Abstract
In response to a number of distinct environmental conditions, the fungal pathogen Candida albicans undergoes a morphological transition from a round, yeast form to a series of elongated, filamentous forms. This transition is believed to be critical for virulence in a mouse model of disseminated candidiasis. Here we describe the characterization of C. albicans ASH1, a gene that encodes an asymmetrically localized transcriptional regulatory protein involved in this response. We show that C. albicans ash1 mutants are defective in responding to some filament-inducing conditions. We also show that Ash1p is preferentially localized to daughter cell nuclei in the budding-yeast form of C. albicans cell growth and to the hyphal tip cells in growing filaments. Thus, Ash1p “marks” newly formed cells and presumably directs a specialized transcriptional program in these cells. Finally, we show that ASH1 is required for full virulence of C. albicans in a mouse model of disseminated candidiasis.
Candida albicans is a common fungal pathogen that causes mucosal infections in healthy individuals and can cause life-threatening disseminated infections in immunocompromised patients. C. albicans is capable of colonizing most tissue types in humans and is therefore able to adapt and thrive in the diverse microenvironments encountered in a host (7, 36). C. albicans responds to changes in its environment by altering its patterns of gene expression, and these responses appear critical to the survival and virulence of this opportunistic pathogen. In the laboratory, C. albicans responds to changes in growth conditions, including starvation, 37°C temperature, neutral pH, exposure to serum, contact with animal cells, or the presence of compounds such as proline and N-acetylglucosamine by switching from a round, single-celled budding-yeast form to elongated filamentous forms. These filamentous forms include a spectrum of morphologies that range from pseudohyphae (chains of elongated cells that remain attached after cell division) to true hyphae (long cylindrical cells separated by septal walls that lack constrictions at sites of cell division) (35). Current evidence is consistent with the idea that the interconversion of these forms is critical for virulence of C. albicans (for reviews, see references 6, 10, 11, 20, 22, 33, 44, and 47). For example, all of the morphological forms are found in infected tissues, and mutations that lock C. albicans into either the yeast or filamentous form produce mutants with significantly reduced virulence when tested in mouse models of disseminated candidiasis. Additional links between filamentous growth and virulence come from studies showing the differential expression of certain cell surface and secreted proteins in filamentous cells compared to budding yeast cells; for example, newly formed filaments adhere better to mammalian cells than do yeast-form cells. It may be that increased adherence and invasion are important for early stages of C. albicans infection; the ability to form yeast cells that bud off from adherent hyphae may subsequently promote the colonization of diverse tissues during disseminated infection (36).
Filamentous growth is common to many species of fungi. For example, Saccharomyces cerevisiae undergoes a process of pseudohyphal growth that is similar in some respects to filamentous growth of C. albicans (17; for reviews, see references 28 and 31). In particular, diploid S. cerevisiae cells grow, in response to nitrogen starvation, as elongated chains of pseudohyphal cells that extend as filaments into solid media. Although S. cerevisiae and C. albicans are closely related evolutionarily, there are important differences between the types of filamentous growth in these two organisms. For example, C. albicans filaments are induced in response to several growth conditions (such as 37°C temperature and exposure to serum) that do not critically affect filamentous growth of S. cerevisiae. Moreover, both S. cerevisiae and C. albicans produce pseudohyphae, yet only C. albicans can make true hyphae, defined by the absence of constrictions at the sites of cell division and by a particular pattern of mitosis and cell division (45). Despite these differences, many of the components that control filamentous growth—particularly those in signaling pathways—are conserved between S. cerevisiae and C. albicans (for example, see references 2, 9, 12, 16, 25, 26, 29, 30, and 40).
The S. cerevisiae Ash1 protein (Ash1p) was originally isolated in screens designed to identify proteins involved in mating-type switching in haploid yeast cells (1, 43). Later, it was discovered that Ash1p is also required for pseudohyphal growth in S. cerevisiae (8). With regards to pseudohyphal growth, Ash1p has been linked to a transcriptional regulatory cascade that, in response to nitrogen starvation, activates expression of the FLO11 gene (37); FLO11 encodes a cell surface protein that is required for pseudohyphal growth. For these reasons, we hypothesized that an ASH1-related gene in C. albicans has a role in filamentous growth, and in this paper, we test this idea.
First, we describe the isolation and characterization of a C. albicans gene that is homologous to the Ash1p gene of S. cerevisiae. C. albicans Ash1p can complement an S. cerevisiae ash1 mutant, indicating that C. albicans Ash1p has biochemical activities (for example, its ability to function as a transcriptional repressor) (32) similar to those of S. cerevisiae Ash1p. C. albicans strains with ASH1 deleted show defects in filamentous growth in vitro and have reduced virulence in a mouse model of systemic candidiasis. We also show than when C. albicans proliferates in the budding-yeast form, Ash1p is asymmetrically localized to the daughter cell nuclei following each cell division, a pattern previously observed for Ash1p from S. cerevisiae (1, 43). When C. albicans enters the hyphal form of growth, Ash1p is localized preferentially to the nuclei of the hyphal tip cells: that is, to the nuclei of only the most recently formed hyphal cells. Given that Ash1p is a transcriptional regulator, it seems likely that its presence in hyphal tip cells endows them with a specialized transcriptional program. Indeed, a number of observations in the literature indicate that C. albicans hyphal tip cells have properties distinct from those cells that make up the internal (subapical) portions of the hyphae. For example, hyphal tip cells are known to selectively secrete phospholipase B (reviewed in references 14 and 21); it has been proposed that the secretion of this and other hydrolytic enzymes destroys the material in front of the hyphal tip cell and thereby enables it to invade host tissues, as reviewed in reference 18. The hyphal tip cells of the filamentous fungus Aspergillus niger are also known to have specialized properties; for example, the enzyme glucomylase is selectively secreted from the tips of mycelia (46). In C. albicans, we propose that Ash1p directs, at least in part, the specialization of the hyphal tip cells.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. G. Fink and colleagues generously provided the cph1/cph1 strain, JKC19 (29). The S. cerevisiae strains YAS242-10B, YAS243-1A, and YAS204 were obtained from A. Sil and colleagues (University of California—San Francisco [UCSF]; unpublished data). Strains YAS242-10B and YAS243-1A have the ADE2 gene integrated at the HO locus and the HO promoter integrated upstream of the CAN1 locus. Both of these promoter-gene fusions cause reporter gene expression to be controlled by the HO promoter. Deletion of S. cerevisiae ASH1 has been described previously (43). The strains CGX69, YAS204, and YDI-56 are of the S. cerevisiae Σ1278b background (16). YDI-56 is a derivative of YAS204 (ash1::HisG-URA3-HisG/ash1::HisG-URA3-HisG, ura3Δ/ura3Δ), which was selected for Ura auxotrophy on plates containing 5-fluoroorotic acid (5-FOA).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| C. albicansa | ||
| CAF2-1 | URA3/ura3::imm434 | 13 |
| CAI4 | ura3Δ/ura3Δ | 13 |
| RM1000 | ura3Δ/ura3Δ his1::HisG/his1::HisG | 34 |
| YDI-1 | ura3Δ/ura3Δ ASH1/ash1::HisG-URA3-HisG | This study |
| YDI-7 | ura3Δ/ura3Δ ash1::HisG/ash1::HisG-URA3-HisG | This study |
| YDI-11 | ura3Δ/ura3Δ ash1::HisG/ash1::HisG | This study |
| YDI-129 | ura3Δ/ura3Δ cph1::HisG/cph1::HisG ash1::HisG/ash1::HisG-URA3-HisG | This study |
| YDI-154 | ura3Δ/ura3Δ ash1::HisG/ash1::HisG::ASH1-URA3 (2d) | This study |
| YDI-157 | ura3Δ/ura3Δ ash1::HisG/ash1::HisG::ASH1-URA3 (6d) | This study |
| YDI-199 | ura3Δ/ura3Δ ash1::HisG/ash1::HisG-URA3-HisG::myc6-ASH1 | This study |
| JKC19 | ura3Δ/ura3Δ cph1::HisG/cph1::HisG-URA3-HisG | 29 |
| S. cerevisiaeb | ||
| CGX69 | MATa/MATα ura3/ura3 | 17 |
| YDI-56 | MATa/MATα ura3/ura3 ash1::HisG/ash1::HisG | This study |
| YAS242-10B | MATα ASH1 can1::HO-CAN1 ho::HO-ADE2 ade2-1 his3-11,15 trp1-1 ura3 leu2-3,112 | Sil, unpublished |
| YAS234-1A | MATα ash1::TRP1 can1::HO-CAN1 ho::HO-ADE2 ade2-1 his3-11,15 trp1-1 ura3 leu2-3,112 | Sil, unpublished |
All C. albicans strains listed as ura3Δ/ura3Δ are homozygous for the ura3::lmm434 mutation, except for CAF2-1, which is heterozygous for the mutation.
