Abstract
Prions are “infectious” proteins. When Sup35, a yeast translation termination factor, is aggregated in its [PSI+] prion form its function is compromised. When Rnq1 is aggregated in its [PIN+] prion form, it promotes the de novo appearance of [PSI+]. Heritable variants (strains) of [PSI+] with distinct phenotypes have been isolated and are analogous to mammalian prion strains with different pathologies. Here, we describe heritable variants of the [PIN+] prion that are distinguished by the efficiency with which they enhance the de novo appearance of [PSI+]. Unlike [PSI+] variants, where the strength of translation termination corresponds to the level of soluble Sup35, the phenotypes of these [PIN+] variants do not correspond to levels of soluble Rnq1. However, diploids and meiotic progeny from crosses between either different [PSI+], or different [PIN+] variants, always have the phenotype of the parental variant with the least soluble Sup35 or Rnq1, respectively. Apparently faster growing prion variants cure cells of slower growing or less stable variants of the same prion. We also find that YDJ1 overexpression eliminates some but not other [PIN+] variants and that prions are destabilized by meiosis. Finally, we show that, like its affect on [PSI+] appearance, [PIN+] enhances the de novo appearance of [URE3]. Surprisingly, [PSI+] inhibited [URE3] appearance. These results reinforce earlier reports that heterologous prions interact, but suggest that such interactions can not only positively, but also negatively, influence the de novo generation of prions.
Prions are best known as the infectious agents proposed to be responsible for the mammalian transmissible spongiform encephalopathies including scrapie in sheep, mad cow disease in cattle, and Creutzfeldt-Jakob disease in humans (1). The prion form of the PrP protein is proposed to propagate its abnormal form to other “normal” PrP protein molecules with the same primary sequence (2, 3). Evidence suggesting that self-propagating prion proteins are not limited to PrP was presented in 1994 by Wickner (4). Three yeast proteins with self-perpetuating, alternate conformations have now been well described: [PSI+] (5), the prion form of the translational termination factor Sup35 (6–8); [URE3] (9), the prion form of the nitrogen catabolite repressor Ure2 (4, 8); and [PIN+] (10, 11), the prion form of Rnq1 (12, 13). In [PSI+] and [URE3] cells, Sup35 and Ure2 are respectively inactivated by aggregation, causing the same phenotypes as mutations in the SUP35 and URE2 genes. No phenotype has yet been associated with inactivation of RNQ1.
The de novo appearance of each of these yeast prions, [PSI+], [URE3], and [PIN+], is enhanced by overproducing the corresponding prion domains (12, 14, 15). The increased number of protein molecules presumably enhances the chance that a prion seed will form de novo. However, de novo appearance of some prions depends on the presence of other prions or prion-like aggregates (13, 16). We first described [PIN+] as a prion-like element having the phenotype of allowing overproduction of Sup35 to convert [psi−] cells to [PSI+] (10, 11), and later showed that [PIN+] is equivalent to the prion form of Rnq1 (13). The presence of [URE3] (13) or the artificial fusion prion [NU+] (16) also permitted overexpression of Sup35 to induce the appearance of [PSI+].
The existence of different heritable forms or strains of prions is a fascinating chapter in the biology of prions. Prion diseases exhibit variable incubation times, neurodegenerative patterns, and PrP prion deposits, all of which remain distinct on transmission in inbred mammals (17). Recent evidence supports the idea that prion strain variation is a result of the PrP protein's ability to propagate in different heritable prion forms (18–20). Others, however, view the existence of prion strains as more compatible with a viral model for prion disease (21). The finding of [PSI+] strains (14), and more recently [URE3] strains (22), in yeast, where the viral hypothesis is unreasonable, supports the idea that prion strains result from multiple prion protein forms.
Distinct strains of [PSI+] have different mitotic stabilities (frequencies of [PSI+] loss), translational termination activities as measured by suppression of nonsense codons (14), and levels of nonaggregated Sup35 (23). Weak [PSI+] are less stable than strong [PSI+] in mitotic division (14), and the levels of nonaggregated Sup35 and accompanying translational termination are higher in weak [PSI+] cells than in strong [PSI+] cells (23). Strains of [PSI+] have also been distinguished by their differential responses to mutations in the SUP35 gene (24, 25) and by their responses to chaperones (26). Several [PSI+] strains have been shown to be dominant, non-Mendelian traits when crossed with [psi−] (5, 27). In addition, the strong [PSI+] phenotype appears in diploids made from mating strong and weak [PSI+] cells (23, 24).
Several in vitro studies support the hypothesis that strains of [PSI+] result from distinct protein conformations of Sup35. Purified Sup35 prion domain (Sup35NM) forms fibers in vitro with either wavy or straight structures (28). Also, a purified chimeric Sup35NM was shown to form aggregates with distinct conformations and distinct in vitro seeding activities (29). Most convincingly, protein extracts from strong [PSI+] cells converted purified Sup35NM into fibers more efficiently than did protein extracts from weak [PSI+] cells (30, 31).
The continued propagation of prions depends on normal chaperone protein levels. The finding that either deleting or overexpressing the HSP104 chaperone gene causes the elimination of [PSI+] (32) supported the prion model for [PSI+] because it suggested that [PSI+] formation involved a conformational change. Deleting, but not overexpressing, HSP104 eliminates [URE3] (33) and [PIN+] (10, 34), and overexpressing YDJ1, which encodes an Hsp40 family chaperone, promotes the loss of [URE3] (33) and a weak strain of Pichia methanolica [PSI+] in Saccharomyes cerevisiae (26). Hsp104 functions together with Hsp40 and Hsp70 (35) to promote the renaturation of denatured or aggregated proteins (36). The effect of Hsp104 on [PSI+] is modified by levels of, and alterations in, the Hsp70 family members Ssa and Ssb (26, 37–39). Maintaining the prion form of Rnq1 requires specific domains of the Hsp40 family member Sis1 (34). In addition, a specific mutation of SIS1 caused an altered aggregation pattern of the Rnq1 prion that appeared to be heritable even in the absence of the SIS1 mutation (34).
Here, we demonstrate the existence of different strains of [PIN+] with distinct phenotypes. We determine the relative competitiveness of these [PIN+] prion strains and of [PSI+] prion strains and find that one factor foretells the outcomes of competitions between two variants of the same prion. We also find that, whereas the [PIN+] prion enhances the de novo appearance of [URE3], the presence of the [PSI+] prion inhibits [URE3] appearance. The non-Mendelian segregation of prions has been reported to deviate from the classical 4:0 ratio (4, 5, 9, 27, 40). Here, we carefully document this effect for [PSI+] and [URE3], and show that it is due to meiosis and not the conditions used to stimulate sporulation. Finally, we show that overproducing the chaperone Ydj1 promotes the elimination of some [PIN+] strains.
Materials and Methods
Cultivation Procedures.
