Abstract
Heterochromatin protein 1 (HP1) is a conserved chromosomal protein that participates in chromatin packaging and gene silencing. A loss of HP1 leads to lethality in Drosophila and correlates with metastasis in human breast cancer cells. On Drosophila polytene chromosomes HP1 is localized to centric regions, telomeric regions, in a banded pattern along the fourth chromosome, and at many sites scattered throughout the euchromatic arms. Recently, one mechanism of HP1 chromosome association was revealed; the amino-terminal chromo domain of HP1 interacts with methylated lysine nine of histone H3, consistent with the histone code hypothesis. Compelling data support this mechanism of HP1 association at centric regions. Is this the only mechanism by which HP1 associates with chromosomes? Interest is now shifting toward the role of HP1 within euchromatic domains. Accumulating evidence in Drosophila and mammals suggests that HP1 associates with chromosomes through interactions with nonhistone chromosomal proteins at locations other than centric heterochromatin. Does HP1 play a similar role in chromatin packaging and gene regulation at these sites as it does in centric heterochromatin? Does HP1 associate with the same proteins at these sites as it does in centric heterochromatin? A first step toward answering these questions is the identification of sequences associated with HP1 within euchromatic domains. Such sequences are likely to include HP1 “target genes” whose discovery will aid in our understanding of HP1 lethality in Drosophila and metastasis of breast cancer cells.
In eukaryotes, there are two major types of chromatin: heterochromatin and euchromatin (1). Heterochromatin corresponds to the relatively gene-poor, late-replicating, repetitious sequences found near centric and telomeric locations. In contrast, euchromatin replicates relatively early in the cell cycle and contains single copy sequences, including the majority of genes. Both euchromatin and heterochromatin are packaged into nucleosomes, the fundamental packaging unit consisting of a histone octamer. Euchromatin and heterochromatin can be distinguished by specific histone tail modifications. In general, the histone tails in heterochromatin are relatively hypoacetylated; however, acetylation of lysine twelve of histone H4 is a distinguishing mark for heterochromatin (2–4). In contrast, histone H3 and H4 tails found in euchromatin are generally acetylated (4). Histone H3 acetylation is often linked to H3 phosphorylation and is likely to represent a two-component code for high levels of gene expression (5, 6).
In addition to distinct differences in histone modification, euchromatin and heterochromatin show differences in nonhistone chromosomal protein constituents. One of the best-studied examples is heterochromatin protein 1 (HP1) first discovered in Drosophila and named for its predominant localization to centric heterochromatin (7) (Fig. 1A). Consistent with this localization, the gene encoding HP1, Su(var)2–5, was isolated as a dominant suppressor of position effect variegation (PEV) (8, 9). PEV is the mosaic pattern of expression exhibited by genes placed near centric heterochromatin by chromosomal rearrangements or transposition events (10). Overexpression of HP1 leads to enhanced silencing of variegating genes. Conversely, a decreased level of HP1 leads to reduced silencing of variegating genes. A complete loss of HP1, as in homozygous Su(var)2–5 null mutants, results in lethality. Larvae survive until the late third instar stage because of maternally supplied HP1 (11, 12). The cause of lethality is unknown. Given the centric localization of HP1, and the interaction between the Schizosaccharomyces pombe HP1-like protein Swi6 and a cohesion protein, chromosome segregation might be affected (13, 14). Thus, HP1 levels are critical for regulating the extent of heterochromatization within centric regions that is required for proper chromosome segregation.
Figure 1.
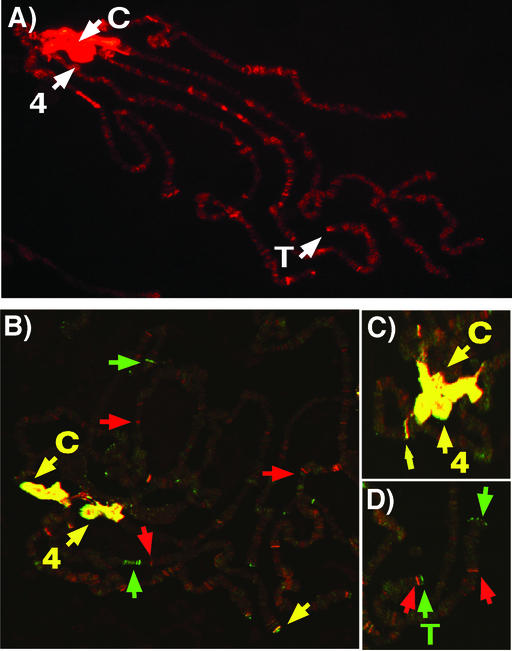
(A) Pattern of HP1 distribution on Drosophila polytene chromosomes. D. melanogaster larval polytene chromosomes were stained with mouse monoclonal C1A9 antibodies against HP1 (gift of Sarah C. R. Elgin) and a secondary antibody conjugated with rhodamine. The chromocenter (C), the fourth chromosome (indicated by 4), telomeres (T), and euchromatic sites associated with HP1. (B) The pattern of HP1 and methylated lysine nine of histone H3 on Drosophila polytene chromosomes. D. melanogaster larval polytene chromosomes were stained with mouse monoclonal C1A9 antibody against HP1 and a rabbit polyclonal antibody that recognizes methylated lysine nine of histone H3 (gift of C. David Allis, University of Virginia, Charlottesville). A Cy5-conjugated rabbit secondary antibody and a FITC-conjugated mouse secondary antibody were used for detection. The chromocenter (C) and the fourth chromosome (indicated by 4) show strong colocalization (yellow). Example locations enriched in HP1 are denoted by green arrows; example locations enriched in methylated lysine nine of histone H3 are indicated by red arrows. (C) Same as in B, showing a closer view of the chromocenter region. (D) Same as in B, showing a closer view of a telomeric region.
In addition to centric regions, HP1 is observed at other regions of the genome known to be heterochromatic. The small fourth chromosome of Drosophila melanogaster, interspersed with heterochromatic domains, shows a banded pattern of HP1 localization (7, 15). Consistent with HP1 having a packaging function at these locations, transgenes inserted along the fourth chromosome exhibit PEV that is suppressed by Su(var)2–5 mutations (15, 16). HP1 localization is also observed at Drosophila telomeres that terminate in repetitive arrays of retrotransposons (17). Telomeric association, however, appears to be independent of primary DNA sequence as broken chromosomes lacking terminal retrotransposons retain HP1 association (12). Telomere-telomere fusions occur in larval neuralblasts of Su(var)2–5 mutants, suggesting HP1 plays a role in telomere capping (12).
