Abstract
Histone mRNAs accumulate in the S phase and are rapidly degraded as cells progress into the G2 phase of the cell cycle. In Saccharomyces cerevisiae, fusion of the 3′ untranslated region and downstream sequences of the yeast histone gene HTB1 to a neomycin phosphotransferase open reading frame is sufficient to confer cell cycle regulation on the resulting chimera gene (neo-HTB1). We have identified a sequence element, designated the distal downstream element (DDE), that influences both the 3′-end cleavage site selection and the cell cycle regulation of the neo-HTB1 mRNA. Mutations in the DDE, which is located approximately 110 nucleotides downstream of the HTB1 gene, lead to a delay in the accumulation of the neo-HTB1 mRNA in the S phase and a lack of mRNA turnover in the G2 phase. The DDE is transcribed as part of the primary transcript and binds a protein factor(s). Maximum binding is observed in the S phase of the cell cycle, and mutations that affect the turnover of the HTB1 mRNA alter the binding activity. While located in the same general region, mutations that affect 3′-end cleavage site selection act independently from those that alter the cell cycle regulation.
The synthesis of histone proteins is tightly regulated during the cell cycle to ensure maximum expression in the S phase coincident with DNA synthesis. Replication-dependent histone mRNAs accumulate in the S phase and are rapidly degraded as cells enter the G2 phase. This periodic accumulation is controlled by both transcriptional and posttranscriptional events (33). At the level of posttranscriptional processing, both mRNA 3′-end formation and mRNA stability play important roles in controlling this cell cycle regulation (33). The 3′ ends of metazoan histone mRNAs differ from general mRNAs in so far as they are nonpolyadenylated. Instead of a poly(A) tail, histone mRNAs possess a highly conserved stem-loop structure immediately upstream of the 3′ end of the mRNA (10).
The formation of metazoan histone mRNA 3′ ends is controlled by a unique multicomponent process (14). Evidence from studies of sea urchin, rodent, and human cells defines at least three factors required for correct 3′-end cleavage of these mRNAs. The snRNA component of the U7 small nuclear ribonucleoprotein (snRNP) base pairs with a loosely conserved purine-rich element (referred to as the histone downstream element [HDE]), which lies approximately 10 to 15 nucleotides (nt) immediately downstream of the cleavage site (7, 29). A second factor, the stem-loop binding protein (SLBP), also referred to as the hairpin binding protein, interacts with the conserved stem-loop at the 3′ end of the mRNA via a unique RNA binding domain (4, 15, 25). This 32-kDa protein remains associated with the mature mRNA as it relocates from the nucleus to the polysomes in the cytoplasm (19). The third factor, an unidentified protein referred to as heat labile factor (HLF), was discovered through the ability of moderate temperature increases to disrupt mRNA 3′-end formation (17). Both SLBP (hairpin binding protein) and HLF levels fluctuate during the cell cycle; the SLBP accumulates in late G1 just prior to the accumulation of histone mRNAs, while HLF becomes limiting in G1 (23, 36). Although the U7 snRNP is constitutively expressed during the cell cycle, conflicting data exist regarding the cell cycle regulation of the interaction of U7 with the HDE (7, 21).
Paradoxically, unlike their metazoan counterparts, all Saccharomyces cerevisiae histone mRNAs appear to be polyadenylated (16). While the factors influencing yeast histone mRNA 3′-end cleavage have not yet been defined, it is thought that the general 3′-end processing machinery is involved. In this regard, histone pre-mRNAs can be processed in vitro by cleavage and polyadenylation extracts capable of processing general mRNAs (10). Furthermore, degenerate sequence motifs, which are similar to those of the positioning elements and efficiency elements and are located upstream of most yeast mRNAs, can be identified upstream of the 3′ ends of yeast histone mRNAs. Despite the difference in the structures of the 3′ ends of metazoan and yeast histone mRNAs, posttranscriptional events also contribute to the cell cycle regulation of yeast histone mRNAs (24, 37).
The role of posttranscriptional events in the cell cycle regulation of yeast histone mRNAs was demonstrated by experiments in which the 3′ untranslated region (UTR) and sequences downstream of the cleavage sites of the histone gene, HTB1, were fused to a bacterial neomycin phosphotransferase (neo) open reading frame under the control of the GAL1 promoter (37). In the presence of galactose, the resultant chimeric mRNA accumulated in the S phase coincident with the endogenous HTB1 mRNA. Deletion of sequences in the HTB1 3′ UTR abolished the cell cycle regulation of the neo-HTB1 transcript. Since the chimera gene also contained the HTB1 3′-end cleavage sites and sequences downstream, it is not clear whether additional events such as 3′-end processing or transcription termination also contribute to cell cycle regulation.
The formation of nonhistone and histone mRNA 3′ ends has been shown to be tightly coupled to transcription termination (13, 27, 30, 31). In S. cerevisiae, mutations in signals controlling 3′-end cleavage and polyadenylation can reduce transcription termination efficiency (5). Additionally, mutations in a number of the trans-acting cleavage and polyadenylation factors, including Pcf11p and Rna15p, result in the disruption of both 3′-end processing and transcription termination (5). Coupling between cleavage and termination appears to be mediated through the C-terminal domain of RNA polymerase II (3, 26). Studies of mice have also revealed a link between mRNA 3′-end processing and transcription termination of histone genes. Removal of the 3′-end processing signals (consisting of the stem-loop structure at the 3′ end of the RNA and the U7 snRNA binding site) of the mouse H2A-614 gene leads to a disruption of transcription termination (12). Interestingly, a physical and genetic linkage between the cleavage and polyadenylation factor CstF-64 (Rna15p) and the transcription factor Res2p, a factor involved in cell cycle regulation of a number of genes, has been demonstrated (2).
In this study, we examined the role of downstream sequences in the 3′-end processing and the cell cycle regulation of the yeast histone HTB1 mRNA. Using a neo-HTB1 chimera gene as a model system, we carried out a mutagenesis study of a region of DNA lying approximately 110 nt downstream of the cleavage sites of the HTB1 gene. We identify a sequence element that lies in the region of transcription termination which influences, in vivo, both mRNA 3′-end cleavage site selection and cell cycle regulation of the upstream mRNA. We show that RNA transcribed from this region binds a protein factor(s) and that maximum binding is observed during the S phase of the cell cycle.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The 10-kb centromeric URA shuttle vector pLJ31-HTB1, containing a neomycin phosphotransferase gene fused to the 3′ untranslated and downstream sequences of the yeast HTB1 gene, was constructed by Xu et al. (37). The gene is under the control of a Gal promoter. The HTB1 region consists of a 1,168-nt SphI-HindIII fragment containing the last 17 amino acids of the coding sequence of HTB1 and 1,113 nt of 3′ UTR and downstream sequences. This fragment was inserted at the BamHI-HindIII site immediately downstream of the neo gene (Fig. 1A). The plasmid pLJ31-HTB1 was kindly provided by Michael Grunstein (University of California at Los Angeles). Sequence analysis revealed the presence of a 157-base-pair (bp) fragment of DNA immediately downstream of the BamHI restriction site at position +848 (Fig. 1A). This sequence represents a duplication of sequences in the 5′ UTR of the neo gene (data not shown).