Standard yeast media and cultivation procedures were used (41). Yeast extract/peptone/dextrose (YPD) with 5 mM guanidine⋅hydrochloride (YPD + GuHCl) was used to eliminate prions (42). YPD with 10 mg/liter cycloheximde (YPD + Cyh) was used to select for cycloheximide-resistant mutations (cyhR) and for random spores in [PSI+] experiments. Synthetic medium with 3 mg/liter cycloheximide was used to select for cytoductants. Growth on YPD plates containing 40 μg/ml of ethidium bromide converted strains to [rho−] (43). Casamino acid (CA) medium contained 0.13% yeast nitrogen base, 0.5% ammonium sulfate, 1% casamino acids, and 2% glucose or 2% glycerol. Uracil, adenine, and tryptophan were added to CA when necessary. CA with 5 mg/liter cycloheximide (CA + Cyh) was used to select for random spores in [PIN+] experiments. Synthetic medium containing galactose and raffinose (SGal + Raf) was used to overexpress YDJ1 or URE2 from the galactose-inducible promoter GAL1. Copper sulfate (Cu; 50 μM) was added to medium to induce expression of RNQ1 or SUP35 under the control of the inducible CUP1 promoter.
Strains.
Opposite mating type yeast strains that are isogenic, sporulate efficiently when intercrossed, and contain the ade1–14 allele, which permits weak and strong [PSI+] to be distinguished, were constructed by mating 74-D694 (MATa ade1–14 trp1–289 his3-Δ200 leu2–3,112 ura3–52) (44) with an efficiently sporulating strain, NKY292 (MATα lys2 ura3 leu2∷hisG ho∷LYS2) (45) (kindly supplied by D. Bishop, University of Chicago) and backcrossing MATα meiotic progeny to 74-D694 four times. Progeny from the final backcross were diploidized by transforming them with pGAL-HO (46) (kindly provided by R. Esposito, University of Chicago). The diploids were sporulated and dissected to obtain MATa and MATα segregants that otherwise had the same genotype as 74-D694. L1842 (MATa) and L1843 (MATα) are one pair of opposite mating type progeny from a single tetrad, as are L1844 (MATa) and L1845 (MATα). L2176 is a spontaneous cyhR derivative of L1845 in which the cyhR mutation was shown to be recessive.
To induce weak and strong [PSI+], L1842, L1843, L1844, and L1845 were transformed with pEMBL-SUP35. Transformants were grown on plasmid selective medium for ≈14 generations and subsequently on YPD for ≈14 generations to promote plasmid loss before plating the cultures for individual colonies on YPD. L2010 and L2012 are, respectively, weak and strong [PSI+] derivatives of L1844.
YHE711 (MATα ura2 leu2 [psi−] [ure-o] [PIN+]) (47) was scored as [PIN+] because expression of an RNQ1-GFP fusion in the strain gave rise to discrete aggregates characteristic of [PIN+] (12, 13). In addition, as expected in a [PIN+] background, overexpression of SUP35NM-GFP in YHE711 led to the appearance of ribbon and curve aggregates characteristic of newly induced [PSI+] (48). Derivatives of YHE711 grown in YPD + GuHCl failed to give rise to either of these types of aggregates and are therefore [pin−]. An RNQ1 deletion derivative of the [PIN+] version of YHE711 was constructed by transformation with a PCR product of the RNQ1∷kanMX4 insertion from yeast strain [American Type Culture Collection (ATCC) no. 4003435; ref. 49] amplified with DNA primers HE230 (RNQ1 5′ UTR, 5′-CACGTATTTCAGTTGTCC-3′) and HE231 (RNQ1 3′ UTR, 5′-CCACTCTTACATTGTCATT-3′). Transformants were selected on YPD containing 300 μg/ml G418, after a recovery period in YPD. To confirm the disruption of RNQ1, candidate mutants were analyzed by PCR using primers HE265 (RNQ1 5′ UTR, 5′-GAATGATCCATCGTTCTTAC-3′), HE266 (RNQ1 3′ UTR, 5′-GATGGCTTATATCCTGCTC-3′), HE267 (kanMX4 pointing to 5′ end, 5′-CTGCAGCGAGGAGCCGTAAT-3′), and HE268 (kanMX4 pointing to 3′ end, 5′-TGATTTTGATGACGAGCGTAAT-3′).
GuHCl-treated versions of yeast strains A3099 (MATα ade2-1 SUQ5 lys1-1 his3–11,15 leu1 kar1-1 ura3∷KanMX4 [psi−][rho−]) (12), c10B-H49 (MATα ade2-1 SUQ5 lys1-1 his3–11,15 leu1 kar1-1 cyhR [psi−][rho−]) (50), BY4741 (MATa his3-Δ1 leu2-Δ met15-Δ ura3-Δ [psi−] [PIN+]) (from Research Genetics), and 3385 (MATa ura2 leu2 kar1 his− [psi−] [ure-o][PIN+]) (4) were used in cytoduction experiments.
Plasmids.
A 2μ plasmid (pEMBL-SUP35) with URA3 leu2-d markers and SUP35 under its native promoter (51) was used to overproduce Sup35 at moderate levels on synthetic medium lacking uracil (−Ura) or high levels (on −Leu). The defective LEU2 promoter present in the leu2-d allele on this plasmid selects for overamplification of the plasmid on −Leu. Moderate overexpression of SUP35 was used to induce [PSI+]; high-level overexpression was used to distinguish different variants of [PIN+] on the basis of growth inhibition. YDJ1 under the control of the GAL1 promoter is present on a CEN LEU2 plasmid (p901); the parent plasmid without YDJ1 is pH316 (33). Plasmids pRNQ1-GFP and pSUP35NM-GFP, which respectively contain the fusions of either RNQ1 or the NM domains of SUP35 to green fluorescent protein (GFP) under the control of the CUP1 promoter, were used to score for [PIN+] as described previously (13). Plasmids used for the [URE3] induction experiments were 2μ-based, with a LEU2 marker and the GAL1 promoter to express URE2 (pH376), URE2 (1–65) (pH382), URE2Δ151–158 (pH377), or no expression of URE2 as a negative control (pH317) (52).
Cytoduction.
Cytoductions were performed by crossing [RHO+] donors to [rho−] recipients. Either the donor or the recipient carried the kar1-1 allele, which inhibits nuclear fusion (53). When the recipient was cycloheximide-resistant (cyhR), cytoductants were selected on synthetic glycerol medium containing cycloheximide. Otherwise, diploids and cytoductants were selected on synthetic glycerol medium deficient in a nutrient required by the donor strain for growth. Cytoductants were then identified by subcloning the population and screening colonies for the recipient mating type and auxotrophic markers.
Analyses of [PSI+] Variants.
After inducing weak and strong [PSI+] in L1842, L1843, L1844, and L1845, the L1842 and L1843 derivatives (shown in Fig. 1) and the L1844 and L1845 derivatives were crossed in all possible combinations. We assayed diploids for [PSI+] strength by color on YPD and level of growth on synthetic medium lacking adenine (−Ade). The diploids were sporulated after propagating for approximately 42 generations. Meiotic progeny from each diploid were assayed for [PSI+] strength, mating type, and curability of [PSI+] by growth on YPD + GuHCl.
