Abstract
Telomeric position effect in Saccharomyces cerevisiae is a chromatin-mediated phenomenon in which telomere proximal genes are repressed (silenced) in a heritable, but reversible, fashion. Once a transcriptional state (active or silenced) is established, however, there is a strong tendency for that state to be propagated. Twenty-five years ago, H. Weintraub and colleagues suggested that such heritability could be mediated by posttranslational modification of chromatin [Weintraub, H., Flint, S. J., Leffak, I. M., Groudine, M. & Grainger, R. M. (1977) Cold Spring Harbor Symp. Quant. Biol. 42, 401–407]. To identify potential sites within the chromatin that might act as sources of “memory” for the heritable transmission, we performed a genetic screen to isolate mutant alleles of the histones H3 and H4 genes that would “lock” telomeric marker genes into a silenced state. We identified mutations in the NH2-terminal tail and core of both histones; most of the amino acid changes mapped adjacent to lysines that are known sites of acetylation or methylation. We developed a method using MS to quantify the level of acetylation at each lysine within the histone H4 NH2-terminal tail in these mutants. We discovered that each of these mutants had a dramatic reduction in the level of acetylation at lysine 12 within the histone H4 tail. We propose that this lysine serves as a “memory mark” for propagating the expression state of a telomeric gene: when it is unacetylated, silent chromatin will be inherited; when it is acetylated an active state will be inherited.
In the beginning of the 20th century, the concept that genotype controlled inheritance of phenotype was defined and developed. However, in 1930, H. J. Muller described a mutant in Drosophila that was in apparent contradiction to the accepted dogma (1). The normal Drosophila compound eye is made of hundreds of red ommatidia, and typical eye-color mutants have a complete change in color of all of the ommatidia. However, Muller's “ever-sporting displacement” mutant resulted in the mosaic expression of red and white pigments in the Drosophila eye. This mosaic expression correlated with a specific chromosomal rearrangement that caused the white locus to be located near the centromere. In some eye cells of an individual, the white gene was expressed (red) whereas in others it was not (white). This phenomenon came to be known as position effect variegation and can now be heralded as the dawning realization that genotype did not necessarily predict phenotype (reviewed in refs. 2 and 3).
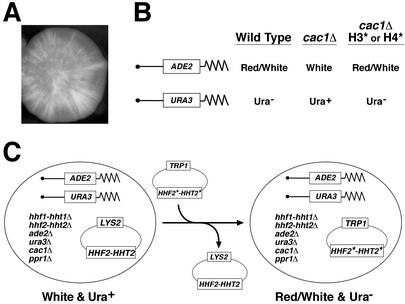
Telomeric position effect in the budding yeast Saccharomyces cerevisiae is very similar to position effect variegation in Drosophila (4). When a normal yeast gene is placed near a telomere, the gene undergoes transcriptional repression, or silencing, that is heritable over many cell generations. However, silencing is reversible, as the gene can become transcriptionally active. For example, when yeast cells with the ADE2 gene placed near a telomere form a colony on solid medium, the colony is composed of subpopulations in which the ADE2 gene is either expressed (white sectors) or repressed (red sectors, see Fig. 1A). The different phenotypes of the sectors in a colony reflect the ability of genetically identical cells to switch between phenotypic states. However, the fact that large sectors are phenotypically uniform reflects the ability of each state to be heritably propagated for multiple generations.
Figure 1.
A genetic screen for histone H3 or H4 mutants that increase telomeric silencing. (A) Photograph of a colony of S. cerevisiae with a telomeric ADE2 gene. Red (gray in photograph) sectors indicate cells in which the gene is silenced. (B) Outline of the principle of a screen for mutants in histone H3* or H4* that acted as bypass suppressors of the cac1Δ phenotype of telomeric silencing (see text for details). (C) Schematic representation of the “plasmid shuffle” method used to screen for histone H3 or H4 mutants that increased silencing. A mutagenized plasmid library of histone H3 and H4 genes (HHT2*-HHF2*-TRP1; pMP3) was transformed into strain UCC1371 (genotype is indicated). Those transformants that regained silencing (red and white sectored colonies that grew poorly, or not at all, in the absence of uracil) and lost the wild-type histone H3 and H4 genes plasmid (HHT2-HHF2-LYS2; pMP9) were chosen for further analysis.
Telomeric silencing is the result of a repressive chromatin structure that initiates from the telomere and extends inward along the chromosome, rendering the enveloped DNA refractory to interaction with factors such as those of the transcriptional machinery (reviewed in ref. 5). The key structural components of telomeric silent chromatin are well defined. They include telomeric DNA sequence, the telomere sequence DNA-binding proteins Rap1p and Ku, nucleosomal core histones H3 and H4, and nonhistone chromatin components Sir2p, Sir3p, and Sir4p (Silent Information Regulators). In a simple view of telomeric silencing, the Sir proteins are recruited to the telomeres through their interactions with Rap1p and Ku at the end of the chromosome, and with each other. They then “polymerize” along telomere-adjacent chromosome regions by binding the NH2-terminal tails of histones H3 and H4 of the associated nucleosomes.
In addition to telomeres, the three Sir proteins act to repress the silent mating loci, HML and HMR. At these loci, additional factors are required that seem to be primarily involved in recruiting the Sir complex and helping to stabilize the silent state (6). In part because of this lack of redundancy, telomeric silencing is semistable and more sensitive to perturbation, particularly as a result of mutations in the NH2 termini of histones H3 and H4. Mutations within residues 16–29 of histone H4 result in a defect in telomeric silencing (7). Deletion analysis of histone H3 implicates residues 4–20 as also being important for telomeric silencing (8).
