Abstract
Bacteria have developed an epigenetic phase variation mechanism to control cell surface pili–adhesin complexes between heritable expression (phase ON) and nonexpression (phase OFF) states. In the pyelonephritis-associated pili (pap) system, global regulators [catabolite gene activator protein (CAP), leucine-responsive regulatory protein (Lrp), DNA adenine methylase (Dam)] and local regulators (PapI and PapB) control phase switching. Lrp binds cooperatively to three pap DNA binding sites, sites 1–3, proximal to the papBA pilin promoter in phase OFF cells, whereas Lrp is bound to sites 4–6 distal to papBA in phase ON cells. Two Dam methylation targets, GATCprox and GATCdist, are located in Lrp binding sites 2 and 5, respectively. In phase OFF cells, binding of Lrp at sites 1–3 inhibits methylation of GATCprox, forming the phase OFF DNA methylation pattern (GATCdist methylated, GATCprox nonmethylated). Binding of Lrp at sites 1–3 blocks pap pili transcription and reduces the affinity of Lrp for sites 4–6. Together with methylation of GATCdist, which inhibits Lrp binding at sites 4–6, the phase OFF state is maintained. We hypothesize that transition to the phase ON state requires DNA replication to dissociate Lrp and generate a hemimethyated GATCdist site. PapI and methylation of GATCprox act together to increase the affinity of Lrp for sites 4–6. Binding of Lrp at the distal sites protects GATCdist from methylation, forming the phase ON methylation pattern (GATCdist nonmethyated, GATCprox methylated). Lrp binding at sites 4–6 together with cAMP-CAP binding 215.5 bp upstream of the papBA transcription start, is required for activation of pilin transcription. The first gene product of the papBA transcript, PapB, helps maintain the switch in the ON state by activating papI transcription, which in turn maintains Lrp binding at sites 4–6.
Cis- and Trans-Acting Pap Switch Components
Bacteria have developed phase variation mechanisms to control cell surface pili–adhesin complexes between expression (phase ON) and nonexpression (phase OFF) states. Pili phase variation can occur by site specific (1, 2) and homologous recombination (3) mechanisms. In addition, a large group of pili operons including pyelonephritis-associated pili (pap) are regulated by an epigenetic switch directly controlled by DNA methylation pattern formation (4, 5). The focus of this paper is on the mechanisms by which the phase OFF and phase ON states are perpetuated, the transition between phase OFF and phase ON states, and the external inputs that control phase switching.
The expression of pap is positively controlled by the local regulators PapI (8 kDa) and PapB (12 kDa) in concert with the global regulators leucine-responsive regulatory protein (Lrp) and catabolite gene activator protein (CAP). DNA adenine methylase (Dam) is also required for pap transcription (Table 1). Knockout mutations in each of the genes encoding these regulatory proteins inhibit the pap phase OFF to phase ON switch (Table 1). In addition, histone-like nucleoid structuring protein (H-NS) modulates pap phase switching because hns mutants show a decreased OFF to ON switch rate (6, 7).
Table 1.
Trans-acting factors that regulate Pap phase variation
| Genotype | Description | Switch rates (OFF to ON)* |
|---|---|---|
| Wild type | 7 × 10−4 per cell per generation | |
| lrp− | Leucine-responsive regulatory protein | Locked OFF |
| crp− | Catabolite gene activator protein | Locked OFF |
| dam− | DNA adenine methylase | Locked OFF |
| papI− | Local regulatory protein | Locked OFF |
| papB− | Local regulatory protein | Locked OFF |
| hns− | Histone-like nucleoid structuring protein | 2 × 10−4 per cell per generation |
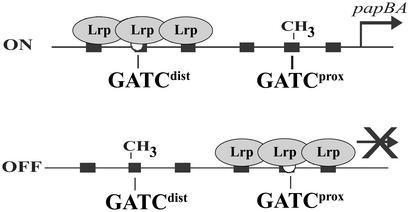
The pap regulatory region encompasses the divergently transcribed papI and papB genes together with the 416-bp intergenic region (Fig. 1). There are six pap DNA Lrp binding sites, designated sites 1–6, within the pap regulatory region spaced three helical turns apart which control transcription at the papBA promoter. Two DNA GATC sites designated GATCprox and GATCdist (proximal and distal with respect to the papBA promoter) are located within Lrp binding sites 2 and 5, respectively. DNA GATC sites are target sites for Dam, which methylates the adenosine of the GATC sequence.
Figure 1.