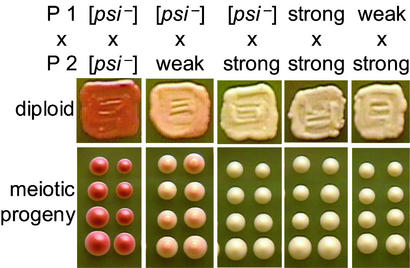
Figure 1.
Meiotic inheritance of [PSI+] variants. Isogenic haploid yeast, parent 1 (P1; L1842 derivatives) and parent 2 (P2; L1843 derivatives) carrying the indicated [PSI+] variants were mated. One representative diploid and two tetrads from each cross are shown.
To perform the random spore analyses of [PSI+] loss frequencies, weak and strong [PSI+] derivatives of L1844 (L2010 and L2012, respectively) were mated to a GuHCl-treated cyhR derivative of L1845 (L2274) to produce diploids SL-1142 and SL-1143, respectively. The resulting diploids were sporulated, and frequencies of [PSI+] loss were determined in at least three independent trials by counting red vs. total number of colonies after suspending cells in 100 μl of 10% gluculase, vortexing for 20 s, diluting 1 × 10−4 in water, and plating to YPD + Cyh.
Analyses of [PIN+] Variants.
Phenotypes of [PIN+] variants in 74-D694, L1844, and L1845 were determined by overexpressing SUP35 from the pEMBL-SUP35 plasmid. Transformants were patched to −Ura, where the plasmid is present in moderate copy number, and then spotted to −Ura, −Leu, and −Ade. The induction of [PSI+] was identified as GuHCl-curable nonsense suppression of ade1–14 that was independent of the plasmid used to induce the appearance of [PSI+]. Phenotypes of [PIN+] variants in c10B-H49 were determined by overexpressing SUP35 and spreading transformants to −Ade.
To determine the relative competitiveness of the [PIN+] variants, we first generated isogenic opposite mating type yeast carrying each of the [PIN+] variants. Independent [PIN+] derivatives of 74-D694 were cytoduced into a GuHCl-treated derivative of c10B-H49 and from there into a GuHCl-treated [pin−] derivative of L1844. Diploids made from crosses of the final cytoductants to a GuHCl-treated [pin−] cyhR derivative of L1845 were transformed with pEMBL-SUP35 and sporulated. The [PIN+] phenotypes of cyhR progeny that maintained pEMBL-SUP35 (obtained by plating sporulated cultures to CA–Ura + Cyh) were determined by growth on −Leu and −Ade. MATα progeny carrying the different [PIN+] variants were backcrossed to the original [PIN+] derivatives of 74-D694. Diploids were tested for the [PIN+] phenotypes and sporulated after propagating for approximately 28 generations. Random spores were selected on CA–Ura + Cyh, and their [PIN+] phenotypes were determined.
Comparison of Rnq1 Among [PIN+] Variants.
Derivatives of 74-D694 were grown in liquid YPD to mid-log (OD600 ≈ 1.0). Harvested cells were resuspended in lysis buffer [50 mM Tris, pH 8.0/150 mM NaCl/0.2% Triton X-100/1.9 μg/ml aprotinin/3.5 μg/ml E-64/5 μg/ml leupeptin/5 μg/ml pepstatin/400 μg/ml 1,10 phenanthroline/500 μg/ml PMSF/50 μg/ml N-(p-tosyl)lysine chloromethyl ketone (TLCK)], and mixed with 750 μl of glass beads/lysis buffer slurries. Total protein lysates were obtained by vortexing each tube eight times for 10 s, with intermittent incubations on ice, and removing cell debris at 10,000 × g for 10 min. The protein concentrations of the cleared lysates were measured (Bio-Rad Protein Assay), and lysis buffer was added to normalize the samples. Total protein lysate (1–2 mg) was fractionated at 280,000 × g for 30 min in a Sorvall TLA100.1 rotor. Pelleted proteins were resuspended in 200 μl of lysis buffer. Rnq1 was detected with a polyclonal antibody (kind gift from S. Lindquist, University of Chicago).
Influence of [PIN+] and [PSI+] on [URE3] Appearance.
[PIN+] and [PSI+] derivatives from 74-D694 were cytoduced into a GuHCl-treated version of c10B-H49, and from there into a GuHCl-treated version of 3385, and finally from 3385 into a GuHCl-treated version of YHE711. The [URE3] prion induction assay was performed as described (52). Briefly, [ure-o] strains were transformed with pH317, pH376, or pH382 and transformant colonies were individually grown to saturation in SGal + Raf-Leu. Starting from 107 cells per plate, serial dilutions were plated on synthetic, dextrose-based medium containing 100 μg/ml ureidosuccinate (USA). Colonies appearing after 5 days at 30°C were recorded.
Elimination of [PIN+] by YDJ1 Overexpression.
Various [psi−] [PIN+] derivatives of 74-D694, as well as one [psi−] [pin−] control, were transformed with p901 (33) or the control plasmid pH316 (lacking YDJ1 but containing the GAL1 promoter). Transformants were grown on media containing galactose to induce YDJ1 expression. Two transformants for each strain and plasmid combination were subcloned on SGal + Raf-Leu 2–3 times consecutively by picking 3–4 same-sized colonies for each successive colony purification step. Colonies underwent an average of 20 cell doublings before being purified again or tested for [PIN+].
The elimination of [PIN+] was scored by checking for the loss of aggregated Rnq1. Colonies were patched to YPD and crossed to tester strains: GuHCl-treated versions of 64-D697 (MATα ade1–14 trp1–289 lys9-A21 leu2–3,112 ura3–52) or SL1010–1A (MATα ade1–14 met8–1 trp1–1 his5–2 leu2–1 ura3–52) transformed with pRNQ1-GFP (13). Diploids from these crosses were selected by complementation on medium (SC-His,Lys,Ura + Cu) that selects for maintenance of pRNQ1-GFP and contains 50 μM Cu to induce expression of the fusion protein. In this assay, [PIN+] colonies have bright green aggregates in the majority of cells, and [pin−] colonies have evenly distributed green fluorescence.
Results
Genetic Analysis of Strains of [PSI+].
To avoid confusion of yeast strains with prion strains we sometimes refer to the latter as variants. Yeast bearing strong or weak [PSI+] prion variants were previously distinguished (14) by the efficiency with which they could suppress the ade1–14 nonsense allele, which contains a premature stop codon that prevents the protein from being completely translated. Yeast that are [psi−] do not grow on −Ade because translation termination at the premature ade1–14 stop codon is efficient, and they are red on YPD due to the accumulation of a metabolic intermediate of the adenine biosynthesis pathway. Yeast strains with weak [PSI+] grow poorly on −Ade and are pink on YPD because they maintain lower levels of functional Sup35 compared with [psi−]; consequently, they partially suppress termination at the premature ade1–14 stop codon and produce some functional Ade1. Yeast strains with strong [PSI+] grow well on −Ade and are white on YPD because they maintain even less functional Sup35 than weak [PSI+], and they efficiently suppress termination at the premature ade1–14 stop codon (23, 30).