In contrast to these chromosomal domains rich in repetitive DNA sequences, HP1 is present at approximately 200 sites within the euchromatic arms of polytene chromosomes that are relatively poor in repetitious DNA sequences. Do these sites represent small domains of repressive chromatin? Are there genes at these sites that are regulated by HP1? These questions are currently under investigation.
Here we describe current studies on the role of HP1 in gene regulation at both euchromatic and heterochromatic domains. We summarize the results from reports that have identified HP1 partner proteins and discuss implications for these findings. Last, we hypothesize about multiple mechanisms of HP1 chromosome association and their impact on gene expression.
HP1 Follows Code
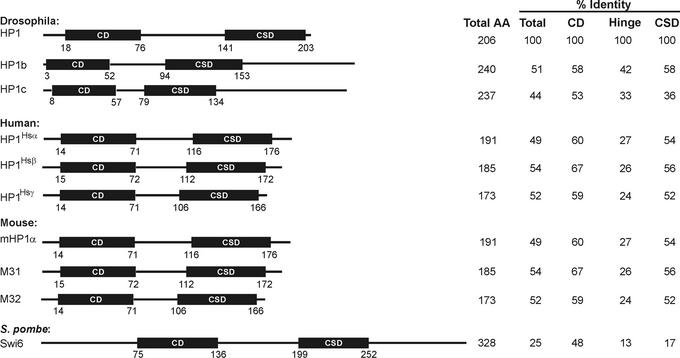
HP1 is a highly conserved protein with family members found in a variety of eukaryotic organisms ranging from S. pombe to humans (18–21). In Drosophila, two additional HP1-like proteins, HP1b and HP1c, sharing amino acid sequence similarity and domain structure, have been identified (22) (Fig. 2). Whereas HP1b shows localization to both euchromatin and heterochromatin just as HP1, HP1c is found only in euchromatin (22). Mice and humans each have three HP1-like proteins that possess similarities in amino acid sequence, domain structure, and centric chromosomal localization properties as Drosophila HP1 (Fig. 2). Although there are minor differences in chromosomal localization and protein interaction partners for HP1-like proteins within a given species (23), it is not clear whether these proteins have specific or redundant functions. In flies, mice, and humans, the HP1-like proteins are small in size, ranging from 173 to 240 aa (Fig. 2). Overall the percent identity of HP1-like proteins to Drosophila HP1 is approximately 50% for mammalian HP1-like proteins. The majority of conserved amino acids are concentrated in two domains. The structure of HP1-like proteins can be simplified as two conserved domains separated by a less conserved hinge region (Fig. 2). The conserved amino-terminal region of HP1-like proteins is termed the chromo domain (CD) (24). This domain is present in 20 proteins in Flybase (www.ebi.ac.uk/proteome/DROME/interpro/stat.html), many of which play roles in gene regulation. The conserved carboxyl-terminal region, termed the chromo shadow domain (CSD), is related to the CD in primary amino acid sequence (25). Both the CD and the CSD have been the subject of extensive structural analysis (26–30). Each domain forms a hydrophobic pocket. The CSD dimerizes (18, 27, 28, 31) as well as interacts with a wide variety of nuclear proteins (see below). Cross-species functional studies in which the mouse HP1-like protein M31 was expressed in S. pombe indicate that species-specific functions of HP1 reside within the CSD (32). The CD is required for chromosome association (33).
Figure 2.
Diagram of HP1 proteins in Drosophila, mouse, human, and S. pombe. Total length of each protein is indicated and drawn to relative scale. Percent identity when compared with Drosophila HP1 over the full length (total), or the CD, CSD, or the hinge region was calculated according to ref. 85.
The mechanism(s) by which HP1 establishes the complex localization pattern on chromosomes remained a mystery for over a decade since its discovery. For many chromosomal proteins, localization is achieved through direct interaction with DNA sequences. Attempts to identify specific interactions between HP1 and DNA sequences, particularly repetitive DNA sequences found within heterochromatin, were not particularly revealing (34). For some chromosomal proteins, localization is achieved through interactions with DNA binding proteins. Therefore, a search for HP1 partner proteins might reveal the “missing link” between HP1 and the chromosome. A phage display assay was performed to identify peptides that interact with the CD and CSD (31). This assay revealed peptide sequences that showed a specific interaction with the CSD. Comparison of the peptide sequences allowed a consensus pentapeptide to be generated (31). Supporting these results, the consensus pentapeptide was found in several proteins shown to interact with HP1 by other types of assays (35–37). To date, the localization pattern of candidate interacting proteins cannot explain the entire localization pattern observed for HP1. In contrast to the results obtained for the CSD, no peptides were identified from the phage display assay that specifically interacted with the CD. These results were surprising because a point mutation within the CD of Drosophila HP1 eliminates the majority of chromosome association, suppresses PEV, and is homozygous lethal (9).
The mystery surrounding interactions of the CD was solved by studies of the murine Suv39h1 protein, a homologue of the Drosophila SU(VAR)3–9 protein (38, 39). A comparative genomic approach in combination with biochemical studies revealed that the SET [a conserved motif in Drosophila Su(rar)3-9, Enhancer of Zeste, and trithorax] domain of Suv39h1 contains methyltransferase activity specific for lysine nine of histone H3. This methylation mark on the histone H3 tail serves as a specific recognition code for the CD of HP1. This discovery supports the histone code hypothesis that proposes histone tail modifications serve as specific recognition motifs for chromatin proteins (40). The HP1 CD, but not the CD of several other proteins, binds methylated lysine nine of histone H3 (41). Therefore, the substrate specificity is likely caused by minor differences in the amino acid sequences of CDs from different proteins. The connection between Suv39h1 and HP1 is consistent with Drosophila research showing that the genes encoding HP1 and SU(VAR)3–9 genetically interact with the heterochromatin silencing system (8) and that the proteins physically interact (42). The relationship between HP1 and Suv39h1 has been maintained by the S. pombe homologues, Swi6 and Clr4, respectively (43, 44), suggesting evolutionary conservation in this mechanism of chromosome association and heterochromatic gene silencing.
