Abstract
The formation of insoluble protein aggregates in neurons is a hallmark of neurodegenerative diseases caused by proteins with expanded polyglutamine (polyQ) repeats. However, the mechanistic relationship between polyQ aggregation and its toxic effects on neurons remains unclear. Two main hypotheses have been put forward for how polyQ expansions may cause cellular dysfunction. In one model neurotoxicity results from the ability of polyQ-expanded proteins to recruit other important cellular proteins with polyQ stretches into the aggregates. In the other model, aggregating polyQ proteins partially inhibit the ubiquitin–proteasome system for protein degradation. These two mechanisms are not exclusive but may act in combination. In general, protein misfolding and aggregation are prevented by the machinery of molecular chaperones. Some chaperones such as the members of the Hsp70 family also modulate polyQ aggregation and suppress its toxicity. These recent findings suggest that an imbalance between the neuronal chaperone capacity and the production of potentially dangerous polyQ proteins may trigger the onset of polyQ disease.
Expansions of CAG trinucleotide repeats encoding polyglutamine (polyQ) tracts in otherwise unrelated proteins are responsible for at least nine different neurodegenerative diseases (1–3). These diseases include Huntington's disease (HD), spinobulbar muscular atrophy, dentatorubral pallidoluysian atrophy, and spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7, and 17. With the exception of spinobulbar muscular atrophy, these neurodegenerative diseases are inherited in an autosomal dominant manner. All nine disorders show late onset of neurological symptoms with progressive neuronal dysfunction and eventual neuronal loss, although the susceptible regions in the nervous system differ among the various disorders. Generally, the pathologic length of the polyQ repeat is ≈40 or greater, whereas healthy individuals have polyQ repeats with fewer residues. Longer CAG repeats result in an earlier age of onset and a more severe pathology, consistent with a disease mechanism by gain of function. These fundamental observations point to a common molecular mechanism underlying the pathology of polyQ diseases. Exactly how polyQ expansions cause neuronal dysfunction is still obscure, however.
A characteristic feature of polyQ diseases is the formation of insoluble, granular, and fibrous deposits in affected neurons termed neuronal inclusions, which have been studied extensively in HD (4). HD is characterized by selective neuronal loss, primarily in the cortex and striatum, leading to motor impairment, personality changes, and dementia. The disease is caused by the expansion of a polyQ segment located within the first exon of the gene encoding huntingtin, an ≈350-kDa protein of unknown but essential function (5, 6). The neuronal inclusions in HD have fibrillar morphology and contain aggregated amino-terminal fragments of huntingtin (7). Similar inclusions containing aggregated polyQ proteins are detected also in other polyQ diseases (8, 9), suggesting a causal relationship between these neurodegenerative disorders and amyloid fibrillogenesis (10). However, it remains unclear whether the aggregates themselves are pathogenic, epiphenomenal, or even beneficial. For example, large polyQ aggregates may provide an advantage over small oligomers by exposing less potentially dangerous protein surfaces.
It is thought that the aggregates result from the ability of long polyQ stretches to form self-associating β-sheets (11, 12). Interestingly, certain transcription factors containing polyQ segments in the nonpathological range, such as TATA-binding protein (TBP) and CREB-binding protein (CBP), are detected in neuronal inclusions (13–15). It has been proposed that sequestration of these essential proteins via polyQ–polyQ interactions may cause neuronal toxicity. Additional proteins detected in neuronal inclusions are ubiquitin, the 19S and 20S proteasome complexes, and several molecular chaperones (8, 16–19). Importantly, the cellular components involved in protein folding and degradation are associated also with intracellular inclusions in other neurodegenerative diseases not caused by polyQ expansion, including Alzheimer's disease, Parkinson's disease, and the prion diseases (20), which suggests that common pathomechanistic principles may underlie these misfolding diseases in general.
In this review, we focus mainly on the molecular mechanism of polyQ aggregation and its cellular toxicity. The functional relationship between molecular chaperones, the ubiquitin–proteasome system, and polyQ aggregation will be discussed.
Mechanism of polyQ Aggregation
In 1994, Perutz proposed that long sequences of polyQ might be able to form stable β-hairpins (11). These structures, also called “polar zippers,” consist of polyQ-containing β-strands held together by hydrogen bonds between both main-chain and side-chain amides. PolyQ-containing hairpins may self-associate, forming stable β-sheet aggregates with fibrillar morphology. In a recent, refined structural model, expanded stretches of polyQ are proposed to form a cylindrical (helical), parallel β-sheet rather than an antiparallel β-sheet (12). In this model, the number of Q repeats per turn in polyQ fibrils is estimated at ≈20, but a single 20-residue helical turn would be unstable. In contrast, a polyQ segment with ≈40 residues could be stabilized by amide hydrogen bonds between successive turns and could act as a nucleus for further growth of a helical fibril. The external and internal diameters of a cylindrical fibril are supposed to be ≈30 and 10 Å, respectively. Thicker amyloid fibers may consist of two or more cylindrical β-sheet fibrils wound around one another (10). A parallel β-sheet structure has been suggested also for the amyloid fibrils formed by the yeast prion protein Sup35 and the Alzheimer's Aβ peptide (21–23).
