Abstract
The isomerization of up to 100 uridines to pseudouridines (Ψs) in eukaryotic rRNA is guided by a similar number of box H/ACA small nucleolar RNAs (snoRNAs), each forming a unique small nucleolar ribonucleoprotein particle (snoRNP) with the same four core proteins, NAP57 (also known as dyskerin or Cbf5p), GAR1, NHP2, and NOP10. Additionally, the nucleolar and Cajal body protein Nopp140 (Srp40p) associates with the snoRNPs. To understand the role of these factors in pseudouridylation, we established an in vitro assay system. Short site-specifically 32P-labeled rRNA substrates were incubated with subcellular fractions, and the conversion of uridine to Ψ was monitored by thin-layer chromatography after digestion to single nucleotides. Immunopurified box H/ACA core particles were sufficient for the reaction. SnoRNPs associated quantitatively and reversibly with Nopp140. However, pseudouridylation activity was independent of Nopp140, consistent with a chaperoning role for this highly phosphorylated protein. Although up to 14 bp between the snoRNA and rRNA were required for the in vitro reaction, rRNA pseudouridylation and release occurred in the absence of ATP and magnesium. These data suggest that substrate release takes place without RNA helicase activity but may be aided by the snoRNP core proteins.
Pseudouridine (Ψ) is the most abundant modified nucleotide in RNA. In vertebrate rRNA, ∼100 uridines are converted to Ψs (for a review, see reference 36). The modifications are catalyzed by a similar number of small nucleolar ribonucleoprotein particles (snoRNPs), each consisting of a unique small nucleolar RNA (snoRNA) and a common set of four core proteins (for reviews, see references 11, 20, 47, and 49). The snoRNAs consist of a common hairpin-hinge-hairpin-tail secondary structure and contain the conserved sequence ANANNA in the hinge region (box H) and an ACA trinucleotide three positions from the 3′ end (box ACA). They are therefore referred to as box H/ACA snoRNAs. Both sides of a bulge in the first and/or second hairpin of these snoRNAs harbor a 3- to 10-nucleotide (nt) long sequence complementary to both sides of the target uridine in rRNA. Thus, box H/ACA snoRNAs determine the site of the pseudouridylation reaction by framing the target uridine in a snoRNA-rRNA hybrid (12, 32). The isomerization of uridine to Ψ is apparently catalyzed by NAP57 (Cbf5p in yeast), one of the snoRNP core proteins. This is supported by genetic evidence in yeast (22, 54) and the crystal structure of TruB, the bacterial homolog of NAP57 (26, 35), bound to tRNA (16). The other box H/ACA snoRNP core proteins are GAR1, NHP2, and NOP10 (3, 13, 15, 24, 51).
Most of the details of the guide mechanism and the composition of box H/ACA snoRNPs are based on genetic and biochemical analyses in yeast. In nuclear extracts of mammalian cells, the four core proteins can assemble with in vitro-synthesized box H/ACA snoRNAs (9, 41). The human NAP57, also known as dyskerin, is mutated in the X-linked bone marrow failure disorder, dyskeratosis congenita, suggesting a role for box H/ACA snoRNPs in this often deadly disease (14). In eubacteria, pseudouridylation of rRNA is both guided and catalyzed by single protein enzymes (for a review, see reference 36). These pseudouridylases, although related to the snoRNP component NAP57, do not require snoRNAs, additional proteins, or other cofactors for their site-specific catalysis (35, 52). Despite this wealth of information, little is known about the requirements and mechanism of snoRNP-mediated pseudouridylation in eukaryotes.
It is not clear if the four core proteins and the snoRNA are sufficient for the conversion of uridine to Ψ or if additional proteins are required. For example, NAP57 was identified as a Nopp140-associated protein (26), raising the question of involvement of Nopp140 in the reaction. Nopp140 is a highly phosphorylated protein located in the nucleolus and the Cajal (coiled) bodies (26, 27). Uniquely, Nopp140 interacts with both box H/ACA and box C/D snoRNPs (18, 53). Box C/D snoRNPs form another major class of snoRNPs that mainly guide the 2′-O-methylation of rRNA. Similar to box H/ACA snoRNPs, they consist of a unique box C/D snoRNA and a set of four common core proteins, the methylase fibrillarin (Nop1p in yeast), NHP2L1/15.5-kDa protein (Snu13p), NAP65 (Nop5/58p), and NOP56 (Nop56p) (for a review, see reference 11). Nopp140 appears to bind more tightly to box H/ACA snoRNPs than box C/D snoRNPs (53). Although these interactions have been observed in vivo and in vitro, it remains to be determined if Nopp140 is an integral part of both classes of snoRNPs and what the nature of its snoRNP interaction is.
Here we show Nopp140 to quantitatively but reversibly associate with snoRNPs in a phosphorylation-dependent manner. As determined by a novel in vitro assay for snoRNP-guided pseudouridylation of rRNA, this Nopp140-snoRNP interaction has no apparent effect on their activity. Pseudouridylation of rRNA solely requires the cognate box H/ACA snoRNP but no other factors, such as magnesium or ATP.
MATERIALS AND METHODS
Site-specifically labeled rRNA substrates.
rRNA substrates used in this study are listed in Fig. 5A. Their sequences are identical in humans and rats except for the penultimate nucleotide in substrate 1. Site-specific labeling of the target uridine was achieved by two-way RNA ligation (see Fig. 3A) (30). RNA oligomers were chemically synthesized and purified as described previously (31), except for the 66-nt-long 3′ fragment of rRNA substrate 1. The latter was transcribed in vitro and therefore required a guanosine at its 5′ end instead of the naturally occurring cytidine. To generate the 32P-labeled Ψ 3′-monophosphate standard, a 5′-RNA fragment was synthesized ending in a Ψ by using a Ψ-CPG support (Chemgenes, Ashland, Mass.). For protection from RNA exonucleases, the ends of most short substrates were appended with two deoxynucleotides. After each 3′ fragment was 32P labeled at its 5′ end, the two corresponding fragments were joined together by oligodeoxynucleotide-mediated ligation (30). The ligation products were purified by urea polyacrylamide gel electrophoresis (PAGE).
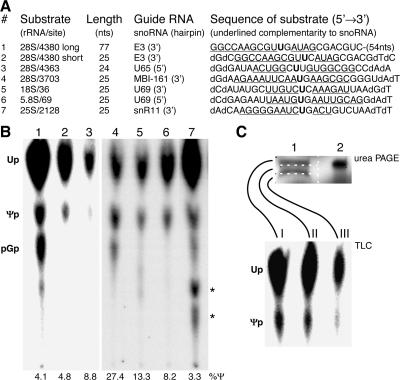
FIG. 5.
The assay works with multiple substrates and extracts. (A) The rRNA substrates used in this study are listed by number, the position of the uridine they represent in the respective human (1-6) and yeast (7) rRNAs, their length in nucleotides, the snoRNA guiding their pseudouridylation (specifying which of the two hairpins contains the complementarity to rRNA), and the nucleotide sequence with the target uridine (bold) and the complementary nucleotides (underlined). Note the replacement of cytidine 4381 by a guanosine in substrate 1 without an effect on activity. The sequences of the human rRNA are identical in rats, except that the numbering of the target uridines differs in substrates 1 and 2 (4141), 3 (4124), and 4 (3459). (B) Autoradiograph of the separated nucleotides liberated from substrates 1 to 7 after incubation in BRL nuclear lysates (1-3), rat liver nucleolar extracts (4-6), and yeast whole-cell extracts (7). Asterisks denote additional products that were detectable when incubations were performed in complex cell fractions and likely represented transphosphorylation products after substrate degradation. (C) Uridine is isomerized in the full-length substrate. After incubation in HeLa nuclear extract, substrate 2 was reisolated, analyzed by denaturing urea PAGE (top panel, lane 1), and compared to half of the substrate before incubation (lane 2). The gel was sectioned into three slices (white dotted boxes), and the RNA was eluted from each slice and digested for analysis of the single nucleotides by TLC (bottom panel).