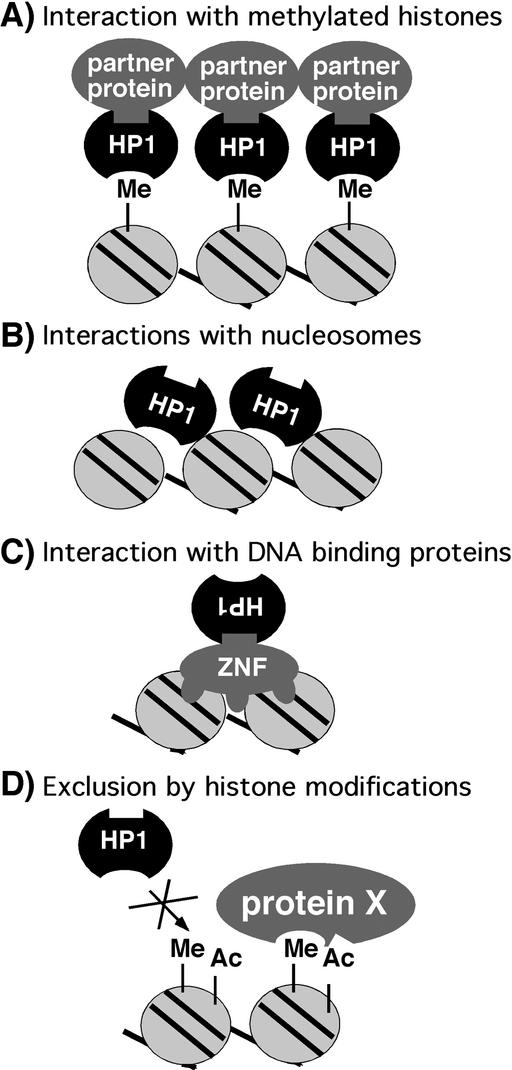
In summary, HP1 serves as a bridging protein, connecting histones, through interactions with the CD, to nonhistone chromosomal proteins, through interactions with the CSD (Fig. 3A). In this case, Suv39h1 sets the histone code for HP1 association. Based on these findings, mechanisms for heterochromatin spreading have been proposed to involve recruitment of Suv39h1 by HP1 and propagation of the methylation mark along the chromosome (39). Details of such spreading mechanisms remain to be elucidated.
Figure 3.
Models for HP1 association and nonassociation with chromosomes. (A) Interaction between the HP1 CD with the methylated lysine nine of histone H3. HP1 serves as a bridge for partner proteins. (B) HP1 interacts with histones in a nonmethylated-dependent fashion. (C) HP1 associates with chromosomes through interactions of the CSD and DNA binding proteins, such as zinc-finger proteins (ZNF). (D) HP1 does not associate with methylated histones that have additional modifications such as acetylation or phosphorylation.
Are There Multiple Mechanisms for HP1 Association?
HP1 localizes to distinctly different environments throughout the genome. Has the discovery of the interaction with the methylated lysine nine of histone H3 cracked the code, or are there alternative mechanisms of HP1 chromosomal association? The importance of this question is evident when reviewing data on the Su(var)2–502 allele of the gene encoding HP1. This allele contains the amino acid substitution of a highly conserved valine to a methionine in the CD. Structural analysis indicates that this residue plays a critical role in the formation of the hydrophobic pocket of the CD (30). In most genetic silencing assays this allele behaves as an HP1 null; however, an important distinction between this allele and null alleles was revealed by a cytological analysis of HP1 staining on chromosomes from HP1 mutants. Whereas null alleles show no HP1 chromosome association, the Su(var)2–502 allele shows diminished HP1 localization to centric regions, but retains association at euchromatic and telomeric sites (12). These data suggest an alternative mechanism of HP1 association might be operating at noncentric locations.
Further evidence for alternate mechanisms of association comes from cytological experiments on polytene chromosomes in wild-type flies. The pattern of staining observed by antibodies to HP1 and methylated lysine nine of histone H3 is not completely coincident. Both antibodies show colocalization to the chromocenter and along the fourth chromosome, but not throughout the euchromatic arms and at telomeric regions (Fig. 1 B–D) (45). One technical explanation for incomplete colocalization is that the epitopes recognized by either antibody are masked by fixation at specific genomic locations. However, if this is not the case, sites within the euchromatic arms that stain with only the HP1 antibody could be generated by HP1 associations through mechanisms independent of SU(VAR)3–9. Interactions of HP1 with unmodified histone tails, the histone-fold domain, and histone H1 might account for the staining pattern observed (34, 46). Such possibilities are diagrammed in Fig. 3B. Alternatively, interactions with nonhistone chromosomal proteins might serve as an additional mechanism of association (Fig. 3C). Interactions between HP1 and transcriptional corepressors that associate with DNA binding proteins (see below) (47, 48) support this hypothesis. The double staining also revealed sites within the euchromatic arms that are detected only by the methyl lysine nine histone H3 antibody. These sites could correspond to different degrees of methylation because the antibody recognizes only dimethylated lysine (Upstate Biotechnology, Lake Placid, NY); HP1 is thought to recognize both methylated states with relatively equal affinity (30). Another explanation for lack of complete colocalization of HP1 and the methyl lysine nine histone H3 antibody is that additional histone modifications might be present that do not permit HP1 association (Fig. 3D). Clearly the code for chromosomal protein association might have multiple components.
HP1 Interacts with a Myriad of Proteins
Does the identification of proteins that associate with HP1 provide clues about the mechanisms of silencing? Genetic analysis of PEV in Drosophila provided a collection of mutations that encode candidate HP1 interaction partners: for example, SU(VAR)3–9, the histone methylase discussed above interacts with HP1 by two-hybrid analysis (42) (Table 1). A second example is SU(VAR)3–7, a zinc finger protein that associates with satellite DNA sequences (49). HP1 and SU(VAR)3–7 colocalize in the Drosophila embryo and on polytene chromosomes (50, 51) and interactions between the two proteins have been demonstrated by yeast two-hybrid analysis and coimmunoprecipitation from embryonic extracts (35, 50). More specifically, the CSD of HP1 interacts with multiple regions of SU(VAR)3–7, but it is not yet clear how these two proteins collaborate to form and/or spread heterochromatin.
Table 1.