CGX69 and YDI-56 are of the Σ1278b strain background; YAS242-10B and YAS243-1A are of the W303 strain background.
S. cerevisiae strains were transformed by lithium acetate transformation (15) with selection on SD −Leu or SD −Ura medium. C. albicans transformations were performed by the lithium acetate method described in reference 4. Strains were routinely grown on YPD plates and liquid medium at 30°C or on SD −Ura medium unless otherwise noted. Counterselections against the C. albicans URA3 gene were performed by overnight growth at 30°C in YPD liquid medium followed by selection on 5-FOA and uridine.
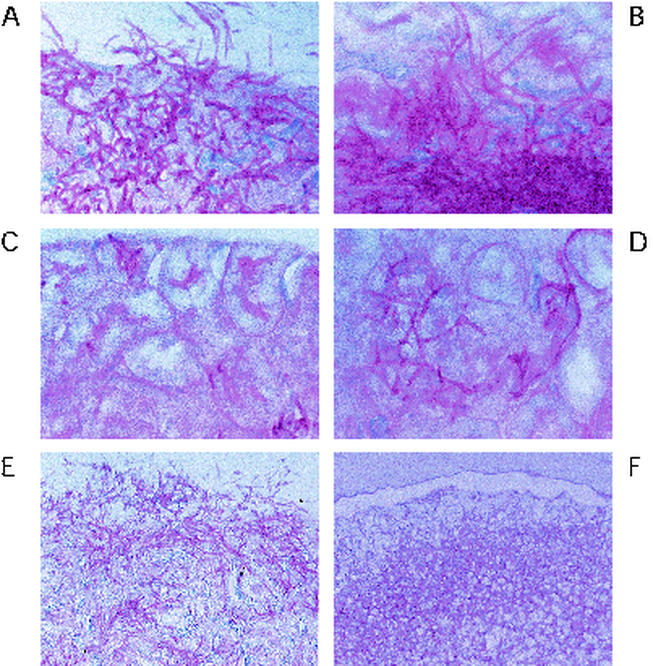
Phenotype testing and induction of C. albicans filamentous growth were done with solid Spider medium as described previously (29); with Lee's medium as described previously (27), at pH 6.8 with the addition of 2% agar; and with YPD with 10% fetal calf serum. Either cells were streaked onto plates, or overnight liquid cultures were diluted in water or YEP medium and plated for single colonies. Plates were incubated for 5 to 6 days at 30°C in plastic sleeves and photographed at a ×7.5 magnification under a Nikon SMZ-U microscope (Fig. 3A) or a Leica M420 microscope (Fig. 3B and C). Sabouraud dextrose agar (Difco), used for tissue fungal burden assays, was prepared according to the manufacturer's directions.
FIG. 3.
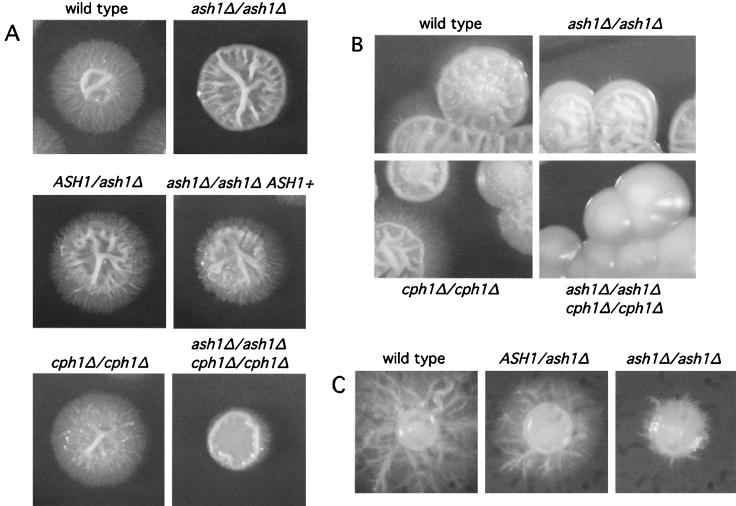
ASH1 is required for filamentous growth of C. albicans. Heterozygous or homozygous strains of the indicated genotypes were grown on various types of filament-inducing solid media: Spider medium (A), YPD medium plus 10% serum (B), and Lee's medium (pH 6.8) with 2% agar (C). Plates were incubated for 5 to 6 days at 30°C. In panel A, the strain depicted in the panel in the second row of the second column is an ash1/ash1 strain into which an intact copy of ASH1 has been introduced.
Cloning the C. albicans ASH1 gene.
The S. cerevisiae Ash1p sequence (GenBank accession no. CAA82028.1) was used in a tBlastn search of the Candida albicans sequence database (http://alces.med.umn.edu/gbsearch/ybc.html), which yielded three overlapping sequence tags with 63% identity to the C terminus of S. cerevisiae Ash1p. With these sequence data, the oligonucleotides DI-1FW (5′-TCTACACACAAATTCATTCC-3′) and DI-2RV (5′-ATTTAGGAAGTACTTCAA CT-3′) (Operon) were designed and used to amplify a 357-bp fragment from C. albicans SC5314 genomic DNA. The resulting PCR fragment was gel purified (Qiaquick gel extraction kit), labeled with 32P in a random prime reaction (Amersham), and used to probe a size-selected C. albicans genomic library made in the Lambda ZapII vector (kindly provided by B. Braun; www.sacs.ucsf.edu/home/JohnsonLab/). Eleven strongly hybridizing clones were obtained. Eight clones were sequenced through the open reading frame (ORF)-containing region and through the vector junctions. Six of these clones contained a full-length ASH1 ORF, and two were N-terminal truncations. One full-length clone, pDI-16, was sequenced to completion and found to contain 1.3 kb upstream and 335 bp downstream, including part of another ORF, DSK2, which begins 185 bp after the stop codon of ASH1. Alignment of the Ash1p proteins was done with GCG Pileup program (Wisconsin Package, version 8.0; Genetics Computer Group, Madison, Wis.) and presented for viewing with Seqvu 1.1 (Garvan Institute of Medical Research, Sydney, Australia).
Expression of C. albicans ASH1 in S. cerevisiae.