Regulatory sequences of the E. coli pap operon. The pap regulatory between the divergently transcribed papBA and papI promoters is depicted. The two GATC sites subject to methylation by Dam are GATCprox and GATCdist, which are located within Lrp binding sites 2 and 5, respectively. The Lrp sites are shown as filled circles and the Lrp binding sites are shown as boxed regions on the expanded DNA sequence. The orientation of the Lrp sites [using a consensus sequence 5′-Gnn(n)TTTt-3′] is indicated with arrows above the sequence. The distance between sites 2 and 5 is 102 bp, and the distance between sites 1 and 6 is 32 bp, measured between conserved base-pairs within the Lrp binding sites (Fig. 6). The CAP and PapB binding sites are shown as open and hatched boxes, respectively. Substitution, deletion, and insertion mutations are shown below the wild-type sequence. Mutant switch phenotypes are indicated in parentheses.
Mutational analyses showed that disruption of Lrp binding sites 2 or 3 resulted in a higher OFF to ON switch rate or a phase locked ON phenotype, respectively. In contrast, disruption of Lrp sites 4 or 5 resulted in a phase locked OFF phenotype (Fig. 1) (8). These results suggested that binding of Lrp proximal to the papBAp promoter inhibits transcription whereas binding of Lrp at the distal site activates transcription. In vitro DNA footprint analyses indicated that Lrp binds with highest affinity to Lrp sites 1–3, and with lower affinity to sites 4–6 (8–10). Insertion or deletion of a single base pair between Lrp binding sites 1 and 2 locks transcription in the ON phase, consistent with the observation that proper spacing between Lrp binding sites is necessary for the high cooperativity in binding (N. Kozak and D.L., unpublished results). Examination of the pap DNA methylation patterns showed that, in phase OFF cells, GATCprox is nonmethylated and GATCdist is methylated, whereas the converse pattern exists in phase ON cells (GATCdist nonmethylated, GATCprox methylated) (Fig. 2). These methylation patterns depend on Lrp (9, 11, 12). Moreover, addition of Lrp to pap DNA in vitro blocks methylation of the pap regulatory GATC sequences (12). Together, these data indicate that in phase OFF cells Lrp is bound at sites 1–3, blocking methylation of GATCprox within site 2. In contrast, Lrp bound to sites 4–6 in phase ON cells blocks methylation of GATCdist within site 5 (Fig. 2). Translocation of Lrp from sites 1–3 to sites 4–6 requires PapI. Lrp thus plays dual roles in regulating pap transcription. Lrp acts as a repressor when bound proximal to the pap BA pilin promoter. Lrp activates pilin transcription when, together with PapI, it is bound distal to the pilin promoter (13). Activation of pap transcription also requires binding of cAMP-CAP at a site 60 bp upstream of Lrp binding site 4 (14–16) and binding of the PapB regulatory protein at a site proximal to the papI promoter (17, 18) (Fig. 1).
Figure 2.
DNA methylation states of phase ON and phase OFF cells. Binding of Lrp at sites 4–6 in phase ON cells and sites 1–3 in phase OFF cells controls the DNA methylation pattern by blocking methylation of the bound GATC site.
Pap Phase Variation Model
A model for Pap phase variation is shown in Fig. 3. Specific details and supporting data for the model are discussed below.
Figure 3.
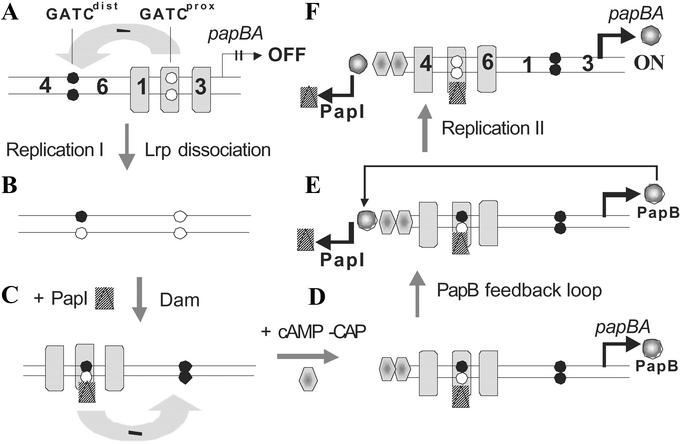
Pap phase variation model. A model for the transition from phase OFF to phase ON is shown in panels A–F. In A, the OFF state is shown in which Lrp binds to sites 1–3, blocking methylation of GATCprox and inhibiting papBA transcription. Nonmethylated pap GATC sites are depicted as open circles, and methylated sites are depicted as closed circles. The binding of Lrp at sites 1–3 reduces the affinity of Lrp for sites 4–6 (“mutual exclusion,” see Fig. 5), and is depicted by an arrow with a negative sign. Transition to the phase ON state is hypothesized to require DNA replication, which dissociates Lrp from pap DNA generating two daughter duplexes of which one is shown (B). PapI and Dam work together to increase the affinity of Lrp for sites 4–6 as discussed in the paper (C). Activation of pap transcription requires cAMP-CAP (D), which stimulates PapB transcription (E). Because PapB activates papI transcription, the phase ON state is self-perpetuating (F).