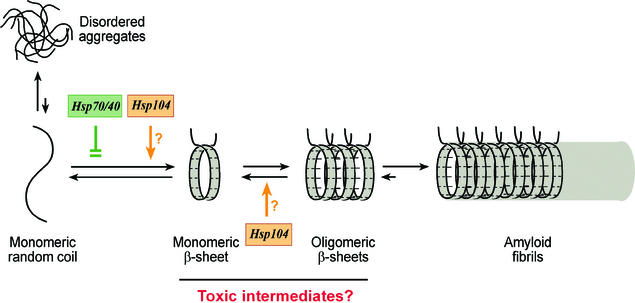
The predicted length dependence of polyQ aggregation was confirmed by in vitro experiments. Wanker and coworkers showed that amino-terminal huntingtin fragments with polyQ tracts exceeding a critical length of ≈40 residues form SDS-resistant aggregates with a fibrillar morphology (24), similar to that of Aβ amyloid and yeast prion protein Sup35 fibrils. It has been proposed that fibril formation in general occurs by a mechanism of nucleated polymerization (10, 25). This mechanism is characterized by the rate-limiting formation of an oligomeric nucleus from monomers that have undergone a (transient) conformational change followed by rapid recruitment of further monomers or oligomers into highly ordered fibrils. A model of how such a mechanism may apply to polyQ fibrils is shown in Fig. 1. In this model, the first step in the aggregation process is proposed to be the repeat length-dependent conformational change of polyQ monomers from random coil to a parallel, helical β-sheet (12). This structural conversion then results in the formation of ordered polyQ oligomers that could function as nuclei for the rapid polymerization of amyloid-like fibrils. Either the initial conversion or the subsequent oligomerization may be the rate-limiting step in polyQ fibrillogenesis, dependent on protein concentration. The intramolecular formation of β-sheet structure is likely to be rate-limiting only at high concentrations of polyQ protein, as reported for the Sup35 protein (26). In the case of huntingtin exon 1, proteolytic cleavage of the polyQ-containing segment from a nonaggregating precursor construct in vitro initiates aggregation, presumably by relieving steric restrictions and facilitating both intramolecular conversions and formation of oligomeric nuclei (24, 27). Thus, after cleavage, the concentration-dependent formation of soluble polyQ oligomers is likely to be rate-limiting in the aggregation of this protein. Indeed, both full-length huntingtin (350 kDa) and the SCA3 protein (ataxin-3, 42 kDa) are cleaved by as-yet-unidentified proteases in vivo (7, 28–30). The resulting production of polyQ-containing fragments dramatically enhances the formation of intracellular inclusions in both transgenic animals and cell-culture systems (28, 31, 32).
Figure 1.
Hypothetical model for the pathway of polyQ aggregation and its modulation by molecular chaperones. The first step in the aggregation process is thought to be the structural conversion of polyQ monomers from a random coil to β-sheet conformation, followed by oligomerization. Once oligomers have formed, polymerization into amyloid-like fibrils occurs rapidly. Monomers or soluble oligomers with β-sheet conformation may be toxic intermediates in the aggregation process, whereas amyloid fibrils as the end product of aggregation may be nontoxic. The Hsp70/Hsp40 chaperone system is proposed to prevent the initial conformational conversion and eventually chaperone-associated disordered aggregates form. In contrast, Hsp104 is proposed to promote conversion into β-sheet conformation at low relative concentration to polyQ protein but at high concentrations may dissociate soluble polyQ oligomers, thereby slowing the aggregation process.
Relationship Between polyQ Aggregation and Toxicity
There are several lines of evidence that suggest a causal link between polyQ aggregation and the disease process. The numbers of neuronal inclusions in patient brains and the severity of neurological symptoms correlate with the polyQ repeat length of the expressed protein (33). Furthermore, the inclusions are present primarily in those neurons that are particularly vulnerable to the disease (7–9). These phenomena have been reproduced in both transgenic animals and cell-culture systems. In transgenic mice expressing polyQ-expanded HD exon 1, neuronal inclusions containing aggregated HD protein form before the onset of neurological symptoms (16). Regulated expression of HD exon 1 in a conditional transgenic mouse model resulted in a progressive neurological phenotype with polyQ inclusions in the striatum and cortex (34). Unexpectedly, when expression of HD exon 1 protein was switched off after polyQ aggregates had formed, neurological symptoms disappeared along with the intracellular inclusions. Although these observations do not establish a causal relationship between the inclusions and either disease initiation or progression, they link the process of polyQ protein aggregation to cellular dysfunction and disease.
