem. 272:19801-19807. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Garcia, A. B., R. Sendra, M. Pamblanco, and V. Tordera. 1997. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 403:186-190. [DOI] [PubMed] [Google Scholar]
- 50.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 51.Sekito, T., J. Thornton, and R. A. Butow. 2000. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 11:103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 53.Sobel, R. E., R. G. Cook, and C. D. Allis. 1994. Non-random acetylation of histone H4 by cytoplasmic acetyltransferase as determined by novel methodology. J. Biol. Chem. 28:18576-18582. [PubMed] [Google Scholar]
- 54.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferases stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 57.Struhl, K., and Z. Moqtaderi. 1998. The TAFs in the HAT. Cell 94:1-4. [DOI] [PubMed] [Google Scholar]
- 58.Syntichaki, P., and G. Thireos. 1998. The Gcn5.Ada complex potentiates the histone acetyltransferase activity of Gcn5. J. Biol. Chem. 273:24414-24419. [DOI] [PubMed] [Google Scholar]
- 59.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Berg, M. A., P. de Jong-Gubbels, and H. Y. Steensma. 1998. Transient mRNA responses in chemostat cultures as a method of defining putative regulatory elements: application to genes involved in Saccharomyces cerevisiae acetyl-coenzyme A metabolism. Yeast 14:1089-1104. [DOI] [PubMed] [Google Scholar]
- 61.Wang, L., L. Liu, and S. L. Berger. 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12:640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston, F., C. Dollard, E. A. Malone, J. Clare, J. G. Kapakos, P. Farabaugh, and P. L. Minehart. 1987. Three genes are required for transactivation of Ty transcription in yeast. Genetics 115:649-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winston, F., and P. Sudarsanam. 1998. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:553-561. [DOI] [PubMed] [Google Scholar]
- 64.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22:5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]