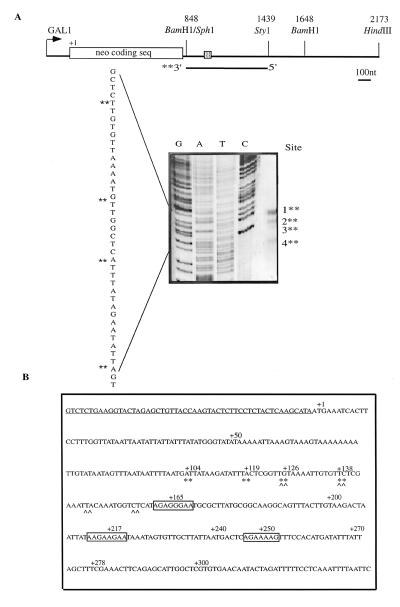
FIG. 1.
(A) S1 nuclease protection of the neo-HTB1 gene: schematic representation of the gene. The gene contains the bacterial neomycin phosphotransferase open reading frame under the control of the GAL1 promoter. The SphI-HindIII fragment of the HTB1 gene was inserted at the BamHI and HindIII sites as shown. The shaded box represents the 17 amino acids of the coding sequence of HTB1. The positions of the restriction enzyme sites are numbered relative to the position of the first base of the neomycin translation start codon (+1). The region between the BamHI site (at position +848) and the shaded box contains 157 bp of extraneous DNA. BLAST analysis of this sequence revealed it to be a repeat of the region surrounding the neo transcription start site. The line below BamHI and StyI sites shows the region used as a DNA probe for S1 nuclease mapping. The probe was labeled with [α-32P]dATP at the BamHI site. Total RNA (isolated from yeast cells grown in galactose and transformed with the plasmid pLJ31-HTB1) was hybridized with the DNA probe. Following digestion with S1 nuclease, the samples were electrophoresed through a 6% denaturing polyacrylamide gel alongside a sequencing ladder. Dideoxy sequencing of the BamHI-StyI fragment was carried out by using a primer from the BamHI region (Table 1). The same S1 protection pattern was observed in three independent experiments. (B) Sequences surrounding the HTB1 mRNA 3′ end. The nucleotide immediately after the stop codon of the HTB1 gene is labeled as +1. ∗∗, neo-HTB1 cleavage sites; ^^, endogenous HTB1 cleavage sites. The underlined sequence represents the 17 amino acids of HTB1 coding sequence. The numbers above the sequence are designated relative to the position of the stop codon of the HTB1 gene. The boxed regions show the purine-rich sequences downstream of the HTB1 gene.
To prepare mutants in the HTB1 downstream region, an 800-nt BamHI fragment was subcloned into the bacterial plasmid pBC (Stratagene Inc.). Mutations were created by using the QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's specifications. Synthetic oligonucleotides serving as mutagenic primers are listed in Table 1. Plasmids isolated from selected colonies were sequenced to confirm the mutagenesis. The mutated BamHI fragment was subsequently subcloned back into the yeast vector, pLJ31-HTB1.
TABLE 1.
Olignucleotides used in this study
| Olignucleotide | Sequence (5′ to 3′) | Use(s) |
|---|---|---|
| Actin1 for | GGCTGCAGGTCGACTCTAGA | Act1 transcripts |
| Actin3rev | TAGATTTTTCACGCTTACTGC | Act1 transcripts |
| Neofor | TACCTGCCCATTCGACCACCAAGC | neo-HTB1 transcripts |
| Neorev | GTCATTTCGAACCCCAGAGTCCCGCT | neo-HTB1 transcripts |
| Neorevcomp | AGCGGGACTCTGGGGTTCGAATGAC | neo-HTB1 3′-end transcripts |
| Htbcodrev | GAGTAGAGGAAGAGTACTTGG | neo-HTB1 3′-end transcript |
| Htbcodfor | GAAAAGAAACCAGCCTCC | Endogenous HTB1 transcript |
| Htbcodrev | GAGTAGAGGAAGAGTACTTGG | Endogenous HTB1 transcript |
| pSAC10a | CTTATTAATGACTCTCTAAAGTTTCCACATG | HTB1 mutagenesis |
| pSAC14b | TAATGACTCAGAAAACAATCCACATGATATTTA | HTB1 mutagenesis |
| pSAC15 | GTTGCTTATTAATGAGAGAGAAAAGTTTCCACA | HTB1 mutagenesis |
| pSAC16 | TTATAAGAAGAATAACTAGTGTTGCTTATT | HTB1 mutagenesis |
| pSAC17 | CTAGTAAGGCTCTTTTCAGAGCCACA | HTB1 mutagenesis |
| Probe 1for | CCGGAATTCGTCTTACAAGTAAAC | Run-on analysis |
| Probe 1rev | CGGGGTACCCTTCCTTTGGTTATA | Run-on analysis |
| Probe 2for | CCGGAATTCACAGGAAGTTAATCA | Run-on analysis |
| Probe 2rev | CGGGGTACCTCAGAAAAGTTTCCA | Run-on analysis |
| Probe 3for | CCGGAATTCGAGAAAAATTTAGGG | Run-on analysis |
| Probe 3rev | CGGGGTACCAATACGCACGTATAA | Run-on analysis |
| Probe 4for | CCGGAATTCATCCAATGGCCATAT | Run-on analysis |
| Probe 4rev | CGGGGTACCCATCTCATCATACGA | Run-on analysis |
| BamH1 | GGATCCTTCTTATTCAAATGT | Sequencing of HTB1 3′ end |
| T7/htb3end | 5′-TAATACGACTCACTATAGGGGAAGAATAAATAGTGTTGCTTA-3′ | In vitro transcription |
| Htbl3endrev | CTAGTATTGTTCACACGAGCC | In vitro transcription, mobility shift assay |
pSAC11 and -13 were derived from the same mutagenesis.
pSAC20 and -21 are further derivatives of pSAC14.
Strains and media.
The S. cerevisiae strain S150-2B (MATa leu2-3,112ura3-52 trp1-289 his3D) was used as the parental strain in this study. The yeast organisms were routinely cultured at 30°C in yeast extract-peptone-dextrose (YEPG) medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] galactose) to a midlogarithmic phase (107 cells/ml, or an optical density at 600 nm [OD600] of 0.6). Typically, 1 μg of plasmid DNA was used to transform S150-2B by using the lithium acetate procedure (9). Transformants were selected on synthetic complete medium containing 1× yeast synthetic dropout medium lacking lysine, histidine, leucine, tryptophan, and uracil (Sigma Chemical Co.), 0.7% (wt/vol) yeast nitrogen base without amino acids (Sigma Chemical Co.), 0.5% (wt/vol) ammonium sulfate, and 2% (wt/vol) galactose and supplemented with 10 mg each of lysine, tryptophan, and histidine/ml and 20 mg of leucine/ml.
Preparation and analysis of RNA.
Total RNA was isolated by the hot-phenol method (9). RNA (30 μg per lane) was separated on 1.0% formaldehyde agarose gels and subsequently blotted to Nytran membranes (Schleicher & Schuell). Digoxygenin-UTP (Roche Biochemicals)-labeled DNA probes were generated by PCR amplification using the primer pairs listed in Table 1, as previously described (9). The membranes were prehybridized for 1 to 3 h in high sodium dodecyl sulfate (SDS) buffer (7% sodium dodecyl sulfate [SDS], 50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% blocking reagent, 50 mM sodium phosphate [pH 7.0], 0.1% N-lauroylsarcosine) at 50°C and hybridized for 15 to 20 h at 50°C, with approximately 25 ng of labeled probe per milliliter of hybridization buffer. Hybridization was detected with a chemiluminescent substrate, CSPD or CDP-Star (both from Roche), as previously described (9).
S1 nuclease analysis.