For expression studies, the C. albicans ASHI ORF was amplified by PCR with Pfu Turbo polymerase (Stratagene) by using primers DI-5FW (5′-CAAAACTACCGTGATACAC-3′) (Operon) and DI-25RV (5′-GGATCCGAGCTCATTGTTGATTATTCGGTATAGAG-3′) (UCSF BRC) with plasmid pDI-16 as the template. A 2.6-kb fragment that includes sequence 1 kb upstream and 185 bp downstream of ASH1 was cloned into the vector pRS425 (42) as an XhoI-SacI fragment to construct the LEU2-marked plasmid pDI-24. This PCR-generated insert was sequenced to verify its accuracy. The XhoI-SacI fragment was digested from pDI-24 and cloned into pRS426 to create the URA3-marked plasmid pDI-25. The S. cerevisiae ASH1 expression plasmid, pAS174, was described previously (43). pAS199 (A. Sil, unpublished observations) contains the same S. cerevisiae ASH1 fragment as pAS174, except pRS425 (LEU2) is the vector backbone. For the HO repression assays, strains were streaked onto SD −Leu plates containing 10 mg of adenine per ml and 0.03% canavanine (1), incubated for 5 days at 30°C, and then transferred to 4°C for 3 days to enhance color differences between Ade+ and Ade− strains before photographing. Pseudohyphal growth assays were performed by incubating cells at 30°C for 5 days on SLAD medium containing 50 μM ammonium sulfate (17). Photographs were taken at a magnification of ×38 on a Nikon SMZ-U microscope.
Disruption of the C. albicans ASH1 gene.
To disrupt the C. albicans ASH1 gene, a modified Ura blaster method was used (5, 13). Heterozygous strains were first constructed by replacing ASH1 coding sequences with the URA3 selectable marker flanked by HisG repeats. Ura+ strains that were deleted for one copy of ASH1 were grown on nonselective medium and plated on 5-FOA and uridine medium to select for loss of the URA3 gene. Two independent Ura− ASH1 heterozygous strains were used to disrupt the second allele of ASH1 with another HisG-URA3-HisG cassette, pDI-23. Sequences from the 5′ and 3′ regions of ASH1 were amplified by PCR, digested with restriction enzymes, and cloned into the disruption vector pBB510, a derivative of pMB7 (13) described in reference 5, to create the disruption plasmid pDI-03. The 5′ flank was amplified with primers 5′ KO-HIII (5′-TACATTAAGCTTCGTGCTGGTCATTACAGCC-3′) and 3′ KO-PstI (5′-ATGTAACTGCAGTTCGGAGTTTGGTTGTAGG-3′), digested with HindIII and PstI, and then cloned into the HindIII-PstI sites of pBB510 to create the cloning intermediate p510-5′. The 3′ flank was amplified with the primers 5′ KO2-NsiI (5′-ATTGAAAATGCATAGCTAAATAACCATCATCATCACGCACC-3′) and 3′ KO2-KpnI (5′-AATACTGGTACCAACTCAAGATTTAGGAAGT-3′), digested with NsiI and KpnI, and cloned into the NsiI-KpnI sites of p510-5′. The resulting plasmid, pDI-03, contains the HisG-URA3-HisG disruption cassette flanked by sequences in the ASH1 promoter and within the 3′ end of ASH1. To avoid disruption of DSK2 promoter sequences at the 3′ untranslated region (UTR) of ASH1, the 3′ flank contains ORF sequence from the 3′ end of ASH1. A second disruption plasmid, pDI-23, was made by digesting pDI-03 with BamHI and BglII to release the HisG-URA3-HisG cassette, which was then gel purified and cloned back into the pDI-03 backbone in the reverse orientation.
Strains with ASH1 deleted were created by homologous integration of a HindIII-XmnI fragment from pDI-03 (to disrupt the first allele) or pDI-23 (to disrupt the second allele). Isolates were screened by PCR and by Southern blotting for the presence of both disruption constructs at the ASH1 locus and for the absence of the disrupted ORF sequence (data not shown). Four independent isolates homozygous for deletions in ASH1 were obtained by this method. Because ash1 mutants obtained by the Ura blaster method retained 93 codons (although no in-frame start codons) at the C terminus, a fifth isolate with the complete ASH1 coding sequence deleted was obtained by the PCR product method (48, 49) in the Ura− His− strain RM1000 (34). The primers DI-55DR (5′-CCGAAGAACCTAAAAAAAAAAAGTAGTCAACATTGTCGAAGCTACCAAATAACCACAGTAGTTTTCCCAGTCACGACGTT-3′) and DI-56DR (5′-TAACAGATATCTAATCCTATATAAATGAAGCTTCCTTTACAATACTTTTCTAAACTCAAGTGTGGAATTGTGAGCGGATA-3′) (vector hybridizing sequence is underlined) were used in separate PCRs with the templates pGEM-HIS1 and pDDB57 to generate the PCR disruption products. Whole-cell PCR, using primers internal to the HIS1 (5′HIS1-RV, 5′-TTGACTATACCTTCGCTGTC-3′; 3′ HIS1-FW, 5′-GCAATAAACCCCTTGTGGAC-3′) or URA3 (5′URA3-RV, 5′-TGGTGAGGCATGAGTTTC-3′; 3′URA3-FW, 5′-GAGATGCTGGTTGGAATGC-3′) selectable markers and outside the flanking region of homology to the ASH1 locus (upstream DI-8-FW, 5′-TCAAGACAAATCACAATTCC-3′; downstream DI-9RV, 5′-ATCCTTCAACACCTTTCC-3′), was used to identify an ash1::URA3/ash1::HIS1 isolate. The phenotype of this strain, with a complete deletion of the ASH1 ORF, was identical to those of ash1::HisG/ash1::His-URA3-HisG strains obtained by the URA blaster method.
Reintroduction of the wild-type ASH1 gene into C. albicans ash1/ash1 mutants.
A KpnI-SacI fragment from pDI-25 was cloned into the KpnI-SacI sites of pDI-26 (D. Inglis, unpublished), a derivative of the maltose-inducible expression plasmid pAU15 (kindly provided by M. A. Uhl), to create the reintegration construct, pDI-29, which expresses ASH1 under the control of its own promoter. pDI-29 was linearized with PacI and transformed into strain YDI-11 (ura3Δ/ura3Δ, ash1::HisG/ash1::HisG). Ura+ transformants were screened by whole-cell PCR with primers DI-20FW (5′-CTGATTTAGTCTACACTACCCAC-3′) and DI-46RV (5′-AGATCTAGTGTGTGAAGGG-3′) (Operon) to verify correct insertion of ASH1 at the 5′ region of ash1::HisG. Forty-five Ura+ transformants that yielded positive results by PCR were tested on Spider plates to compare the filamentous growth of these strains with that of the ash1/ash1 parent strain and the ASH1/ash1 heterozygote, which also carries one copy of ASH1. Surprisingly, the integration of wild-type ASH1 with plasmid pDI-29 failed to restore filamentous growth to all but 2 of the 45 transformants that screened positive by PCR for correct integration of the plasmid. Of these two transformant strains, one (YDI-154) was slightly more filamentous than the wild-type CAF2-1 strain on Spider medium (data not shown). The other transformant (YDI-157) produced filaments similar to those of the ASH1/ash1 heterozygote (YDI-1) and was also restored for virulence comparable to that of YDI-1.
Localization studies of Ash1p.
A PCR fragment generated by using Pfu Turbo polymerase (Stratagene) and containing a multimerized epitope of myc (Glu-Gln-Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu-Asn), flanked by BglII restriction sites, was cloned into the BglII site 8 amino acids downstream of the third potential start methionine at the N terminus of the ASH1 coding sequence of the 2μm CaASH1 plasmid, pDI-25. The insertion was sequenced for accuracy, and the plasmid complemented the pseudohyphal growth defects of the S. cerevisiae ash1/ash1 mutant YDI-56. An XhoI-HindIII fragment containing myc-ASH1 and 1 kb of the C. albicans ASH1 promoter sequence from pDI-25 was cloned into the XhoI-HindIII sites of pDI-29 to generate the plasmid pDI-30, which is identical to the reintegration construct pDI-29, except for the sequence encoding the myc epitope. pDI-30 was digested with PacI and integrated into the ASH1 promoter region of YDI-11 to generate the myc-tagged ASH1 strain YDI-199.