The Self-Perpetuating Phase OFF State.
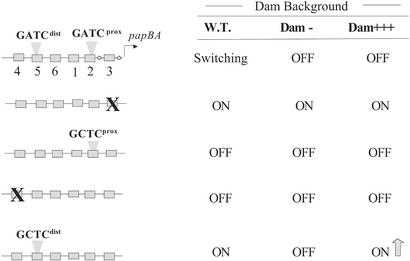
Only the distal GATC site of the pap regulatory region is methylated in phase OFF cells (GATCdist methylated, GATCprox nonmethylated). Initial analysis indicated that methylation of GATCdist was required for maintenance of the OFF transcription state because introduction of an A to C transversion within GATC results in a phase-locked ON phenotype (Fig. 1) (10). Although the adenosine within GATC is obviously required for methylation of this site by Dam, the affinities of Lrp for wild-type pap DNA and DNA containing the GCTCdist mutation appear similar (10), and dimethyl sulfate footprint analyses indicate that Lrp does not closely contact the adenosine of GATC (8). Further studies (10) showed that overproduction of Dam prevented the phase OFF to ON transition in cells containing a wild-type pap sequence but not in cells containing the GCTCdist mutation (Fig. 4). Thus, overmethylation of GATCdist prevents the phase OFF to ON transition, consistent with the hypothesis that methylation of this distal GATC site helps maintain cells in the OFF state.
Figure 4.
The effects of DNA methylation on Pap phase variation. Wild-type and mutant pap DNA regulatory regions are shown at the left. Substitutions in Lrp binding sites 3 and 4 are depicted by an “X” and point mutations in GATCdist and GATCprox that disrupt DNA methylation are shown as “GCTC” sequences. The Pap switch phenotypes under variation levels of Dam are shown at the right. “Switching” indicates reversible phase variation, “OFF” indicates locked OFF, “ON” indicates locked ON, and “ON” with an arrow indicates up-regulation of pap pilin transcription. At the upper right, “W.T.” indicates wild-type levels of Dam, “Dam−” indicates a deletion of the dam gene, and “Dam+++” indicates overproduction of Dam (>4-fold). Data are from refs. 8 and 10.
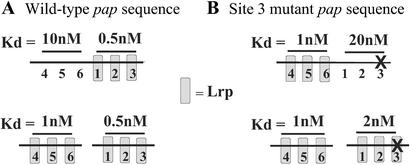
Another factor that may contribute to maintenance of the phase OFF state is a mutual binding exclusion phenomenon. The affinity of Lrp is about 2 times higher for sites 1–3 compared with sites 4–6 when the sites are separated (Fig. 5A Lower). However, when the sites are linked (intact pap regulatory region) the affinity of Lrp for sites 4–6 is reduced 10-fold (Fig. 5A, compare Upper and Lower). These results indicate that binding of Lrp at sites 1–3 exerts a negative effect on Lrp binding at sites 4–6. This mutual binding exclusion effect is reduced from 10-fold to only about 2-fold when unsupercoiled DNAs are used (unpublished data), showing a strong dependence on DNA topology. Because Lrp is known to form higher oligomers under certain conditions (19, 20) and bend DNA (21), one possible mechanism is that the conformation of pap sites 4–6 might be altered as a result of a binding of Lrp at sites 1–3 located 102 bp away (measured from site 2 to site 5) (Fig. 1). Because the binding of Lrp to pap DNA is highly cooperative (8), this could serve as a signal amplification mechanism. As discussed below, when the affinity of Lrp for sites 1–3 is reduced by mutation of Lrp binding site 3, mutual exclusion works in reverse, lowering the affinity of Lrp at sites 1–3 as a result of binding of Lrp at sites 4–6 (Fig. 5B).
Figure 5.