Several lines of evidence indicate that posttranslational modifications (especially acetylation) of lysines in the NH2-terminal tails of histone H3 and H4 are critical to telomeric silencing. For instance, the histone tails in silenced chromatin are hypoacetylated compared with the rest of the genome (9, 10). [However, there are conflicting reports with respect to the acetylated state of lysine 12 (K12) of histone H4.] Most likely this hypoacetylated state is achieved by the deacetylase activity of Sir2p, which can remove the acetyl groups from acetylated histone tail peptides in vitro (reviewed in ref. 11). The hypoacetylated state may be critical for Sir3p binding to chromatin. In vitro, a fragment of the Sir3 protein binds with greater affinity to histone H4 tail peptides that are completely unacetylated at K5, K8, K12, or K16, compared with those that are fully acetylated at these positions (12). Of these four lysines, in vivo analysis has implicated K16 as the “key” lysine in telomeric silencing (reviewed in ref. 13). From these studies, it has been interpreted that acetylation of K16 prevents silencing, i.e., precludes binding of Sir3p to chromatin. However, peptides with K16 as the sole site of acetylation still bind exceptionally well to Sir3p in vitro (12).
For histone H3 less is known and the situation is even more complex, in part because methylation and phosphorylation also occur within the tail (reviewed in ref. 13). The lysine residues in its NH2-terminal tail are hypoacetylated when in silent chromatin as well, and when K9, K14, K18, and K23 all are mutated to arginine or glycine, telomeric silencing is reduced, although not to the extent that the same change at K16 on histone H4 reduces silencing (8). Thus, there is still ambiguity as to what posttranslational histone modification “code” is required for telomeric silencing.
Although a basic understanding of telomeric silent chromatin structure has been developed, its epigenetic character remains a mystery. In particular it is unclear how the heritable propagation of a state occurs within the context of a certain level of switching between expression states. The switching can be explained in part as the result of shifts in a competition between silencing components and transcriptional-activating factors for assembly onto telomere-proximal DNA (14). Despite this competition, examination of a colony of cells with ADE2 at a telomere indicates that under normal circumstances the preexisting transcriptional state is most often inherited (see Fig. 1A). This finding indicates the existence of some mechanism to favor the status quo through successive cell cycles. In particular, assembly (or reassembly) of the silent chromatin must occur during, or shortly after, each round of DNA replication. Consistent with this idea, a number of unrelated mutations or drug treatments that lengthen S phase, and presumably affect the kinetics and coordination of molecular events in S phase, are able to suppress defects in silencing (15, 16). Furthermore, telomeric silencing is sensitive to mutations in subunits of chromatin assembly factor 1 (CAF-1) or in ASF1; both facilitate assembly of newly replicated DNA into nucleosomes in vitro (17–20). Hence there appears to be an intimate coordination between silent chromatin assembly and DNA replication.
The role of histone modification as a means of “marking” chromatin to perpetuate the molecular memory of an expression state after DNA replication was suggested 25 years ago (21) and is likely to be germane to telomeric position effect in S. cerevisiae. Gene activation proceeds by a series of steps that includes recruitment of histone acetylases, which help produce the hyperacetylated state, and presumably prevents Sir3p binding (reviewed in ref. 22). Conversely, silent chromatin contains the Sir2p deacetylase and helps maintain Sir3p binding (5). Thus the acetylated state of histones could serve as the molecular mark that serves as “memory” for propagating the transcriptionally active state against a competing silencing complex, which once established, would maintain a deacetylated histone (reviewed in ref. 23). Other models for the heritable transmission of a chromatin state that include DNA methylation are likely irrelevant, as such modifications have not been detected in S. cerevisiae (24, 25). Similarly, those models that include methylation of specific histone residues such as K9 of histone H3 (a modification that is presumably irreversible), which then serve as binding sites for “silencing factors” such as HP1, are also unlikely; neither this histone modification, nor HP1 homologs, have been identified in S. cerevisiae telomeric chromatin (26).
Given the apparent complexity of interactions with histone H3 and H4 in the formation of telomeric chromatin, we sought to identify potential sites of reversible modification that might be particularly critical in imparting memory to a telomeric gene. That is, those residues and modifications that are responsible for the heritable bias of a telomeric gene's transcriptional state to be passed on from one cell to its progeny. Here we present our initial findings that combine a genetic screen and development of mass spectrometric methods to analyze these modified sites.
Materials and Methods
Plasmids, Oligos, and Strains.
A 2.7-kb PstI fragment of HHT2–HHF2 was inserted into PstI sites of pRS317 and pRS314 to create pMP9 and pMP3, respectively (27). Histone H3 and H4 mutants were generated in three ways (see Supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org for details).
The yeast strain BY4705 was modified to create UCC1373 [MATa ade2Δ∷hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 adh4∷URA3-TEL(VII-L) ADE2-TEL(VR) hhf2- hht2Δ∷MET15 hhf1-hht1Δ∷LEU2 ppr1Δ∷KanMX]. UCC1371 is isogenic with UCC1373 but also contains cac1Δ∷HIS3. Both strains use pMP9 (LYS2 CEN ARS)-HHF2-HHT2 as a covering plasmid to provide functional copies of histones H3 and H4.
Screening Procedure.
Mutant histone H4 plasmids (pMP3) were transformed into UCC1371 as described (18). Pink/red colonies were picked and restreaked onto yeast complete media (YC)-Trp and replica-plated onto YC-Trp and YC-Lys to identify colonies that had lost pMP9. Plasmids were rescued from Lys−/Trp+ colonies and transformed into UCC1371 and UCC1373 to retest ADE2 and URA3 silencing. Plasmids were then sequenced to identify mutations in both HHF2 and HHT2.
All silencing assays were carried out as described (28).
Transcriptional Microarrays.
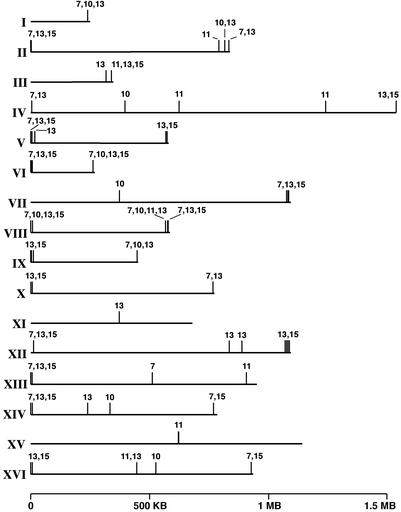
Isolation of mRNA, labeling, hybridization, and data analysis were carried out as described (29). genespring software (Silicon Genetics, Redwood City, CA) was used to analyze all transcript array data. A two-step process identified those loci indicated in Fig. 3. First, for each mutant allele, all genes that were calculated to be significantly different (>2 SD) than wild type were selected. Then the genes were ranked and those that were down-regulated the most were identified.