Elucidating the mechanism(s) by which polyQ aggregation exerts cellular toxicity now represents one of the most challenging problems in the field. Interestingly, there are several reports that the neuronal inclusions themselves are not necessary for the initiation of symptoms and may even help to protect against cellular dysfunction (35, 36). Saudou et al. showed that expression of a dominant-negative ubiquitin-conjugating enzyme in cell culture reduces huntingtin aggregation but enhances its toxicity (36). Zoghbi and coworkers reported similar results in SCA1 transgenic mice (37). These studies strongly indicate that microscopically detectable inclusions are not necessary for disease initiation. As mentioned above, the critical step in the aggregation process is thought to be an intramolecular conformational change in the polyQ protein that precedes the formation of an oligomeric nucleus (Fig. 1). It is possible that these nuclei or other early, i.e., prefibrillar, oligomeric intermediates in the aggregation process are the toxic agents. This idea parallels the recent demonstration by two groups that early stages in the process of protein misfolding and amyloid fibril formation are important for cellular toxicity, whereas the fibrils themselves are not toxic (38, 39). Specifically, soluble dimers and trimers of the Aβ peptide impair neuronal functions in rat brains but not Aβ monomers, protofibrils, and fibrils (39). The toxic aggregation intermediates of Aβ peptide accumulate extracellularly and may damage cell surface structures. Similarly, the unprotected β-sheets in intracellular polyQ oligomers or even in the monomeric polyQ fragments generated from full-length precursor proteins may interact unfavorably with the surfaces of other proteins, thereby impairing various cellular functions (Fig. 1). To critically test this “toxic-intermediate” hypothesis of polyQ disease, it will be important to define the biochemical and biophysical properties of the exposed β-sheets in the soluble states of polyQ proteins.
Transcriptional Dysregulation by polyQ Aggregation
Analyzing the cellular components contained in neuronal polyQ inclusions may offer valuable clues as to how polyQ expansions cause cellular dysfunction. To date, numerous proteins have been shown to be sequestered into the inclusions using immunohistochemical and biochemical approaches. One class of recruited proteins comprises essential transcription factors with nonpathological length polyQ tracts, such as TATA-binding protein and CBP (13–15, 40). For example, Ross and coworkers reported that CBP is depleted from its normal nuclear location and becomes sequestered into polyQ aggregates in HD cell-culture models, HD transgenic mice, and human HD postmortem brains (15). The CBP homolog p300, lacking a substantial polyQ stretch, is not recruited to neuronal inclusions, suggesting that sequestration occurs via polyQ–polyQ interactions. Importantly, the expression of expanded polyQ proteins specifically interfered with CBP-activated gene transcription, causing cellular toxicity. Gene-array studies also showed that expression of genes controlled by cAMP-response elements (CREs) is down-regulated in HD transgenic mice or cell-culture models (41, 42). More recently, transgenic mice with disruptions in two CRE-binding proteins (CREB1 and CREM) in the adult forebrain were shown to develop progressive neurodegeneration in the dorsolateral striatum reminiscent of HD (43). Taken together, at least in HD, dysregulation of CREB-mediated transcription by sequestration of CBP into neuronal inclusions may cause cellular toxicity.
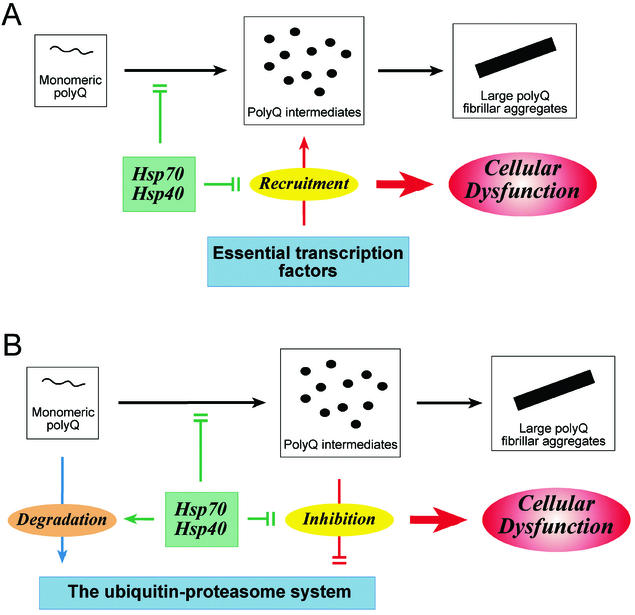
Considering that alterations in gene expression have been reported in several other polyQ disorders, including SCA1, SCA3, and dentatorubral pallidoluysian atrophy (44, 45), sequestration of transcription factors into neuronal inclusions provides an attractive explanation for the toxicity common to these diseases. Because transcription factors act in the nucleus, this model also would explain the greater toxicity of intranuclear versus cytosolic polyQ protein aggregation. Whether recruitment is mediated by (toxic) intermediates in the aggregation process or the final polyQ fibrils is still unclear. On the other hand, additional, alternative mechanisms also may be involved in polyQ-related transcriptional dysregulation. For example, TAFII130, another important positive regulator of CREB-mediated transcription, was shown to bind directly to long polyQ stretches in SCA3 and dentatorubral pallidoluysian atrophy, even though TAFII130 lacks a polyQ tract (45). Long polyQ stretches also may cause a pronounced inhibition of the histone acetyltransferase (HAT) activities of CBP and other proteins such as P/CAF (p300/CBP-associated factor) by binding to their acetyltransferase domains, not their polyQ tracts (46). Interestingly, administration of histone deacetylase inhibitors had remarkable beneficial effects in a fly model of HD (46). Although it is unclear at the moment which mechanism(s) described above is primarily responsible for alterations in gene expression by polyQ expansion proteins, transcriptional dysregulation is clearly one important element of polyQ toxicity (Fig. 2A).