Diploids resulting from pairwise matings of isogenic yeast that were either [psi−], weak [PSI+], or strong [PSI+] displayed nonsense suppression levels equal to that of the [PSI+] parent with the “strongest” phenotype (Fig. 1). Tetrad analyses revealed that strong [PSI+] diploids always segregated strong [PSI+] in a 4:0 ratio and weak [PSI+] diploids usually segregated weak [PSI+] in a 4:0 ratio (Fig. 1 and Table 1).
Table 1.
Tetrad analysis of [PSI+] variants
| Parent 1 | Parent 2 | Tetrads dissected | Viable progeny | [PSI+] phenotypes of meiotic progeny
|
||
|---|---|---|---|---|---|---|
| [psi−] | Weak | Strong | ||||
| [psi−] | [psi−] | 14 | 56 | 56 | 0 | 0 |
| Weak | [psi−] | 59 | 200 | 10 | 190 | 0 |
| Weak | Weak | 33 | 118 | 2 | 116 | 0 |
| Strong | [psi−] | 33 | 121 | 0 | 0 | 121 |
| Strong | Strong | 16 | 60 | 0 | 0 | 60 |
| Weak | Strong | 18 | 67 | 0 | 0 | 67 |
| Strong | Weak | 21 | 77 | 0 | 0 | 77 |
Each row represents the sum of progeny obtained from two to four independent diploids. At least one diploid was from a cross between derivatives of L1842 and L1843, and one was from a cross between derivatives of L1844 and L1845. Weak and strong refer to the [PSI+] variants.
Meiosis Eliminates [PSI+] and [URE3] Prions.
Approximately 2–5% of the spores from weak [PSI+] diploids were [psi−] (Table 1, rows 2 and 3), which is significantly more than the 0.7% loss of weak [PSI+] among the mitotic progeny from one of the parents of these diploids (average of ≈2,800 colonies from three independent, equally represented trials). This, together with an earlier finding that a weak [PSI+] (then called [ETA+]) was very unstable in meiosis (27), led us to investigate this phenomenon further.
Haploid cells did not exhibit an enhanced loss of weak [PSI+] when exposed to the same conditions that induce meiosis in an isogenic weak [PSI+] diploid. We incubated three yeast strains on sporulation medium: a weak [PSI+] haploid, L2010; a weak [PSI+] diploid isogenic to L2010 but heterozygous for cyhR, SL-1142; and a weak [PSI+] cyhR meiotic segregant from this diploid, SL1142–1A. Random spores were selected from the diploid culture on YPD + Cyh. SL1142–1A was also plated on this medium, and L2010 was plated on YPD lacking cycloheximide. The frequency of [psi−] among the random spore colonies was 5.7% (average of ≈2,400 colonies from three independent, equally represented trials). The frequency of [psi−] among mitotic colonies from either of the haploid controls was only 0.4% (7 [psi−] of 1,670 colonies from L2010, and 15 [psi−] of 3,530 colonies from SL1142–1A). Therefore, the observed effect was not a result of the conditions used to induce sporulation. The effect was also not due to heightened instability in the diploid phase because the frequency of weak [PSI+] loss from mitotic diploid progeny was only 0.07% (average of ≈3,450 colonies from three independent, equally represented trials). Thus, some aspect of meiosis interferes with the inheritance of [PSI+].
Likewise, although [URE3] is highly stable during mitotic growth, it is frequently lost in meiotic segregants (4, 9, 40). We examined the effect of meiosis-inducing conditions on the stability of [URE3]. [URE3] was stable in mitotic growth: it was efficiently cytoduced from strain 1735 to 1019 (16 of 16 cytoductants examined were USA+). Growing 1735 on sporulation medium did not decrease the stability of [URE3]. Furthermore, a diploid made by crossing 1735 with 1019 stably maintained [URE3] (100 of 100 colonies examined after growth on YPD were USA+). However, sporulation of this diploid produced mostly USA− spores (39 of 48 USA− spores). Thus, the process of meiosis causes loss of [URE3].
[PIN+] Variants.
We previously described the isolation of spontaneously appearing [PIN+] colonies after prolonged incubation of a [pin−] [psi−] derivative of 74-D694 (11). Rare [PIN+] cells were detected by selecting for the appearance of [PSI+] after overexpression of SUP35. We eliminated [PSI+], but not [PIN+], by overexpressing HSP104. Later, the [PIN+] status of these isolates was shown to be a consequence of the prion form of Rnq1 because the loss of [PIN+] correlated with the loss of Rnq1 aggregates (13).
The phenotypes of [PIN+] were originally described as allowing moderate overproduction of Sup35 to convert [psi−] cells to [PSI+] and as inhibiting growth in the presence of extreme overproduction of Sup35 (10). We now distinguish [PIN+] variants with different levels (low, medium, high, and very high) of [PSI+] induction and growth inhibition (Fig. 2).
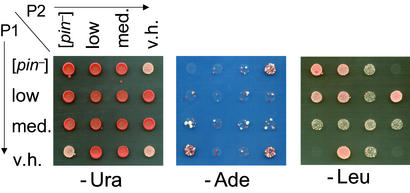
Figure 2.
Characterization of [PIN+] variants and their inheritance through cytoduction. Independent derivatives of the original 74-D694 are shown (Originals). These were each cytoduced into a [pin−] version of c10B-H49, a kar1–1 yeast strain, and from there back into a [pin−] version of 74-D694 (Cytoductants). Both the originals and cytoductants carry the pEMBL-SUP35 plasmid. When the plasmid is maintained at moderate level on −Ura distinct levels of [PSI+] induction are observed on transfer to −Ade. When the plasmid is amplified on −Leu, different levels of growth inhibition are observed. The different [PIN+] variants are cytoplasmically inherited because cytoductants and donors exhibit the same [PIN+] phenotypes. Row 1 is the [pin−] control; rows 2, 3, and 5 are the low, medium (med.), and very high (v.h.) spontaneous [PIN+] variants obtained in [pin−] 74-D694; row four is high [PIN+] from the original 74-D694.
Because [PIN+] is cytoducible (12, 13), cytoduction should transfer the distinct phenotypes if they result from heritable [PIN+] variants, but not if they are the result of Mendelian modifier mutations. Derivatives of 74-D694 carrying the different [PIN+] isolates and a [pin−] control were cytoduced into a [pin−] derivative of c10B-H49, and from there back into a [pin−] derivative of 74-D694. The cytoductants displayed the donor's [PIN+] phenotypes (Fig. 2). This result proves that Mendelian mutations do not cause the variable phenotypes.
Genetic Analysis of Variants of [PIN+].