HP1 interacting partners and candidate partners
| Protein | Organism | HP1 variant | Methodology | HP1 domain | Ref(s). |
|---|---|---|---|---|---|
| Transcription regulation/chromatin modifying proteins | |||||
| H1 | Drosophila | HP1 | rPD | nd | 46 |
| HP1-BP74 H1-like | Mouse | mHP1α | Y2H, FW, rPD | Hinge region | 46, 86 |
| H3 | Mouse | mHP1α, M31, M32 | FW, rPD, exIP | CD | 46, 54 |
| H3 | Drosophila | HP1 | rPD | nd | 46 |
| Methylated K9 of H3 | S. pombe | Swi6 | rPD, ChIP | CD | 38, 43 |
| Methylated K9 of H3 | Drosophila | HP1 | IF, FAITC, NMR | CD | 26, 41 |
| Methylated K9 of H3 | Mouse | mHP1α, M31, M32 | rPD | CD | 39 |
| Methylated K9 of H3 | Human | HP1Hsα, HP1Hsβ, HP1Hsγ | rPD, SPRA | CD | 38 |
| H4 | Mouse | M31 | rPD | nd | 54 |
| H4 | Drosophila | HP1 | In vitro cross-linking | CSD | 34 |
| MacroH2A1.2* | Mouse | M31 | IF | nd | 87 |
| SUVAR3–9 | Drosophila | HP1 | IF, Y2H, exIP | CSD | 42 |
| Suv39h1 | Mouse | M31 | IF, exIP, SED | nd | 88, 89 |
| SUV39H1 | Human | HP1Hsβ | IF, exIP, SED | nd | 88, 89 |
| Suvar3–7 | Drosophila | HP1 | IF, Y2H, exIP | CSD | 35, 50 |
| KAP-1/TIF1β | Human | HP1Hsα, HP1Hsγ | IF, rPD, exIP, SPRA, GFC | CSD | 28, 46–48 |
| KAP-1/TIF1β | Mouse | mHP1α, M31, M32 | IF, rPD, Y2H, exIP, GFC | CSD | 28, 37, 47, 86, 90 |
| TRF1/PIN2 | Mouse | M31 | IF | nd | 91 |
| TAFII130 | Human | HP1Hsα, HP1Hsγ | Y2H, transPD, exPD | CSD | 36 |
| TIF1α | Mouse | mHP1α, M31, M32 | Y2H, rPD | CSD | 86, 90 |
| mSNF2β | Mouse | mHP1α | Y2H | CSD | 86 |
| Rb | Human | HP1, HP1Hsγ | Y2H, exPD, exIP, ChIP | nd | 57, 82 |
| Rb | Maize | HP1γ | rPD, Y2H | nd | 82 |
| Dnmt3a | Mouse cells | mHP1α | IF | nd | 92 |
| Dnmt3b | Mouse cells | mHP1α | IF | nd | 92 |
| ATRX/HP1-BP38 | Mouse | mHP1α, M31 | Y2H, IF | CSD | 86, 93 |
| Pim-1 | Human | HP1Hsγ | Y2H, exIP, rPD | CSD | 65 |
| CKII | Drosophila | HP1 | In vitro phosphorylation | nd | 63 |
| dAF10 | Drosophila | HP1 | transPD | CSD | 94 |
| DNA replication and repair | |||||
| CAF-1 p150 | Mouse | mHP1α, M31 | IF, Y2H, rPD, GFC, NMR | CSD | 28, 37 |
| CAF-1 p150 | Human | HP1Hsα | rPD | CSD | 48 |
| Ku70 | Human | HP1Hsα | Y2H, rPD, exIP | CSD | 95 |
| BRCA-1* | Human | HP1Hsα | IF | nd | 96 |
| ORC1 | Drosophila | HP1 | transIP | CD, CSD | 58 |
| ORC2 | Drosophila | HP1 | IF, exPD, exIP, transIP | CD, CSD | 58 |
| ORC3 | Drosophila | HP1 | transIP | CD, CSD | 58 |
| ORC4 | Drosophila | HP1 | transIP | CD, CSD | 58 |
| ORC5 | Drosophila | HP1 | exIP, transIP | CD, CSD | 58 |
| ORC6 | Drosophila | HP1 | exIP, transIP | CD, CSD | 58 |
| Xorc1 | Xenopus | XHP1α, XHP1γ | Y2H | nd | 58 |
| HOAP | Drosophila | HP1 | IF, exIP | nd | 62 |
| Nuclear architecture | |||||
| Lamin B receptor | Human | HP1Hsα, HP1Hsβ, HP1Hsγ | Y2H, rPD, exPD, transPD, exIP | CSD | 48, 52–54 |
| HP1-BP84 | Mouse | mHP1α, M31 | Y2H | CSD | 86 |
| Lamin B | Mouse | M31 | BA | CD | 55 |
| LAP2β | Mouse | M31 | BA | CD | 55 |
| Nuclear envelope | Mouse | mHP1α, M31, M32 | IF, BA | CD | 55 |
| Other chromosome-associated proteins | |||||
| Psc3 | S. pombe | Swi6 | IF, Y2H, exPD, ChIP | CD+Glu-rich | 13 |
| DDP1 | Drosophila | HP1 | IF | nd | 97 |
| Arp4/dArp6 | Drosophila | HP1 | IF | nd | 98, 99 |
| INCENP | Human | HP1Hsα, HP1Hsγ | Y2H, transPD | Hinge region | 100 |
| Ki-67 | Human | mHP1α, M31, M32 | Y2H, exPD, IF | CSD | 101 |
| SP100B | Human | HP1Hsα, HP1Hsβ, HP1Hsγ | IF, Y2H, rPD, transPD | CSD | 48, 102, 103 |
| EST AA153281 | Mouse | mHP1α, M31 | Y2H, rPD | CSD | 37 |
| EST AA003533 | Mouse | mHP1α, M31 | Y2H, rPD | CSD | 37 |
BA, binding assay; ChIP, chromatin immunoprecipitation; exIP, co-immunoprecipitation using extract; exPD, pull-down assay using extracts; FAITC, fluorescence anisotropy, isothermal titration calorimetry; FW, far Western analysis; GFC, gel filtration chromatography; IF, immunofluorescence colocalization; nd, not determined; rIP, coprecipitation using recombinant proteins; rPD, pull-down assay using recombinant proteins; transIP, immunoprecipitation with in vitro-translated protein; transPD, pull-down assay using in vitro translated protein; SED, sedimentation assay; SPRA, surface plasmon resonance analysis; Y2H, yeast two-hybrid assay.