Figure 2.
Two models for polyQ-mediated toxicity and its suppression by the Hsp70/Hsp40 chaperone system. (A) Transcriptional dysregulation. Intermediates of polyQ aggregation recruit essential transcription factors, thereby inhibiting their transcriptional activities and causing cellular dysfunction. The Hsp70/Hsp40 chaperone system prevents this recruitment and thus mitigates polyQ toxicity. (B) Inhibition of the ubiquitin–proteasome system. PolyQ aggregation intermediates trap the 19S regulatory complex (and other components of the degradative machinery), resulting in a partial inhibition of proteasome-dependent proteolysis and eventually cellular dysfunction. The Hsp70/Hsp40 chaperone system may prevent or reduce this effect by stabilizing polyQ protein in a soluble, degradation-competent state and shielding aggregates.
Role of Molecular Chaperones in polyQ Aggregation and Toxicity
Components involved in protein folding and degradation represent another group of proteins frequently recruited to polyQ inclusions. These factors include several molecular chaperones such as Hsp70 and Hsp40 as well as ubiquitin and the 20S and 19S proteasome complexes (8, 16–19, 40). Their presence in aggregates suggests that expanded polyQ tracts are recognized as misfolded conformers and that cellular quality-control mechanisms are activated in an attempt to prevent their accumulation (20).
Hsp70 chaperones promote protein folding by an ATP-dependent process of binding and release of extended polypeptide segments enriched in hydrophobic residues that are typically exposed by nonnative, i.e., fully or partially unfolded, proteins (47, 48). These structural features often give rise to intermolecular association (aggregation) mediated by hydrophobic interactions and β-sheet formation (49). Hsp70 binding may prevent protein aggregation directly by shielding the interactive surfaces of nonnative polypeptides and indirectly by inhibiting or reversing intramolecular misfolding. Hsp70 cooperates in this function with members of the Hsp40 family. These cochaperones are homologs of bacterial DnaJ and contain a so-called J domain. They have a critical role in mediating substrate binding to Hsp70 but alone are inefficient in preventing aggregation. Most Hsp40s recognize nonnative polypeptide segments and target them to Hsp70 by a direct interaction. In addition, the J domain of Hsp40 activates the Hsp70 ATPase, thereby catalyzing the formation of the ADP-state of Hsp70, which binds protein substrate tightly (48). This Hsp40 effect may be particularly important for the binding by Hsp70 of extended sequences not containing hydrophobic amino acid residues (50), such as polyQ. Expression of Hsp70 and Hsp40 chaperones is induced under various conditions of cell stress such as heat shock, which result in unfolding and aggregation of certain proteins.
There are several reports that increased expression of the Hsp70/Hsp40 chaperone system can suppress polyQ-induced neurotoxicity in fly models (51–54) and a mouse model of polyQ disease (ref. 55; Table 1) [reviewed by Bonini in this issue (ref. 56)]. These studies identified the Hsp40 protein dHdj1, the Drosophila homolog of human Hdj1, and the J-domain cochaperone dTPR2 as well as dHsp70, a Drosophila homolog of human Hsp70, as active components (51–54). Suppression of toxicity by expression of Hdj1 or dTPR2 alone is most likely caused by the ability of these cochaperones to activate the endogenous Hsp70 for polyQ protein binding, an effect that may be less pronounced with other Hsp40 homologs. In contrast, expression of a dominant negative mutant form of Hsp70 increased polyQ toxicity (52). The finding by Cummings et al. that overexpression of Hsp70 suppresses polyQ-induced toxicity in SCA1 transgenic mice (55) validates molecular chaperones and the components involved in regulating their expression as promising targets in developing a possible therapy for polyQ diseases (57).
Table 1.