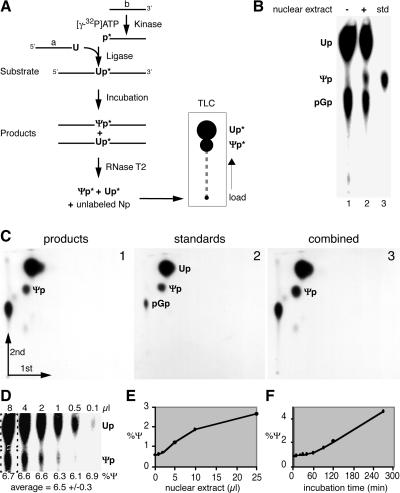
FIG. 3.
In vitro assay for pseudouridylation of rRNA. (A) Schematic of the assay outlining the synthesis of the site-specifically labeled rRNA substrate by oligodeoxynucleotide-mediated ligation of the two independently synthesized 5′ (a) and 3′ (b) halves after labeling of b with 32P (p*). The reaction products are analyzed by nucleolytic liberation of the uridine (Up*) and Ψ 3′ monophosphates (Ψp*) and their separation by TLC followed by autoradiography. (B) Autoradiograph of a TLC demonstrating the production of Ψ only in the presence of HeLa nuclear extract (lane 2). Ψ 3′ monophosphate was loaded as standard (std, lane 3). Guanosine 5′,3′-diphosphate (pGp) was generated by the digest of incompletely ligated 3′-half substrate (b in panel A). (C) Analysis of the reaction products by two-dimensional TLC. The identity of the product Ψp (panel 1) is confirmed by the generation of a single spot when standard nucleotides (panel 2) are mixed with the product prior to chromatography (panel 3). (D) Quantitation by phosphorimager analysis varies minimally over an 80-fold difference in product loading on TLC. The amounts of Up and Ψp were each determined in rectangles (dotted lines) that had been background corrected, and the amount of Ψp was expressed as a percentage of total Up and Ψp (%Ψ). The production of Ψp increased with the amount of HeLa nuclear extract (E) and with increasing incubation time (F). Note that the higher activity in panel D than in panels E and F was caused by incubation in a different extract, BRL nucleolar extract.
Subcellular fractions.
Nuclear lysate was prepared from five subconfluent 150-cm-diameter dishes of buffalo rat liver (BRL) cells. The cells were scraped off, washed once with 10 ml of cold phosphate-buffered saline (PBS), and swollen for 15 min in 1.25 ml of hypoosmotic homogenization buffer (10 mM HEPES-KOH [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol [DTT]). The cells were disrupted with 12 to 15 strokes of a tight-fitting pestle in a Dounce homogenizer. After a 10-min centrifugation at 1,000 × g, the pellets (crude nuclei) were resuspended in 0.5 ml of a solution containing 10 mM Tris-HCl (pH 7.5), 3.3 mM MgCl2, 1 mM DTT, and 0.25 M sucrose and disrupted by tip sonication to produce nuclear lysate. To prepare nucleolar extract, the nuclear lysate was sedimented at 3,000 × g for 10 min and the pellet was resuspended in an equal volume of solution containing 20 mM HEPES (pH 7.5), 2 mM MgCl2, 1 mM DTT, and 10% glycerol using a Dounce homogenizer. After adjustment to 0.5 M NaCl, the mixture was incubated on ice for 30 min and centrifuged at 47,000 × g for 10 min to produce the nucleolar extract in the supernatant. Nucleolar extracts were prepared analogously from 2 × 109 rat liver nuclei isolated as described previously (4, 28). Rat liver low-salt nuclear extracts corresponded to the second nuclear extract described in reference 28. Yeast whole cell extracts (48) and HeLa nuclear extracts (8) were prepared as described previously.
Plasmids and in vitro transcription.
The 66-nt 3′ fragment of rRNA substrate 1 and the rat E3 and E2 snoRNAs (45) were transcribed using a T7-MEGA shortscript transcription kit (Ambion, Austin, Tex.). The transcription templates were generated by PCR amplification of plasmids pGEM28Sp, pTM105, and pTM125, respectively. For pTM105 (rat E3 snoRNA in pGEMm), the cDNA of rat E3 snoRNA was isolated by reverse transcription from BRL whole-cell RNA, amplified by PCR as described elsewhere (53), and cloned into the EcoRI and SacI sites of pGEMm. The primers used to amplify E3 cDNA were 5′-TAATACGACTCACTATAGTAAGCAGGATTTTACTACAATAT-3′ and 5′-TCATGTATGAGACCAAGCGT-3′, generating a T7 promoter sequence (underlined) and an extra guanosine (bold) at the 5′ end of the transcript for better transcription efficiency. pGEMm was generated from pGEM-9Zf(−) (Promega, Madison, Wis.) by digestion with XbaI and SpeI and religation. For pTM125 (E2 in pIIIA/MS2-1), the rat E2 snoRNA cDNA was generated as described above for E3 and cloned into the SmaI site of pIIIA/MS2-1 (46). The primers used to amplify the E2 cDNA were 5′-TAATACGACTCACTATAGGTGTGCACATTGTTAGA-3′ and 5′-TACTTGTAGCAAGAATTACT-3′, generating two extra guanosines (bold) at the 5′ end of the transcript. pGEM28Sp [nt 2701 to 4204 of rat ribosomal DNA in pGEM-9Zf(−)] was a kind gift from Paul McDermott, Medical Center, Charlton, S.C. The 66-bp fragment of rat 28S ribosomal DNA (nt 4142 to 4207) was amplified from pGEM28Sp using the primers 5′-TCGAATTCTCGAGTAATACGACTCACTATAGATAGCGACGTCGCTTT-3′ and 5′-GGCGAATTCTGCTTCAC-3′, replacing cytidine 4142 with a guanosine at the 5′ end of the transcript (bold). The transcription products were all gel purified.
Pseudouridylation assay.
A standard 25-μl reaction contained 10 μl of HeLa nuclear extract or BRL nuclear lysate, 5 μl of 5× reconstitution buffer (100 mM HEPES-KOH [pH 7.5], 600 mM KCl, 10 mM MgCl2), 1 pg of tRNA, 20 U of RNasin (Panvera, Madison, Wis.), and approximately 10 to 20 fmol of rRNA substrate. Reaction mixtures were incubated for 2 h at 30°C and terminated on ice, and the RNA was phenol-chloroform extracted, ethanol precipitated, and digested with 5 U of RNase T2 (Sigma, St. Louis, Mo.). The resulting single-nucleotide 3′-monophosphate mixture was separated on thin-layer chromatography (TLC) cellulose polyethyleneimine plates (Selecto Scientific, Suwanee, Ga.) using isopropanol-HCl-water (70:15:15 [vol/vol/vol]) as a solvent and, in the case of two-dimensional TLC, isobutyric acid-0.5 M NH3 (5:3 [vol/vol]) in the first dimension (33). TLC plates were quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). High-salt nucleolar extracts were diluted with a solution containing 20 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 2 mM MgCl2, and 5% glycerol before activity measurement. Identical buffer conditions were used for the immunopurified snoRNPs. For the RNA depletion experiments, 1 μl of micrococcal nuclease (100 to 300 U/μl; Fermentas MBI, Hanover, Md.) was added to 10 μl of BRL nuclear lysate containing 1 mM CaCl2 and inactivated by the addition of 6 mM EGTA after a 30-min incubation at 37°C. To reconstitute activity, 0.6 pmol of in vitro-transcribed snoRNAs were added to the nuclease-treated nuclear lysates and incubated at 30°C for 1 h prior to activity determination.
RNA-RNA gel shifts.
Gel shifts were performed as described previously (42). Briefly, ∼20 fmol of 32P-labeled substrate 2 and 1 pmol of in vitro-transcribed E3 snoRNA were mixed in 20 μl of hybridization buffer (50 mM Tris glycine [pH 8.1], 250 mM KCl, 10 mM MgCl2) and incubated at 30°C for 1.5 h. For competition experiments, increasing amounts of RNA oligonucleotides (1, 10, and 160 pmol) were added to the hybridization mixture prior to the substrate. Before the mixture was loaded onto a native 8% polyacrylamide gel, 50 μg of tRNA/ml and 10% glycerol were added.