To determine the relative competitiveness of the different [PIN+] variants, we crossed pairs of opposite mating type yeast harboring each of the different [PIN+] variants in all possible combinations and determined the [PIN+] phenotypes of the resulting diploids by measuring [PSI+] induction and growth inhibition levels. High [PIN+] outcompetes low and medium [PIN+] (data not shown), and medium [PIN+] outcompetes low (Fig. 3); however, very high [PIN+] was outcompeted by high, medium, and low [PIN+] (Fig. 3 and data not shown). Therefore, the winner in these competitions is not always the one with the “strongest” [PIN+] phenotype. Meiotic progeny always exhibited the same phenotype as the diploid parent (Table 2). This result was true even when two different [PIN+] variants were crossed. For example, crosses between low [PIN+] and very high [PIN+] produced low [PIN+] diploids whose meiotic progeny always inherited low [PIN+].
Figure 3.
Genetic analysis of variants of [PIN+]. Independent diploids carrying the pEMBL-SUP35 plasmid reveal the outcome of crosses between haploid parents (P1 and P2) carrying the [pin−], low, medium (med.), or very high (v.h.) [PIN+] variants. The [PIN+] phenotypes are scored as in Fig. 2.
Table 2.
Meiotic inheritance of [PIN+] variants
| Parent 1 | Parent 2 | [PIN+] phenotypes of meiotic progeny
|
||||
|---|---|---|---|---|---|---|
| Total | Low | Med. | V.H. | [pin−] | ||
| [pin−] | [pin−] | 26 | 0 | 0 | 0 | 26 |
| [pin−] | Low | 30 | 29 | 0 | 0 | 1 |
| [pin−] | Med. | 29 | 0 | 29 | 0 | 0 |
| [pin−] | V.H. | 30 | 0 | 0 | 30 | 0 |
| Low | Low | 30 | 29 | 0 | 0 | 1 |
| Med. | Med. | 30 | 0 | 30 | 0 | 0 |
| V.H. | V.H. | 30 | 0 | 0 | 30 | 1 |
| Low | V.H. | 62 | 60 | 0 | 0 | 2 |
| Med. | V.H. | 60 | 0 | 60 | 0 | 0 |
| Med. | Low | 60 | 0 | 58 | 0 | 2 |
[PIN+] phenotypes were scored using the SUP35 overexpression assays shown in Fig. 2. For each row, ≈10 progeny were obtained from 3 or 6 independent diploids. Low, medium (Med.), and very high (V.H.) variants of [PIN+] were used.
Because very high [PIN+] was outcompeted by low, medium, and high [PIN+], it was possible that the latter phenotypes were caused by a combination of [PIN+] and an additional “modifier” prion distinct from [PIN+]. In this case, diploids formed from crosses of very high [PIN+] to, e.g., low [PIN+] would contain both the [PIN+] prion and the modifier prion, resulting in the low [PIN+] phenotype. To test this possibility, we cytoduced low and medium [PIN+] derivatives of c10B-H49 into a Δrnq1 derivative of BY4741 that, while unable to maintain [PIN+], should be able to maintain other cytoduced modifiers. The Δrnq1 recipient was then cytoduced into a very high [PIN+] derivative of c10B-H49 to test whether the hypothesized prion modifier would convert very high into low or medium [PIN+]. Because most cytoductants remained very high [PIN+] regardless of whether [pin−] control (11 of 12 independent cytoductants) or presumptive modifier-containing donor cytoplasm (11 of 12 independent cytoductants) was used, the data do not support the prion modifier hypothesis. In control experiments, we cytoduced weak or medium [PIN+] derivatives of BY4741 into a very high [PIN+] derivative of c10B-H49 and found that the majority of the recipients (10 of 15) were indeed converted into the phenotype of the donor (low or medium). Thus, it appears that the distinct phenotypes result from heritable differences in the [PIN+] aggregates themselves.
Comparison of Rnq1 Aggregation Among [PIN+] Variants.
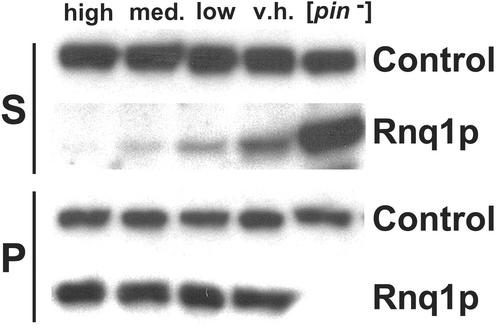
We compared the amounts of soluble and aggregated Rnq1 in the high, medium, low and very high [PIN+] variants. Each maintained indistinguishable amounts of aggregated Rnq1, but the levels of soluble Rnq1 showed strain-specific differences (Fig. 4). Strikingly, the hierarchy of [PIN+] variants determined by the competition experiments described above exactly corresponded to the gradient, from least to most, of soluble Rnq1 exhibited by these strains. For example, the variant with the most soluble Rnq1, very high, was lost when crossed with each of the other [PIN+] variants, whereas the variant with the least soluble Rnq1, high, outcompeted all of the other [PIN+] variants.
Figure 4.
Comparison of levels of soluble and aggregated Rnq1 among [PIN+] variants. Lysates were fractionated into soluble (S) and pellet (P) fractions by ultra-centrifugation. Rnq1 was detected by Western blotting with polyclonal Rnq1 antibody (kind gift from S. Lindquist). Stripped blots were then re-probed with monoclonal Sup35 antibody (Control) as a loading control. Note, the soluble Rnq1 in this figure were exposed twice as long as the Rnq1 in the pellet. The gradient depicted was generally reproducible; however, in three of eight independent protein isolations, the level of soluble Rnq1 in the v.h. and low variants appeared similar.
Effect of [PIN+] and [PSI+] on [URE3] Appearance.
Because [PIN+] facilitates the appearance of [PSI+] (10, 11), we asked whether it has a similar effect on the appearance of another prion, [URE3]. We compared the frequency with which overexpression of URE2 can induce the appearance of [URE3] in the [PIN+] yeast strain YHE711 and a [pin−] derivative of this strain obtained by growth on GuHCl. The original YHE711 strain had enhanced [URE3] induction relative to the [pin−] derivative, which had a drastically reduced frequency of [URE3] appearance (Table 3). Because deleting RNQ1 from the original [PIN+] strain also abolished the ability to induce [URE3], YHE711 does not harbor other elements in addition to [PIN+] that independently allow for [URE3] induction (Table 3). Clearly, [PIN+] facilitates [URE3] appearance. We further showed that [PIN+] did not cause greater overproduction of Ure2, nor did it stabilize newly appearing [URE3] (data not shown).
Table 3.