Two probes were used to map the 3′ ends of the neo-HTB1 mRNAs. A 591-nt BamHI-StyI fragment was isolated from the plasmid pLJ31-HTB1 (Fig. 1A). A second 458-nt StyI-ScaI DNA fragment (see Fig. 3B) was used to determine the 3′ end closer to the neo sequences. These DNA fragments were 3′ end labeled at the BamHI and StyI sites, respectively, by using a Klenow fragment of DNA I polymerase and incorporating [α-32P]dATP (specific activity, 3,000 Ci/mmol).
FIG. 3.
S1 nuclease mapping of the 3′ end of neo-HTB1 generated by mutant genes. Total RNA was isolated from yeast cells grown in galactose-containing medium and transformed with the WT or mutant plasmids. RNA was hybridized with a DNA probe labeled with [α-32P]dATP. (A) Schematic representation of the neo-HTB1 transcript, showing the position of the DNA probe. The values above the BamHI and StyI sites represent their positions relative to that of the neomycin translation start codon. Following digestion with S1 nuclease, samples were electrophoresed through a 6% denaturing polyacrylamide gel. Lane 1, pLJ31-HTB1 grown in glucose; lane 2, pLJ31-HTB1 grown in galactose; lane 3, pSAC10; lane 4, pSAC11; lane 5, pSAC17; lane 6, pSAC13; lane 7, pSAC14; lane 8, pSAC20; lane 9, pSAC21. All mutants were grown in galactose. The four protected fragments are labeled 1∗∗, 2∗∗, 3∗∗, and 4∗∗. The position of a 311-nt molecular mass marker is shown on the left of the figure. The S1 cleavage pattern was observed in at least two independent experiments. (B) S1 nuclease mapping of mutants pSAC10, -11, and -16. Total RNA was hybridized with an antisense DNA probe labeled at the 3′ end with [α-32P]dATP. Diagrammatic representation of the neo-HTB1 transcript showing the region of the StyI-ScaI DNA probe. The probe was labeled at the StyI site. The multiple arrows show the position of the normal cleavage sites, while the single arrow shows the position of the new cleavage sites observed with mutants pSAC10, -11, and -16. Protected fragments were electrophoresed through a 6% denaturing polyacrylamide gel. Lane 1, WT pLJ31-HTB1-transformed cells grown in glucose; lane 2, WT pLJ31-HTB1-transformed cells grown in galactose; lane 3, pSAC10; lane 4, pSAC11; lane 5, pSAC16; lane P, probe alone; lane M, HaeIII-cut φX174 DNA ladder. All mutants were grown in galactose. The S1 cleavage pattern was observed in at least two independent experiments. (C) Sequences surrounding the new cleavage sites in mutants pSAC10, -11, and -16 are shown. The schematic representation of the neo-HTB1 gene is shown as outlined in the upper half of panel B. The single vertical arrow shows the location of the new cleavage site immediately downstream of the BamHI site. The sequence in this region is shown in expanded form below the schematic representation. The BamHI site is represented by underlined bold letters. The final nucleotide represents the cleavage site.
The hybridization reactions and S1 digestions were carried out as described previously (8). For all S1 nuclease assays, the probe concentration was always in excess of that of cellular RNA. Total RNA (100 μg) was precipitated in 70% EtOH and 0.3 M NaOAc, pH 5.3, with the labeled DNA probes (5 × 105 cpm). The mix was then resuspended in 30 μl of S1 hybridization buffer [80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.4), 10 mM EDTA, 400 mM NaCl]. The samples were heat denatured at 85°C for 10 min and hybridized at 42°C for 16 h. After the hybridization, 300 μl of S1 digestion buffer (50 mM sodium acetate, 4.5 mM ZnSO4, 280 mM NaCl, 20 μg of salmon sperm DNA/ml) containing S1 nuclease (Promega) (100 U) was added to the RNA-probe mix. S1 digestion was carried out for 1 h at 37°C. The reaction was stopped by the addition of 80 μl of S1 nuclease stop buffer (4 M ammonium acetate, 50 mM EDTA [pH 8.0], 50 μg of tRNA/ml). The samples were extracted with phenol-chloroform and then ethanol precipitated before electrophoresis on a 6% acrylamide-7 M urea gel in 1× Tris-borate-EDTA buffer.
TRO assays.
Transcription run-on (TRO) assays were carried out as previously described (1), with the following modifications: yeast cultures (100 ml each), transformed with the plasmid pLJ31, were grown to an OD600 of approximately 0.1 to 0.15. Cells were harvested and washed with ice-cold distilled water (dH2O). Cells were rendered more permeable by resuspension in 0.95 ml of ice-cold dH2O and 50 μl of 10% (wt/vol) sodium N-lauryl sarcosine sulfate. The mix was incubated on ice for 20 min and then centrifuged for 1 min. The pellet was resuspended in 100 μl of transcription buffer (50 mM Tris-HCl [pH 7.9], 5 mM MgCl2, 100 mM NaCl, 1 mM MnCl2, 2 mM dithiothreitol, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 100 μCi [α-32P]UTP [3,000 Ci/mmol]). Transcription was allowed to proceed for 5 min at 30°C, and the reaction was stopped by the addition of 1 ml of ice-cold H2O. Pelleted cells were resuspended in 500 μl of TES buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.1% [wt/vol] SDS), and total RNA was extracted as described previously. RNA was partially hydrolyzed for 5 min on ice (0.2 M NaOH), neutralized (0.2 M Tris-HCl [pH 7.2]), and used for hybridization to DNA fragments corresponding to the untranslated and downstream regions of the HTB1 gene.
To generate HTB1 DNA fragments, DNA in 200-bp increments corresponding to the regions downstream of the neo open reading frame was amplified by PCR using oligonucleotides which include recognition sites for EcoRI (upstream oligonucleotide) or KpnI (downstream oligonucleotide) (Table 1). The amplified DNA was cloned into the M13 phage. Single-stranded M13 DNA corresponding to each 200-bp fragment was immobilized onto nylon filters. Filters were prehybridized for 2 h in a solution containing 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10× Denhardt solution, 50% formamide, and 0.2% SDS. Hybridization was carried out in the same solution overnight. Filters were washed four times with 2× SSC-0.1% SDS at room temperature and 0.2× SSC-0.1% SDS at 37 to 42°C for 10 min. Run-on signals were then visualized with an Instant Imager (Packard) or an Image Reader FLA 3000 (Fujifilm).
Synchronization of yeast cells.
Strain S150-2B MATa cells containing either the pLJ31-HTB1 plasmid or its mutant derivatives were grown in selective medium (synthetic complete medium without uracil) to early log phase and were then grown overnight in YEPG medium. α1-mating factor (TRP-HIS-TRP-LEU-GLN-LEU-LYS-PRO-GLY-GLN-PRO-MET-TYR) was added to the yeast culture at a concentration of 2 μg/ml. The culture was then incubated for another 3 h at 30°C until the cells were arrested in the G1 phase of the cell cycle. Arrested cells showing the characteristic peanut shape (shmooed) appearance were identified by microscopic examination. When at least 90% of the cells had shmooed, the cells were centrifuged at 3,000 × g for 5 min at 4°C. The α-factor was removed by washing the cells twice with 150 ml of ice-cold sterile distilled water, followed by centrifugation at the same speed as before. After the last wash, the pelleted cells were resuspended in 300 ml of prewarmed YEPG medium. Samples (15 ml) were then taken at 5-min intervals, spun at 3,000 × g for 5 min, and frozen at −70°C.
In vitro transcription of RNA.