Immunopurification of snoRNPs.
Immunoprecipitations were performed as described previously (26) except under more stringent conditions, i.e., in the presence of 0.5 M NaCl-1% Triton X-100-0.2% sodium dodecyl sulfate (SDS) (unless indicated otherwise), with affinity-purified antipeptide antibodies against Nopp140 (27) and NAP57 (26). The immunoprecipitates were washed repeatedly with equal stringency and finally with pseudouridylation buffer. The precipitated proteins were analyzed by 9% standard (21) and 15% Tricine SDS-PAGE (44) followed by Coomassie blue and/or silver staining (29) or transfer to nitrocellulose and antibody detection. The following primary antibodies were used for immunodetection: anti-NAP57, RL11 affinity-purified rabbit immunoglobulin Gs (IgGs) (0.4 μg/ml) (26); anti-Nopp140, RF11 affinity-purified rabbit IgGs (0.1 μg/ml) (27); anti-GAR1, Rab2B rabbit serum (1:1,000) (9); anti-NHP2 (P15) and anti-NOP10 (P13), affinity-purified rabbit IgGs, (0.15 μg/ml) (41); and anti-fibrillarin, D77 mouse monoclonal IgGs (0.2 μg/ml) (1). The immunoprecipitated snoRNPs were assayed for pseudouridylase activity while bound to protein A-Sepharose (Pharmacia, Piscataway, N.J.) by the antibodies.
Glycerol gradients and phosphatase treatment.
SnoRNPs were fractionated on 10 to 30% glycerol gradients as described previously (50). Briefly, 500 μl of rat liver low-salt nuclear extracts were adjusted to 10 mM Tris (pH 7.5)-60 mM KCl in the presence or absence of 500 mM potassium acetate and fractionated on 10 ml gradients by centrifugation at 25,000 rpm for 15 h in a SW-41Ti rotor. The gradients were fractionated and the proteins were precipitated by 5% trichloroacetic acid, separated by SDS-PAGE, and analyzed by Western blotting. Bovine serum albumin (4.4S) and thyroglobulin (20S) were used as sedimentation markers in parallel gradients.
For phosphatase treatment, 50 U of calf intestinal alkaline phosphatase (New England Biolabs, Beverly, Mass.) was added to 0.5 ml of rat liver nuclear extracts or to 0.2 ml of high-salt nucleolar extracts after dilution to 0.1 M salt and incubated at 37°C for 30 min. Twenty-five microliters of treated extracts were analyzed by Western blotting for the phosphorylation status of Nopp140, and the remainder was used for immunoprecipitation experiments to test the Nopp140-snoRNP interaction.
RESULTS
Nopp140 associates reversibly with snoRNPs.
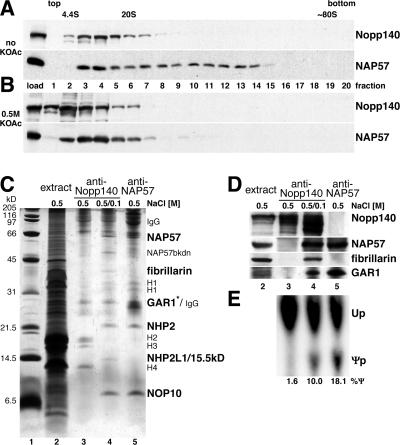
We demonstrated previously that Nopp140 interacts with box H/ACA and box C/D snoRNPs in vivo and in vitro (18, 26, 53). To further investigate the Nopp140-snoRNP interaction, nuclear extracts were fractionated on glycerol gradients under physiological salt conditions and analyzed by Western blotting (Fig. 1A). Nopp140 (upper panel) and NAP57 (lower panel) cosedimented at ∼10S (fractions 3 and 4) as expected for single snoRNPs (10, 13, 50). Neither protein was detectable in the top fraction of the gradient, indicating that these proteins quantitatively associated with snoRNPs or other higher-molecular-weight structures. Moreover, both proteins extended farther into the gradient, likely due to higher-order complexes with each other and/or with higher-molecular-weight structures in these extracts, e.g., preribosomes and chromatin. In yeast, box H/ACA snoRNPs can be separated from box C/D snoRNPs by 0.5 M potassium acetate (13). Under this condition, Nopp140 dissociated from snoRNPs and sedimented as a single species at the top of the gradient (Fig. 1B, upper panel). In addition, NAP57 no longer extended into the gradient but formed a homogenous peak at ∼10S, indicating the disruption of the higher-order complexes (lower panel). Thus, Nopp140 interacted with box H/ACA snoRNPs in a salt-sensitive fashion.
FIG. 1.
Reversible association of Nopp140 with snoRNPs. Western blots of fractions separated on 10 to 30% glycerol gradients under physiological (A) or high-salt (B) conditions (0.5 M KOAc) and probed with anti-Nopp140 antibodies (upper panels) and anti-NAP57 antibodies (lower panels) are shown. The top and bottom of the gradient are indicated, as are the migrating positions of bovine serum albumin (4.4S) and thyroglobulin (20S). The 80S position was estimated by comparison of published gradients. (C) Silver-stained SDS-PAGE of immunoprecipitations from 0.5 M salt nucleolar extracts (lane 2) with anti-Nopp140 (lanes 3 and 4) and anti-NAP57 antibodies (lane 5). Before precipitation in lane 4, the extracts were diluted to 0.1 M salt. The specifically precipitating proteins (bold), the immunoglobulins (IgG), contaminating histones (H1 to H4), and a NAP57 breakdown product (NAP57bkdn) are indicated (see Fig. 6E for a detailed explanation). The asterisk draws attention to the fact that GAR1 is visible only in lane 5, not in lane 4, in which it comigrates with the anti-Nopp140 IgG light chains (see Fig. 1D and 6E) (53). Lane 2 contains 1/100 of the input used for the precipitations. (D) Western blots of the same samples as in panel C (lanes 2 to 5). Respective migration regions were probed with anti-Nopp140, -NAP57, -fibrillarin, and -GAR1 antibodies as indicated on the right-hand side and detected by enhanced chemiluminescence. (E) Autoradiograph of nucleotides separated by TLC after liberation from substrate 1, which was incubated with the same immunoprecipitates shown in panels C and D (lanes 3 to 5) to determine their pseudouridylase activity in vitro as described in the legend to Fig. 3.
Using coimmunoprecipitation, we tested whether this Nopp140-snoRNP interaction was reversible. To avoid the interference of immunoglobulin heavy and light chains, samples were not boiled before loading, which largely prevented the reduction of the disulfide bonds and caused migration of the immunoglobulins at 80 kDa and above. As expected from our glycerol gradient analysis, immunoprecipitation of Nopp140 from nucleolar extracts (Fig. 1C, lane 2) in the presence of 0.5 M salt failed to coprecipitate snoRNP core proteins (lane 3). In contrast, dilution of the salt from 0.5 to 0.1 M in the extracts led to the specific coprecipitation with Nopp140 of all known box H/ACA snoRNP core proteins (lane 4). For comparison, box H/ACA snoRNPs were precipitated with anti-NAP57 antibodies in the presence of 0.5 M salt (lane 5; see also Fig. 6E). In addition to the four box H/ACA snoRNP core proteins, Nopp140 also coprecipitated fibrillarin and a protein that likely corresponded to the NHP2L1/15.5-kDa protein of box C/D snoRNPs (lane 4) (5, 34, 43). The precipitation of Nopp140, NAP57, fibrillarin, and GAR1, or lack thereof, under the various salt concentrations was confirmed by Western blotting (Fig. 1D). These data illustrate the highly specific reassociation of Nopp140 with only the snoRNPs in the complex mixture of nucleolar extracts (compare lanes 2 and 4), demonstrating a reversible electrostatic interaction between Nopp140 and both major classes of snoRNPs.