Effects of molecular chaperones on polyQ aggregation and toxicity in vivo
| Chaperones | Organisms | PolyQ disease model | Aggregation | Toxicity | References |
|---|---|---|---|---|---|
| Hsp70↑ | Mouse | SCA-1 | → | ↓ | 55 |
| dHsp40 (dHdj1)↑ | Drosophila | HD | → | ↓ | 51 |
| dTPR2↑ | Drosophila | HD | → | ↓ | 51 |
| dHsp70↑ | Drosophila | SCA-3 | → | ↓ | 52 |
| dHsp70↓ | Drosophila | SCA-3 | n.d. | ↑ | 52 |
| dHsp40 (dHdj1)↑ | Drosophila | SCA-3 | n.d. | ↓ | 53 |
| dHsp40 (dHdj1)↓ | Drosophila | SCA-3 | n.d. | ↑ | 53 |
| dHsp40 (dHdj2)↑ | Drosophila | SCA-3 | n.d. | ↓ | 53 |
| dHsp70↑ + dHdj1↑ | Drosophila | SCA-3 | → | ↓↓ | 53 |
| dHsp40 (dHdj1)↑ | Drosophila | HD | n.d. | ↓ | 53 |
| dHsp40 (dHdj2)↑ | Drosophila | HD | n.d. | → | 53 |
| dHsp40 (dHdj1)↑ | Drosophila | SCA-1 | → (more compact) | ↓ | 54 |
| dVCP/Cdc48↓ | Drosophila | SCA-3 | → | ↓ | 63 |
| dVCP/Cdc48↑ | Drosophila | SCA-3 | → | ↑ | 63 |
| Hsp104↑ | C. elegans | DRPLA | ↓ | ↓ | 66 |
| Hsp70 (SSA1)↑ | S. cerevisiae | HD | ↓ | (−) | 62 |
| Hsp104↑ | S. cerevisiae | HD | ↓ | (−) | 62 |
| Hsp104↓ | S. cerevisiae | HD | ↓↓ | (−) | 62 |
| Hsp104↓ | S. cerevisiae | SCA-3 | ↓ | (−) | 67 |
n.d., not determined.
Surprisingly, in all cases studied, overexpression of Hsp70/Hsp40 chaperones did not prevent the formation of polyQ aggregates, although polyQ toxicity was suppressed. Experiments by Muchowski et al. (58) helped to resolve this puzzle. They demonstrated that these molecular chaperones, when present at sufficient levels, profoundly modulate the aggregation process and the physical properties of the resulting polyQ inclusions without significantly changing their appearance in the fluorescence microscope. Purified mammalian Hsp70 and Hsp40 (Hdj1) suppressed, in an ATP-dependent manner, the in vitro assembly of polyQ-expanded HD exon 1 constructs into ordered, SDS-insoluble amyloid fibrils and instead allowed the formation of amorphous, SDS-soluble aggregates that were associated with Hsp70 (ref. 58; Fig. 1). This effect of the chaperones was reproduced in HD exon 1-expressing yeast (58) and mammalian COS-1 cells (57) by coexpressing Hsp70 and Hsp40 homologs. In addition, Bonini and coworkers demonstrated in their Drosophila disease model that overexpression of Hsp70/Hsp40 strongly increases the SDS-solubility of polyQ aggregates (53). Considering that Hsp70 (in concert with Hsp40) binds to extended polypeptide segments, it may inhibit the formation of intramolecular β-sheet conformation and thus block ordered oligomerization and fibril growth (Fig. 1). On the other hand, binding of Hsp70 to the polyQ segments must be transient and of relatively low affinity, because Hsp70 cycles its substrates in an ATP-dependent manner, and glutamine is not a preferred residue in Hsp70-binding peptides (59). Consequently, the formation of irregular hydrogen bonds between polyQ sequences, resulting in amorphous aggregation, might not be suppressed efficiently except at a high molar excess of Hsp70. Assuming that this proposed mechanism is correct in outline, one would predict that Hsp70 and Hsp40 inhibit the recruitment of other polyQ-containing proteins such as CBP and TATA-binding protein into polyQ aggregates, explaining in part how the chaperones may mitigate polyQ-induced neurotoxicity (Fig. 2A). In addition, the observed association of Hsp70 with the aggregates may result in the coating of potentially dangerous surfaces. Indeed, the normal cellular levels of Hsp70/Hsp40 (and other chaperones) may be sufficient to control the damaging effects of polyQ-expanded proteins for decades, but eventually aging processes may result in a reduction of the available chaperone capacity (20, 60). Thus, a shift in the balance between cellular chaperone capacity and production of polyQ-expanded protein may be crucial in triggering the onset of disease (“chaperone hypothesis of polyQ disease”; ref. 58). Longer polyQ sequences would require more chaperone binding to avoid a toxic aggregation pathway, and therefore patients expressing such sequences would develop neuronal dysfunction earlier in life.
Interestingly, increased expression of Hsp70 also suppresses the toxicity induced by the non-polyQ-containing protein α-synuclein in a fly model for Parkinson's disease, again without altering the microscopic appearance of the neuronal inclusions formed (61). This observation points to the exciting possibility that the Hsp70/Hsp40 chaperone system has a general potential in mitigating the toxicity caused by misfolding proteins.