Influence of [PIN+] on [URE3] appearance
| Experiment | Frequency of [URE3] as % of [pin−] control
|
|||||
|---|---|---|---|---|---|---|
| [pin−] | Δrnq1 | [PIN+] Variants
|
||||
| Original | Very high | Medium | Low | |||
| 1 | 100 ± 59 | 4,600 ± 1,100 | ||||
| 2 | 100 ± 49 | 4,300 ± 2,400 | ||||
| 3 | 100 ± 50 | 37,000 ± 12,500 | 3,500 ± 1,200 | |||
| 4 | 100 ± 39 | 9,200 ± 4,600 | 550 ± 80 | |||
| 5* | 100 | 19 | 23,000 | 3,500 | ||
| 6* | 100 | 23 | 1,100 | 1,100 | ||
| 7* | 100 | 830 | 650 | 720 | ||
| 8 | 100 ± 50 | 800 ± 360 | 2,100 ± 1,100 | 1,200 ± 650 | ||
| 9 | 100 ± 142 | 15,00 ± 6,400 | 2,800 ± 1,300 | 1,000 ± 1,100 | ||
| 10 | 100 ± 143 | 39,000 ± 19,000 | 510 ± 230 | 5,700 ± 3,800 | ||
| Average | 100 | 21 | 16,700 | 1,700 | 2,400 | 960 |
Full-length Ure2 was overproduced in derivatives of YHE711. The frequency of [URE3] colonies appearing in the [pin−] strain per 106 cells plated is normalized to 100, and other values were normalized accordingly. The YHE711 derivatives were: GuHCl-treated ([pin−]), RNQ1 deletion (Δrnq1), not GuHCl-treated (Original), GuHCl-treated and cytoduced with very high, medium, or low [PIN+]. Averages and standard deviations are shown when three or more transformants were assayed. Induction of [URE3] by overproducing just the Ure2 prion domain (1-65 aa) was also facilitated by the presence of [PIN+] (data not shown). Each experiment also included controls in which URE2 was not overexpressed, where the numbers of [URE3] colonies was very low (not shown).
Indicates that only two measurements were performed, the average is shown.
To determine whether the different variants of [PIN+] that were characterized above in terms of their distinct [PSI+] induction frequencies could also be distinguished on the basis of their effect on [URE3] induction, we cytoduced several different variants of [PIN+] into a GuHCl-treated derivative of YHE711. Surprisingly, the original YHE711 [PIN+] facilitated [URE3] appearance more efficiently than even the very high [PIN+] (Table 3), yet the original YHE711 [PIN+] was less efficient than very high [PIN+] in the Sup35-based [PSI+] induction phenotypes (data not shown). Whereas the presence of the low, medium, or very high [PIN+] elements enhanced the frequency of [URE3] induction relative to the [pin−] control, we could not consistently observe any differences in [URE3] induction levels among strains with the low, medium and very high [PIN+] variants (Table 3).
Although it generally appears that prions enhance the appearance of other prions (13), we find here that [PSI+] does not facilitate [URE3] induction, but rather inhibits its appearance. By overexpressing the URE2Δ151–158 allele, which efficiently induces [URE3] (15, 54), we found that a [PSI+] [pin−] derivative of YHE711 is less inducible to [URE3] compared with a [psi−] [pin−] derivative (averages of 346 ± 107 vs. 989 ± 394 [URE3]/106 cells from six independent experiments).
Effect of YDJ1 Overexpression on [PIN+].
Various [psi−] [PIN+] derivatives of 74-D694, as well as a [psi−] [pin−] control, were transformed with p901, carrying YDJ1 under the control of the GAL1 promoter. Transformants were grown on media containing galactose to induce YDJ1 expression, after which Rnq1 aggregation was used to detect [PIN+]. Some [PIN+] variants were readily cured by overproducing Ydj1, whereas other [PIN+] variants were not cured (Table 4). [PIN+] was never lost in control experiments by using the pH316 empty vector (Table 4). The experiment gave similar results when repeated with two of the curable and three of the incurable [PIN+] variants (Table 4). Thus, we find that overexpressing YDJ1 promotes the loss of some [PIN+] variants.
Table 4.
YDJ1 cures some [PIN+] variants
| Strain | Expt. 1
|
Expt. 2
|
||||||
|---|---|---|---|---|---|---|---|---|
| Control
|
YDJ1
|
Control
|
YDJ1
|
|||||
| [pin−] | [PIN+] | [pin−] | [PIN+] | [pin−] | [PIN+] | [pin−] | [PIN+] | |
| [pin−] | 24 | 0 | 24 | 0 | 35 | 0 | 48 | 0 |
| V.H. | 0 | 24 | 0 | 24 | ||||
| Low | 0 | 24 | 0 | 24 | ||||
| Med. | 0 | 24 | 8 | 16 | ||||
| High | 0 | 24 | 2 | 22 | 0 | 52 | 0 | 64 |
| L1941 | 0 | 24 | 3 | 21 | ||||
| L1952 | 0 | 24 | 14 | 10 | 0 | 61 | 29 | 32 |
| L1947 | 0 | 24 | 22 | 2 | 0 | 66 | 11 | 56 |
| L1949 | 0 | 24 | 0 | 24 | 0 | 49 | 4 | 62 |
| L1956 | 0 | 24 | 0 | 24 | 0 | 46 | 0 | 66 |
Strains are derivatives of 74-D694. The very high (V.H.), low, medium (Med.), and high [PIN+] variants are indicated as such. Other independent [PIN+] isolates are also indicated (L1941–1956). Colonies checked from the control (pH316) and YDJ1 bearing (p901) transformants are shown.
Discussion
According to the protein-only prion model, two explanations of the prion strain phenomena are possible. Prion variants may result from inherent flexibility of the tertiary structure allowing one chain of amino acids to have two or more self-perpetuating conformations that are stably inherited. Alternatively, variants might result from a single tertiary conformation arranged into two or more different quaternary arrangements that are stable and self-perpetuating. Here, the existence of distinct heritable variants of the [PIN+] prion is described, and the phenotypic differences between them are shown not to be due to either nuclear or cytoplasmic modifiers. It is now clear that each of the well characterized yeast prions—[PSI+], [URE3], and [PIN+]—can exist as different distinct heritable variants.
Although different prions, e.g., [PSI+] and [PIN+], or [PSI+] and [URE3], can be maintained together in a single cell (11, 13, 16), such coexistence may not be possible for two variants of the same prion. Indeed, diploids formed by mating cells with different [PSI+] variants that could be distinguished from each other by Sup35-GFP staining retained only one of the [PSI+] variants (25). Furthermore, if prion variants could coexist, one might expect diploids formed from crosses between isogenic cells bearing weak and strong [PSI+] to exhibit a phenotype more extreme than strong [PSI+], and to occasionally segregate out both strong and weak [PSI+] in mitotic and meiotic growth. We show here that this is not the case. Rather, except for rare cases of loss of [PSI+], the diploids and all mitotic and meiotic progeny were indistinguishable from the strong [PSI+] parent. Furthermore, weak [PSI+] did not emerge from the diploids created by crossing weak and strong [PSI+] even after stimulation of [PSI+] loss by short-term growth in medium containing GuHCl (M.E.B. and S.W.L., unpublished work).
Although one could argue that strong and weak [PSI+] coexist, but that the phenotype of strong [PSI+] cannot be made any stronger, similar results obtained for crosses between cells bearing the different [PIN+] variants cannot easily be explained in this way. This result is because, unlike crosses between weak and strong [PSI+], where the variant with the strongest phenotype prevails, [PIN+] variants with less dramatic phenotypes (low, medium, and high [PIN+]) prevailed over very high [PIN+], which has the most dramatic phenotype. Thus, if the different [PIN+] variants coexisted, the very high [PIN+] phenotype could not reasonably be expected to be hidden by the [PIN+] variants with milder phenotypes.