RNA transcripts were generated by T7 RNA polymerase runoff transcription as previously described (8). The sequences to be transcribed were cloned downstream of the T7 promoter. Runoff transcription products were obtained by linearizing the plasmid with a restriction enzyme downstream of the sequence to be transcribed. Alternatively, templates were obtained by amplification of a DNA fragment by using a primer containing the T7 promoter fused to sequences complementary to the 5′ start of the RNA transcript and a reverse primer complementary to the end of the transcript. Where radiolabeled in vitro-transcribed RNA was required, a molar ratio of 26:1 of cold rUTP-[α-32P]UTP (3,000 Ci/mmol) was added.
Preparation of yeast cell extracts and protein-RNA binding assays.
Cell pellets from either synchronous or asynchronous cultures were prepared as previously described (9). Various concentrations of total protein extract were added to 5 × 105 cpm of [α-32P]UTP-labeled RNA probe. Then, 1× binding buffer [10 mM Tris-HCl (pH 7.5), 5 mM Mg(OAc)2, 100 mM KOAc, 2 mM dithiothreitol, 0.1 mM spermine, 100 ng of bovine serum albumin/μl, 8U of RNasin, 0.2 μg of tRNA/μl, 10% (vol/vol) glycerol, 5 mg of heparin/ml] was added to the protein-probe mix and the volume was adjusted to 20 μl with double-distilled water (ddH2O). The reaction mixture was incubated on ice for 30 min and then loaded onto 4% (40:1 acrylamide-bisacrylamide) nondenaturing gel containing 1× Tris-borate-EDTA buffer and 5% (vol/vol) glycerol. After electrophoresis, the gel was dried and exposed to X-ray film.
Quantification of mRNA transcripts.
The level of neo-HTB1 mRNA transcripts during the cell cycle was quantified by using the densitometric program Gel Works 1D Advanced, version 3.01 (Nonlinear Dynamics Ltd.).
neo-HTB1 mRNA transcripts levels were normalized with respect to the level of actin transcripts in each lane. For each data set, the value at every time point was accessed relative to the value at 20 h after the end of log-phase growth (T20) and the results were plotted accordingly.
RESULTS
The model system we used to examine the influences of downstream sequences on the 3′-end formation and cell cycle regulation of yeast histone mRNAs was based on the plasmid pLJ31-HTB1, in which the coding region of the neomycin phosphotransferase gene was fused to a fragment of the HTB1 gene. The HTB1 fragment contains the last 17 amino acids of the open reading frame, the stop codon, and 1,113 bp of 3′ untranslated and downstream sequences. The chimera gene is under the control of a GAL1 promoter (see Materials and Methods). Using this same system, Xu et al. (37) have shown that the HTB1 sequences present in the chimera gene are sufficient to confer cell cycle regulation on the neo-coding sequence. The salient features of the plasmid are outlined in Fig. 1A.
To ensure that the chimera neo-HTB1 mRNA was correctly processed, the 3′ end of the mRNA was mapped by using S1 nuclease protection experiments (Fig. 1A). Four cleavage sites were identified and mapped to positions +104, +119, +126, and +138 relative to the position of the stop codon of the HTB1 gene (Fig. 1B). These cleavage sites correspond closely to those mapped in the endogenous HTB1 gene (data not shown and Fig. 1B), indicating that the chimera gene is authentically processed at the correct cleavage sites.
To identify potentially conserved sequences required for 3′-end processing and/or cell cycle regulation, we initially carried out a bioinformatic analysis of the sequences in the region encompassing the 3′ ends of yeast histone mRNAs. The 3′ ends of five of the eight yeast histone mRNAs have been mapped (20, 32, 35). These histone mRNAs were aligned with respect to their 3′-end cleavage sites by using the ClustalW multiple sequence alignment program. Using parameters that identify the features which are most conserved between members of a set of sequences, we observed that each of the histone genes contained stretches of purine-rich sequences downstream of the 3′-end cleavage sites (Fig. 1B and data not shown). Based on their locations, these purine stretches can be defined as being proximal or distal to the 3′-end cleavage sites. While not all of the histone genes contained identifiable proximal purine-rich sequences, they all contained one or more purine-rich stretches in the region of nt +60 to +120 downstream of their 3′ ends (data not shown). The HTB1 gene contains three such purine rich clusters at positions +21, +73, and +106 relative to the position of the strongest cleavage site (Fig. 1B). The presence of purine-rich stretches, or “words,” downstream of yeast genes is quite unusual. Two recent bioinformatics studies have shown that the most common words downstream of yeast genes are U-rich stretches, while the least common words are GA-rich stretches (18, 34). Given the known role of downstream purine-rich sequences in metazoan histone mRNA processing and the unusual distribution of GA stretches downstream of the yeast histone genes, we first focused our analysis on one of the HTB1 distal purine-rich sequences.
Mutations in the downstream sequences of the HTB1 mRNA alter 3′-end cleavage site selection and the steady-state levels of the neo-HTB1 transcript.
The distal purine-rich sequence of the HTB1 gene that we have examined lies approximately 110 nt downstream of the last cleavage site of the mRNA and contains the sequence AGAAAAG (Fig. 1B). To examine the role of this sequence in the 3′-end processing and cell cycle regulation of the HTB1 gene, mutants were prepared in which purines were replaced by pyrimidines. Since the purine-rich HDE of metazoan histone mRNAs is always found in the context of the stem-loop of the histone mRNA, we also prepared a mutant (pSAC17) in which a stem-loop structure was inserted upstream of the purine-rich sequence. Additional mutations were also prepared in the sequences surrounding the purine stretch (Fig. 2A). Following transformation of the mutated plasmids into yeast cells, Northern blotting was performed on RNA extracted from cultures grown in galactose-containing medium. The blots were hybridized with a probe containing neo sequences only (Fig. 2A). As shown in Fig. 2B, a number of alterations are apparent in the neo-HTB1 transcript.
FIG. 2.
Northern blot analysis of neo-HTB1 transcripts from mutants in the downstream sequences of the HTB1 gene. (A) Schematic representation of the neo-HTB1 gene. The shaded box represents the 17 amino acids of the HTB1 gene. The region of the neo gene used as a DNA probe for Northern blot hybridization is indicated. The cleavage sites are indicated by arrows. The open box shown downstream of the cleavage sites indicates the location where mutations were made. The region is shown in expanded form in the lower panel and numbered relative to the position of the stop codon of the HTB1 gene. The WT sequences where mutations took place are underlined. The introduced mutations and the names of the mutants are shown below the sequence. (B) Northern blot hybridization of total RNA from each mutant probed with the neo-specific probe. Lane 1, pLJ31-HTB1 grown in glucose; lane 2, pLJ31-HTB1 grown in galactose. All mutants were grown in galactose. The full-length neo-HTB1 transcript is marked with a solid arrow, while the minor transcript, which was detected with the neo probe only, is indicated by an open arrowhead. Transcripts detected in mutants pSAC10, -11, and -16 are labeled Transcript A and Transcript B and are indicated by a double solid arrowhead and single solid arrowhead, respectively. The Northern blot patterns were independently observed in at least three independent experiments.
Growth of cells in galactose-containing medium results in the induction of an mRNA of approximately 1.2 kb that hybridizes to a probe containing neo sequences only (Fig. 2B, lane 2). This transcript also hybridizes to a probe containing sequences downstream of the neo region and including the HTB1 region of the chimera, indicating that this transcript represents the neo-HTB1 chimera RNA (Table 1 and data not shown). A second minor transcript of approximately 1.0 kb is also detected by the neo probe alone (Fig. 2B, lane 2). This species is most likely a degradation product or a transcript emanating from the opposite strand, since when using a variety of primers along the neo and HTB1 sequences, we observed only a single product by reverse transcription-PCR (data not shown), and no additional protected fragments were observed by using S1 nuclease mapping (see Fig. 3A and B).