Figure 3.
Mutations in the histone H4 tail result in a general down-regulation of telomere proximal genes. Transcriptional changes between strains with mutant and wild-type histone H4 were determined by competitive hybridization to DNA microarrays. Each array contained >6,000 S. cerevisiae ORFs, and each hybridization was carried out a minimum of two times. A schematic of each S. cerevisiae chromosome is presented. Vertical lines above each chromosome mark loci that were transcriptionally down-regulated in the mutant strain. Numbers above these lines correspond with residue position number (e.g., G11T) in the mutants that showed an effect. For alleles L10F and G11T, the 10 loci that were down-regulated the most are indicated, for G7I and A15T ≈20 loci and for G13L ≈30 loci are shown.
Histone Purification.
Histones were isolated according to Waterborg (30). Histones were then separated on a Zorbax reverse-phase C-18 column with a Waters HPLC system using a 0.5%/min gradient from 40% to 60% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA). Histone H4 coeluted with H2A at ≈43% ACN/0.1%TFA and histone H3 eluted at ≈50% ACN/0.1%TFA. Purified histones were then dried with a speed-vac and stored at either 4° or −20°.
Chemical Acetylation/Trypsin Digestion.
Purified and dried histone samples were resuspended in 50 μl deuterated acetic acid (Acros, Fair Lawn, NJ). Five microliters deuterated acetic anhydride was added, and the samples were left at room temperature for 6 h. Samples were then dried in a speed-vac, resuspended in 20 mM NH4HCO3 and 100 ng trypsin (Worthington), and incubated at 37° overnight. Digestion buffer was removed by speed-vac before mass spectrometric analysis.
MS and Analysis.
MS and analysis are described in additional Materials and Methods, which are published as supporting information on the PNAS web site.
Results
Genetic Screen to Identify Histone H3 and H4 Mutants That Increase Telomeric Silencing.
As noted above, histones H3 and H4 are known to play a critical role in telomeric silencing, and their NH2-terminal tails appear to be hypoacetylated in silenced chromatin (9). Conversely, the tails become hyperacetylated when a telomeric gene becomes transcriptionally active. We wanted to identify mutations in histones H3 and H4 that would increase the probability that a silenced state would occur, with the idea that these mutants might “lock” the histone into a posttranslational modification that was critical for silencing. The corresponding modified residue would be a candidate for a site on the histone in which the memory of a silenced or active transcriptional state might be stored.
We created and used S. cerevisiae strains in which the two chromosomal copies of the histone H3 and H4 genes were deleted and replaced with a wild-type copy of the histone genes (HHT2-HHF2) on a centromere plasmid containing a LYS2 gene (pMP9; Fig. 1C) (27). In addition, the ADE2 and URA3 genes were integrated adjacent to telomeres on the right arm of chromosome V and left arm of chromosome VII, respectively (Fig. 1B) (18). In cells that were otherwise wild type, these markers served as independent reporters of telomeric position effect (28). The cells give rise to colonies that are red and white sectored because of variegated expression of ADE2 and that grow poorly, or not at all, in the absence of uracil. [The transcriptional activator of the URA3 gene, PPR1, is deleted, effectively crippling the URA3 promoter and preventing it from being expressed when it is proximal to the telomere (14).]
To screen for histone mutants that would increase the frequency of telomeric silencing, the strain was “sensitized” by deletion of the CAC1 gene (19, 20). This gene encodes a subunit of the chromatin assembly factor (CAF-I), which is a chaperone for histones H3 and H4 and likely helps in chromatin assembly. Loss of the gene product results in a loss of silencing. We hypothesize that in the absence of Cac1p, histones H3 and H4 are still deposited on the chromatin, but that they may be inappropriately modified on their way to deposition. Such modifications then prevent proper formation of silent chromatin. Thus we created a strain (UCC1371) that was defective in telomeric silencing (white colonies that could grow in the absence of uracil); this strain was used to screen for histone mutants that bypass the cac1 defect and reinstate telomeric silencing (red and white sectored colonies and poor growth in the absence of uracil; Fig. 1 B and C).
Libraries of histone H3 and H4 mutants were transformed into the yeast strain described above (UCC1371), and the plated colonies were screened for increased redness (silencing of the telomeric ADE2 gene) compared with colonies containing unmutagenized wild-type histones. Only colonies that had lost the wild-type histone/LYS2 plasmid, which indicated that the mutant histones could replace the wild-type histones, were selected and examined further (see Fig. 1C). These strains were then tested for their ability to grow in media lacking uracil to address whether the telomeric URA3 was also silenced. After verifying that the increase in telomeric silencing was plasmid-linked, the plasmids were sequenced to identify changes in histone H3 or H4. Those plasmids that had a single point mutation are presented (Table 1 and Fig. 2A).
Table 1.
Mutations in the core region of histone H3 and H4 that increase telomeric silencing
| Histone | Wild-type residue | Mutant residues isolated |
|---|---|---|
| H3 | D77 | A, G, N, V |
| D81 | G | |
| H4 | ||
| R39 | K | |
| H75 | Y |
Figure 2.