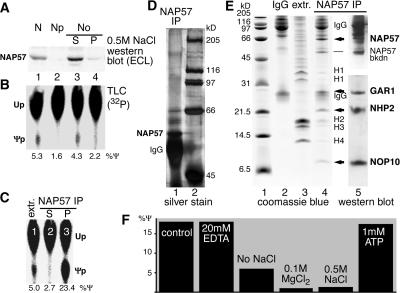
FIG. 6.
Minimal requirements for in vitro pseudouridylation of rRNA. (A and B) Subfractionation of BRL nuclear lysate (lane 1) into nucleoplasm (lane 2) and 0.5 M NaCl nucleolar extract supernatant (lane 3) and pellet (lane 4) is shown. The fractions were analyzed by Western blotting with anti-NAP57 antibodies (A) and for their pseudouridylase activity towards substrate 1 by TLC (B). Note that the activity cofractionated with the putative pseudouridylase NAP57 in the nucleolar extract (lane 3). (C) The pseudouridylase activity was precipitated from rat liver nucleolar extracts (extr.) with anti-NAP57 antibodies (lane 3) and removed from the supernatant (lane 2). Fivefold less extract was assayed (lane 1) than was used in the immunoprecipitation experiments. (D) Analysis of the active NAP57 immunoprecipitate by SDS-9% PAGE and silver staining (lane 1). NAP57, the immunoglobulin heavy chains (IgG), and the molecular size markers (lane 2) are indicated. (E) Comparison on Coomassie blue-stained Tricine SDS-15% PAGE of the active NAP57 immunoprecipitate (lane 4) to nucleolar extract (lane 3) and anti-NAP57 antibodies (lane 2). Note that 3,000-fold more extract was used for the precipitation than was loaded in lane 3. The only specifically precipitated protein bands (arrows and dash), the immunoglobulin chains (IgG), the histones (H1 to 4), and the molecular size markers (lane 1) are indicated. The identity of the specifically precipitated bands was confirmed by consecutive probing of a Western blot with anti-GAR1, -NHP2, and -NOP10 antibodies (lane 5, lower half) and -NAP57 antibody (upper half). (F) The pseudouridylation of substrate 1 by the immunopurified snoRNPs bound to protein A-Sepharose was determined under the indicated conditions relative to the control reaction containing 2 mM MgCl2, 100 mM NaCl, 20 mM HEPES (pH 7.5), and 5% glycerol.
Nopp140 phosphorylation controls its association with snoRNPs.
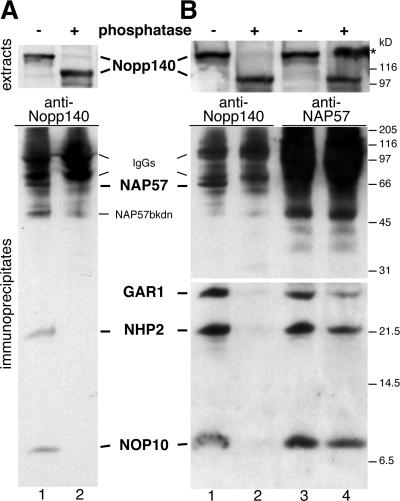
The major contributor to charge in Nopp140 is its high degree of phosphorylation of ∼80 phosphates/molecule (25, 27). When rat liver nucleolar extracts were analyzed by SDS-PAGE and Western blotting, Nopp140 showed a characteristic mobility shift from 140 to 100 kDa after phosphatase treatment, indicating complete dephosphorylation (Fig. 2A, top panel) (25, 27). Immunoprecipitation from these extracts with anti-Nopp140 antibodies followed by Western blotting showed a coprecipitation of all box H/ACA snoRNP core proteins tested from mock-treated extracts but not from phosphatase-treated extracts (Fig. 2A, lanes 1 and 2), whereas precipitation of both forms of Nopp140 was confirmed by Western blotting (not shown). Consistent with this, box H/ACA snoRNAs were coprecipitated with Nopp140 from nucleolar extracts exclusively in the absence of phosphatase pretreatment (not shown). These data suggest that box H/ACA snoRNPs interacted only with phosphorylated Nopp140.
FIG. 2.
Nopp140 dephosphorylation releases box H/ACA snoRNPs. Western blots of immunoprecipitates from high-salt nucleolar extracts diluted to 0.1 M salt (A) and from low-salt nuclear extracts (B) probed consecutively with antibodies against the antigens, indicated in bold between the panels, are shown. Prior to immunoprecipitations, the rat liver extracts were incubated in the absence (odd lanes) or presence (even lanes) of alkaline phosphatase. The dephosphorylation of Nopp140 in the extracts was documented by its mobility shift from 140 to 100 kDa (upper panels). The extracts were incubated with anti-Nopp140 antibodies (lanes 1 and 2) and anti-NAP57 antibodies (lane 3 and 4), and the coprecipitating antigens were visualized by Western blotting (lower panels). The lower half was exposed longer for ECL detection (B) (bottom panels). The asterisk refers to Nopp140 spillover from an adjacent lane on the gel, and additional labels are as described for Fig. 1.
The nucleolar fractions used in the above-described experiment were generated by 0.5 M salt extraction and dilution to 0.1 M salt to reassociate Nopp140 with the snoRNPs as described above (Fig. 1C and D). To ascertain whether Nopp140-snoRNP complexes formed in vivo would also be dissociated by phosphatase treatment, we repeated the experiments with low-salt rat liver nuclear extracts (28). The results were identical (Fig. 2B, compare lanes 1 and 2). Although no mobility shift of any of the box H/ACA snoRNP core proteins was observed upon phosphatase treatment (not shown, but see Fig. 2B, lanes 3 and 4), the possibility existed that dephosphorylation did not disrupt the Nopp140-snoRNP interaction but destabilized the snoRNP itself. However, anti-NAP57 antibodies coprecipitated all four box H/ACA snoRNP core proteins from low-salt nuclear extracts in the presence and absence of phosphatase pretreatment, demonstrating that the snoRNPs remained intact (Fig. 2B, compare lanes 3 and 4). Consistent with this, the immunoprecipitated snoRNPs remained active regardless of phosphatase treatment (see below). In summary, Nopp140 associated with intact box H/ACA snoRNPs in its phosphorylated form but not in its dephosphorylated form.
In vitro assay for pseudouridylation.
To test the effect of the Nopp140-snoRNP interaction on snoRNP activity and to study the molecular mechanism of this reaction, we developed an in vitro assay system for snoRNP-mediated pseudouridylation of rRNA. The assay consisted of three major steps: first, the synthesis of a site-specifically labeled rRNA substrate; second, its incubation with cellular fractions; and third, the analysis of the product rRNA for pseudouridylation (Fig. 3A). Site-specific labeling and synthesis of the rRNA substrate enabled the selection of any of the ∼100 target uridines in rRNA as the substrate and the sensitive detection of the isomerized uridine. Specifically, we separately synthesized a 5′-half RNA ending in the target uridine (Fig. 3A) and a 3′ half of a rRNA substrate (Fig. 3A), phosphorylated the 3′ half with 32P, and joined the two fragments by oligodeoxynucleotide-mediated ligation. In the second step, the substrate rRNA containing the 3′-32P-labeled target uridine was incubated with cellular extracts to produce the pseudouridylated rRNA products. The third step took advantage of the specific labeling of the target uridine by its liberation from the product rRNA by RNase T2 digestion and by the separation of the resulting uridine and Ψ 3′ monophosphates by TLC followed by autoradiographic detection.
The specific rRNA substrate used in our studies, unless stated otherwise, corresponded to uridine 4380 of human 28S rRNA with 10 nt upstream and 66 nt downstream (see Fig. 5A, 1). Incubation of this substrate generated, in addition to uridine 3′ monophosphate (Up), a new spot by TLC in the presence but not in the absence of HeLa nuclear extract (Fig. 3B, lanes 1 and 2). The new spot comigrated with the Ψp standard (lane 3). The incubation products were further analyzed by two-dimensional TLC and compared to uridine and Ψ standards (Fig. 3C, panels 1 and 2). When the products and standards were mixed prior to chromatography, they produced a single spot in the position of Ψp, confirming its identity (panel 3).