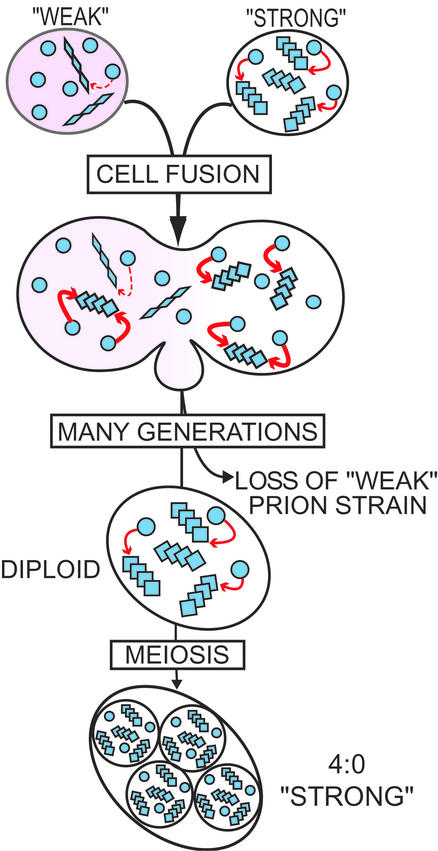
Because two variants of the same prion compete for the same pool of newly synthesized protein to reproduce and be heritable, faster replicating variants should eventually outcompete slower or less stable variants by starving them for convertible protein (Fig. 5). Indeed we found that [PIN+] variants, which maintain little soluble Rnq1 but abundant aggregated Rnq1, indicating fast reproduction, did take over when combined with [PIN+] variants that maintain more soluble Rnq1 and less abundant aggregated Rnq1. Previous observations indicate that [PSI+] variants show a similar pattern (23, 24).
Figure 5.
Cartoon depicting a competition for soluble protein between variants of the same prion. Haploids carrying soluble protein (free circles) and aggregated-prion protein as either “weak” (rhomboids) or “strong” (squares) variants are mated. Mitotic growth of the diploid results in loss of the weak variant, presumably because it is starved for convertible soluble protein. When the diploid sporulates, each spore inherits the strong prion variant.
Recent work has demonstrated that certain prions facilitate the appearance of other prions: [PIN+] and [URE3] (13), or the artificial fusion protein prion [NU+] (16), permit overexpression of SUP35 to induce the appearance of [PSI+]; and [PSI+] and [URE3] facilitate the appearance of [PIN+] (13). Here, we show that [PIN+] facilitates, but [PSI+] inhibits, the de novo appearance of [URE3].
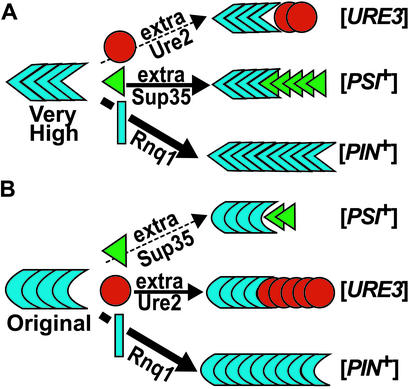
Two mechanisms of prion-facilitated prion appearance have been proposed (13, 16). According to the seeding model, heterologous prions provide a template for initial cross-seeding of a de novo prion aggregate. The titration model hypothesizes that preexisting heterologous prion aggregates sequester a protein that normally inhibits prion appearance, thereby allowing other prions to appear more easily. Our current finding of no correlation between the efficiencies with which the different [PIN+] variants promote the appearance of [PSI+] and the efficiencies with which they promote the appearance of [URE3] can most easily be explained by the cross-seeding model. We propose that some [PIN+] variants cross-seed Sup35 better than Ure2, whereas others exhibit the opposite preference (Fig. 6). More complicated scenarios involve combinations of the seeding and titration models, or multiple inhibitors with distinct binding properties.
Figure 6.
Models illustrating the different seeding preferences proposed for two [PIN+] variants. (A) The very high [PIN+] variant inefficiently seeds Ure2 (red circles), but efficiently seeds Sup35 (green triangles) converting them into the [PSI+] shape. (B) The original YHE711 [PIN+] variant inefficiently seeds Sup35, but efficiently seeds Ure2, converting them into the [URE3] shape. Both [PIN+] variants propagate their forms by converting Rnq1 (blue rectangles) with the highest efficiency.
The [PIN+] variants described here cannot easily be distinguished by the amounts of aggregated Rnq1 (Fig. 4). There is also no correlation between the levels of soluble Rnq1 and the phenotypes of the [PIN+] variants: the order of increasing soluble Rnq1 levels is high, medium, low, then very high. One possibility to explain this conundrum is that different prion conformations of Rnq1 are better at influencing [PSI+] appearance, and these conformations are only coincidentally distinguishable by soluble Rnq1 levels. Another possibility is that accessory proteins, such as Sis1 (34), are associated in different amounts with each of the [PIN+] variants. The presence of such proteins may hinder the action of [PIN+] or, if these are chaperone proteins, they might be essential for creating the action of [PIN+]. It is also possible that other variants of [PIN+] that do not facilitate the induction of [PSI+] or [URE3] may exist.
We have unexpectedly found that the presence of one prion can inhibit the de novo appearance of another, because [PSI+] inhibited the appearance of [URE3]. Whereas the effect of [PSI+] on the induction of [URE3] is inhibitory, it still suggests that heterologous prions interact. [PSI+] may inhibit de novo [URE3] appearance by occasionally joining and “poisoning” [URE3] seeds thereby inhibiting [URE3] propagation as previously proposed to explain the [PSI+] curing effect of certain SUP35 mutants and the [URE3] curing effect of URE2-GFP (55–57). Alternatively, [PSI+] may sequester Ure2Δ151–158 thereby reducing the amount of protein available to form [URE3] seeds. That [PSI+] stimulates [PIN+] appearance but inhibits [URE3] appearance is inconsistent with the inhibitor model and more compatible with the idea that heterologous prions directly interact—sometimes causing cross-seeding and sometimes causing inhibition.
Prions are stable, heritable elements. Yeast cells should therefore acquire multiple different prions by mating with other cells—even though the prions may be disadvantageous under some circumstances. Our finding that prions are occasionally eliminated by meiosis may be an indication that yeast have evolved mechanisms of ridding themselves of prions. Several possibilities might explain how prions are eliminated by meiosis. Because overexpressing HSP104 is known to eliminate [PSI+] (32), the elevated HSP104 levels associated with sporulating cultures (58) might disrupt the inheritance of [PSI+]. Alternatively, because meiotic progeny receive less cytoplasm than mitotic daughter cells (59), they might lose [PSI+] more frequently as a result of inheriting fewer [PSI+] seeds. Because most Ure2 amyloid filaments are found in a single cytoplasmic network in [URE3] cells (60), it is not surprising that [URE3] is often lost in meiosis.
