Abstract
We have developed a series of programs, collectively packaged as Array File Maker 4.0 (AFM), that manipulate and manage DNA microarray data. AFM 4.0 is simple to use, applicable to any organism or microarray, and operates within the familiar confines of Microsoft Excel. Given a database of expression ratios, AFM 4.0 generates input files for clustering, helps prepare colored figures and Venn diagrams, and can uncover aneuploidy in yeast microarray data. AFM 4.0 should be especially useful to laboratories that do not have access to specialized commercial or in-house software.
Rationale
DNA microarray experiments quickly generate millions of data points, a flood of information that can overwhelm biologists. Extracting useful information from transcriptional profiles can be a major challenge. To this end, some freeware has been developed that clusters and displays data, but there is clearly a need for more user-friendly computer resources for DNA-microarray analysis, particularly for labs that do not specialize in genomics and/or have limited access to computer programmers. In our experience, we have found an urgent need for programs that manage large data sets quickly and easily. When facing a data set consisting of more than 1,000 individual microarray experiments, there are endless questions to be asked and hypotheses to be checked. Querying the data set often becomes rate-limiting, as sifting through and rearranging millions of data points is a monotonous and laborious task.
We have developed a series of programs, collectively packaged as Array File Maker 4.0 (AFM), that rapidly manipulate data, prepare files for use in freeware clustering programs, and make figures. The programs are written in Microsoft Visual Basic [1] and are used within the familiar confines of Microsoft Excel, allowing the researcher to use standard Excel functions in addition to AFM-specific ones. We have primarily used AFM to manage array data from yeast, but the program can be easily tailored for use with data sets from any organism, including large mammalian array files (D. Wigle and J. Rossant, personal communication).
Figure 1.
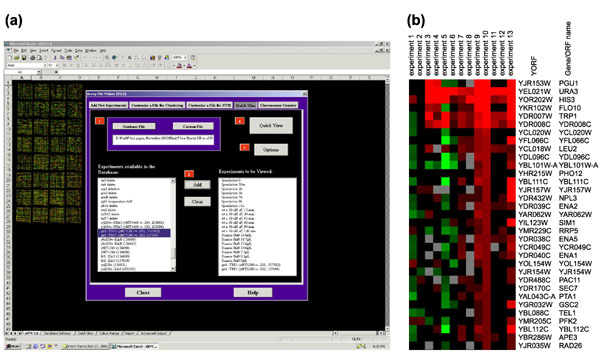
(a) The AFM 4.0 Quickview function page. (b) Expression ratio data that have been replaced with colors, as generated by the AFM Quickview function.
Array File Maker 4.0
The input file for AFM 4.0 is any database of expression ratios. AFM 4.0 is thus compatible with data from spotted cDNA, spotted oligonucleotide, or Affymetrix microarrays. All steps required to determine the expression ratios from microarray experiments - for instance spot finding, spot quantitation, background subtraction, normalization, and so on - must be done prior to using AFM 4.0.
Clustering
Clustering is a powerful method for discovering patterns in array data. Two excellent clustering freeware programmes are available: Cluster and Support Vector Machine [2,3]. Each of these freeware packages requires data to be in a distinct file format. Although these are not difficult to prepare, a user can waste hours reformatting his/her data into these set file formats. Starting with a database of microarray experiments, AFM quickly prepares input files for either program.
Quickview
Huge columns of numbers are mind-numbing to most users. Microarray data are much easier to visualize as colored squares, where the color and color intensity indicate whether a gene is induced or repressed, and the extent of any induction and repression, respectively [2]. AFM's Quickview function replaces expression ratios with a corresponding range of colors, thereby transforming an array database into a color representation that can be easily manipulated as an Excel file. We frequently use Quickview to prepare figures for presentation.
Venn diagrams
When analyzing microarray data, the Venn diagram is a useful visualization tool to indicate the extent of overlap between multiple experiments. AFM 4.0 generates Venn diagrams that report the number of genes, induced or repressed above a user-defined threshold that share differences or similarities in between two and four transcriptional profiles.
Chromosome Counter
As recently revealed by Hughes and colleagues [4], aneuploidy is a common phenomenon in lab strains of yeast. Approximately 8% of the strains in the complete set of yeast deletions contain extra chromosomes. Moreover, even strains without high chromosome loss or gain can accidentally obtain, and then retain, extra chromosomes (P.J. and M.T., unpublished observations). In yeast, aneuploidy is easily observed as a global increase or decrease in transcript levels along the entire duplicated or deleted chromosome [4]. For example, a median expression ratio of 2:1 for any given chromosome in a haploid strain would strongly indicate that that strain had an extra copy of that chromosome. AFM's Chromosome Counter calculates the mean or median expression ratios across each yeast chromosome. Checking every yeast microarray experiment for aneuploid starting strains is a necessary precaution, as aneuploid yeast may yield false correlations between otherwise unrelated transcriptional profiles.
Additional programs
In addition to AFM, we have also written beta versions of programmes that, first, transform intensity output files from the GSI Lumonics Quantarray program into normalized expression-ratio data and, second, automatically file yeast microarray data with details of the particular experiment. Quantarray Data Handler 3.0 accepts intensity data from Quantarray and does the following: it subtracts slide background from all data points; it normalizes Cy3 and Cy5 signal intensities; it eliminates spots below a threshold intensity; and it median-centers the final data set, leaving the user with expression-ratio data. Array Database 1.0 is a databank for array data that allows labs to store images from microarray experiments with the full experimental details. As a single person can perform 30-40 microarray experiments a week, keeping track of cell-culture conditions and other experimental details is essential.
Downloading files
AFM 4.0, Quantarray Data Handler 3.0, and Array Database 1.0 can be downloaded at the Tyers Lab Home Page http://www.mshri.on.ca/tyers/ and are copyrighted against commercial gain. These programs run on PCs (Windows 95 or later) and require only Microsoft Office (97 or later) software as an operating platform. Full help manuals are also available at the website. AFM 4.0 and associated files are also available for download with the complete version of this article, online. For further information or to provide feedback please contact the authors.
AFM 4.0 and associated files are also available for download from the AFM 4.0 index page. AFM 4.0 should be downloaded to a local hard drive before use. The individual files are: the AFM 4.0 user manual as a Microsoft Word file or a pdf file, the AFM 4.0 software as an Excel file, an instructions file and a test database available as an Excel file. AFM 4.0 and the test database are available as ZIP files from the AFM 4.0 index page.
Supplementary Material
To open the AFM 4.0 software and associated files, click on the links below. To download the files to disk, right click on the link and choose 'Save Target As' (on a PC) or press 'Control' and click the link (on a Mac)
AFM 4.0 user manual - Microsoft Word file
Disclaimer: Whilst every effort is made by the authors, editors and publishers to ensure that the software performs as described, they accept no liability whatsoever for the consequences of downloading or using it. This information has not been peer-reviewed. Responsibility rests solely with the author(s).
References
- Microsoft Visual Basic http://msdn.microsoft.com/vbasic/default.asp
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Jr, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To open the AFM 4.0 software and associated files, click on the links below. To download the files to disk, right click on the link and choose 'Save Target As' (on a PC) or press 'Control' and click the link (on a Mac)
AFM 4.0 user manual - Microsoft Word file