Denotes cell cycle-dependent association.
In addition to a gene silencing function, HP1 is thought to play a role in nuclear organization. This hypothesis is based on the discovery that HP1 interacts with lamin B receptor, either directly (52, 53) or indirectly through interactions with histones (54). In addition, experimental data support an interaction between HP1 and B-type lamin and Lap2β, lamin-associated protein, located within the nuclear envelope. In vitro, these interactions foster nuclear envelope assembly, suggesting HP1 plays a role in organizing nuclear architecture (55). Given that heterochromatin localizes to the nuclear periphery in many eukaryotic cell types, HP1 might tether heterochromatin to the nuclear envelope, leaving active regions of the genome free to coalesce into transcription factories within the interior of the nucleus (56).
The localization of HP1 to many sites throughout the Drosophila euchromatic arms brings to question the role of HP1 in the regulation of gene expression. Supporting a role for HP1 in transcriptional regulation, HP1 has been shown to interact with numerous proteins involved in modulating chromatin structure and gene expression (Table 1). In mammals, association of HP1, the retinoblastoma (Rb) protein and SUV39H1 with the cyclin E promoter correlates with gene silencing (57). Furthermore, HP1 has been implicated in gene repression mediated by Krüppel-associated box (KRAB) zinc finger proteins (47, 48). Taken together, these findings suggest that HP1 is recruited to specific genes by protein–protein interactions, resulting in gene silencing by an unknown mechanism.
In addition to transcriptional regulators, HP1 interacts with proteins involved in DNA replication and repair. Chromatin assembly factor 1 (CAF1) is a three-subunit complex that assembles histones H3 and H4 onto newly replicated DNA in both euchromatic and heterochromatic regions of the genome. In mammals, the large subunit, p150, contains a domain that interacts with the CSD of HP1 (37). Deletion of this domain does not alter CAF1-mediated chromatin assembly after replication in vitro or targeting of HP1 to heterochromatin in vivo during DNA replication. However, deletion of this domain reduces the amount of CAF1 present in heterochromatin outside of S phase. Although the significance of CAF1-HP1 interaction is not clear, the data suggest that CAF1 might stabilize heterochromatin structure during times of chromosome decondensation and transcription.
HP1 associates with origin recognition complex (ORC) proteins (58, 59). This presents an intriguing parallel to the situation in Saccharomyces cerevisiae where ORC proteins associate with silent information regulator (SIR) proteins to generate silent chromatin (60, 61). A high molecular weight complex isolated from Drosophila embryos was recently shown to contain HP1/ORC-associated protein (HOAP) in addition to HP1 and ORCs. HOAP has sequence similarity to high mobility group proteins and binds to satellite sequences in vitro (62). Mutations in the genes encoding ORC proteins and HOAP are suppressors of PEV, suggesting a role in heterochromatin formation (62).
What regulates HP1 association with protein partners? Posttranslational modifications are likely to govern interactions between HP1 and partner proteins and/or itself. In Drosophila, HP1 is multiply phosphorylated giving rise to at least eight differently charged isoforms (63). Drosophila embryonic extracts possess HP1-containing complexes that differ in HP1 phosphorylation status (58, 62). For example, hypophosphorylated HP1 is found in a complex containing ORC and HOAP (58, 62). In Drosophila, casein kinase II (CKII) is credited for the phosphorylation of serine residues at the amino and carboxyl termini of HP1. Mutation of these serine residues to alanine reduces the amount of HP1 localized to centric heterochromatin and reduces gene silencing, suggesting phosphorylation plays a role in chromosome association and/or complex stability (63, 64).
In mammals, HP1 phosphorylation changes through the cell cycle. HP1Hsα and HP1Hsγ exhibit increased levels of phosphorylation during mitosis (23). HP1Hsγ is a substrate for Pim-1 kinase that phosphorylates a serine cluster in the center of the protein (65). Phosphorylation of HP1Hsα is thought to disrupt protein–protein interactions that are necessary to maintain most of the centric localization. In G2, phosphorylated HP1Hsα shifts from a centric location to being dispersed throughout the nucleus. Clearly, the role of phosphorylation needs further investigation to understand the biological significance of this dynamic process.
HP1 Regulates Gene Expression
Effects on Gene Expression Near Centric Heterochromatin.
The gene encoding HP1, Su(var)2–5, was isolated in a screen for suppressors and enhancers of PEV of the white+ gene brought into juxtaposition with heterochromatin through a chromosomal rearrangement (8). To determine the effects of HP1 on chromatin packaging, stocks containing the well-characterized Drosophila hsp26 gene inserted within centric heterochromatin were used for chromatin structure analysis (16). These transgenes exhibit less accessibility to nucleases and are packaged into a more regular nucleosome array than euchromatic insertions. In an HP1 mutant background the transgenes become more accessible to restriction enzyme digestion, indicating a more “open” chromatin configuration (66). To determine the transcriptional mechanism impaired by packaging with HP1, high-resolution chromatin structure analysis was performed (67). The results indicated that general transcription factors such as TFIID and RNA polymerase II are not associated with heterochromatic transgenes exhibiting silencing. Thus, association of HP1 correlates with a “closed” chromatin configuration that limits the accessibility of regulatory sites to trans-acting factors. This is similar to the mechanism of X-chromosome inactivation in mammals where transcription factors are also absent from genes on the inactive X (68), but contrasts the mechanism of silencing at the mating type loci in S. cerevisiae and Polycomb-mediated silencing in Drosophila in which general transcription factors and RNA polymerase II are found in association with silenced promoters (69–71). Differences between these systems and HP1 silencing could reflect fundamentally different properties of the silencing systems or developmentally different stages of silent chromatin maturation.