Thus far only two other chaperones structurally unrelated to Hsp70/Hsp40 have been identified as modulators of polyQ aggregation (Table 1). Both belong to the Hsp100/Clp family of AAA proteins (ATPases associated with various cellular activities; refs. 62 and 63). The yeast chaperone Hsp104 and its bacterial homolog, ClpB, can solubilize small protein aggregates in concert with Hsp70/Hsp40 (64, 65). Coexpression of Hsp104 with expanded polyQ protein reduced the formation of polyQ aggregates in yeast (62) and Caenorhabditis elegans (66). In C. elegans expression of polyQ protein in body wall muscle cells strongly impairs the ability of the animals to move, an effect that was shown to be alleviated by the expression of yeast Hsp104 (66). Interestingly, deletion of Hsp104 in yeast also prevented the aggregation of expanded polyQ proteins (62, 67), thus recapitulating the effects of Hsp104 on the aggregation of the yeast prion Sup35, which contains a Q/N-rich amino-terminal domain that mediates aggregation (68). It is proposed that Hsp104 catalyzes a structural change of Sup35 into an aggregation-prone conformation (68). Similarly, Hsp104 may facilitate the conversion of a polyQ random coil into β-sheet conformation, perhaps by relieving a steric block exerted by sequences adjoining the polyQ repeat (Fig. 1). Considering that overexpression of Hsp104 also suppresses polyQ aggregation, it is possible that excess amounts of the chaperone, in cooperation with Hsp70/Hsp40, effectively dissociate polyQ oligomers that nucleate the aggregation process (Fig. 1). A mammalian counterpart of Hsp104 has not been identified yet, but VCP (valosin-containing protein, the mammalian homolog of yeast Cdc48) is a distantly related AAA protein involved in membrane fusion, protein disassembly, and degradation (69–72). Higashiyama et al. recently reported that loss-of-function mutants of VCP suppress polyQ-mediated toxicity in SCA3 transgenic flies without inhibiting the formation of visible polyQ aggregates (63). Further in vitro and in vivo studies will be required to investigate whether VCP/Cdc48 affects polyQ aggregation by a mechanism similar to that of Hsp104.
Role of the Ubiquitin–Proteasome System in polyQ Aggregation and Toxicity
The ability of molecular chaperones to prevent (or reverse) protein aggregation is also important in aiding the proteolytic degradation of proteins that cannot be refolded (48). Most cytosolic proteins destined for degradation are marked by covalent attachment of a polyubiquitin chain at lysine residues (73). In this process, ubiquitin is activated first by the ubiquitin-activating enzyme (E1) and then transferred to a ubiquitin-conjugating enzyme (E2). The latter then links the activated ubiquitin to the protein substrate in functional cooperation with an E3 ubiquitin ligase, which acts as a specificity factor. Polyubiquitinated protein substrate is recognized and degraded by a large molecular machine, the 26S proteasome, which consists of a barrel-shaped proteolytic core complex of 20S, capped at both ends by 19S regulatory complexes (74). The 19S cap can be divided further into two subcomplexes, the “lid” and the “base.” The lid forms the distal part of the cap and functions in recognition and binding of polyubiquitinated substrate proteins. The base contacts the 20S core and contains a ring of six AAA ATPases that mediate unfolding and translocation of the substrate into the proteolytic chamber of the 20S complex. Concomitantly with translocation, ubiquitin molecules are released from the substrate and recycled. Importantly, the ubiquitin–proteasome system does not only participate in normal protein turnover, but its activity is required also for essential regulatory functions in a variety of cellular processes (74). As shown recently, these functions also include a role of the 19S complex in transcriptional regulation (75), which may impact on the transcriptional dysregulation in polyQ disease described above.
The finding that polyQ inclusions stain positively for ubiquitin and the 20S and 19S complexes suggested that the ubiquitin–proteasome system may be involved in polyQ pathogenesis (17–19). There are several reports that formation of polyQ inclusions is accelerated when proteasome inhibitors are added to transfected cells (18, 19, 37). Indeed, soluble HD exon 1 proteins are degraded in a proteasome-dependent process, as demonstrated by pulse–chase experiments in a Chinese hamster ovary cell-culture model (P.B., unpublished results). Unexpectedly, in these experiments the half-lives of polyQ constructs were found to be similar, independent of polyQ repeat lengths. Comparable results were obtained after expression of polyQ proteins in spinobulbar muscular atrophy cells (76). On the other hand, a polyQ-expanded ataxin-1 protein translated in vitro was shown to be more resistant to proteasome-dependent degradation than versions with polyQ repeats in the normal range, although both short and long polyQ proteins seemed to be ubiquitinated with similar efficiencies (37). Further experiments will be necessary to resolve the differences between these studies. Given that soluble polyQ proteins are degraded by the proteasome, it is likely that this step is preceded by ubiquitination, although this has not been demonstrated yet in vivo. Interestingly, in the cellular system mentioned above (76), Hsp70 and Hsp40 were observed to enhance the degradation of an expanded polyQ protein. Transfer of polyQ-expanded proteins to the degradation machinery may be mediated by Hsp70 together with the newly discovered protein CHIP (carboxy-terminus of Hsp70-interacting protein), which is thought to act as an E3 ligase in the ubiquitination of nonnative proteins in cooperation with Hsp70 and Hsp40 (77, 78). Because Hsp70 is likely to recognize an unstructured polyQ monomer (58), it will be interesting to investigate whether CHIP is involved in the ubiquitination of soluble polyQ proteins.