For cells in the budding-yeast form of growth, cultures were grown to mid-log phase (optical density at 600 nm [OD600] of ∼ 6) in M199 culture medium (pH 4.5) at 23, 25, and 30°C or in YPD at 25 or 30°C. Chains of connected budding yeast cells (pseudohyphae) were produced by growth in M199 (pH 7.0) at 23°C. Hyphal forms of C. albicans were induced by growing late-log (OD600 of 20) to stationary-phase cultures (OD600 of 40) and diluting cells to OD600 of 0.8 to 1.0 in prewarmed YPD with 10% serum at 30, 35, and 37°C or in YPD at 35°C with 20% serum in a shaking water bath. Cells were harvested by sterile pouring or by transfer with a wide-bore pipette tip into a 15-ml conical tube and fixed at room temperature in 4.5% formaldehyde for 1 h. Cells were prepared for antibody hybridization and microscopy by methods described in reference 43 with modifications for C. albicans based on reference 45. Cells were resuspended in a total volume of 0.5 ml in SP (1.2 M sorbitol, 0.1 M potassium phosphate) buffer with 1 to 2 μl of β-mercaptoethanol and 40 μl of Zymolase-20T (ICN Pharmaceuticals) and incubated at 37°C with gentle shaking for a total of 10 to 15 min for yeast-form cells and 15 to 20 min for hyphal cells. Sixteen microliters of cells was transferred to a polylysine-coated slide well, and cell wall digestion was continued for 10 min at room temperature, while the cells settled onto the slide wells. Excess solution was gently aspirated from the wells, and the cells were allowed to dry. To flatten cells, slides were submerged in −20°C methanol for 5 min, followed by −20°C acetone for 30 s, and then dried completely. The 9E10 myc monoclonal antibody (a gift from Joachim Li, UCSF) was used at a 1:6,000 dilution for the yeast and hyphal experiments shown. Previous batches of 9E10 from the same source have been used at 1:300 for yeast-form cells and at 1:800 for hyphal cells. A Cy3-conjugated goat anti-mouse antibody (obtained from Jackson ImmunoResearch Laboratories, West Grove, Pa.) was used at a 1:200 dilution to detect the myc epitope. A rabbit polyclonal antibody (38) raised against S. cerevisiae Tup1p cross-reacts with C. albicans Tup1p and produces a staining pattern that is tightly associated with the nucleus in both mother and daughter cells of C. albicans yeast and filamentous forms. A purified version of this antibody (S. Green, unpublished observations) was used at 1:400 as a positive control for nuclear stain and detected with a 1:200 dilution of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody (SC2012; Santa Cruz Biotechnology). Cells were blocked at room temperature for 1 to 2 h in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA) and 0.1% Tween 20. Sixteen microliters of the 9E10 and anti-Tup1p primary antibodies (diluted in PBS with 1% BSA) was hybridized to cells for 1 h at room temperature. Sixteen microliters of secondary antibodies was hybridized to cells in the dark for 1 h. Excess primary and secondary antibodies were removed by washing the slide wells four times with PBS. To visualize cell nuclei, cells were stained after the first secondary antibody wash by applying 16 μl of 1-μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (diluted in PBS) to slide wells for 2 min. Slide wells were then washed four times with PBS and aspirated before the slides were treated with Fluormount-G (Southern Biotechnology Associates, Inc.). Digital images were captured by a charge-coupled device (CCD) camera on a Leica DMLB microscope through either a ×100 oil objective lens for yeast cell images or through a ×40 objective lens for hyphal cell images.
Virulence studies and histological sections.
Groups of six female (18 to 20 g) BALB/c mice (Charles Rivers) were each injected by tail vein with C. albicans cells and monitored daily for survival and other signs of infection. Cells were prepared for injection by diluting overnight cultures (grown at 30°C in liquid YPD) into fresh YPD to an OD600 of 0.1 to 0.2, growing the cells for approximately 4 h, and then washing and resuspending the cells in sterile saline (0.9% NaCl2) and counting them with a hemacytometer. A total of 106 cells per mouse (0.5 ml of 2 × 106 cells per ml of solution) were injected by tail vein. Animals were sacrificed by CO2 and handled according to UCSF Committee on Animal Research (CAR) guidelines.
Whole kidneys were dissected from infected mice and prepared for histological staining (41) by being fixed overnight in 4% neutral buffered formalin (NBF), followed by serial dehydration in alcohol and then toluene. Samples were embedded in paraffin, cut into 5-μm sections, and stained with a periodic acid-Schiff base (Sigma Diagnostics), followed by a light green counterstain (Harleco). Images were photographed through a Leica DMLB microscope. Colony counts were obtained by removing kidneys from infected mice, weighing them, and homogenizing the tissue in 1 to 3 ml of sterile distilled water. Serial dilutions were plated onto Sabouraud dextrose agar, and plates were incubated at 30°C for 1 to 2 days before CFU were counted.
Nucleotide sequence accession number.
The complete C. albicans ASH1 sequence has been submitted to the GenBank database under accession no. AF237674.
RESULTS
Identification and sequence of C. albicans ASH1.
To identify an ASH1 homolog in C. albicans, we searched the partial C. albicans sequence database available in July 1998 (http://alces.med.umn.edu/gbsearch/ybc.html) for sequence traces similar to those of S. cerevisiae Ash1p. A 577-nucleotide sequence was identified that, when translated, matched the zinc finger region located in the C terminus of S. cerevisiae Ash1p. Oligonucleotides were designed to PCR amplify a 358-nucleotide-pair fragment from genomic DNA, and this PCR product was used to probe a lambda library containing C. albicans genomic fragments (5). Several full-length clones were obtained and sequenced, and the complete C. albicans ASH1 sequence was submitted to the GenBank database (accession no. AF237674). This sequence is identical to that now found in assembly 6 of the C. albicans genome sequence provided by the Stanford DNA Sequencing and Technology web site (http://www-sequence.stanford.edu/group/candida/).
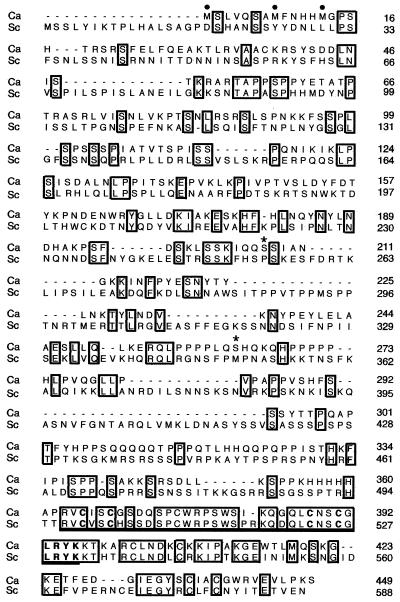
C. albicans ASH1 is predicted to encode a 449-amino-acid protein with 36% identity overall to the S. cerevisiae protein (Fig. 1). The zinc finger region is 81% identical over 34 amino acids. The N-terminal regions are much less similar, with only 23% identity over 359 amino acids. Like the S. cerevisiae protein, C albicans Ash1p has three potential consensus sites for phosphorylation by Cdc28/CDK (1) and two consensus sites in the zinc finger for acetylation, a modification that alters the activity of the related GATA-1 transcriptional regulator in mammals (3).