To quantitate the Ψ produced in the assay, the TLCs were analyzed by phosphorimaging (Fig. 3D). Although the recovery of RNA after incubation was somewhat variable, the quantitation was unaffected by the amount of product loaded for TLC analysis. Over an 80-fold difference in product loading, the calculated activity fluctuated only marginally and, more importantly, independently of the amount loaded (Fig. 3D, 8 μl to 0.1 μl). In contrast, the pseudouridylation activity was dependent on the concentration of the nuclear extract and on the duration of the incubation (Fig. 3E and F).
In vitro pseudouridylation requires snoRNA-rRNA hybridization.
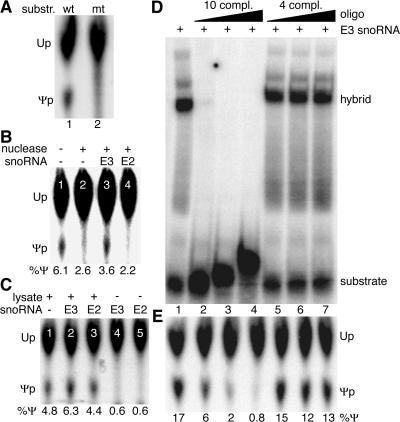
Pseudouridylation of uridine 4380 in our synthetic rRNA substrate 1 (see Fig. 5A) was predicted to be guided by the E3 snoRNA based on its 10- and 4-nt complementarities flanking the uridine residue (12). To test whether complementarity on both sides of the target uridine was required for in vitro pseudouridylation of substrate 1, its 5′ half was replaced with that of another substrate (no. 3; see Fig. 5A). Indeed, unlike the case of the wild-type substrate 1 (Fig. 4A, lane 1), no Ψ was formed during incubation of this hybrid substrate with nuclear lysate (lane 2). As determined in vivo (12, 32), therefore, complementarity of the substrate rRNA on both sides of the target uridine to the same snoRNA was important for its pseudouridylation in vitro, supporting the site specificity of the reaction.
FIG. 4.
In vitro pseudouridylation requires snoRNP-rRNA hybridization. (A) Autoradiograph of a TLC of uridine and Ψ liberated from the substrate after incubation with rat liver nucleolar extract. Ψ is produced from the wild-type rRNA substrate 1 (lane 1) but not a mutant substrate consisting of the 3′ half of substrate 1 but the 5′ half of substrate 3 (lane 2). (B) Pseudouridylation of substrate 1 in nuclear lysate pretreated with micrococcal nuclease (lanes 2 to 4) or not pretreated (lane 1). Addition of the cognate E3 snoRNA (lane 3) but not the noncomplementary E2 snoRNA (lane 4) partially restored the activity. Note that “trailing” of uridine often artificially elevated the numerical value of Ψ production under negative conditions (e.g., see lanes 2 and 4). (C) Ψ production in substrate 1 (lane 1) is enhanced in untreated nuclear lysates by E3 snoRNA (lane 2) but not E2 snoRNA (lane 3). Addition of either snoRNA alone failed to produce Ψ (lanes 4 and 5). (D) RNA-RNA gel shift analysis. Autoradiograph of a native polyacrylamide gel loaded with a mixture of the 32P-labeled rRNA substrate 2 and its cognate, unlabeled snoRNA E3 (lane 1). Addition of 10-fold-increasing concentrations of an unlabeled RNA oligonucleotide corresponding to the 5′ half of the rRNA substrate with 10 nt complementarity to E3 competed effectively for the gel shift (lanes 2 to 4), but addition of an oligonucleotide corresponding to the 3′ half with only 4-nt complementarity (lanes 5 to 7) did not. Note that the 5′-half oligonucleotide also had the ability to hybridize to and shift the substrate when present in sufficiently high concentrations (lanes 3 and 4). (E) Addition of equal amounts of substrate 2 to immunopurified snoRNPs (see Fig. 6) produced Ψ efficiently (lane 1). This activity was competed by the 5′-half oligonucleotide (lanes 2 to 4) but not the 3′-half oligonucleotide (lanes 5 to 7).
To further investigate the RNA requirements of the reaction, nuclear lysate was pretreated with micrococcal nuclease and the nuclease was subsequently inactivated by the addition of EGTA. Such pretreatment abolished pseudouridylation, demonstrating a requirement for RNA in the lysate (Fig. 4B, compare lanes 1 and 2). The activity was partially restored by the addition of in vitro-transcribed E3 snoRNA (Fig. 4B, lane 3), the guide RNA for rRNA substrate 1, but not by the addition of the unrelated E2 box H/ACA snoRNA (lane 4). Thus, the in vitro reaction required the cognate snoRNA, and exogenous E3 was able to assemble into a functional snoRNP with proteins in the lysate. Indeed, E3, but not E2 snoRNA, similarly enhanced the amount of Ψ produced in rRNA substrate 1 when added to untreated lysates (Fig. 4C, lanes 1 to 3). Although the addition of E3 snoRNA increased or restored the activity only marginally, the effect was reproducible in several independent experiments. The snoRNAs E3 and E2 in the absence of nuclear lysate were insufficient for the production of Ψ, consistent with a snoRNP requirement for the pseudouridylation of rRNA (Fig. 4C, lanes 4 and 5).
To determine if the cognate snoRNA indeed hybridized to the synthetic substrates and if such hybridization was required for the activity, we performed RNA-RNA gel shift and in vitro pseudouridylation assays in the presence of cRNA oligonucleotides. Addition of unlabeled E3 snoRNA to labeled rRNA substrate 2 (Fig. 5A) caused a characteristic shift, suggesting that the RNAs indeed hybridized with each other (Fig. 4D, lane 1). E3 snoRNA potentially forms 10 bp 5′ and four bp 3′ of the target uridine in substrate 2. Addition of increasing amounts of an unlabeled oligonucleotide corresponding to the 5′ half of rRNA substrate 2 efficiently competed for hybrid formation (Fig. 4D, lanes 2 to 4). However, addition of an unlabeled oligonucleotide corresponding to the 3′ half of substrate 2 (with only 4-nt complementarity) failed to interfere (Fig. 4D, lanes 5 to 7), as did addition of an oligonucleotide of unrelated sequence (not shown). Similarly, in vitro pseudouridylation of the same rRNA substrate was inhibited by the addition of the rRNA oligonucleotide with 10-nt complementarity (Fig. 4E, lanes 2 to 4) but not the one with only 4-nt complementarity (lanes 5 to 7) or of unrelated sequence. Although identical amounts of rRNA substrate and competitor oligonucleotide were used in the gel shift and pseudouridylase assays, the latter was inhibited less, suggesting that the rRNA-snoRNA interaction was stronger when the snoRNA was present in the context of the snoRNP rather than by itself. Collectively, these data demonstrate that the synthetic rRNA substrates hybridize with their cognate snoRNAs and that this hybridization is required for the in vitro pseudouridylation of their target uridine.
The in vitro assay is generally applicable.
We tested a variety of substrates in our assay. For example, the in vitro activity was independent of the length of the substrate, since uridine 4380 of human rRNA was equally isomerized to Ψ if presented in the context of 77 nt or only 25 nt (Fig. 5A and B, compare substrates 1 and 2). Selection of different target uridines tested the functionality of different snoRNPs. Thus, short substrates with uridines corresponding to the nearby positions 4363 and 3702 in human 28S rRNA were also efficiently pseudouridylated in nuclear lysates (Fig. 5A and B, substrates 3 and 4). The pseudouridylation of these last two sites is guided by the snoRNAs, U65 and MBI-161, whose complementarity to the substrate lies in the bulge of their 5′ and 3′ hairpins, respectively (12, 17). Even substrates 5 and 6 were modified in the assay (Fig. 5A and B), representing uridines 36 of human 18S rRNA and 69 of human 5.8S rRNA, both complementary to the same box H/ACA snoRNA, U69 (12). Thus, in vitro pseudouridylation was catalyzed by guide snoRNAs with complementarity in either and/or both hairpins and modified all three rRNAs. Additionally, the assay worked with different cell extracts from various species: in HeLa nuclear extracts (Fig. 3B and C), in nuclear lysates from human lymphoblasts (not shown), in nuclear lysates from buffalo rat liver cells (Fig. 5B, substrates 1 to 3), in rat liver nucleolar extracts (Fig. 5B, substrates 4 to 6), and in yeast whole-cell lysates (Fig. 5B, substrate 7). In summary, the assay is generally applicable as it occurs on multiple bona fide substrates and in extracts from multiple species.