In contrast to the silencing effects HP1 has on euchromatic genes, HP1 is required for the expression of genes that naturally reside within heterochromatin. Over the years genetic and molecular analyses have revealed genes that reside within centric heterochromatin (72, 73). Two well-characterized genes are light, an essential gene encoding a protein involved in the vesicle transport pathway (74, 75), and rolled, an essential mitogen-activated protein kinase (76). Heterochromatic genes are not specific for Drosophila; they also have been discovered in Arabidopsis (77) and are likely to be found in other organisms as genomic sequence analysis becomes more complete. In Drosophila, heterochromatic genes appear to be unrelated to each other in function, however, they do share some common properties. Structurally, many heterochromatic genes have introns containing middle repetitive DNA sequences (74) (D. E. Cryderman and L.L.W., unpublished data). Heterochromatic genes are inefficiently expressed and sometimes exhibit PEV when translocated to euchromatin (78, 79). In addition, heterochromatic genes require heterochromatin proteins such as HP1 for expression (11).
How does HP1 establish a chromatin configuration that hinders the expression of euchromatic genes while fostering the expression of heterochromatic genes? This question will be better addressed as the promoter regions of heterochromatic genes are analyzed. Assuming a role for HP1 in chromatin compaction, HP1 might bring distant regulatory elements in association with the promoter region of heterochromatic genes. Alternatively, HP1 may be required to set up a favorable chromatin configuration within the promoter proximal region and/or be involved in the recruitment of general transcription factors as suggested by a recent report showing an interaction between HP1 and the general transcription factor TFII130 (36).
Effects of Tethering HP1.
What are the effects of HP1 on gene expression at locations other than centric heterochromatin? One approach taken to address this question has been to generate HP1 fusion proteins containing heterologous DNA binding domains. In mammalian cell transient transfection assays, effects of HP1 fusion proteins on the expression of reporter genes possessing the appropriate DNA binding sequences are assayed. In these experiments both murine and human HP1 family members repress transcription when tethered to a small number of sites located in close proximity to the promoter (36). Transcriptional repression no longer occurs as the binding sites are moved to distances more than 2 kb from the promoter (80). Does this indicate that HP1-mediated repression has only short-range capabilities, perhaps operating on a gene-by-gene basis? If this is the case, HP1 located at euchromatic sites (Fig. 1) might play a role in the regulation of individual euchromatic genes, rather than entire domains.
The effects of tethering HP1 in a chromosomal context, as in transgenic Drosophila, demonstrate the complexities of gene silencing. A Gal4–HP1 fusion protein tethered upstream of a reporter gene caused silencing at only one of six genomic locations tested (51, 81). Interestingly, the site that supported silencing was flanked by middle repetitive DNA sequences, reminiscent of heterochromatin domains. In this case, silencing could spread in trans to a homologue lacking the tethering sites. These results suggest that not all chromosomal contexts will support the formation of silent chromatin by HP1. The inability to silence at certain locations might depend on the chromatin of the neighboring region, including the types of histone modifications, as well as gene density and transcriptional status of the region.
Identification of HP1 Target Genes.
As a second approach to determine the effects of HP1 on gene expression at locations other than centric heterochromatin, investigators have identified potential target genes by their response to HP1 dosage. Representational difference analysis identified two genes that are up-regulated in homozygous HP1 mutant larvae (59). Interestingly, one of the genes misregulated in the HP1 homozygous mutant maps to cytological region 31, a chromosome division that stains intensely with antibodies to HP1 (7). Two additional randomly selected genes within region 31 show up-regulation in HP1 homozygous mutants (59). For all four candidate HP1 target genes, mutations in additional modifiers of PEV, including Su(var)3–9, cause increases in gene expression. These results suggest that HP1 might function to silence genes located within euchromatic domains in a mechanism similar to that operating in centric regions.
A microarray approach has also been used to identify candidate genes regulated by HP1. Several hundred genes mapping within the euchromatic arms are up-regulated in an HP1 mutant background (D. E. Cryderman and L.L.W., unpublished data). In addition, several hundred genes were down-regulated, a pattern similar to that of heterochromatic genes. For all of the candidate target genes identified in Drosophila to date, it remains to be determined whether misregulation is caused by a direct interaction with HP1. It will be of interest to determine whether interactions between HP1 and specific genes are conserved through evolution.
In mammals, it is also likely that HP1 plays a role in the regulation of genes at noncentric locations. HP1 has been identified as a partner protein for many promoter-associated factors involved in control of gene repression. These include the transcription intermediate factor TIF1β that interacts with zinc finger proteins containing Krüppel-associated box (KRAB) domains known to be involved in transcriptional repression. The role of HP1 in KRAB-mediated repression is not clear, but might involve recruitment of histone deacetylases (47, 48).
The first example of a direct association of HP1 with a promoter region came from studies on the cyclin E gene (57). Binding of the Rb protein upstream of the cyclin E promoter causes gene silencing, partly through the recruitment of histone deacetylases. In addition, chromatin cross-linking and immunoprecipitation experiments place Rb at the promoter with HP1 and methylated lysine nine of histone H3 (57). Consistent with the finding, an “Rb binding motif” is present within the amino acid sequences of HP1 from a variety of species (82). It is unclear whether HP1 is recruited to the cyclin E promoter by Rb, methylated lysine nine of histone H3, SUV39H1, or any combination of these interacting molecules. In support of such interactions, HP1 possesses the ability to simultaneously interact with the methylated histone H3 tail and Rb (57). One hypothesis is that Rb recruits histone deacetylases first, because the histone H3 methyltansferase cannot use an acetylated lysine as a substrate for methylation (43), then SUV39H1 methylates the histone tail which serves as the substrate for HP1 binding. Interestingly, Drosophila SU(VAR)3–9 was recently purified in a complex with histone deacetylase HDAC1, suggesting that the two proteins might cooperate to methylate previously acetylated histone tails (83).
The identification of HP1 target genes has implications for understanding breast cancer metastasis in humans. HP1Hsα, but not HP1Hsβ or HP1Hsγ, is down-regulated in highly invasive/metastatic breast cancer cells compared with poorly invasive/nonmetastatic breast cancer cells (84). Introduction of a tagged HP1Hsα into the highly invasive/metastatic cells, which normally have low levels of HP1Hsα, lead to a less in vitro invasive phenotype. These results imply that modulation of the levels of HP1Hsα alters molecular properties of cells needed for invasion. Consistent with the cell culture studies, HP1Hsα is down-regulated in tissues from distant metastatic sites in breast cancer patients (84). One hypothesis is that HP1Hsα normally silences genes required for metastasis, making HP1Hsα a candidate metastasis suppressor. Depending on when HP1Hsα is down-regulated during tumor progression, HP1Hsα could be used as a predictive/prognostic marker for metastatic breast cancer.