Overexpression of YDJ1, which interacts with Hsp104 and Hsp70 to rescue denatured proteins (35), was previously shown to cause the loss of a [URE3] variant (33). Here, we show that overexpressing YDJ1 eliminates some, but not other, [PIN+] variants. [URE3] and [PIN+] are both eliminated by deletion, but not overexpression, of HSP104 (10, 33, 34). The spontaneous [PIN+] variants described in this paper, including those that were not cured by Ydj1 overexpression, were cured by deleting HSP104 (M.E.B. and S.W.L., unpublished work). Possibly, YDJ1 may cure some [PIN+] variants by sequestering Hsp104 (33), but other [PIN+] variants may be less sensitive to the reduction of Hsp104. Alternatively, Ydj1 may stimulate protein refolding and therefore cure [PIN+] by disaggregating it. If this result were true, it could mean that some [PIN+] variants maintain too many seeds, or replicate too quickly to be eliminated by overexpressing just Ydj1. Possibly overexpressing Hsp104 or Hsp70 along with Ydj1 would eliminate some of these incurable [PIN+] variants. Indeed, overexpression of Ydj1 together with Ssa1 or Ssb1 did cause loss of a weak [PSI+] (26).
Our results show that [PIN+] prions exhibit strain variation, as do [PSI+] and [URE3] (14, 22), further supporting the hypothesis that strain variation is compatible with the protein-only model for prions. The recent suggestions that prions affect the appearance of other prions (13, 16) and that prion strain variation and species barrier are related phenomena (29) indicate that elucidating the molecular basis of [PIN+] strain variation may yield important clues about the transmissibility of prion diseases between different species.
Acknowledgments
We thank D. Bishop, R. Esposito, and S. Lindquist for strains, plasmids, and antibodies; I. Derkatch, J. Gavin-Smyth, and Y. Vitrenko for helpful comments about the manuscript; and J. Gavin-Smyth for help with the figures. This work was partially supported by a grant from the National Institutes of Health (GM56350) to S.W.L.
Abbreviations
- GuHCl
guanidine⋅hydrochloride
- Cyh
cycloheximide
- GFP
green fluorescent protein
- USA
ureidosuccinate
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 2.Griffith J S. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 4.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 5.Cox B S. Heredity. 1965;20:505–521. [Google Scholar]
- 6.Liebman S W, Derkatch I L. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 7.Serio T R, Lindquist S L. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 8.Wickner R B, Taylor K L, Edskes H K, Maddelein M L, Moriyama H, Roberts B T. Adv Protein Chem. 2001;57:313–334. doi: 10.1016/s0065-3233(01)57026-6. [DOI] [PubMed] [Google Scholar]
- 9.Lacroute F. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derkatch I L, Bradley M E, Zhou P, Chernoff Y O, Liebman S W. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derkatch I L, Bradley M E, Masse S V, Zadorsky S P, Polozkov G V, Inge-Vechtomov S G, Liebman S W. EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sondheimer N, Lindquist S. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 13.Derkatch I L, Bradley M E, Hong J Y, Liebman S W. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 14.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 16.Osherovich L Z, Weissman J S. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 17.Mabbott N A, Bruce M E. J Gen Virol. 2001;82:2307–2318. doi: 10.1099/0022-1317-82-10-2307. [DOI] [PubMed] [Google Scholar]
- 18.Bessen R A, Kocisko D A, Raymond G J, Nandan S, Lansbury P T, Caughey B. Nature (London) 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 19.Caughey B, Kocisko D A, Raymond G J, Lansbury P T. Chem Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 20.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 21.Mestel R. Science. 1996;273:184–189. doi: 10.1126/science.273.5272.184. [DOI] [PubMed] [Google Scholar]
- 22.Schlumpberger M, Prusiner S B, Herskowitz I. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Derkatch I L, Uptain S M, Patino M M, Lindquist S, Liebman S W. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derkatch I L, Bradley M E, Zhou P, Liebman S W. Curr Genet. 1999;35:59–67. doi: 10.1007/s002940050433. [DOI] [PubMed] [Google Scholar]
- 25.King C Y. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 26.Kushnirov V V, Kryndushkin D S, Boguta M, Smirnov V N, Ter-Avanesyan M D. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 27.Liebman S W, All-Robyn J A. Curr Genet. 1984;8:567–573. doi: 10.1007/BF00395701. [DOI] [PubMed] [Google Scholar]
- 28.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 29.Chien P, Weissman J S. Nature (London) 2001;410:223–227. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- 30.Uptain S M, Sawicki G J, Caughey B, Lindquist S. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochneva-Pervukhova N V, Chechenova M B, Valouev I A, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- 32.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 33.Moriyama H, Edskes H K, Wickner R B. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondheimer N, Lopez N, Craig E A, Lindquist S. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 36.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 37.Chernoff Y O, Newnam G P, Kumar J, Allen K, Zink A D. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung G, Jones G, Wegrzyn R D, Masison D C. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aigle M, Lacroute F. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 41.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 42.Tuite M F, Mundy C R, Cox B S. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldring E S, Grossman L I, Krupnick D, Cryer D R, Marmur J. J Mol Biol. 1970;52:323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 44.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 45.Kane S M, Roth R. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herskowitz I, Jensen R E. Methods Enzymol. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-z. [DOI] [PubMed] [Google Scholar]
- 47.Edskes H K, Hanover J A, Wickner R B. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou P, Derkatch I L, Liebman S W. Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- 49.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 50.Kochneva-Pervukhova N V, Poznyakovski A I, Smirnov V N, Ter-Avanesyan M D. Curr Genet. 1998;34:146–151. doi: 10.1007/s002940050379. [DOI] [PubMed] [Google Scholar]
- 51.Ter-Avanesyan M D, Kushnirov V V, Dagkesamanskaya A R, Didichenko S A, Chernoff Y O, Inge-Vechtomov S G, Smirnov V N. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 52.Edskes H K, Wickner R B. Proc Natl Acad Sci USA. 2000;97:6625–6629. doi: 10.1073/pnas.120168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conde J, Fink G R. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maddelein M L, Wickner R B. Mol Cell Biol. 1999;19:4516–4524. doi: 10.1128/mcb.19.6.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kochneva-Pervukhova N V, Paushkin S V, Kushnirov V V, Cox B S, Tuite M F, Ter-Avanesyan M D. EMBO J. 1998;17:5805–5810. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DePace A H, Santoso A, Hillner P, Weissman J S. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 57.Edskes H K, Gray V T, Wickner R B. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brewer B J, Fangman W L. Proc Natl Acad Sci USA. 1980;77:5380–5384. doi: 10.1073/pnas.77.9.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speransky V V, Taylor K L, Edskes H K, Wickner R B, Steven A C. J Cell Biol. 2001;153:1327–1336. doi: 10.1083/jcb.153.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]