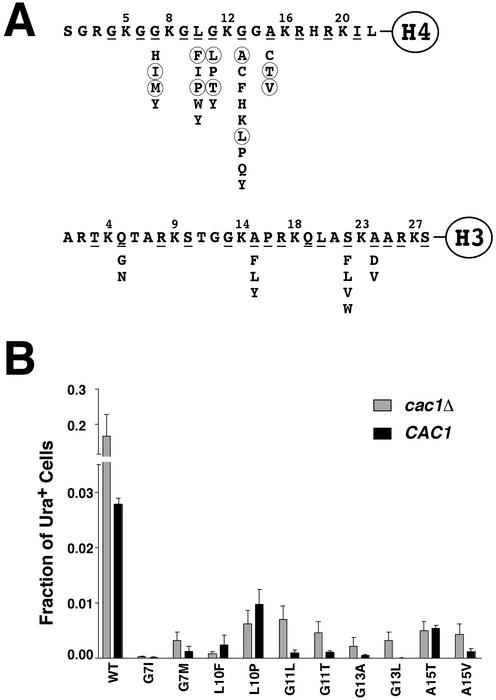
Changes in amino acids adjacent to lysines in the tails of histone H3 and H4 increase telomeric silencing of marker genes. (A) Representation of the NH2-terminal tails of histones H3 and H4. Those residues that were specifically targeted for mutagenesis are underlined. Single amino acid changes that gave rise to increased silencing are indicated below the wild-type sequence. Those changes that were subjected to further analysis are circled. (B) Silencing of URA3 was measured by the fraction of cells in each strain that could form colonies on media lacking uracil for each of the histone H4 tail mutants circled in A. Gray and black bars represent data using cells from UCC1371 (cac1Δ) and UCC1373 (CAC1), respectively. Three independent transformants were used for each measurement.
The mutations were classified into four categories: those mapping to either the tails of histone H3 (Q5, A15, S22, and A24) or histone H4 (G7, L10, G11, G13, and A15) (see Fig. 2), or to the “core” of histone H3 (D77 and D81) or histone H4 (R39 and H75) (see Table 1). The mutant histones still required SIR gene function for silencing (data not shown), indicating that the mutant histone repression was still mediated by silent chromatin. We focused on the mutants within the histone H4 tail because of the documented role that acetylation of lysines within this domain plays in telomeric silencing, and because this tail was amenable to biochemical analysis of the modifications (described below).
Residues Adjacent to Histone H4 Tail Lysines Are Involved in Telomeric Silencing.
We chose two alleles at random from each of the mutated residues in the histone H4 tail (circled residues in Fig. 2A) for further characterization. The ability of these mutants to increase telomeric silencing at the URA3 gene was quantified. Fig. 2B shows the fraction of cells that gave rise to a colony in the absence of uracil, which reflects the level of expression from URA3. As expected, the presence of CAC1 in strains containing wild-type histones caused a significant reduction in the ability of cells to grow in the absence of uracil. However, each of the mutants increased silencing even further, regardless of whether or not CAC1 was present. Mutants G7I, G7M, L10F, G13L, and G13A had the largest increase in silencing (≈50- to 500-fold reduction in uracil growth compared with wild type), whereas L10P, G11T and G11L, A15T, and A15V allowed a little more expression (≈10- to 40-fold reduction compared with wild type). In general, when CAC1 was introduced into a histone mutant strain, silencing was increased further, with the possible exceptions of alleles L10F, L10P, and A15T, where no statistically significant difference was measured between CAC1 and cac1 strains. The two mutants that exhibited the most consistent CAC1-dependent increase in silencing were G11L and G11T (10- and 8-fold, respectively). Taken together these results indicated that these 10 mutant alleles of histone H4 completely bypassed the need for CAC1 in telomeric silencing.
However, it was possible that the histone mutants increased silencing indirectly; for instance, they may have resulted in the misregulation of silencing proteins. Increased dosage of SIR3 causes increased silencing at telomeres and can compensate for some mutants that lose telomeric silencing (31). Currently there are fewer than 200 genes that have been implicated in telomeric silencing, according to the Yeast Proteome Database (www.incyte.com/proteome/databases.jsp). To examine whether these genes were differentially regulated by the mutant histones, a transcriptional microarray analysis was performed on one representative allele from each mutant site. For A15T, G13L, G11T, and L10F alleles, there was no significant (2-fold) increase or decrease in any of these genes (Table 2, which is published as supporting information on the PNAS web site). Only in the G7I allele was an effect observed: a 2-fold increase in expression of the EST2 and SAS4 genes. EST2 encodes the reverse transcriptase of telomerase (32), and SAS4 encodes a subunit of the SAS histone acetylase complex that is required for telomeric silencing (33). The overexpression of EST2 has been determined to have no effect on telomeric silencing (34). The modest increase in SAS4 may impact telomeric silencing if this subunit is normally a limiting component, or if it is critical for targeting of the complex to telomeres.
The transcript microarray data also permitted us to map, on a genomewide scale, loci that were transcriptionally repressed (in addition to the telomeric ADE2 and URA3 genes) in the histone H4 mutants relative to wild type. Quite strikingly, when the genes were ranked by their expression level for each mutant compared with wild type, virtually all of the 20–40 genes that were down-regulated the most in the G7I, G13L, and A15T alleles were located in close proximity (within 20 kbp) to telomeres (Fig. 3 and Table 2). This bias was maintained in the L10F allele, although to a lesser extent; six of 10 genes were telomeric. Although telomeric loci were repressed in the G11T allele as well, they represented fewer of those most affected, only two of the 10 most repressed genes. Taken together, these data indicate that the increase in silencing on the two telomeric marker genes (ADE2 and URA3) caused by these mutant histone H4 alleles reflect an effect that occurred on telomeric chromatin in general.
Acetylation Analysis of Histone H4 Silencing Mutants by MS.