Since the discovery of HP1 over 12 years ago by the laboratory of Sarah C. R. Elgin (Washington University, St. Louis), HP1 has grown in popularity. In part, this has been caused by the fact that HP1 has unexpectedly been identified as an interacting partner for a wide variety of proteins with diverse nuclear functions. Dissecting the function of HP1 in association with its partner proteins lies ahead. These experiments will shed light on the connections between chromatin structure, gene expression, DNA replication and repair, and nuclear organization.
Acknowledgments
We thank Sarah C. R. Elgin for the Drosophila HP1 antibody and C. David Allis for the gift of histone H3 methyl K9 antibodies and the histone code hypothesis. We thank Pamela Geyer and members of the Wallrath lab for suggestions regarding this manuscript. Work in the laboratory of L.L.W. is supported by American Cancer Society Grant GMC-100527 and National Institutes of Health Grant GM61513. Research into the role of HP1 in breast cancer metastasis was supported by a Carver Collaborative Pilot grant to L.L.W. and D.A.K. from the Roy J. and Lucille A. Carver College of Medicine at the University of Iowa.
Abbreviations
- CAF1
chromatin assembly factor 1
- CD
chromo domain
- CSD
chromo shadow domain
- HP1
heterochromatin protein 1
- ORC
origin recognition complex
- HOAP
HP1/ORC-associated protein
- PEV
position effect variegation
- Rb
retinoblastoma
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Richards E J, Elgin S C. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 2.Jeppesen P, Mitchell A, Turner B, Perry P. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 5.Cheung P, Tanner K G, Cheung W L, Sassone-Corsi P, Denu J M, Allis C D. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 6.Lo W S, Trievel R C, Rojas J R, Duggan L, Hsu J Y, Allis C D, Marmorstein R, Berger S L. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 7.James T C, Eissenberg J C, Craig C, Dietrich V, Hobson A, Elgin S C. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 8.Wustmann G, Szidonya J, Taubert H, Reuter G. Mol Gen Genet. 1989;217:520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- 9.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 11.Lu B Y, Emtage P C, Duyf B J, Hilliker A J, Eissenberg J C. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 13.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal S I, Watanabe Y. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 14.Bernard P, Maure J F, Partridge J F, Genier S, Javerzat J P, Allshire R C. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 15.Sun F L, Cuaycong M H, Craig C A, Wallrath L L, Locke J, Elgin S C. Proc Natl Acad Sci USA. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallrath L L, Elgin S C. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 17.Pardue M L, DeBaryshe P G. Genetica. 1999;107:189–196. [PubMed] [Google Scholar]
- 18.Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O. Development (Cambridge, UK) 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 19.Couteau F, Guerry F, Muller F, Palladino F. EMBO Rep. 2002;3:235–241. doi: 10.1093/embo-reports/kvf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eissenberg J C, Elgin S C. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 21.Volpe A M, Horowitz H, Grafer C M, Jackson S M, Berg C A. Genetics. 2001;159:1117–1134. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smothers J F, Henikoff S. Mol Cell Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minc E, Allory Y, Worman H J, Courvalin J C, Buendia B. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 24.Eissenberg J C. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- 25.Aasland R, Stewart A F. Nucleic Acids Res. 1995;23:3168–3174. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs S A, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 27.Cowieson N P, Partridge J F, Allshire R C, McLaughlin P J. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 28.Brasher S V, Smith B O, Fogh R H, Nietlispach D, Thiru A, Nielsen P R, Broadhurst R W, Ball L J, Murzina N V, Laue E D. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ball L J, Murzina N V, Broadhurst R W, Raine A R, Archer S J, Stott F J, Murzin A G, Singh P B, Domaille P J, Laue E D. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen P R, Nietlispach D, Mott H R, Callaghan J, Bannister A, Kouzarides T, Murzin A G, Murzina N V, Laue E D. Nature (London) 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 31.Smothers J F, Henikoff S. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Ma A, Chow C M, Horsley D, Brown N R, Cowell I G, Singh P B. Mol Cell Biol. 2000;20:6970–6983. doi: 10.1128/mcb.20.18.6970-6983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platero J S, Hartnett T, Eissenberg J C. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao T, Heyduk T, Allis C D, Eissenberg J C. J Biol Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
- 35.Delattre M, Spierer A, Tonka C H, Spierer P. J Cell Sci. 2000;113:4253–4261. doi: 10.1242/jcs.113.23.4253. [DOI] [PubMed] [Google Scholar]
- 36.Vassallo M F, Tanese N. Proc Natl Acad Sci USA. 2002;99:5919–5924. doi: 10.1073/pnas.092025499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murzina N, Verreault A, Laue E, Stillman B. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 38.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Nature (London) 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 39.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature (London) 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 40.Jenuwein T, Allis C D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs S A, Taverna S D, Zhang Y, Briggs S D, Li J, Eissenberg J C, Allis C D, Khorasanizadeh S. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama J, Rice J C, Strahl B D, Allis C D, Grewal S I. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 44.Grewal S I, Elgin S C. Curr Opin Genet Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 45.Cowell I G, Aucott R, Mahadevaiah S D, Borgoyne P S, Huskisson N, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Wu R, et al. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen A L, Oulad-Abdelghani M, Ortiz J A, Remboutsika E, Chambon P, Losson R. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 47.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., 3rd Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lechner M S, Begg G E, Speicher D W, Rauscher F J., 3rd Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cleard F, Spierer P. EMBO Rep. 2001;2:1095–1100. doi: 10.1093/embo-reports/kve243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cleard F, Delattre M, Spierer P. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seum C, Delattre M, Spierer A, Spierer P. EMBO J. 2001;20:812–818. doi: 10.1093/emboj/20.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Q, Worman H J. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 53.Ye Q, Callebaut I, Pezhman A, Courvalin J C, Worman H J. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 54.Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos P A, Singh P B, Giannakouros T, Georgatos S D. EMBO Rep. 2001;2:920–925. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kourmouli N, Theodoropoulos P A, Dialynas G, Bakou A, Politou A S, Cowell I G, Singh P B, Georgatos S D. EMBO J. 2000;19:6558–6568. doi: 10.1093/emboj/19.23.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pombo A, Jones E, Iborra F J, Kimura H, Sugaya K, Cook P R, Jackson D A. Crit Rev Eukaryotic Gene Expression. 2000;10:21–29. [PubMed] [Google Scholar]