Intracellular aggregation of polyQ proteins has been proposed to impair the ubiquitin–proteasome system (reviewed in ref. 20). Once an expanded polyQ protein has escaped degradation and β-sheet oligomers are initiated, 19S regulatory complexes, unable to unfold these oligomers, may become trapped in the growing aggregates. This trapping of 19S particles may result in a partial inhibition of proteasomal activity and eventually cellular dysfunction (Fig. 2B). In contrast, large neuronal polyQ inclusions, the final product of the aggregation process, are compartmentalized in so-called aggresomes (ref. 19 and P.B., unpublished observations) and would no longer be able to trap 19S particles efficiently. Two recent reports generally support this hypothesis (79, 80). Using a rapidly degraded version of green fluorescent protein as a reporter, Bence et al. showed that expression of an expanded polyQ protein partially inhibited the ubiquitin–proteasome system in cell culture (79). Navon and Goldberg provided proof of principle that a nondegradable model substrate, unrelated to polyQ proteins, can function as a dominant inhibitor of the unfolding and degradation of otherwise proteasome-degradable proteins in vitro (80). Further biochemical studies will have to reveal whether intermediates in the process of polyQ aggregation indeed inhibit the unfolding and degradation activities of the proteasome. Such a mechanism could explain how protein misfolding may cause cellular toxicity and how the Hsp70/Hsp40 chaperone system may mitigate this toxicity. There are several reports that the Hsp70/Hsp40 chaperone system is not only essential for proper folding but also for the rapid degradation of certain proteins (81–83). This effect may be attributed to the activity of these chaperones in preventing the formation of intermolecular β-sheets by nonnative proteins, maintaining them in a degradation-competent state (Fig. 2B).
Perspectives
Recent years have seen major strides toward understanding the molecular basis of polyQ diseases based on a combination of biochemical studies in vitro and the analysis of numerous animal and cell-culture models in vivo. However, the exact mechanisms by which expanded polyQ proteins exert cellular toxicity remain to be established. In testing current hypotheses, it will be important to dissect the process of polyQ aggregation into distinct steps. Significant progress would result from the development of techniques that allow the accumulation and isolation of aggregation intermediates at different stages of the process. These intermediates then could be tested for their ability to interact with molecular chaperones, to recruit other polyQ-containing proteins, or to inhibit the proteasome system.
It is striking that many other neurodegenerative diseases including Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis, and the prion diseases all involve the assembly of structurally unrelated proteins into intracellular or extracellular amyloid fibrils. Together with the recent finding that intermediates formed early in the aggregation pathway can be inherently highly cytotoxic (38, 39), even for proteins that are not disease-associated (38), it seems likely that common fundamental mechanisms underlie the toxicity of amyloid formation. This notion is strongly supported by the observation that molecular chaperones can suppress the toxicity of amyloidogenic proteins as different as α-synuclein in Parkinson's disease (61), Tau protein in Alzheimer's disease (H. Xu, personal communication), or the polyQ proteins. Searching for ways to pharmacologically induce the expression of molecular chaperones in neurons may open up a promising approach to the treatment of these diseases (57).
Acknowledgments
We thank J. Young for critically reading the manuscript and E. Wanker for discussion. Work in our laboratory is funded by Deutsche Forschungsgemeinschaft Grant SFB 596. H.S. is supported by a research fellowship of the Japan Society for the Promotion of Science.