Finally, we confirmed that the Ψ 3′ monophosphate observed by TLC was generated in the context of the full-length synthetic substrate and did not represent phosphorylation of preexisting single Ψ, e.g., that had been liberated from tRNA in the extracts. For this purpose, rRNA substrate 2 was isolated after incubation in nuclear lysate and analyzed by denaturing urea polyacrylamide electrophoresis (Fig. 5C, top). When the full-length and smaller (degraded) RNA bands were excised by division into equal-size gel slices (Fig. 5C, lane 1, gel slices I to III), digested, and analyzed individually for their Ψ content using TLC, most of the Ψ was found in the full-length or near-full-length RNA (Fig. 5C, bottom). Analysis of substrate 1 after incubation in HeLa nuclear extract produced similar results (not shown). Taken together, the data argue that uridine was faithfully isomerized to Ψ within the context of the full-length rRNA substrate.
Pseudouridylation by purified core snoRNPs is independent of Nopp140 and energy.
Having established faithful in vitro reconstitution of snoRNA-guided rRNA pseudouridylation that fulfills all previously in vivo-demonstrated or postulated criteria for catalysis, we investigated the minimal requirements for the reaction. Nuclei from buffalo rat liver cells were lysed and fractionated into nucleoplasm and nucleoli, and the nucleoli were extracted with 0.5 M sodium chloride, resulting in a nucleolar supernatant and pellet. When the subcellular fractions were analyzed by Western blotting for the presence of the putative rRNA pseudouridylase, NAP57, most NAP57 fractionated with nucleoli and was extracted with 0.5 M salt (Fig. 6A, lane 3). The rRNA pseudouridylase activity cofractionated with NAP57 in the nucleolar salt extract (Fig. 6B, lane 3). We demonstrated previously that anti-NAP57 antibodies specifically precipitate box H/ACA snoRNPs (53). Using these antibodies, box H/ACA snoRNPs were immunoprecipitated from the nucleolar extracts, removing the pseudouridylase activity from the supernatant and recovering it in the immunoprecipitate (Fig. 6C, lanes 2 and 3). The comparable activity of the extracts and the immunoprecipitated snoRNPs bound to protein A-Sepharose via anti-NAP57 antibodies, suggested that the antibodies had no adverse effect on snoRNP activity. This was confirmed by the lack of reduction of pseudouridylase activity of nucleolar extracts when antibodies were added to NAP57 or to any of the other box H/ACA snoRNP core proteins and to Nopp140 (data not shown).
The activity of the immunoprecipitated snoRNPs enabled us to test the functionality of the snoRNPs coprecipitated with anti-Nopp140 antibodies (Fig. 1C and D). Box H/ACA snoRNPs pseudouridylated their rRNA substrate alone and in association with Nopp140 (Fig. 1E, lanes 3 and 4). No activity was observed in the precipitates containing Nopp140 alone (Fig. 1E, lane 3). Although a minor effect of Nopp140 on snoRNP activity cannot be excluded, the increased activity in the absence of Nopp140 (Fig. 1E, compare lane 5 to lane 4) was likely due to the increased amounts of snoRNPs present in that fraction (Fig. 1D, compare the amounts of GAR1 in lanes 4 and 5). Thus, box H/ACA snoRNPs also pseudouridylated rRNA when complexed with Nopp140.
The protein composition of the active NAP57 immunoprecipitates was analyzed by SDS-PAGE and Western blotting. Silver staining revealed NAP57 (above the immunoglobulin heavy chain) but revealed no additional protein band of similar intensity (Fig. 6D, lane 1). Three minor bands corresponded to albumin from the IgG fraction and two unidentified proteins with masses of approximately 60 kDa. These last two proteins could represent NAP65 (Nop5/58p) and NOP56 (Nop56p), which are associated with Cajal body-specific small nuclear RNAs that possess both box H/ACA and box C/D motifs and their corresponding core proteins (7, 19). Apparently, therefore, no protein of slower mobility than NAP57's was present in stoichiometric amounts in the NAP57 precipitates (Fig. 6D).
Faster-migrating proteins were analyzed on a 15% polyacrylamide Tricine gel system (Fig. 6E). Comparison to NAP57 antibodies (lane 2) and to the extract itself (lane 3) revealed the specific bands coprecipitating stoichiometrically with NAP57 as 28-, 23-, and 10-kDa proteins, whereas the remaining bands corresponded to immunoglobulins and contaminating histones (Fig. 6E, lane 4). The specifically precipitated proteins were identified as GAR1, NHP2, and NOP10, respectively, by consecutive probing of a Western blot (Fig. 6E, lane 5). Additionally, a protein band of ∼50 kDa was observed variably in the NAP57 precipitates (Fig. 6E, lane 4). On Western blots of the upper half of the precipitates, anti-NAP57 antibodies specifically recognized, in addition to NAP57, this band and identified it as a NAP57 breakdown product (lane 5, NAP57bkdn). The time-dependent increase of this band during incubation of nucleolar extracts at 37°C (data not shown) and its absence in freshly prepared extracts corroborated this conclusion (e.g., see Fig. 1C, lane 5). The minor precipitation of histones was nonspecific, since they were present even when the precipitation of NAP57 and the other snoRNP core proteins was prevented with competing free NAP57 peptide (not shown). Our data suggest that box H/ACA snoRNPs active in rRNA pseudouridylation are solely composed of the four core proteins, NAP57, GAR1, NHP2, and NOP10, and a box H/ACA snoRNA. Nevertheless, given the presence in our precipitates of ∼100 different box H/ACA snoRNPs, each with a specific snoRNA, we cannot rule out the possibility that certain particles contain additional proteins that are below the detection limit and that contribute to the activity. The fact, however, that the immunoprecipitated particles are active towards five different mammalian rRNA substrates (listed in Fig. 5A; data not shown) suggests that the four core proteins together with the specific snoRNA are sufficient for site-specific pseudouridylation.
This activity was independent of snoRNP phosphorylation, since snoRNPs precipitated from extracts that were phosphatase pretreated or not were comparably active, i.e., 10.4% Ψ and 7.7% Ψ, respectively (see Fig. 2B, lanes 3 and 4). Similarly, direct phosphatase treatment of immunopurified snoRNPs barely affected their pseudouridylase activity, i.e., 19.9% Ψ with treatment and 17.2% Ψ without treatment. These data demonstrated that phosphorylation of box H/ACA snoRNPs was not a major factor in the regulation of their activity.
Using the immunoprecipitated snoRNPs washed with 0.5 M NaCl, 1% Triton X-100, and 0.2% SDS, we determined the factor requirements for snoRNP-mediated pseudouridylation. The standard conditions included 2 mM MgCl2, 100 mM NaCl, 20 mM HEPES (pH 7.5), and 5% glycerol (Fig. 6F, control). Addition of 20 mM EDTA (in the absence of MgCl2) had no effect on the activity, indicating that it was magnesium independent (Fig. 6F, 20 mM EDTA). Omission of NaCl lowered the activity by more than half (Fig. 6F, No NaCl), consistent with a minimal salt requirement to support the snoRNA-rRNA hybridization. However, high magnesium or sodium chloride concentrations abolished the activity (Fig. 6F, 0.1 M MgCl2 and 0.5 M NaCl). Furthermore, snoRNP-mediated pseudouridylation of rRNA was energy independent, since no energy source was added to the washed precipitates. This was supported by the observation that apyrase treatment of nucleolar extracts had no effect on their pseudouridylase activity (not shown) and that the addition of 2 mM creatine phosphate or 1 mM ATP failed to stimulate the activity in nuclear lysates (not shown) or of immunopurified snoRNPs (Fig. 6F, 1 mM ATP). Finally, the lack of an ATP and magnesium requirement extended to the release of the product from the snoRNA, since more than 95% of the Ψ containing product rRNA was routinely harvested from the supernatant of the protein A-Sepharose tethered snoRNPs. In summary, therefore, helicase activity was apparently not involved in the snoRNP-dependent in vitro pseudouridylation cycle, which merely relied on physiological salt conditions but not on other factors.