Experiments with the mutants pSAC10, -11, and -16 resulted in the generation of a smaller transcript (transcript B; Fig. 2B, lanes 3, 4, and 5). The full-length neo-HTB1 transcript is also altered in size in these mutants (transcript A). S1 mapping showed that transcript A contained both neo and HTB1 sequences and had the same 3′-end cleavage sites as the neo-HTB1 wild-type (WT) transcript (see below).
All of the other mutants showed the same transcript pattern as cells harboring the WT neo-HTB1 transcript (Fig. 2B, lanes 6 to 10). We did observe some fluctuations in the steady-state levels of the neo-HTB1 transcript in some of the mutants (mutants pSAC10 and -20; Fig. 2B, lanes 3 and 9). In most cases this fluctuation correlated with the OD600 at which the cells were harvested, since increased transcript levels were observed as the OD600 of the culture increased. However, we consistently observed that the steady-state levels of the neo-HTB1 transcript in mutant pSAC20 were significantly lower than those of the WT transcript (Fig. 2B, lane 9). In three independent experiments in which RNA from cultures harvested at the same OD600 was used, the steady-state levels of the neo-HTB1 transcript in mutant pSAC20 were on average fivefold lower than those of the WT transcript. This mutant contains two sets of mutated triplets (Fig. 2A), including the triplet mutated in pSAC14. Since the steady-state level of neo-HTB1 mRNA in mutant pSAC14 appears normal (Fig. 2B, lane 8), we can deduce that the additional bases mutated in pSAC20 are responsible for the decrease in steady-state levels. Surprisingly, the levels of the neo-HTB1 transcript in mutant pSAC21 are similar to the WT levels (Fig. 2B, lane 10). This mutant contains the same mutated bases as mutant pSAC20 plus an additional triplet base change, suggesting that the additional changes in sequence had compensated for the decrease in transcript levels observed with the pSAC20 mutant.
Transcript B is generated by the use of an alternative 3′-end cleavage site.
To determine whether the shorter transcripts (transcripts A and B) observed with mutants pSAC10, -11, and -16 resulted from the use of alternative 3′-end cleavage sites, S1 mapping using probes encompassing both neo and HTB1 sequences was performed. As shown in Fig. 3A, transcript A results from cleavage at the normal 3′ cleavage sites downstream of the HTB1 sequences (Fig. 3A, lanes 3 and 4, and data not shown). Since no other 3′-end cleavage sites were identified, the size difference between the full-length transcripts generated by the WT and mutant genes may result from the presence of a shorter poly(A) tail or from transcription from an alternative initiation site. The full-length WT neo-HTB1 mRNA and transcripts A and B are all equally retained on oligo(dT) cellulose columns (data not shown). As with mutants pSAC10, -11, and -16, the full-length neo-HTB1 transcripts observed in all other mutants are cleaved at the normal 3′-end cleavage sites (Fig. 3A).
To identify the 3′ ends of transcript B, S1 nuclease mapping was carried out by using a DNA probe encompassing the 3′ end of the neo gene and the 5′ end of the HTB1 sequences (Fig. 3B). We observed that all three mutants (pSAC10, -11, and -16) contain a new 3′-end cleavage site which lies approximately 45 nt downstream of the neo stop codon and 12 nt downstream of the BamHI site (Fig. 3B, lanes 3, 4, and 5, and C). Therefore, transcript B does not contain any HTB1 sequences. This new cleavage site was not observed in the WT mRNA or with any of the other mutants (Fig. 3B, lanes 1 and 2, and data not shown).
Mutations downstream of the HTB1 gene lie in a region of RNA polymerase II termination.
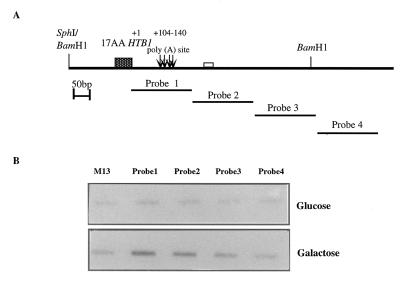
The area where the mutations were made (hereafter referred to as the distal downstream element [DDE]) lies quite distal to the 3′-end cleavage sites, and yet these mutants affect both the steady-state levels and 3′-end cleavage site selection of the neo-HTB1 mRNA. We therefore wanted to test whether the region in which the mutations were made was in fact transcribed as part of the primary pre-mRNA transcript. TRO experiments were carried out as previously described (1). This assay quantifies the density of RNA polymerase molecules along a transcript by incorporating [α-32P]UTP into the elongating RNA transcript and is most effective for analysis of highly transcribed genes. Single-stranded DNA probes, corresponding to sequences located in approximately 200-nt increments downstream of the HTB1 stop codon, were prepared by the cloning of these fragments into the replicative form of the phage M13. Single-stranded DNA from the phage was immobilized onto nylon membranes and hybridized to 32P-labeled nascent RNA isolated from cells transformed with the WT neo-HTB1 plasmid (Fig. 4). To distinguish between transcripts emanating from the endogenous gene and the neo-HTB1 gene, we compared the levels of nascent transcripts in cells grown in glucose and galactose. The first DNA fragment (probe 1 [nt +9 to +196]; Fig. 1B) contains the entire 3′ UTR and the 3′-end cleavage sites (nt +104, +119, +126, and +138), while probe 2 (+235 to +409) contains the DDE, which lies close to the 5′ end of the fragment (nt +250; Fig. 1B). As shown in Fig. 4B, we observed transcription proceeding from the position of probe 1 to probe 2. Taking the transcription level to be 100% for probe 1 and having subtracted the background hybridization, in our analysis of galactose-containing medium we observed that transcription decreased to 63% in the probe 2 region, 24% in the probe 3 region, and 0% in the probe 4 region. Similarly, in glucose-containing medium, there was a decrease in transcription to 51% in the probe 2 region, 16% in the probe 3 region, and 0% in the probe 4 region. These results suggest that transcription termination was occurring somewhere in the region of probes 2 and 3. Analysis of run-on transcription in cells transformed with the mutated plasmids indicated that all mutant transcripts were transcribed as far as the region of probe 2, and there appeared to be no significant quantitative differences in the levels of transcription of the mutant mRNAs (data not shown).
FIG. 4.
The DDE is transcribed as part of the primary transcript. (A) Schematic representation of the HTB1 region of the neo-HTB1 gene, showing the position of the 200-nt DNA fragments to which nascent RNA was hybridized. The fragments are labeled Probe 1 to Probe 4. The values above the poly(A) sites show their positions relative to that of the stop codon of HTB1 (+1). The shaded box represents the HTB1 coding sequences, while the open box downstream of the poly(A) sites (arrows) represents the DDE. (B) Hybridization of nascent transcripts from yeast cells transformed with the WT plasmid pLJ31-HTB1 grown in glucose or galactose. M13, single-stranded phage DNA with no insert. Probes 1 to 4 were as described for panel A.
Mutations in the DDE alter the cell cycle periodicity of the neo-HTB1 transcript.