FIG. 1.
Sequence alignment of the predicted amino acid sequences from C. albicans and S. cerevisiae Ash1p. Boxes indicate identical residues. Three possible start codons for C. albicans Ash1p are indicated by dots. Two predicted serine residues encoded by nonstandard CUG codons of C. albicans (39) are indicated by asterisks. The highly conserved GATA-like zinc finger region is underlined. Amino acid numbers are indicated on the right.
C. albicans Ash1p can regulate expression of the HO promoter and promote pseudohyphal growth in S. cerevisiae.
As reviewed in the introduction, S. cerevisiae Ash1p regulates both expression of the HO gene (which encodes the endonuclease that initiates mating-type switching) (1, 43) and pseudohyphal growth under conditions of nitrogen starvation (8). To determine whether C. albicans Ash1p is a functional homolog of the S. cerevisiae protein in both respects, we asked whether C. albicans Ash1p could regulate (in this case, repress) expression of HO and promote pseudohyphal growth in S. cerevisiae ash1 mutant strains. For these experiments, C. albicans ASH1 and S. cerevisiae ASH1 were each cloned into a high-copy-number 2μm vector and transformed into S. cerevisiae ash1 mutant strains.
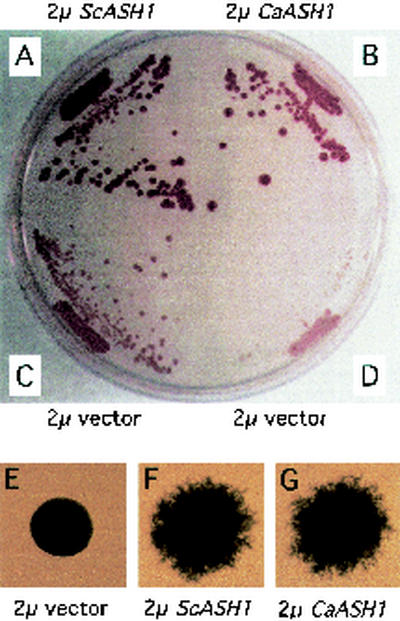
To detect HO repression, we used a reporter strain with ASH1 deleted and which carries the CAN1 and ADE2 reporter genes, each under the control of the HO promoter (Fig. 2A, B, and D). A control strain (Fig. 2C) carries both reporter constructs, the wild-type genomic copy of ASH1, and a vector control plasmid. These strains allow growth of colonies on medium that contains canavanine only when HO is repressed, because expression of CAN1 is lethal in the presence of canavanine. When ASH1 is deleted from this strain, colonies fail to grow in the presence of canavanine, indicating that HO-CAN1 is expressed (Fig. 2D). The HO-ADE2 gene provides a second test of HO expression: expression of HO-ADE2 produces white colonies in low-adenine medium, whereas repression of HO-ADE2 results in red colonies. When the C. albicans ASH1 plasmid was introduced into S. cerevisiae ash1 strains, healthy red colonies grew on low-adenine medium containing canavanine (Fig. 2A and B), indicating that C. albicans ASH1 can repress both the HO-CAN1 and HO-ADE2 reporter genes.
FIG. 2.
C. albicans Ash1p represses HO and promotes pseudohyphal growth in S. cerevisiae ash1 mutant strains. The HO-ADE2 HO-CAN1 ash1 reporter strain produces red colonies on 0.03% canavanine and 10 mg of adenine per ml of SD −Leu medium when transformed with a high-copy-number plasmid that carries either S. cerevisiae (A) or C. albicans (B) ASH1. The same strain transformed with vector (D) fails to grow on this medium; the few surviving colonies are white, indicating HO-ADE2 is not repressed. The HO-ADE2 HO-CAN1 ASH1 control strain (C) has the genomic copy of ASH1 intact and contains the vector control plasmid. Strains were incubated for 5 days at 30°C. For the experiments shown in panels E to G, diploid ash1/ash1 strains of the Σ1278b background were transformed with the indicated plasmids and then incubated on nitrogen-limiting SLAD medium for 5 days at 30°C. Colonies were photographed at a magnification of ×38.
To test whether C. albicans ASH1 can stimulate pseudohyphal growth in S. cerevisiae, ash1/ash1 strains of the S. cerevisiae Σ1278b background (17) were transformed with plasmids expressing either the C. albicans or the S. cerevisiae ASH1 gene and grown on low-nitrogen medium. We found that C. albicans ASH1 restored pseudohyphal growth as efficiently to the S. cerevisiae ash1/ash1 strain as did the S. cerevisiae ASH1 (Fig. 2F and G). Based on these results, we believe that C. albicans Ash1p is a functional homolog of S. cerevisiae Ash1p in that it can replace the function of Ash1p in S. cerevisiae both for repression of HO and for promotion of pseudohyphal growth.
C. albicans ash1/ash1 mutants have defects in filamentous growth.
C. albicans is diploid, and to determine whether ASH1 has a role in regulating filamentous growth in C. albicans, strains lacking both copies of ASH1 were constructed in the SC5314 background by replacing coding sequences with the URA3 selectable marker (13). ASH1/ASH1 (wild type), ASH1/ash1, and ash1/ash1 strains were tested on several types of solid media that induce filament formation. Both ash1 heterozygous and homozygous mutants show significantly reduced filamentous growth on Spider medium (Fig. 3A). The reintroduction of one copy of wild-type ASH1 to the ash1/ash1 strain partially restores filamentous growth (Fig. 3A), indicating that the original filamentous growth defects are attributable to the loss of ASH1. On YPD with 10% serum at 30°C, ash1/ash1 homozygous mutants show a slight reduction in hyphal growth compared with the parental strain (Fig. 3B). ash1/ash1 mutants showed no obvious defects in forming germ tubes, regarded as precursors to hyphal growth, in response to serum in liquid media (data not shown). On solid Lee's medium at neutral pH, ash1/ash1 strains were severely reduced for filamentous growth (Fig. 3C). These results show that ASH1 is important for filamentous growth under several specific environmental conditions.
Analysis of ash1 cph1 double mutants.
In S. cerevisiae, Ash1p and Ste12p regulate parallel but independent pathways of filamentous growth in response to Ras2p signaling. To determine the relationship between C. albicans ASH1 and the C. albicans Ste12p homolog, CPH1, C. albicans strains lacking both ASH1 and CPH1 were constructed by the method described above. On Spider medium, the ash1/ash1 cph1/cph1 double-mutant strain exhibited less filamentous growth overall than did either of the single-mutant strains, although the double mutant did produce low levels of peripheral hyphae (Fig. 3A). On YPD with 10% serum at 30°C, each of the single mutants formed filamentous colonies, whereas the ash1/ash1 cph1/cph1 strain formed round, smooth afilamentous colonies under the same conditions (Fig. 3B). These data suggest that Ash1p and Cph1p contribute additively to filamentous growth in C. albicans.
Ash1p is asymmetrically localized to daughter cells.
When S. cerevisiae proliferates by budding, Ash1p is asymmetrically localized to daughter cell nuclei, where it represses transcription of the HO gene (1, 43). To test whether this asymmetric localization also occurs in C. albicans, Ash1p was tagged at its N-terminal end with six tandem copies of the 11-amino-acid myc epitope (23). This altered form of Ash1p complements the filamentous growth defect when reintegrated into the C. albicans ash1/ash1 strain, YDI-11; moreover, C. albicans myc6-Ash1p successfully promotes pseudohyphal growth when introduced into the S. cerevisiae ash1 mutant strain, YDI-56 (data not shown).