DISCUSSION
We describe an in vitro assay for snoRNP-mediated pseudouridylation of eukaryotic rRNA. Using this assay, purified mammalian box H/ACA snoRNPs consisting of only the four core proteins, NAP57, GAR1, NHP2, and NOP10, and a snoRNA were apparently sufficient for site-specific pseudouridylation of synthetic rRNA substrates. Except for physiological concentration of salt, no exogenous factors were required for catalysis. The ATP and magnesium independence of this reaction ruled out the need for helicase activity. The box H/ACA snoRNPs associated with phosphorylated but not dephosphorylated Nopp140 and were active in rRNA pseudouridylation independently of their Nopp140 association. These findings are consistent with a phosphorylation-dependent role for Nopp140 in the organization of the hundreds of snoRNPs during the modification of rRNA.
A major challenge in this study was the development of a sensitive method to detect the pseudouridylation in rRNA. Unlike 2′-O-methylation, the other major snoRNP-guided modification of rRNA, pseudouridylation is an isomerization and does not involve addition of a molecule that could be labeled for detection purposes. Several methods have been developed to detect the pseudouridylation of RNA, in particular, the release of 3H from position 5 of the pyridine ring (6), the strong stops generated during reverse transcription of RNA after specific derivatization of Ψs (2), and the separation of Ψ from uridine by TLC after digestion of the RNA to single nucleotides (33). This last method has been used extensively to study in vitro the pseudouridylation of 32P-labeled spliceosomal small nuclear RNAs (snRNAs) (for a review, see reference 39), which appears to depend on at least partial RNP assembly (38, 40). We adapted this assay, which also supports snoRNA-guided pseudouridylation of snRNA (19), for the pseudouridylation of rRNA. To increase the sensitivity and to monitor the isomerization of uridines in different contexts, we site-specifically labeled the target uridine with 32P in short synthetic rRNAs.
The instability of the short rRNA substrates when incubated with whole-cell lysates or subcellular fractions, but not with immunopurified snoRNPs, posed another important problem. While the terminal addition of two deoxynucleotides increased the stability, it did not abolish the degradation of a significant fraction of the substrate rRNAs (e.g., see Fig. 5C). Similarly, the elongation of the rRNA substrate or the addition of a stem-loop consisting of the MS2 binding site (data not shown) helped stabilize the substrate only marginally. This may be caused by the fact that for ease of synthesis the sequence upstream of the target uridine consisted of only 12 nt or less. Therefore, the increased stability of a full-length snRNA, compared to our short synthetic rRNA substrates, may explain its enhanced in vitro pseudouridylation potential (19). It will be interesting to test in the future how a longer, more physiological rRNA substrate will behave in pseudouridylation and, in particular, if its modification will also occur in the absence of any cofactors.
The lack of a cofactor requirement for catalysis by box H/ACA snoRNPs is reminiscent of that for pseudouridylation by TruB, the closest bacterial homolog to NAP57 (26, 35). Catalysis by TruB solely requires the enzyme and the target tRNA, whereas we demonstrated a requirement for hybridization of the rRNA substrate with the box H/ACA snoRNA of the snoRNP. The fact that the isomerized rRNA product can be harvested from the supernatant after incubation with the immunoprecipitated, protein A-Sepharose-bound snoRNPs demonstrates that the product is released after pseudouridylation. Therefore, the snoRNP must allow the energy-independent rRNA-snoRNA association and dissociation, in the case of the E3 snoRNA and its substrate rRNA, a stretch of 4 and 10 consecutive base pairs flanking the target uridine. The lack of a requirement for RNA helicase activity is remarkable considering the intimate protein-RNA contacts predicted between NAP57 and the snoRNA-rRNA three-helix junction that cradles the target uridine, based on the cocrystal structure of TruB with the T stem-loop of tRNA (16). The physiological concentration of salt in the in vitro reaction is likely required to promote the hybridization of the two RNAs, whereas higher salt concentrations may hyperstabilize the snoRNA-rRNA hybrid, preventing its dissociation, and thereby inhibit the reaction. Alternatively, increased salt concentration could lead to minor structural rearrangements within the snoRNP. Regardless, snoRNP-mediated pseudouridylation of synthetic rRNA substrates occurs in the absence of helicase activity in vitro. This is in agreement with the mechanism for box C/D snoRNP-mediated 2′-O-methylation of rRNA in vitro (37).
Genetic and biochemical experiments with yeast have identified Cbf5p/NAP57, GAR1, NHP2, and NOP10 as the four core proteins of box H/ACA snoRNPs (3, 13, 15, 22, 24, 51). These findings were supported by in vitro studies with mammalian cells (9, 41, 53). We now demonstrate that indeed these four proteins alone, together with a box H/ACA snoRNA, form the core snoRNP particles in mammalian cells. Moreover, these core snoRNPs are apparently sufficient for site-specific pseudouridylation of synthetic rRNA substrates in vitro. At present it is not clear if all four proteins are required or if a smaller complex would suffice. Nevertheless, these data provide the first biochemical evidence for NAP57 as the pseudouridylase of rRNA, because this predicted enzyme is one of only four proteins in the minimal active complex.
Pseudouridylase activity could only be partially rescued after nuclease pretreatment of nuclear extracts and addition of the in vitro-transcribed cognate snoRNA (Fig. 4B). A similar lack of stimulation was observed when rescuing the in vitro pseudouridylation of uridine 46 in human U5 snRNA by the addition of U85 snoRNA to nuclease-pretreated extracts (19). This suggested that the digested or nicked snoRNAs in the snoRNPs could not be replaced by an exogenous snoRNA. Alternatively, nuclease treatment could have caused the irreversible disassembly of the snoRNPs. The latter, however, was not the case, since even after micrococcal nuclease treatment, all four snoRNP core proteins could be precipitated with NAP57 antibodies in the presence of 0.5 M salt or 2 M urea (data not shown). The fact that addition of exogenous snoRNA to untreated extracts increased their activity to a similar extent suggested that these extracts contained either free snoRNP proteins or RNA-free snoRNPs. Indeed, this property of nuclear extracts has previously been exploited for the in vitro reconstitution of box H/ACA snoRNPs (9). The similar enhancement of pseudouridylase activity by the addition of in vitro-transcribed snoRNA in extracts and in snoRNPs immunoprecipitated by anti-NAP57 antibodies (data not shown) suggests that the snoRNA associated with RNA-free snoRNPs rather than individual snoRNP proteins. Therefore, the biogenesis of snoRNPs may involve an independent assembly of all or some of the core proteins followed by the integration of the specific snoRNA.
We demonstrate that Nopp140 associates with and dissociates from intact snoRNPs in a salt-dependent manner. At physiological concentration and in the cell, snoRNPs associate with Nopp140, while at increased salt concentrations they dissociate, suggesting an electrostatic interaction between Nopp140 and the snoRNPs. This is not surprising given the high positive and negative charge density on Nopp140. Nopp140 is phosphorylated by casein kinase 2 to the unusually high degree of ∼80 phosphates/molecule (25, 27). Although we showed previously that this phosphorylation was important for the binding of nuclear localization signals in vitro, there is no evidence for a Nopp140-nuclear localization signal interaction in vivo. However, Nopp140 clearly interacts with snoRNPs in vivo, and we now demonstrate that this interaction is dependent on its phosphorylation in vitro. Thus, phosphorylation and dephosphorylation of Nopp140 may govern the association of Nopp140 with snoRNPs in the cell. Nopp140 appears to be predominantly in its fully phosphorylated state in the cell (unpublished data), consistent with its physical association with casein kinase 2 (23). The dissociation of the snoRNPs from Nopp140, therefore, appears to require the action of a phosphatase that remains to be identified.