To test whether mutations in the DDE had any effect on cell cycle regulation of the neo-HTB1 transcript, cells transformed with the mutated plasmids were synchronized at the G1 border of the cell cycle by using the α1-mating factor. Once the cells were synchronized, the α-factor was removed and RNA was prepared from samples taken at 5-min intervals. The RNA was hybridized with a DNA probe specific to the neo region of the gene (Fig. 2A). The pattern of the endogenous histone mRNA was monitored by hybridizing the RNA samples with a probe specific to the coding region of HTB1 (endogenous HTB1 primers; Table 1). To allow for adjustment for the amount of RNA loaded in each lane, the blots were also hybridized to an actin DNA probe. As shown in Fig. 5A, the endogenous HTB1 mRNA was detected 20 min after α-factor removal and peaked at 35 to 40 min and levels of the mRNA decreased at 45 to 50 min, with little or none detected at 55 min. The WT neo-HTB1 transcript (Fig. 5B) shows an accumulation pattern similar to that of the endogenous HTB1; however, the rate of decline in mRNA levels appears slower than that of the endogenous HTB1. These results confirm that the sequences in the 3′ UTR and downstream of the cleavage sites can confer cell cycle periodicity on a neo mRNA (37).
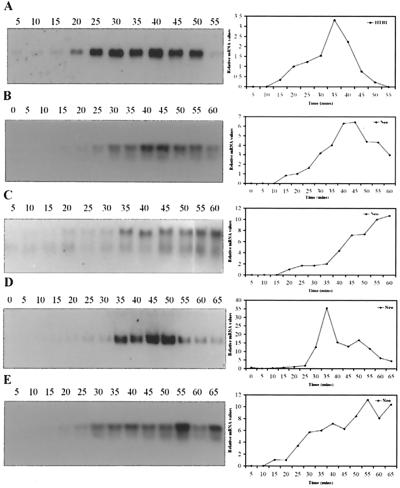
FIG. 5.
Cell cycle regulation of the neo-HTB1 transcript. Yeast cells transformed with the plasmid pLJ31-HTB1 or mutant plasmids were synchronized to the G1 phase of the cell cycle by the addition of the α1-mating factor. RNA was extracted at 5-min intervals and hybridized to a probe specific for the coding region of the HTB1 gene (A) (Table 1) or a neo-specific probe (B to E) (Fig. 2A and Table 1). To account for slight variations in RNA loading, the blots were stripped and rehybridized with an actin DNA probe. The levels of hybridization to the three probes were quantified, normalized to the actin mRNA levels, and expressed as mRNA levels at each time point relative to the level of hybridization at 20 min following removal of α1-mating factor (right panels). (A) Levels of endogenous HTB1 mRNA; (B) mRNA levels of WT neo-HTB1; (C) mRNA levels of neo-HTB1 from cells transformed with mutant pSAC14 plasmid; (D) mRNA levels of neo-HTB1 from cells transformed with mutant pSAC20 plasmid; (E) mRNA levels of neo-HTB1 from cells transformed with mutant pSAC21 plasmid. The numbers above each lane indicate the time (min) after release from α1-factor. The hybridization patterns shown in each panel were independently observed in at least three experiments.
Of the mutants analyzed, only pSAC14 and pSAC21 show an altered cell cycle pattern of neo-HTB1 expression (Fig. 5C and E, respectively). In both cases, there appears to be a delay in the accumulation of the neo-HTB1 mRNA. Once induced, both transcripts continued to accumulate, and there appeared to be no turnover of the mRNA, with levels remaining high at 55 and 60 min. Other mutants, such as pSAC10, -11, and -16, showed accumulation patterns identical to that of the WT neo-HTB1 mRNA (data not shown). Interestingly, the pSAC20 mutant showed a normal pattern of cell cycle accumulation (Fig. 5D); however, the peak of accumulation is narrower than that observed with the endogenous HTB1 or with the WT neo-HTB1 mRNA. This mutant contains the same base changes as mutant pSAC14 plus an additional three base changes (Fig. 2A), while mutant pSAC21 contains the same base changes as pSAC20 but with a further three base changes (Fig. 2A). Since mutant pSAC20 appeared to show normal cell cycle regulation, the additional three base changes in this mutant must compensate for the alterations caused by the three base changes in mutant pSAC14. This compensation appears to be overridden by the additional base changes in mutant pSAC21.
Sequences in the DDE bind protein factor(s).
The results obtained from the analysis of mutations in the DDE suggest that these sequences influence the accumulation and subsequent turnover of HTB1 mRNA during the cell cycle. Since two of the mutants showed a delay in the accumulation of and a lack of turnover of neo-HTB1 mRNA, we reasoned that a protein factor might be required to stabilize the pre-mRNA in the S phase or destabilize it immediately thereafter. To examine whether this region was capable of binding protein factors, a 100-nt [32P]dUTP-labeled RNA probe, consisting of the DDE and 80 nt of surrounding sequences (Fig. 6A), was generated by using T7 RNA polymerase. The RNA was incubated with cell extracts prepared from yeast cells harvested at different time points following release of cells from α-factor synchronization. In the presence of cell extracts, the labeled RNA bound to one or more protein factors, as indicated by the retardation of the mobility of the RNA on nondenaturing gels (Fig. 6B). Interestingly, maximum binding was observed in extracts prepared at 35 min following α-factor release (Fig. 6B, lanes 5 and 6), the time point corresponding to the peak of neo-HTB1 mRNA during the cell cycle. Binding is also observed at 30 min (Fig. 6B, lanes 2 to 4) and 40 min (lanes 7 to 9), with a marked decrease in binding occurring at 45 min (lanes 10 to 12).
FIG. 6.
A protein factor binds to the DDE. (A) Schematic representation of the neo-HTB1 gene. The shaded box represents the region of the last 17 amino acids of the HTB1 gene. The arrows indicate the locations of the cleavage sites. The open box represents the location of the DDE. The sequences within the DDE are shown in expanded form below the schematic representation. Underlined sequences represent the forward and reverse primers used to amplify the DDE sequences. T7 promoter sequences are shown in bold letters. (B) RNA corresponding to the sequence shown in panel A was radiolabeled by incorporation of [α-32P]UTP. The RNA was incubated with cell extracts prepared at the times (min) indicated following release from α1-factor synchronization. Following the incubation, the samples were electrophoresed through a 4% nondenaturing acrylamide gel. Lane 1, RNA probe alone; lanes 2 to 4, cell extract prepared 30 min after release from α-factor arrest; lanes 5 and 6, 35-min extract; lanes 7 to 9, 40-min extract; lanes 10 to 12, 45-min extract. Increasing concentrations (10, 20, and 40 μg) of cell extracts were used per 20 μl of reaction mixture. Lanes 2, 5, 7, and 10, 10 μg of extract; lanes 3, 8, and 11, 20 μg; lanes 4, 6, 9, and 12, 40 μg. The band shift pattern was observed in at least three independent experiments. (C) RNA probes corresponding to the WT, pSAC14, pSAC20, and pSAC21 sequences were in vitro transcribed and labeled with [α32P]UTP. The RNA was incubated with cell extract prepared 35 min after release from α1-factor. Lanes 1 to 3, WT probe with 10, 20, and 40 μg of extract, respectively; lanes 4 to 6, pSAC14 probe with 10, 20, and 40 μg of extract, respectively; lanes 7 to 9, pSAC20 with 10, 20, and 40 μg of extract, respectively; lanes 10 to 12, pSAC21 with 10, 20, and 40 μg of extract, respectively. The arrow shows the position of band-shifted RNA. The band shift pattern was observed in at least three independent experiments.