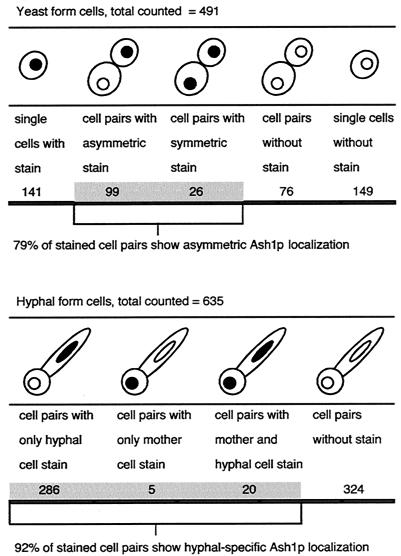
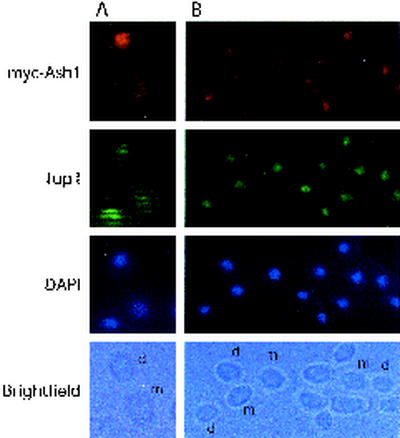
To localize Ash1p, a strain containing one copy of myc6-ASH1 as the sole source of Ash1p was stained with monoclonal antibodies against the myc epitope and visualized by indirect immunofluorescence (23). When C. albicans was grown as a budding-yeast form (in M199 [pH 4.5] and YPD media at 23 and 30°C, respectively), Ash1p was specifically localized to daughter cell nuclei: of 125 postanaphase cell pairs stained for Ash1, 79% showed localization of Ash1p to the nucleus of the daughter cell, but not to the nucleus of the mother cell (Fig. 4 and 5A). C. albicans Ash1p was not observed in mother-daughter cell pairs undergoing mitosis (not shown), as is also the case for S. cerevisiae (1, 43). C. albicans yeast-form cells that are exposed to mild filament-inducing conditions bud in a unipolar fashion, similar to pseudohyphal cells of S. cerevisiae, and produce chains of cells. Under these conditions, C. albicans Ash1p is also localized to daughter cells, seen as cells that bud from the growing chain (Fig. 5B). This pattern of Ash1p localization is similar to that seen in S. cerevisiae pseudohyphal cells (8).
FIG. 4.
Summary of the characteristics of the yeast- and hyphal-form cells examined in this study.
FIG. 5.
Ash1p is localized to daughter cells of C. albicans growing in the yeast form. (A) Cells expressing myc-Ash1p (YDI-199) were grown in M199 (pH 4.5) at 23°C (conditions that favor the budding-yeast form) and processed for indirect immunofluorescence (see Materials and Methods). Cells were stained for myc-Ash1p with 9E10 mouse antibodies and Cy3-conjugated secondary antibodies that recognize the mouse 9E10 antibody. Tup1p was stained with rabbit polyclonal antibodies and FITC-conjugated secondary antibodies. Cell nuclei were visualized with DAPI stain, and whole cells were examined by bright-field imaging. Cells are stained as described in panel A. Yeast cells grown in M199 (pH 7.0) at 23°C appear as chains of attached budding cells. In panels A and B, selected mother (m) and daughter (d) cells are labeled.
Ash1p is localized to hyphal tip cells.
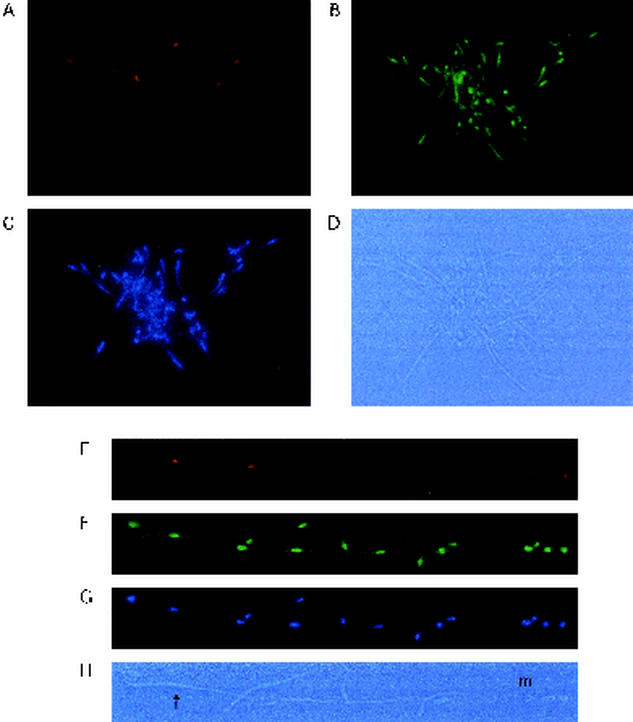
When C. albicans cells are grown for short periods of time in medium that strongly induces hyphal growth (YPD plus 20% serum at 35°C, for example), the mother cells appear rounded, and the daughter cells appear highly elongated, because the latter are beginning to form hyphae. In such mother-daughter pairs, Ash1p is localized specifically in the daughter cell nuclei (Fig. 4 and Fig. 6). Of the stained mother-daughter pairs observed in this study, 92% showed Ash1p in the daughter cell nucleus and not in the mother cell nucleus. If these mother-daughter pairs are incubated for longer times (YPD plus 20% serum at 35°C for 6 to 8 h), mature hyphae form in which multiple cells are joined end to end. In such hyphae, Ash1p is observed in the nuclei of apical hyphal cells (hyphal tip cells), but not in any of the other cells in the hyphae (Fig. 6). The apical cell is the site of active hyphal growth and constitutes the newest cell of the growing hypha.
FIG. 6.
Ash1p localizes to hyphal tip cells. (A to D) Ash1p is observed in the hyphal daughter cell nucleus of mother-daughter hyphal cell pairs grown for 2 h at 35°C in YPD plus 20% serum (A, anti-myc stain; B, anti-Tup1p stain; C, DAP1 stain; D, bright-field image). (E to H) Hyphal cells grown for 4 to 6 h show chains of cells in a hyphal filament with Ash1p located only in the nucleus of the hyphal tip (apical) cell (indicated by the arrow). The original mother cell (m) has also produced budding cells. The images are as described for panels A to D.
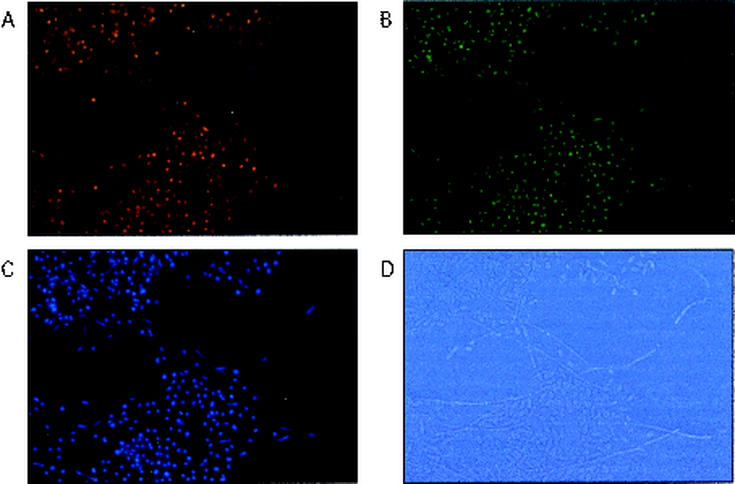
Upon longer exposures to YPD plus 20% serum at 35°C, hyphae begin to generate blastospores, which bud off from multiple positions along the hyphae. It also appears that the nuclei of these newly released blastospores stain positively for Ash1p (Fig. 7), but in this experiment, it is difficult to rigorously distinguish cells that have recently budded from hyphae from those that have undergone additional cell divisions.