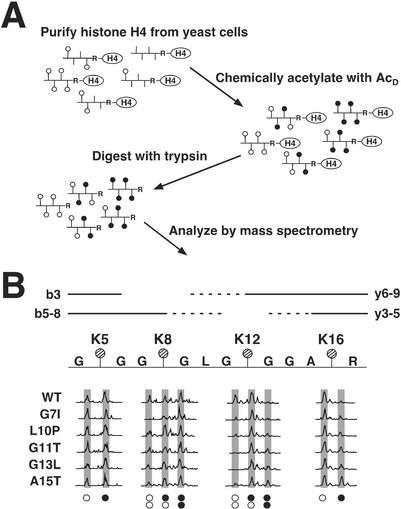
Because the results of our screen pinpointed residues in proximity to the histone H4 lysines at positions 5, 8, 12, and 16 as being important in controlling telomeric silencing, we suspected that acetylation of these residues in the mutants might be altered. Although antibodies to specific acetylated lysines have been used to determine levels of acetylation (9), we found these reagents to be unsuitable for our analysis. The mutations in histone H4 we created changed the recognition epitope for these antibodies. Furthermore, it is unclear how acetylation at a nearby lysine changes the affinity of the antibody for its epitope, thus making it difficult to truly quantify the level of acetylation at each lysine. Therefore we sought to eschew these issues by developing a mass spectrometric approach to measure the level of acetylation at each individual lysine residue in the tail of histone H4. Presented in Fig. 4A is an outline of this method. After initial isolation by standard procedures (30), purified histone H4 was prepared by reverse-phase HPLC (see Materials and Methods). The primary structure of the resulting population of histone H4 molecules was likely complex because any or all of the four tail lysine residues at positions 5, 8, 12, and 16 might be acetylated. To prepare a homogeneous, chemically identical population, purified histone H4 was treated with deuterated acetic anhydride, which labeled each unacetylated lysine with a deuterated acetyl moiety (35).
Figure 4.
Mass spectrometric analysis to measure acetylation at specific lysine residues in the histone H4 tail. (A) A schematic of the method used to determine site-specific acetylation of histone H4. Vertical lines represent lysines that can be acetylated, ○ represents those lysine residues acetylated in vivo (AcH), and ● represents lysine residues chemically acetylated by deuterated acetic anhydride in vitro (AcD); the mass difference between ○ and ● is 3 Da (42 versus 45, respectively). Tryptic digestion of purified and chemically modified histones produced a chemically identical population of tail peptides containing K5, K8, K12, and K16, beginning at G4 and terminating at R17. These peptides were then analyzed by MS. (B) Mass spectra showing altered levels of protonated versus deuterated acetylation in mutant and wild-type histone H4. A schematic of the histone H4 tryptic peptide is shown with each of the four lysine residues marked with a hatched circle. The lines (solid and dashed) above the peptide represent b and y ions. The b3 ion provided acetylation information for K5 and b ions 5-8 (b5–8) provided information for K8; similarly, y ions 3–5 (y3–5) were used to assess acetylation at K16 and y ions 6–9 (y6–9) were used for K12. (The b4 and y2 ions were indistinguishable in the spectra and were therefore omitted from analysis; the mass of the b2 ion was below the detection range.) Spectra of various b and y ions are shown for wild-type histone H4 along with mutants G7I, L10P, G11T, G13L, and A15T. The x axis and y axis of the mass spectra represent mass-to-charge ratio and relative abundance, respectively. Peaks are highlighted by shaded boxes and are aligned above symbols denoting whether the lysines are acetylated with a protonated (○) or deuterated (●) acetyl. ○○ represents fragment ions containing two lysine residues that are both modified with protonated acetyls; ○● represents fragments with one protonated and one deuterated acetyl; ●● represents fragments in which both acetyls are deuterated.
Trypsin efficiently cleaves after lysine or arginine in a peptide; however, acetylation of lysine protects these residues from cleavage (36). With each of the histone H4 lysines masked by either a protonated acetyl from endogenous acetylation or a deuterated acetyl from the chemical acetylation, digestion with trypsin produced peptides for which cleavage occurred only after arginine residues. For histone H4, the four acetylatable lysine residues within the NH2 terminus, K5, K8, K12, and K16, remained together in a single peptide (residues 4–17) (data not shown).
Tandem MS was used to determine the endogenous level of site-specific acetylation at each lysine within the mutant histone H4 tail peptides (see Fig. 2A). This approach capitalized on the fact that the acetyl groups added in vivo, in the yeast cell, have a mass 3 Da less than the acetyl groups added in vitro by the reaction of the purified proteins with deuterated acetic anhydride. Tryptic products of histone H4 that corresponded to the fully acetylated peptide containing residues 4–17 were identified by HPLC electrospray ionization MS. Isotope patterns of this peptide indicated the extent of in vivo and in vitro acetylation in this segment (36, 37). Aided by “data-dependent” scanning, these molecular ions were fragmented by low-energy collision-induced dissociation (38), and the resulting b and y fragment ions were analyzed (39).
The level of in vivo acetylation at each lysine residue was determined from the relative intensities of isotope peaks caused by in vivo acetylation (AcH) and in vitro acetylation (AcD) for specific b and y ions. Because the peptide fragments used for these measurements were formed from chemically identical parent ions, fragmentation occurred independent of where in vivo acetylation occurred. The intensities of protonated and deuterated fragments ions were determined from the maximum ion currents of the appropriate isotope peaks of specific b and y ions (see Materials and Methods).
Measuring the level of endogenous acetylation at K16 was straightforward because it involved fragment ions (specifically, y3, y4, or y5 ions) that contained only this site of acetylation. Similarly, the level of acetylation at K5 was directly determined by using the fragment, b3. Quantification of acetylation at the internal lysine residues, however, was more complicated because b and y ions that include K8 and K12 also include K5 and K16, respectively (see Fig. 4B). Thus for fragments containing two sites of acetylation, there were three possible mass states: the ion can have two protonated acetylations (e.g., AcH at K12 and AcH at K16), one protonated acetylation and one deuterated acetylation (AcH at K12 and AcD at K16 or AcD at K12 and AcH at K16), or two deuterated acetylations (AcD at both K12 and K16). Thus, although we could determine the total relative number of protonated acetylations for a particular fragment, we could not tell directly how much acetylation is at either of the internal lysines. Given that we knew the acetylation level at one site, we could calculate the level at the other. By subtracting the contribution of the external site, information we obtained by analyzing fragments y3–5 or b3, the level of acetylation exclusively at the internal lysine was determined (see Materials and Methods).