To determine whether binding was affected by the mutations made in this region, the binding experiments were repeated by using RNA incorporating the mutations. We observed that binding was slightly reduced in the pSAC14 mutant RNA (Fig. 6C, lanes 4 to 6), while no binding was observed in the pSAC21 mutant RNA (lanes 10 to 12). Mutant pSAC20 showed binding similar to that of the WT RNA; however, the kinetics of binding was altered somewhat, with reduced binding observed at the lower concentrations of extract and greater binding at the highest extract concentration (Fig. 6C, lanes 7 to 9). These results suggest that the DDE, as defined by the mutants described here, influences accumulation of HTB1 during the cell cycle through the binding of specific protein factors. The correlation between the continued accumulation of the mRNA in pSAC14 and -21 and the decrease in binding activity in the cell extracts suggests that the protein factors function to destabilize the pre-mRNA.
DISCUSSION
We have identified a novel sequence element lying approximately 110 nt downstream of the 3′ end of HTB1 that influences at least two cellular processes, namely, mRNA 3′-end processing and cell cycle regulation. The element is referred to as the DDE. To analyze this sequence element, we generated a series of mutations within the DDE. Experiments using three of the mutants, pSAC10, -11, and -16, resulted in the selection of an alternative 3′-end cleavage site just downstream of the neo sequences of the neo-HTB1 chimera, generating a new transcript referred to here as transcript B. In addition, the full-length neo-HTB1 transcript generated in mutants pSAC10, -11, and -16, referred to here as transcript A, is approximately 50 nt shorter than the WT transcript. Since transcript A is generated through the use of the same cleavage sites as the WT transcript, the difference in sizes between the WT and transcript A is most likely accounted for by the presence of a shorter poly(A) tail on the mutant-derived transcripts or by the use of an alternative transcription initiation site. Further experiments will be required to distinguish between these two possibilities.
We observed that experimentation using one of the mutants, pSAC20, resulted in a fivefold or greater decrease in the steady-state levels of the neo-HTB1 mRNA. Since the mutated sequences are not contained in the final transcript, this reduction in steady-state levels of neo-HTB1 may be a result of a decrease in efficient 3′-end processing or may affect the stability of the final transcript. Interestingly, Kim et al. (22) have identified a 54-nt element downstream of the distal poly(A) site of the Schizosaccharomyces pombe uv15+ gene that can influence the stability of the mature mRNA. Like the DDE described here, the uv15+ element is not included in the mature mRNA. Analysis of the mutant pSAC20 RNA during the cell cycle revealed that the RNA, while induced in the S phase, appears to be more rapidly degraded as cells enter the G2 phase (Fig. 5D). Thus, the inherent instability of the mutant pSAC20 RNA may be a consequence of its rapid turnover in the G2 phase of the cell cycle. Taken together, the data suggest that sequences downstream of the HTB1 gene, though not part of the mature mRNA, can affect the steady-state levels of the final transcript.
It is surprising that mutant pSAC21 mRNA, which contains the same mutated bases as mutant pSAC20 (Fig. 2A) plus an additional triplet base pair change, shows steady-state transcription levels similar to those of the WT mRNA. Furthermore, mutant pSAC14 shares three of the six base pair changes of pSAC20 and it too shows normal steady-state levels of neo-HTB1 transcript. These findings suggest that the presence of a specific sequence is not alone sufficient to determine the overall function of this region and that a secondary structure may also play a role (see below).
Of the other mutants generated, pSAC13 and -17 showed normal steady-state levels (Fig. 2A). Mutant pSAC13 (AAA to TTT) includes three of the bases changed in mutant pSAC11 (AGAAA to TCTCT) (Fig. 2A). Unlike pSAC11, this mutant does not result in the generation of transcript B. Therefore, we can conclude that transcript B is generated as a result of the introduction of bases (TCT) that are common between mutants pSAC10 and -11 or, alternatively, that it is the introduction of C residues that accounts for the generation of transcript B. The latter argument is supported by findings for mutant pSAC16 in which a single base change from A to C resulted in the generation of transcript B (Fig. 2A). Finally, it is surprising that the introduction of a stem-loop structure (pSAC17) at the same location as mutant pSAC16 caused the transcript pattern to revert to that of the WT. This may indicate that the overall sequence context in this region is important to its function.
Sequences located at a distance from the 3′-end cleavage sites are known to influence transcription termination. To determine whether the DDE was transcribed as part of the pre-mRNA, TRO experiments were carried out. While we have not defined the exact position of termination, the results of this study show that transcription does indeed proceed as far as the DDE and that the majority of transcripts terminate no more than 150 nt downstream of the element.
The processes of 3′-end cleavage and polyadenylation and transcription termination are intimately linked (5). A bipartite signal consisting of a functional polyadenylation element and a downstream transcription pause element are required for efficient transcription termination (30). Mutations in the former element have been shown to reduce the efficiency of termination. Among the components of cleavage factor 1A in yeast, the most notable proteins required for efficient termination are Rna15, Rna14, and Pcf11 (30). These proteins appear to influence termination through their interaction with the C-terminal domain of RNA polymerase II (2, 11). A connection between transcription termination and 3′-end processing of the nonpolyadenylated mouse histone H2A gene has also been demonstrated (12). Removal of cis-acting sequences that are required for 3′-end processing of this gene disrupts transcription termination. The DDE described here is located just upstream of the region of transcription termination. However, according to the findings for the mutations examined in this study, it appears that these cis-acting sequences are not directly required for transcription termination (data not shown). Given the location of the element and the known connection between transcription termination and 3′-end cleavage, further mutations may uncover a link between these two processes.
Cell cycle regulation is altered by mutations in the DDE.
Our data also reveal that two of the mutants in the DDE, pSAC14 and pSAC21 (Fig. 5C and E), can alter the periodic accumulation of neo-HTB1 mRNA during the cell cycle. Interestingly, these mutations, while in the same general location as those altering 3′-end cleavage, are in fact distinct. Mutations altering 3′-end cleavage do not alter cell cycle regulation, and mutations altering cell cycle regulation do not alter 3′-end cleavage. The mutations in pSAC-14 and -21 lie adjacent to one another (Fig. 2A). Using the RNA folding program MFOLD (http://www.bioinfo.rpi.edu/), we found that the sequences surrounding the DDE can be folded into a putative stem-loop structure (Fig. 7). Both mutant pSAC14 and mutant pSAC21 lie at the top end of the stem structure, while the mutants that affect 3′-end cleavage lie further down the stem. Using a band shift assay, we showed that an RNA molecule encompassing this stem-loop structure binds a protein factor or factors that are maximally present in the S phase of the cell cycle. The fact that maximum binding is observed in the S phase of the cell cycle may indicate the binding of a cell cycle-regulated factor. Both pSAC14 and -21 mutants showed a reduction and an absence of protein binding, respectively, and also showed a lack of turnover of the mRNA following the S phase, suggesting that the protein factor is required for destabilization of the mRNA following the S phase. Furthermore, mutant pSAC20, which exhibited more rapid turnover of the RNA, showed a greater degree of binding to proteins present in extracts from the S phase of the cell cycle. Using the MFOLD program, we observed that the putative secondary structure of these RNAs, in the region of the mutations, was altered (data not shown), strengthening the argument that a specific factor is interacting with this region. The sequences in the DDE that regulate 3′-end processing and cell cycle periodicity appear to act independently of each other; however, since they are so closely linked spatially, it is possible that further mutations may reveal an overlapping link between 3′-end processing and cell cycle regulation.
FIG. 7.