FIG. 7.
Mature hyphae produce budding daughter cells that express Ash1p. Cells were grown overnight at 35°C in YPD plus 20% serum and stained as described in the legend to Fig. 6.
An important control for all of the localization experiments discussed in this and the previous section is the demonstration that an antibody directed against a different nuclear protein effectively stains all of the nuclei in the cell population. This is particularly important for growing hyphae, because the susceptibility of cells to the staining procedure could vary along the hypha. For this control, we used antibodies against the nuclear protein Tup1 (Fig. 5 to 7). Tup1p is observed in all nuclei (compare with the DAPI-stained images), and this observation rules out the possibility that the daughter cell and hyphal tip-cell-specific staining of Myc-Ash1p is due to the greater susceptibility of these cells to antibody.
ash1 mutant strains are reduced for virulence.
A mouse model of systemic candidiasis is a sensitive assay for determining differences in virulence between C. albicans strains, and in this way, we tested whether Ash1p is important for virulence in vivo. For this analysis, 106 wild-type or mutant C. albicans cells were injected by tail vein into groups of six mice. We performed two separate experiments with two independently derived ash1/ash1 mutant strains.
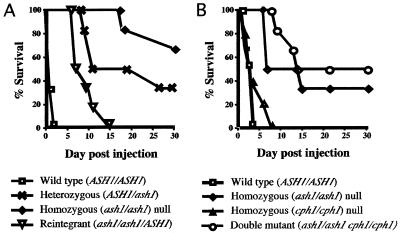
In the first experiment, we compared four strains: the ASH1/ASH1 parental strain (CAF2-1), an ASH1/ash1 heterozygous strain (YD-01), an ash1/ash1 (YDI-7) fully mutant strain, and a homozygous mutant strain (YDI-157) that carries an intact ASH1 gene reintegrated into the genome. As shown in Fig. 8A, disruption of one copy of ASH1 results in decreased virulence, and disruption of both copies reduces virulence even further. When an intact copy of the ASH1 gene is reintroduced into the ash1/ash1 strain, virulence is increased to approximately that of the ash1/ASH1 heterozygote, a result consistent with the fact that these two strains each have a single intact copy of ASH1. This result confirms that the virulence defects of the ash1/ash1 mutant strain are due to the absence of ASH1 and not to some other change resulting from transformation or other manipulations used in the construction of mutant strains. The doubling times of ash1/ash1 mutant strains grown at 30 and 37°C in YPD medium were comparable to that of the wild type when measured in vitro (data not shown), suggesting that its defect in virulence is not due simply to a nonspecific growth defect.
FIG. 8.
ASH1 is important for virulence in a mouse model of systemic candidiasis. Shown are survival curves of mice systemically infected with URA3+ C. albicans strains of the genotypes ASH1/ASH1 (CAF2-1), ASH1/ash1 (YDI-1), ash1/ash1 (YDI-7), and ash1/ash1::ASH1 (YDI-157) (A) or ASH1/ASH1 (CAF2-1), ash1/ash1 (YDI-27), ash1/ash1 cph1/cph1 (YDI-129), and cph1/cph1 (JKC19) (B).
In a second virulence experiment (Fig. 8B), we compared an independently constructed ash1/ash1 mutant strain (YDI-27), the wild-type strain (CAF2-1), a cph1/cph1 strain (JKC19), and an ash1/ash1 cph1/cph1 strain (YDI-129). The cph1 mutants are included in this experiment, because, as described above, the ash1 cph1 double mutant shows a greater defect in filamentous growth than do the single mutants. As in the first virulence experiment, we found that deletion of ASH1 leads to a marked reduction in virulence. In contrast, the cph1/cph1 mutant strain appears fully virulent, as reported by Lo et al. (30). The additional deletion of CPH1 from the ash1/ash1 strain led to only small differences in survival of mice infected with these two strains. One notable difference between the ash1/ash1 strain and the ash1/ash1 cph1/cph1 double-mutant strain was that animals injected with the double-mutant strain showed fewer symptoms of infection, such as weight loss, reduced activity, or roughened coat appearance, compared to the ash1/ash1 single-mutant strain, even though the survival times were only slightly different.
ash1 mutants colonize tissues and produce hyphae in vivo.
To observe the morphology of C. albicans cells in vivo, kidneys of mice systemically infected with 106 C. albicans cells were removed, sectioned, and stained with periodic acid-Schiff base for the presence of C. albicans. Mice injected with 106 wild-type C. albicans cells of the SC5314 strain background succumb to infection within 2 to 4 days (Fig. 8) and at 2 days show a large number of Candida cells in the kidneys (Fig. 9A and B). Both hyphae and blastospores are visible at the sites of infection. At the same time point, very few ash1/ash1 cells are observed in the kidney (Fig. 9C and D). At later time points (>7 days), ash1/ash1 cells could easily be detected in the kidney, and both blastospores and hyphae were present (Fig. 9E and F). Thus, although most mice infected with 106 ash1/ash1 cells survive past 18 days, ash1/ash1 strains were nonetheless observed in the kidney as yeast and filamentous forms after 7 days and even after 30 days (the longest time point at which kidneys infected with the ash1/ash1 strain were examined; Fig. 9E and F). The tissue fungal burden of animals that succumbed to infection was quantified by homogenizing the kidneys and brains and plating on Sabouraud agar. Both ash1/ash1 and ash1/ash1 cph1/cph1 mutant strains achieved similar levels of colonization (3 × 107 CFU/g of kidney) to those reached by the wild type. However, the wild-type strains achieved this density within a day or 2, while the ash1/ash1 mutant strains required several weeks. These observations indicate that the virulence defect of the ash1/ash1 mutants is not simply due to the inability to survive or to grow as filaments in the mouse. It is possible, for example, that the attenuated virulence of ash1/ash1 mutants is due to defects in the regulation of filamentous growth or in the proper specialization of hyphal tip cells.
FIG. 9.
ash1/ash1 mutants colonize kidneys much more slowly than do ash1/ash1/ASH1+ reintegrants, although they do produce hyphae in vivo. Histological sections of mouse kidneys were prepared as described in Materials and Methods. (A, B) The ash1/ash1/ASH1+ strain 24 h after infection. Panel A shows a locus of infection on the edge of the kidney and a large colony inside the kidney (B). (C to F) Shown are the ash1/ash1 mutant strains after 2 (C and D), 15 (E), and 30 (F) days. Panels A to D were photographed at ×40, and panels E and F were photographed at ×20.
DISCUSSION
Asymmetric cell division is critical for the development of nearly all organisms, ranging from unicellular bacteria and yeasts to multicellular plants and animals. The asymmetric localization of gene products to one of the two daughter cells formed during cell division leads to genetically identical cells with the potential for dramatically different cell fates.
In this paper, we show, by staining the transcriptional regulator Ash1p, that the human fungal pathogen C. albicans undergoes asymmetric cell division in all three of its morphological forms—budding-yeast-form cells, pseudohyphae, and hyphae. All three forms are found in infected tissues, and the ability of C. albicans to switch between them is thought to be crucial for its pathogenesis (for a recent review, see reference 19). In budding-yeast-form cells, C. albicans Ash1p is observed in daughter cell nuclei, but not in mother cell nuclei. It is also observed only in daughter cell nuclei in chains of pseudohyphal cells. Finally, in mature hyphae, Ash1p is observed only in the nuclei of hyphal tip cells—that is, to the cells active in hyphal growth. Thus, as far as can be seen by immunofluorescence, hyphae consist of long, branched chains of elongated cells, with Ash1p absent from all but the growing tip cells.