Although the interaction between Nopp140 and snoRNPs is charge mediated, the specificity of their interaction is striking. Thus, despite the abundance of many highly charged proteins in nucleolar 0.5 M salt extracts, Nopp140 exclusively associates with snoRNPs after dilution of the extracts to 0.1 M salt (Fig. 1C, lane 4). This specificity may be generated by multiple interactions between Nopp140 and the snoRNPs, e.g., the phosphate groups of Nopp140 with the positively charged NAP57 carboxy-terminal tail and the lysine-rich repeats of Nopp140 with the snoRNA. Both interactions would be required for their association, explaining why dephosphorylation alone is sufficient for dissociation. Additionally, such a dual interaction could explain why, despite much effort, no single interaction between Nopp140 and a snoRNP component could be identified, e.g., by using two- or three-hybrid interaction approaches (unpublished results). The fact that binding of Nopp140 to snoRNPs does not interfere with their activity as pseudouridylases indicates that Nopp140 binding and rRNA hybridization to snoRNPs can occur simultaneously. Thus, Nopp140 may aid in the organization of the snoRNPs along the pre-rRNA before and during their function as modifying enzymes. Although it remains to be determined what the stoichiometry of the Nopp140-snoRNP interaction is, the 10 alternating phosphorylated and basic repeats of Nopp140 could serve as a scaffold for several snoRNPs. Taken together, our data point to Nopp140 as an electrostatic chaperone for snoRNPs.
Acknowledgments
We are grateful to the following investigators for reagents and advice: John Aris, Francois Dragon, Witek Filipowicz, Tao Huang, Paul McDermott, Catherine Newnham, Vanda Pogacic, Marvin Wickens, and Yunfeng Yang. Critical reading of the manuscript by Susan Smith and Jon Warner is appreciated.
This study was supported by grants from the American Cancer Society (U.T.M.) and the NIH (C.C.Q.).
REFERENCES
- 1.Aris, J., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakin, A., and J. Ofengand. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32:9754-9762. [DOI] [PubMed] [Google Scholar]
- 3.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, G., and V. Potter. 1966. Nuclei from rat liver: isolation method that combines purity with yield. Science 154:1662-1665. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M. S., H. Sasaki, M. S. Campbell, S. K. Kraeft, R. Sutherland, C. Y. Yang, Y. Liu, D. Auclair, L. Hao, H. Sonoda, L. H. Ferland, and L. B. Chen. 1999. HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation. J. Biol. Chem. 274:36544-36549. [DOI] [PubMed] [Google Scholar]
- 6.Cortese, R., H. O. Kammen, S. J. Spengler, and B. N. Ames. 1974. Biosynthesis of pseudouridine in transfer ribonucleic acid. J. Biol. Chem. 249:1103-1108. [PubMed] [Google Scholar]
- 7.Darzacq, X., B. E. Jady, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, P., R. Reddy, and H. Busch. 1984. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry 23:5421-5425. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 12.Ganot, P., M.-L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 13.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 14.Heiss, N. S., S. W. Knight, T. J. Vulliamy, S. M. Klauck, S. Wiemann, P. J. Mason, A. Poustka, and I. Dokal. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19:32-38. [DOI] [PubMed] [Google Scholar]
- 15.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J. P. Gelugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang, C., and A. R. Ferre-D'Amare. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107:929-939. [DOI] [PubMed] [Google Scholar]
- 17.Hüttenhofer, A., M. Kiefmann, S. Meier-Ewert, J. O'Brien, H. Lehrach, J. P. Bachellerie, and J. Brosius. 2001. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 20:2943-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaac, C., Y. Yang, and U. T. Meier. 1998. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 142:319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jady, B. E., and T. Kiss. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, D., U. T. Meier, G. Dobrowolska, and E. G. Krebs. 1997. Specific interaction between casein kinase 2 and the nucleolar protein Nopp140. J. Biol. Chem. 272:3773-3779. [DOI] [PubMed] [Google Scholar]
- 24.Lübben, B., P. Fabrizio, B. Kastner, and R. Lührmann. 1995. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem. 270:11549-11554. [DOI] [PubMed] [Google Scholar]
- 25.Meier, U. T. 1996. Comparison of the rat nucleolar protein Nopp140 to its yeast homolog SRP40: differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 271:19376-19384. [PubMed] [Google Scholar]
- 26.Meier, U. T., and G. Blobel. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 127:1505-1514. (Author’s correction, 140:447.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier, U. T., and G. Blobel. 1992. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 70:127-138. [DOI] [PubMed] [Google Scholar]
- 28.Meier, U. T., and G. Blobel. 1990. A nuclear localization signal binding protein in the nucleolus. J. Cell Biol. 111:2235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merril, C. R., D. Goldman, and M. L. Van Keuren. 1984. Gel protein stains: silver stain. Methods Enzymol. 104:441-447. [DOI] [PubMed] [Google Scholar]
- 30.Moore, M. J., and C. C. Query. 2000. Joining of RNAs by splinted ligation. Methods Enzymol. 317:109-123. [DOI] [PubMed] [Google Scholar]
- 31.Newnham, C. M., and C. C. Query. 2001. The ATP requirement for U2 snRNP addition is linked to the pre-mRNA region 5′ to the branch site. RNA 7:1298-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura, S. 1972. Minor components in transfer RNA: their characterization, location, and function. Prog. Nucleic Acid Res. Mol. Biol. 12:49-85. [PubMed] [Google Scholar]
- 34.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Luhrmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurse, K., J. Wrzesinski, A. Bakin, B. G. Lane, and J. Ofengand. 1995. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA 1:102-112. [PMC free article] [PubMed] [Google Scholar]
- 36.Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514:17-25. [DOI] [PubMed] [Google Scholar]
- 37.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patton, J. R. 1994. Formation of pseudouridine in U5 small nuclear RNA. Biochemistry 33:10423-10427. [DOI] [PubMed] [Google Scholar]
- 39.Patton, J. R. 1994. Pseudouridine formation in small nuclear RNAs. Biochimie 76:1129-1132. [DOI] [PubMed] [Google Scholar]
- 40.Patton, J. R. 1991. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol. Cell. Biol. 11:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pogacic, V., F. Dragon, and W. Filipowicz. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20:9028-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Query, C. C., M. J. Moore, and P. A. Sharp. 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 8:587-597. [DOI] [PubMed] [Google Scholar]
- 43.Saito, H., T. Fujiwara, S. Shin, K. Okui, and Y. Nakamura. 1996. Cloning and mapping of a human novel cDNA (NHP2L1) that encodes a protein highly homologous to yeast nuclear protein NHP2. Cytogenet. Cell Genet. 72:191-193. [DOI] [PubMed] [Google Scholar]
- 44.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 45.Selvamurugan, N., O. H. Joost, E. S. Haas, J. W. Brown, N. J. Galvin, and G. L. Eliceiri. 1997. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 25:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SenGupta, D. J., B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 48.Sorger, P. K., G. Ammerer, and D. Shore. 1989. Identification and purification of sequence-specific DNA-binding proteins, p. 199-223. In T. E. Creighton (ed.), Protein function: a practical approach. IRL Press, Oxford, United Kingdom.
- 49.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 50.Tyc, K., and J. A. Steitz. 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Lührmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all box H/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrzesinski, J., K. Nurse, A. Bakin, B. G. Lane, and J. Ofengand. 1995. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for Ψ746 in 23S RNA is also specific for Ψ32 in tRNAphe. RNA 1:437-448. [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, Y., C. Isaac, C. Wang, F. Dragon, V. Pogacic, and U. T. Meier. 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 11:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]