Presented in Fig. 4B are mass spectra of wild-type and selected mutant histone tryptic peptides; the spectra are placed beneath the lysine residue for which they provide acetylation information. There were two possible mass states for y3–5 ions: one at +42 (a single AcH represented by ○) and one at +45 (a single AcD represented by ●). In wild-type histone H4, the level of acetylation at K16 was very high, with only a small amount of deuterated acetylation. However, mutations close to K16 correlated with an increase in the amount of deuterated acetylation (note the change in peak areas in Fig. 4B). For the A15T mutant, there were equal levels of protonated and deuterated acetylation, indicating the level of endogenous acetylation dropped from ≈90% to ≈50%. Similarly, the amount of protonated acetylation dramatically decreased at K12 and K8, particularly for mutants containing amino acid changes near these positions. (See mutants G11T and G13L for K12 acetylation and mutant G7I for K8 acetylation.)
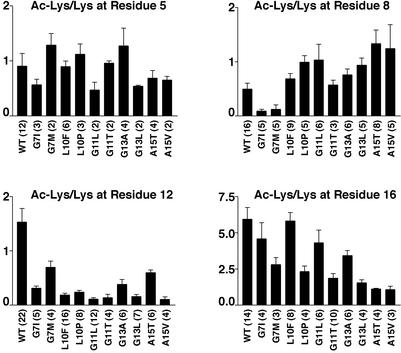
Summarized in Fig. 5 are the results of the quantitative mass spectrometric analysis of histone acetylation for each of the four lysine residues of histone H4. The data, presented as the ratio of acetylated lysine to unacetylated lysine, were based on the analysis of numerous b and y ions from multiple histone preparations; the number of data points that contributed to each set of data are marked in parentheses. The most dramatic observation was that acetylation of K12 was diminished in each of the mutants with the most drastic change occurring for mutants G11L, G11T, and G13L. However, in general, we found that mutation of a residue adjacent to a specific lysine decreased the ability of that lysine to be acetylated. For example, in addition to the decrease in acetylation at K12, G7I and G7M showed a striking decrease in the level of acetylation of K8, and A15T and A15V reduced acetylation at position 16. Acetylation of K5 appeared to be relatively unaffected by the histone tail mutations analyzed.
Figure 5.
Mutant histone H4 proteins exhibit decreased acetylation at specific lysine residues. The ratio of acetylated lysine to unacetylated lysine at positions 5, 8, 12, and 16 was determined for histone H4 mutants compared with wild-type histone H4. Levels of acetylation were calculated by comparing peak intensities of protonated vs. deuterated ions for specific y and b fragments (see Materials and Methods). Multiple b and y ions were used to calculate the fraction of acetylation at each site; the total number of intensity measurements, which is the sum of all ions measured from independent histone preparations, is shown in parentheses.
Discussion
We have identified 13 residues on histones H3 and H4 that are important in telomeric silencing by screening for mutants in these histone genes that bypass the loss of telomeric silencing phenotype of a cac1Δ strain. Single amino acid changes in the NH2-terminal tails of histone H3 and H4, and within their core regions, reinstated telomeric silencing. Interestingly, mutations mapping to the core of these histones suggest another region for potential interaction between nucleosomes and the SIR complex.
In principle, there are several ways in which these mutants may be acting. They may increase the affinity of the SIR complex for the nucleosomes, thus stabilizing the silent chromatin state against competition by gene activation pathways. Such stabilization itself may proceed by several routes. Some may be very direct; a new ionic or hydrogen bond may be formed between the new residue and the SIR complex. This may be true for three of the changed residues, for which single amino acid substitutions were found. However, for the other 10 in which at least two substitutions were identified, each with very different side chains, this scenario is unlikely. Rather these residues may increase silencing by preventing the transcriptional activation pathway from competing effectively. In particular they may prevent the activation-associated, posttranslational modifications that diminish the ability of silent chromatin to form. Indeed, as discussed below, our data suggest that this is the case for the alleles that mapped to the tails of histone H4, where we were able to determine that the acetylation of lysines was reduced. Another possibility is that the amino acid substitutions may increase silencing by stabilizing the nucleosome, perhaps reducing the ability of the nucleosome to be “remodeled” for gene activation (22)—in essence these alleles would not increase telomeric silencing per se, but rather make activation more difficult. Also, it is unclear whether the histone H3 and H4 mutants are critical for increasing silencing by acting in steps of silent chromatin assembly and/or maintenance. But given that they all bypassed the need for CAC1, it is likely that they may influence a step in assembly, at least in part.
How then might these specific alleles be acting? Here we can gain potential insights from studying the structure of the yeast nucleosome (40). For example, R39 of histone H4 is located within the core of the nucleosome at an interface with the DNA. The R39K allele in histone H4 may cause the interaction between histone H4 and the DNA to change within the nucleosome. Thus this allele may increase silencing by enhancing the stability of the nucleosome. H75Y also maps to the core of histone H4; it, too, may be acting to stabilize the nucleosome. However, H75 is also located in proximity to the histone H3 loop that contains K79 and could be acting through this region of the nucleosome (see below).
The mutations in D77 and D81 of histone H3 are particularly intriguing, as they map on either side of K79, a residue we recently identified to be methylated by the methyltransferase Dot1p (41). This residue maps to the top and bottom surfaces of the nucleosome and has the potential to be another binding site for the Sir complex on the nucleosome. The methylation of K79 modulates the level of telomeric silencing; methylation seems to reduce binding of Sir proteins to the nucleosomes. Thus the mutant alleles at D77 and D81 may be acting either by reducing or eliminating Dot1p-mediated methylation of K79. Alternatively, they may change the surface of the nucleosome in a way that mitigates the effects of K79 methylation and instead facilitates Sir binding.