- 57.Nielsen S J, Schneider R, Bauer U M, Bannister A J, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera R E, Kouzarides T. Nature (London) 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 58.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 59.Hwang K K, Eissenberg J C, Worman H J. Proc Natl Acad Sci USA. 2001;98:11423–11427. doi: 10.1073/pnas.211303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross D S. Trends Biochem Sci. 2001;26:685–686. doi: 10.1016/s0968-0004(01)01985-5. [DOI] [PubMed] [Google Scholar]
- 61.Gasser S M, Cockell M M. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- 62.Shareef M M, King C, Damaj M, Badagu R, Huang D W, Kellum R. Mol Biol Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao T, Eissenberg J C. J Biol Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 64.Zhao T, Heyduk T, Eissenberg J C. J Biol Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 65.Koike N, Maita H, Taira T, Ariga H, Iguchi-Ariga S M. FEBS Lett. 2000;467:17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 66.Cryderman D E, Cuaycong M H, Elgin S C, Wallrath L L. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
- 67.Cryderman D E, Tang H, Bell C, Gilmour D S, Wallrath L L. Nucleic Acids Res. 1999;27:3364–3370. doi: 10.1093/nar/27.16.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfeifer G P, Riggs A D. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- 69.Breiling A, Turner B M, Bianchi M E, Orlando V. Nature (London) 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 70.Sekinger E A, Gross D S. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- 71.Sekinger E A, Gross D S. EMBO J. 1999;18:7041–7055. doi: 10.1093/emboj/18.24.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sinclair D A, Schulze S, Silva E, Fitzpatrick K A, Honda B M. Genetica. 2000;109:9–18. doi: 10.1023/a:1026500620158. [DOI] [PubMed] [Google Scholar]
- 73.Carvalho A B, Dobo B A, Vibranovski M D, Clark A G. Proc Natl Acad Sci USA. 2001;98:13225–13230. doi: 10.1073/pnas.231484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devlin R H, Bingham B, Wakimoto B T. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warner T S, Sinclair D A, Fitzpatrick K A, Singh M, Devlin R H, Honda B M. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- 76.Eberl D F, Duyf B J, Hilliker A J. Genetics. 1993;134:277–292. doi: 10.1093/genetics/134.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Copenhaver G P, Nickel K, Kuromori T, Benito M I, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell L D, et al. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 78.Weiler K S, Wakimoto B T. Genetics. 1998;149:1451–1464. doi: 10.1093/genetics/149.3.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wakimoto B T, Hearn M G. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Vlag J, den Blaauwen J L, Sewalt R G, van Driel R, Otte A P. J Biol Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]
- 81.Seum C, Spierer A, Delattre M, Pauli D, Spierer P. Chromosoma. 2000;109:453–459. doi: 10.1007/s004120000101. [DOI] [PubMed] [Google Scholar]
- 82.Williams L, Grafi G. Trends Plant Sci. 2000;5:239–240. doi: 10.1016/s1360-1385(00)01653-8. [DOI] [PubMed] [Google Scholar]
- 83.Czermin B, Schotta G, Hulsmann B B, Brehm A, Becker P B, Reuter G, Imhof A. EMBO Rep. 2001;2:915–919. doi: 10.1093/embo-reports/kve210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirschmann D A, Lininger R A, Gardner L M, Seftor E A, Odero V A, Ainsztein A M, Earnshaw W C, Wallrath L L, Hendrix M J. Cancer Res. 2000;60:3359–3363. [PubMed] [Google Scholar]
- 85.Henikoff S, Henikoff J G. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 87.Turner J M, Burgoyne P S, Singh P B. J Cell Sci. 2001;114:3367–3375. doi: 10.1242/jcs.114.18.3367. [DOI] [PubMed] [Google Scholar]
- 88.Czvitkovich S, Sauer S, Peters A H, Deiner E, Wolf A, Laible G, Opravil S, Beug H, Jenuwein T. Mech Dev. 2001;107:141–153. doi: 10.1016/s0925-4773(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 89.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, et al. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Netzer C, Rieger L, Brero A, Zhang C D, Hinzke M, Kohlhase J, Bohlander S K. Hum Mol Genet. 2001;10:3017–3024. doi: 10.1093/hmg/10.26.3017. [DOI] [PubMed] [Google Scholar]
- 92.Bachman K E, Rountree M R, Baylin S B. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 93.McDowell T L, Gibbons R J, Sutherland H, O'Rourke D M, Bickmore W A, Pombo A, Turley H, Gatter K, Picketts D J, Buckle V J, et al. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Linder B, Gerlach N, Jackle H. EMBO Rep. 2001;2:211–216. doi: 10.1093/embo-reports/kve039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song K, Jung Y, Jung D, Lee I. J Biol Chem. 2001;276:8321–8327. doi: 10.1074/jbc.M008779200. [DOI] [PubMed] [Google Scholar]
- 96.Maul G G, Jensen D E, Ishov A M, Herlyn M, Rauscher F J., 3rd Cell Growth Differ. 1998;9:743–755. [PubMed] [Google Scholar]
- 97.Cortes A, Huertas D, Fanti L, Pimpinelli S, Marsellach F X, Pina B, Azorin F. EMBO J. 1999;18:3820–3833. doi: 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frankel S, Sigel E A, Craig C, Elgin S C, Mooseker M S, Artavanis-Tsakonas S. J Cell Sci. 1997;110:1999–2012. doi: 10.1242/jcs.110.17.1999. [DOI] [PubMed] [Google Scholar]
- 99.Kato M, Sasaki M, Mizuno S, Harata M. Gene. 2001;268:133–140. doi: 10.1016/s0378-1119(01)00420-6. [DOI] [PubMed] [Google Scholar]
- 100.Ainsztein A M, Kandels-Lewis S E, Mackay A M, Earnshaw W C. J Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scholzen T, Endl E, Wohlenberg C, van der Sar S, Cowell I G, Gerdes J, Singh P B. J Pathol. 2002;196:135–144. doi: 10.1002/path.1016. [DOI] [PubMed] [Google Scholar]
- 102.Lehming N, Le Saux A, Schuller J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]