Putative secondary structure for the region encompassing the DDE. The sequence surrounding the DDE was folded into a secondary structure by using the program MFOLD. Arrows show the positions of nucleotides altered in the various mutants. The numbers next to the structure represent the positions of the bases relative to that of the stop codon of the HTB1 gene (Fig. 1B).
Given the location of the DDE and the myriad of proteins associated with RNA polymerase II, it is not surprising that this RNA fragment binds to proteins present in cell extracts. Aranda and Proudfoot (2) have uncovered a physical and genetic link between the C-terminal domain of CstF-64 (Rna15p), a protein required for 3′-end processing and transcription termination, and the transcription factor Res2p. This protein is a component of the G1/S transcription factor complex MBF (Mlu1 cell cycle box-binding factor), which regulates the activation of a set of genes at the G1/S boundary of the cell cycle (38). While histone genes are not regulated by Res2p, taken together, the data suggest that 3′-end processing and transcription termination events play a role in the control of cell cycle-regulated genes. This argument is strengthened by recent data showing that the phosphorylation and ubiquitination state of poly(A) polymerase, a component of the cleavage and polyadenylation machinery, exhibits variation during the cell cycle (6, 28). Interestingly, it has been shown that transcription of the human histone gene, H3.3, is regulated by a transcription termination site located within an intron in the gene. This gene belongs to the class of replication-independent, basally expressed histone genes that do not accumulate during the S phase of the cell cycle. Rather, members of this class of histone genes are constitutively expressed in certain tissues such as spermatids and nondividing higher eukaryotic cells. Basally transcribed histone genes differ from replication-dependent histone genes, as they often possess introns and are polyadenylated. Transcription elongation of the H3.3 gene is blocked in cells arrested at the initiation of the S phase. DNA binding studies revealed the presence of protein factors in HeLa cell nuclear extracts that bind specifically to the region of the transcription elongation block. The block in transcriptional elongation was shown to correlate with the protein binding activity (33). The data presented here, showing that sequences close to the site of termination of a yeast histone gene play a role in cell cycle regulation, may provide an insight into the evolutionary process that resulted in such divergent mechanisms for the regulation of histone genes in lower and higher eukaryotes.
Acknowledgments
We thank Tharappel C. James for helpful discussions and experimental advice. Thanks also to Stephan Keegan, who provided technical assistance. We thank Agustin Aranda and Aurora Marco for helpful contributions in the TRO analyses.
This work was supported by a grant to U.B. from Enterprise Ireland (SC/97/326) and as part of the National Development Plan (grant SC/01/398) and was partly funded by the European Community-European Regional Development Plan. M. del Olmo is funded by Generalitat Valenciana (GV99-105-1-13) and the Ministerio de Educación y Ciencia (CICYT ALI99-1224-C02-02).
REFERENCES
- 1.Aranda, A., J. E. Perez-Ortin, C. Moore, and M. del Olmo. 1998. Transcription termination downstream of the Saccharomyces cerevisiae FBP1 poly(A) site does not depend on efficient 3′ end processing. RNA 4:303-318. [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda, A., and N. Proudfoot. 2001. Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell 7:1003-1011. [DOI] [PubMed] [Google Scholar]
- 3.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battle, D. J., and J. A. Doudna. 2001. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA 7:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, K. Keller, and N. Proudfoot. 1998. Coupling transcriptional termination to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 6.Bond, G., C. Prives, and J. L. Manley. 2000. Poly(A) polymerase phosphorylation is dependent on novel interactions with cyclins. Mol. Cell. Biol. 20:5310-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond, U., and T. Yario. 1994. The steady-state levels and structure of the U7 snRNP are constant during the human cell cycle: lack of cell cycle regulation of histone mRNA 3′ end formation. Cell. Mol. Biol. Res. 40:27-34. [PubMed] [Google Scholar]
- 8.Bond, U. M., T. A. Yario, and J. A. Steitz. 1991. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 5:1709-1722. [DOI] [PubMed] [Google Scholar]
- 9.Bracken, A., and U. Bond. 1999. Reassembly and protection of small nuclear ribonucleoprotein particles by heat shock proteins in yeast cells. RNA 5:1586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler, J. S., P. P. Sadhale, and T. Platt. 1990. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol. Cell. Biol. 10:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo, O., and J. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 12.Chodchoy, N., N. B. Pandey, and W. F. Marzluff. 1991. An intact histone 3′-end processing site is required for transcription termination in a mouse histone H2A gene. Mol. Cell. Biol. 11:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Co-ordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. [DOI] [PubMed] [Google Scholar]
- 14.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ ends of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 15.Dominski, Z., L. Zheng, R. Sanchez, and W. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahrner, K., J. Yarger, and L. Hereford. 1980. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 8:5725-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gick, O., A. Kramer, A. Vasserot, and M. L. Birnstiel. 1987. Heat-labile regulatory factor is required for 3′ end processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. USA 84:8937-8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. Genomic detection of new yeast pre-mRNA 3′ end processing signals. Nucleic Acids Res. 27:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson, R. J., J. Sun, D. G. Willis, and W. F. Marzluff. 1996. Efficient extraction and partial purification of the polyribosome-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35:2146-2156. [DOI] [PubMed] [Google Scholar]
- 20.Hereford, L., K. Fahrner, T. Woolford, M. Rosbash, and D. B. Kaback. 1979. Isolation of yeast histone genes H2A and H2B. Cell 18:1261-1271. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman, I., and M. L. Birnstiel. 1990. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature 346:665-668. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M., W. Lee, J. Park, J. B. Kim, Y. K. Jang, R. H. Seong, S. Y. Choe, and S. D. Park. 2000. The stress-activated MAP kinase sty1/spc1 and a 3′ regulatory element mediate UV-induced expression of the gene uv15+ at the post-transcriptional level. Nucleic Acids Res. 28:3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luscher, B., and D. Schumperli. 1987. RNA 3′ end processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lycan, D. E., M. Osley, and L. M. Hereford. 1987. Role of transcriptional and post-transcriptional regulation in the expression of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, F., A. Schaller, S. Eglite, D. Schumperli, and B. Muller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. Patterson, M. Wickens, and D. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 27.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 28.Mizrahi, N., and C. Moore. 2000. Posttranslational phosphorylation and ubiquitination of the Saccharomyces cerevisiae poly(A) polymerase at the S/G2 stage of the cell cycle. Mol. Cell. Biol. 20:2794-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNAs. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 30.Proudfoot, N. J. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 31.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 32.Smith, M. M., and K. Murray. 1983. Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J. Mol. Biol. 69:641-661. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, A., L. Zhang, J. Herrmann, B. Wu, L. Kedes, and D. Wells. 1997. Cell-cycle-specific transcription termination within the human histone H3.3 gene is correlated with specific protein-DNA interactions. Genet. Res. 69:101-110. [DOI] [PubMed] [Google Scholar]
- 34.van Helden, J., M. del Olmo, and J. E. Perez-Ortin. 2000. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 28:1000-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells, D., and D. Brown. 1991. Histone and histone gene compilation: an alignment update. Nucleic Acids Res. 19(Suppl.):2173-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. Hurt, and W. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, H., L. Johnson, and M. Grunstein. 1990. Coding and noncoding sequences at the 3′ end of yeast histone H2B mRNA confer cell cycle regulation. Mol. Cell. Biol. 10:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, Y., T. Takeda, S. Whitehall, N. Peat, and N. Jones. 1997. Functional characteristics of the fission yeast Start-specific transcription factor Res2. EMBO J. 16:1023-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]