In the NH2-terminal tail of histone H3, residues adjacent to all six lysines (K4, K9, K14, K18, K23, and K27) were heavily mutagenized. However, the only mutations that increased silencing were residues adjacent to K4, K14, or K23. Based on our findings at the histone H4 tail, we suspect that posttranslational modification of these lysines was affected. At K14 and K23, a decrease or lack of acetylation is the probable effect, whereas at K4 it is likely to be a decrease in methylation (reviewed in ref. 13). The importance of K14 and K23 in telomeric silencing was also found when cells lacked the histone H4 acetylase Hat1p (27), thus supporting the significance of the results reported here. Given that mutations in SET1 result in a decrease in telomeric silencing (42) and that Set1p mediates methylation of K4 (43–46), it is likely that the ability of Set1p to act on K4 is changed in the Q5G and Q5N alleles.
Directed mutagenesis of the residues adjacent to the five lysines (K5, K8, K12, K16, and K20) in the tail of histone H4 identified mutations only in those residues adjacent to K8, K12, and K16. K5 is reversibly acetylated, but apparently that has no consequence in telomeric silencing. No mutant alleles were identified at positions R17, R19, or I21. Given that virtually any change in the region encompassing residues 16–29 has been shown to result in a loss of telomeric silencing (8, 47, 48), it was not surprising that we did not identify mutant alleles in our screen for increased telomeric silencing.
The MS analysis after chemical acetylation by deuterated acetic anhydride allowed us to determine the relative level of acetylation at the four potentially acetylated lysines in the histone H4 tail. K16 was most dramatically underacetylated when A15 was changed, and K8 was similarly affected when G7 was mutated. This finding indicated that, on a global scale, these mutations prohibited or greatly reduced the efficacy of the major histone acetyltransferase(s) that act on these lysines. Although formally possible, it is doubtful that these mutations increased the efficacy of deacetylases that act on K8 or K16, because a variety of amino acid replacements at these residues (G7M, G7I, G7H, and G7Y; A15C, A15T, and A15V) all had the same phenotype.
However, the most striking result from the histone H4 tail mutants was the dramatic underacetylation of K12 in all of the mutants analyzed (Fig. 5). Returning to the idea that the acetylated status of a lysine might serve as a way for the chromatin state to be inherited, this result indicated that unacetylated K12 increased the probability that a silenced chromatin state would be inherited in a telomeric gene. We interpret this to mean that K12 is the site of a memory mark within the nucleosome for the heritable transmission of an active or silenced state in a telomeric gene.
If this idea is true, it has further implications with respect to histone H4 deposition in silent chromatin. It appears that shortly after synthesis of histone H4, it is acetylated at positions K5 and K12, most likely by the Hat1p histone acetylase (49–51). If an unacetylated K12 is needed for silent chromatin formation, then K12 must be rapidly deacetylated shortly after deposition onto the DNA in a silenced region of the genome. This may represent part of the silent chromatin maturation process that has been invoked to occur during, or shortly after, S phase (14, 52, 54).
Although the mass spectroscopic analysis of bulk histones has provided us with an intriguing insight about K12, the method is limited in that we have not determined the modified state of histones specifically within telomeric chromatin. We envision that the reduction in acetylation at specific lysines seen within total histone H4 mirrors the decreased likelihood that the lysine will be modified within telomeric chromatin as well. The application of this mass spectroscopic technology to isolated telomeric chromatin in the future will provide the greatest amount of information we seek. Nevertheless, by combining the analysis of bulk histones with the transcript microarray data for the five alleles of histone H4, in which we show there were no significant perturbations to expression of genes involved in telomeric silencing, we have alleviated the concern that the histone mutants increased telomeric silencing by indirect effects. Such indirect effects have plagued previous studies reporting the involvement of the RPD3 and HDA1 genes in telomeric silencing (55, 56). Mutants in these histone deacetylase genes changed the expression level of known silencing factors (57). Similarly, the earlier report of the requirement for HAT1 in telomeric silencing (Hat1p acetylates K12 on histone H4) may also be indirect, particularly given that mutations in the histone H3 tail were required to uncover the HAT1 dependency (27). Alternatively, the HAT1 result may be caused by there being more than one pathway for depositing the histone H3/H4 tetramer into silenced chromatin (58) [both ASF1- and CAC1-dependent complexes are involved in telomeric silencing (17, 19, 20)]. Perhaps mutation of the histone H3 tail forces histone deposition to proceed via a pathway that relies on acetylation of K12.
The transcript array data showed a preponderance of telomeric loci in the list of genes down-regulated to the greatest extent in the histone H4 alleles, particularly the G7I, G13L, and A15T alleles (Fig. 3 and Table 2). Because none of the known silencing components were perturbed, this observation fortifies the hypothesis that these histone H4 mutants have a direct involvement in forming silent telomeric chromatin. It is also worth noting that the proportion of telomeric loci silenced was greatest in the A15T allele, which likely reflects that K16 also needs to be unacetylated for telomeric silencing. This finding suggests that both K16 and K12 may serve as key memory marks for silent chromatin. Because Sir-mediated silencing appears to require multiple weak interactions between the nucleosome and Sir complex, there are likely to be several such memory marks within the nucleosome. This finding also implies that there may not be a single place within the nucleosome where memory is stored, and we believe that our mutants will help to identify more of these marks.
One of the key strengths of this mutant hunt was that it allowed us to identify not only key residues that were modified, but also the particular modified state of the residue that appears to be relevant for telomeric silencing (i.e., acetylation states of lysines in histone H4). Previous attempts mutated the lysines and then inferred what the change meant (reviewed in ref. 13). The collection of mutants created in our study should also be useful in identifying those activities that are critical for switching a silenced gene to an active state and/or maintaining it as such. For instance, genetic screens designed to identify unlinked neomorphic alleles that could acetylate K12. Alternatively, the histone mutants could be used as a substrate in biochemical assays with histone acetylases and methylases that have been shown to be involved in telomeric silencing (44–46, 59). Ultimately it will be interesting to determine whether such factors are gene-specific or part of a general system of maintaining gene expression.
The analysis of these mutant histones may also define interdependency of modifications within the histones (reviewed in ref. 60). For instance, mutation of G7 in histone H4 confers reduced acetylation at K8 as well as at K12, which raises the possibility that unacetylated K8 may decrease the chance for K12 acetylation. That is, in vivo, there may be a hierarchy of acetylation along the histone H4 tail.