Abbreviations
- polyQ
polyglutamine
- HD
Huntington's disease
- SCA
spinocerebellar ataxia
- CBP
CREB-binding protein
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Orr H T. Genes Dev. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 4.Tobin A J, Signer E R. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- 5.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 6.Nasir J, Floresco S B, O'Kusky J R, Diewert V M, Richman J M, Zeisler J, Borowski A, Marth J D, Phillips A G, Hayden M R. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J L, Fischbeck K H, Pittman R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 9.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 10.Rochet J C, Lansbury P T., Jr Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 11.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perutz M F, Finch J T, Berriman J, Lesk A. Proc Natl Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, Dawson T M, Ross C A. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 16.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 17.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 18.Chai Y, Koppenhafer S L, Shoesmith S J, Perez M K, Paulson H L. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 19.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker E E. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman M Y, Goldberg A L. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 21.Balbirnie M, Grothe R, Eisenberg D S. Proc Natl Acad Sci USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benzinger T L, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Biochemistry. 2000;39:3491–3499. doi: 10.1021/bi991527v. [DOI] [PubMed] [Google Scholar]
- 23.Antzutkin O N, Balbach J J, Leapman R D, Rizzo N W, Reed J, Tycko R. Proc Natl Acad Sci USA. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 25.Kelly J W. Nat Struct Biol. 2000;7:824–826. doi: 10.1038/82815. [DOI] [PubMed] [Google Scholar]
- 26.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 27.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunkes A, Mandel J L. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler V C, White J K, Gutekunst C A, Vrbanac V, Weaver M, Li X J, Li S H, Yi H, Vonsattel J P, Gusella J F, et al. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Hasegawa H, Tanaka K, Kakizuka A. Cell Death Differ. 2001;8:871–873. doi: 10.1038/sj.cdd.4400901. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Nat Genet. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- 32.Cooper J K, Schilling G, Peters M F, Herring W J, Sharp A H, Kaminsky Z, Masone J, Khan F A, Delanoy M, Borchelt D R, et al. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 33.Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto A, Lucas J J, Hen R. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 35.Klement I A, Skinner P J, Kaytor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 36.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 37.Cummings C J, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr H T, Beaudet A L, Zoghbi H Y. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 38.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson C M, Stefani M. Nature (London) 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 39.Walsh D M, Klyubin I, Fadeeva J V, Cullen W K, Anwyl R, Wolfe M S, Rowan M J, Selkoe D J. Nature (London) 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 40.Suhr S T, Senut M C, Whitelegge J P, Faull K F, Cuizon D B, Gage F H. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luthi-Carter R, Strand A, Peters N L, Solano S M, Hollingsworth Z R, Menon A S, Frey A S, Spektor B S, Penney E B, Schilling G, et al. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 42.Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft T F, Kato K, Rubinsztein D C. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 43.Mantamadiotis T, Lemberger T, Bleckmann S C, Kern H, Kretz O, Villalba A M, Tronche F, Kellendonk C, Gau D, Kapfhammer J, et al. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 44.Lin X, Antalffy B, Kang D, Orr H T, Zoghbi H Y. Nat Neurosci. 2000;3:157–163. doi: 10.1038/72101. [DOI] [PubMed] [Google Scholar]
- 45.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, et al. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 46.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature (London) 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 47.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 48.Hartl F U, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 49.Dobson C M, Karplus M. Curr Opin Struct Biol. 1999;9:92–101. doi: 10.1016/s0959-440x(99)80012-8. [DOI] [PubMed] [Google Scholar]
- 50.Misselwitz B, Staeck O, Rapoport T A. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 51.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 52.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 53.Chan H Y, Warrick J M, Gray-Board G L, Paulson H L, Bonini N M. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 54.Fernandez-Funez P, Nino-Rosales M L, de Gouyon B, She W C, Luchak J M, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner P J, et al. Nature (London) 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 55.Cummings C J, Sun Y, Opal P, Antalffy B, Mestril R, Orr H T, Dillmann W H, Zoghbi H Y. Hum Mol Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 56.Bonini N M. Proc Natl Acad Sci USA. 2002;99, Suppl. 4:16407–16411. doi: 10.1073/pnas.152330499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, Hayer-Hartl M K, Hartl F U, Wanker E E. Hum Mol Genet. 2001;10:1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 58.Muchowski P J, Schaffar G, Sittler A, Wanker E E, Hayer-Hartl M K, Hartl F U. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heydari A R, Takahashi R, Gutsmann A, You S, Richardson A. Experientia. 1994;50:1092–1098. doi: 10.1007/BF01923466. [DOI] [PubMed] [Google Scholar]
- 61.Auluck P K, Chan H Y, Trojanowski J Q, Lee V M, Bonini N M. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 62.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higashiyama H, Hirose F, Yamaguchi M, Inoue Y H, Fujikake N, Matsukage A, Kakizuka A. Cell Death Differ. 2002;9:264–273. doi: 10.1038/sj.cdd.4400955. [DOI] [PubMed] [Google Scholar]
- 64.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Zvi A P, Goloubinoff P. J Struct Biol. 2001;135:84–93. doi: 10.1006/jsbi.2001.4352. [DOI] [PubMed] [Google Scholar]
- 66.Satyal S H, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer J M, Morimoto R I. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kimura Y, Koitabashi S, Kakizuka A, Fujita T. Genes Cells. 2001;6:887–897. doi: 10.1046/j.1365-2443.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 68.Serio T R, Lindquist S L. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- 69.Latterich M, Frohlich K U, Schekman R. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 70.Dai R M, Li C C. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 71.Ye Y, Meyer H H, Rapoport T A. Nature (London) 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 72.Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 73.Pickart C M. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 74.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 75.Gonzalez F, Delahodde A, Kodadek T, Johnston S A. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 76.Bailey C K, Andriola I F, Kampinga H H, Merry D E. Hum Mol Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 77.Demand J, Alberti S, Patterson C, Hohfeld J. Curr Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 78.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 80.Navon A, Goldberg A L. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 81.Lee D H, Sherman M Y, Goldberg A L. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 83.Ohba M. FEBS Lett. 1997;409:307–311. doi: 10.1016/s0014-5793(97)00535-8. [DOI] [PubMed] [Google Scholar]