It has long been appreciated that the hyphal tip (apical) cells of C. albicans differ from internally positioned (subapical) hyphal cells. For example, much of the metabolism and growth of hyphae is concentrated in these cells. Moreover, the organelle composition of the hyphal tip cell appears different from that of other hyphal cells; in particular, hyphal tip cells are vacuole poor and cytosol rich compared to the rest of the hyphal cells (for review, see reference 18). Finally, secretion of at least some hydrolytic enzymes occurs selectively at the hyphal tip cell (reviewed in references 14 and 20). Many studies indicate that the penetration of host epithelial surfaces by C. albicans is carried out by the hyphal tip cells, and it is reasonable to believe that the tip cells specifically secrete hydrolytic enzymes that damage host tissues, providing sites of penetration. (For recent reviews, see references 19 and 21.) Because Ash1p is localized to the nuclei of hyphal tip cells and because it is a transcriptional regulator, it seems likely that Ash1p regulates at least a portion of the specializations that take place in hyphal tip cells.
In addition to specifically “marking” daughter cells, Ash1p is also required for filamentous growth on some types of filament-inducing medium. However, the severity of the defect caused by deleting ASH1 varies, depending on the nature of the medium. These results suggest that one function of Ash1p in daughter cells is to establish or maintain filamentous growth on certain types of media. For example, ash1/ash1 mutant cells show a pronounced defect of filamentous growth on Spider (low-nutrient) medium. It has been proposed that, in response to this medium, C. albicans grows filamentously in a “foraging mode,” seeking out new locations of greater nutritional richness (24). It is possible Ash1p in the hyphal tip cells regulates a process that is required for hyphae to sense or to respond to low-nutrient medium, thus causing the filamentation defect of ash1/ash1 mutants on this medium.
In S. cerevisiae, the best-understood function of Ash1p takes place in budding cells. In daughter cells, it represses transcription of the HO endonuclease gene, which carries out the first step in mating-type interconversion. Because Ash1p is specifically localized to daughter cell nuclei, only mother cells are able to switch mating types. As far as is known, C. albicans is unlikely to carry out mating-type interconversion; its genome lacks silent mating cassettes and a gene closely related to HO. It is possible that the asymmetric localization of Ash1p has a deeply conserved function in S. cerevisiae and C. albicans (perhaps involving filamentous growth) and that its regulation of HO was a relatively recent evolutionary add-on in S. cerevisiae.
Finally, our work shows that ash1/ash1 mutants of C. albicans are significantly attenuated for virulence in a mouse model of disseminated candidiasis. Examination of the kidneys revealed that the ash1/ash1 mutants can still form hyphae in vivo; however the number of hyphal cells observed in the kidney was significantly smaller than that of the wild-type control strain observed at the same time after infection. Although it is premature to make a firm conclusion, it is possible that the virulence defect in the ash1/ash1 strain is due to subtle defects in the regulation of hyphal formation or to a defect in hyphal tip cell specialization.
In conclusion, this study has focused on the role of C. albicans Ash1 protein. We found that this protein is required for filamentous growth under certain laboratory conditions, is required for full virulence in a mouse model of infection, and marks daughter cell nuclei in budding and pseudohyphal forms and tip (apical) cell nuclei in hyphae. We propose that the asymmetric localization of Ash1p is crucial for the proper specialization of the hyphal tip cell and that this specialization is important for virulence.
ADDENDUM IN PROOF
While this paper was under revision, the expression of C. albicans ASH1 in S. cerevisiae was independently reported by Munchow et al. (S. Munchow, D. Ferring, K. Kahlina, and R. P. Jansen, Curr. Genet. 41:73-81, 2002).
Acknowledgments
We thank A. Sil, B. Braun, and C. Hull for critical reading of the manuscript and constructive suggestions; A. Sil, M. Maxon, and P. Sudbery for valuable discussions and sharing of reagents; and L. Prentice for training and use of the histology laboratory facilities at UCSF. G. Fink, A. Sil, and I. Herskowitz generously provided strains.
We also acknowledge the support of the NIDR and Burroughs Wellcome Fund provided to the Candida albicans Genome Sequencing Project and to Stanford University and the University of Minnesota for maintaining the public C. albicans sequence database. This work was supported by a National Science Foundation Graduate Fellowship to D.O.I. and by National Institutes of Health grant GM37049 to A.D.J.
REFERENCES
- 1.Bobola, N., R. P. Jansen, T. H. Shin, and K. Nasmyth. 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84:699-709. [DOI] [PubMed] [Google Scholar]
- 2.Bockmuhl, D. P., and J. F. Ernst. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1, and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. J., and N. A. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 7.Calderone, R. A. (ed.). 2002. Introduction and historical perspectives, p. 3-13. Candida and candidiasis. ASM Press, Washington, D.C.
- 8.Chandarlapaty, S., and B. Errede. 1998. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 18:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csank, C., K. Schröppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 12.Feng, Q., E. Summers, B. Guo, and G. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghannoum, M. A. 2000. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13:122-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 16.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 18.Gow, N. A. R. 2002. Cell biology and cell cycle of Candida, p. 145-158. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 19.Gow, N. A. R., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 20.Hube, B., and J. Naglik. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997-2005. [DOI] [PubMed] [Google Scholar]
- 21.Hube, B., and J. Naglik. 2002. Extracellular hydrolysis, p. 107-122. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 22.Kobayashi, S. D., and J. E. Cutler. 1998. Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol. 6:92-94. [DOI] [PubMed] [Google Scholar]
- 23.Kolodziej, P. A., and R. A. Young. 1991. Epitope tagging and protein surveillance. Methods Enzymol. 194:508-519. [DOI] [PubMed] [Google Scholar]
- 24.Kron, S. J., and N. A. Gow. 1995. Budding yeast morphogenesis: signalling, cytoskeleton and cell cycle. Curr. Opin. Cell Biol. 7:845-855. [DOI] [PubMed] [Google Scholar]
- 25.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 26.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. R. Gow, A. J. P. Brown, and D. Y. Thomas. 1996. Signal transduction through homologues of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 28.Lengeler, K. B., R. C. Davidson, C. D'souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 30.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 31.Madhani, H. D., and G. R. Fink. 1998. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 8:348-353. [DOI] [PubMed] [Google Scholar]
- 32.Maxon, M. E., and I. Herskowitz. 2001. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. USA 98:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687-692. [DOI] [PubMed] [Google Scholar]
- 34.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 35.Odds, F. C. 1988. Candida and candidosis. Baillière Tindall, London, United Kingdom.
- 36.Odds, F. C. 1994. Candida species and virulence. ASM News 60:313-318. [Google Scholar]
- 37.Pan, X., and J. Heitman. 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20:8364-8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redd, M. J., M. B. Arnaud, and A. D. Johnson. 1997. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J. Biol. Chem. 272:11193-11197. [DOI] [PubMed] [Google Scholar]
- 39.Santos, M. A., and M. F. Tuite. 1995. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 41.Sheehan, D. C., and B. B. Hrapchak. 1987. Theory and practice of histotechnology, 2nd ed. Batelle Press, Columbus, Ohio.
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sil, A., and I. Herskowitz. 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84:711-722. [DOI] [PubMed] [Google Scholar]
- 44.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 46.Wessels, J. G. H. 1993. Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 123:397-413. [DOI] [PubMed] [Google Scholar]
- 47.Whiteway, M. 2000. Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol. 3:582-588. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]