The application of mass spectrometric methods to quantitatively evaluate levels of histone modifications, as we have shown here, promises to enhance our understanding of chromatin structure regulation. We have found the combination of chemical isotopic acetylation of histones with trypsin cleavage and MS to be more robust and to require much less sample than other physical methods of examining acetylation at specific residues [i.e., Edman degradation (61)]. Furthermore, it provides greater flexibility in analysis than site-specific antibodies (53), which cannot bind to mutant histones. Future applications of this technology will lie in development of biochemical isolation procedures of locus-specific chromatin.
The combination of further analysis of the histone H3 and H4 mutants isolated in this study along with further development of mass spectroscopic methods will facilitate a greater understanding of how heritable states of telomeric genes in S. cerevisiae are propagated. These methods, and the paradigms that evolve from our studies, are likely to be applicable to a wide variety of systems in which chromatin states are responsible for propagating an epigenetic phenotype.
Supplementary Material
Acknowledgments
We thank R. Gardner, D. Smith, A. Stellwagen, and F. van Leeuwen for comments on the manuscript. National Institutes of Health Grant GM43893 and an Ellison Medical Foundation Senior Scholar Award (to D.E.G.) supported this work.
Abbreviation
- Sir
Silent Information Regulators
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Muller H J. J Genet. 1930;22:299–324. [Google Scholar]
- 2.Henikoff S. Trends Genet. 1990;6:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- 3.Spofford J B. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 1c. New York: Academic; 1976. pp. 955–1018. [Google Scholar]
- 4.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 5.Moazed D. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 6.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 7.Grunstein M. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J S, Ling X, Grunstein M. Nature (London) 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 9.Suka N, Suka Y, Carmen A A, Wu J S, Grunstein M. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 10.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschling D E. Curr Biol. 2000;10:R708–R711. doi: 10.1016/s0960-9822(00)00714-4. [DOI] [PubMed] [Google Scholar]
- 12.Carmen A A, Milne L, Grunstein M. J Biol Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 13.Richards E J, Elgin S C. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 14.Aparicio O M, Gottschling D E. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 15.Laman H, Balderes D, Shore D. Mol Cell Biol. 1995;15:3608–3617. doi: 10.1128/mcb.15.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axelrod A, Rine J. Mol Cell Biol. 1991;11:1080–1091. doi: 10.1128/mcb.11.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler J K, Adams C R, Chen S R, Kobayashi R, Kamakaka R T, Kadonaga J T. Nature (London) 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 18.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 21.Weintraub H, Flint S J, Leffak I M, Groudine M, Grainger R M. Cold Spring Harbor Symp Quant Biol. 1977;42:401–407. doi: 10.1101/sqb.1978.042.01.042. [DOI] [PubMed] [Google Scholar]
- 22.Narlikar G J, Fan H Y, Kingston R E. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 23.Turner B M. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 24.Proffitt J H, Davie J R, Swinton D, Hattman S. Mol Cell Biol. 1984;4:985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 26.Jenuwein T. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 27.Kelly T J, Qin S, Gottschling D E, Parthun M R. Mol Cell Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Leeuwen F, Gottschling D E. Methods Enzymol. 2002;350:165–186. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- 29.Bedalov A, Gatbonton T, Irvine W P, Gottschling D E, Simon J A. Proc Natl Acad Sci USA. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterborg J H. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]
- 31.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 32.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 33.Xu E Y, Kim S, Rivier D H. Genetics. 1999;153:25–33. doi: 10.1093/genetics/153.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans S K, Sistrunk M L, Nugent C I, Lundblad V. Chromosoma. 1998;107:352–358. doi: 10.1007/s004120050318. [DOI] [PubMed] [Google Scholar]
- 35.Riordan J F, Vallee B L. Methods Enzymol. 1967;11:565–570. [Google Scholar]
- 36.Steiner R F, Albaugh S, Fenselau C, Murphy C, Vestling M. Anal Biochem. 1991;196:120–125. doi: 10.1016/0003-2697(91)90127-f. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Smith D L, Griep M A. Protein Sci. 1998;7:1781–1788. doi: 10.1002/pro.5560070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes R N, Gross M L. Methods Enzymol. 1990;193:237–263. doi: 10.1016/0076-6879(90)93418-k. [DOI] [PubMed] [Google Scholar]
- 39.Biemann K. Methods Enzymol. 1990;193:886–887. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]
- 40.White C L, Suto R K, Luger K. EMBO J. 2001;20:5201–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Leeuwen F, Gafken P R, Gottschling D E. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 42.Nislow C, Ray E, Pillus L. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briggs S D, Bryk M, Strahl B D, Cheung W L, Davie J K, Dent S Y, Winston F, Allis C D. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogan N J, Dover J, Khorrami S, Greenblatt J F, Schneider J, Johnston M, Shilatifard A. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 45.Roguev A, Schaft D, Shevchenko A, Pijnappel W W, Wilm M, Aasland R, Stewart A F. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagy P L, Griesenbeck J, Kornberg R D, Cleary M L. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 48.Johnson L M, Fisher-Adams G, Grunstein M. EMBO J. 1992;11:2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 51.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 52.Li Y C, Cheng T H, Gartenberg M R. Science. 2001;291:650–653. doi: 10.1126/science.291.5504.650. [DOI] [PubMed] [Google Scholar]
- 53.White D A, Belyaev N D, Turner B M. Methods. 1999;19:417–424. doi: 10.1006/meth.1999.0878. [DOI] [PubMed] [Google Scholar]
- 54.Kirchmaier A L, Rine J. Science. 2001;291:646–650. doi: 10.1126/science.291.5504.646. [DOI] [PubMed] [Google Scholar]
- 55.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. Nature (London) 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 57.Bernstein B E, Tong J K, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mello J A, Sillje H H, Roche D M, Kirschner D B, Nigg E A, Almouzni G. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marmorstein R, Roth S Y. Curr Opin Genet Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Reinberg D. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 61.Sobel R E, Cook R G, Allis C D. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.