Quantitation of the mutual exclusion effect on LRP binding by UV footprint analysis. DNA fragments containing the intact pap regulatory region (Upper) or unlinked regions containing only Lrp binding sites 1–3 or 4–6 (110 bp each) (Lower) were cloned into plasmid vector pTZ19U (41). Supercoiled plasmids were isolated from a Dam− strain, and the affinity of Lrp for sites 1–3 and 4–6 was measured by UV footprinting as described (42). Briefly, samples were irradiated at 254 nm, and UV-induced pyrimidine dimers were detected by extension of 32P-end-labeled primers with TaqDNA polymerase. The affinities of Lrp for pap DNA sites 1–3 and 4–6 were identical to affinities determined by using electrophoretic mobility shift analysis (unpublished data). The location of a 6-bp substitution mutation in site 3 (see Fig. 1) is depicted by an “X” in B.
Activation of papBA transcription requires binding of Lrp at sites 4–6 (8), and thus, binding of Lrp at sites 1–3 indirectly inhibits transcription because of mutual exclusion of Lrp binding at sites 4–6. In addition, Lrp binding at sites 1–3 appears to directly block pap transcription in vivo (13). Although papBA transcription is low in cells containing either lrp− (3 Miller units, MU) or hns− (59 MU) mutations, cells lacking both Lrp and H-NS display a basal transcription level (528 MU), which is about one-eighth that of phase ON cells (4,200 MU) (13). This transcription activity is similar to that observed for hns− cells in which pap regulatory sequences upstream of the papBA promoter, including Lrp binding sites, have been deleted (486 MU), and thus likely represents basal papBA promoter activity (13). The mechanism by which H-NS represses pap basal transcription is not known, but likely involves specific binding to pap regulatory sequences (6).
Further evidence that Lrp directly represses pap transcription comes from in vitro analysis of pap transcription. Addition of RNA polymerase-σ70 to a supercoiled pap DNA template containing the papBA promoter resulted in Lrp- and cAMP-CAP-independent transcription (intrinsic papBA promoter activity), with a transcription start site identical to that observed in vivo (22). Lrp was titrated while simultaneously monitoring transcription by primer extension and Lrp binding by in vitro methylation protection (IVMP). In IVMP, first applied by van der Woude et al. (12), binding of Lrp to the GATCdist and GATCprox sites is monitored by addition of Dam followed by restriction enzyme MboI, which cuts at fully nonmethylated GATC sites. It was observed that half-maximal inhibition of papBA transcription occurred at the same level of Lrp that gave half-occupancy of the GATCprox site. Thus, binding of Lrp at the GATCprox region (sites 1–3) correlated with repression of pap transcription. Moreover, mutational disruption of Lrp binding site 3, which reduced the affinity of Lrp for sites 1–3 by about 40-fold (from a Kd of 0.5 nM to 20 nM, measured using intact pap DNA, Fig. 5) also abrogated Lrp-dependent inhibition of pap transcription. Together, these results strongly indicate that binding of Lrp at sites 1–3 in phase OFF cells directly blocks the intrinsic papBA promoter activity.
The Phase OFF to Phase ON Transition.
The Pap phase OFF to ON switch rate is about 100-fold lower than the phase ON to OFF rate (23), resulting in a bias in bacterial populations toward the OFF state. This is generally true for most pili operons, including type 1 pili, regardless of their switch mechanisms (4).
We hypothesize that an opportunity for transition to the transcriptionally active ON phase only occurs after DNA replication (Fig. 3B). First, replication should dissociate Lrp from sites 1–3, removing the mutual exclusion effect on binding of Lrp at sites 4–6. Second, GATCdist will become transiently hemimethylated, providing an opportunity for binding of Lrp to sites 4–6 with the aid of PapI, which is required together with Lrp for methylation protection of GATCdist (9) and transition to the phase ON state (Fig. 3C). Third, dissociation of Lrp from sites 1–3 provides an opportunity for methylation of GATCprox by Dam, which is essential for pap transcription (10). These individual steps in the OFF to ON transition are discussed below.
Role of PapI.
PapI is a small (8 kDa) coregulatory protein that shares homology only with other PapI-like genes present in many diverse pili operons in Escherichia coli, Salmonella typhimurium, and likely other enteric bacteria (5). Two functions of PapI have been identified: specific binding to Lrp (24) and specific binding to DNA sequences within pap sites 2 and 5 (A.H. and D.L., unpublished data). Notably, the affinity of PapI for pap DNA alone is very low, and cannot be detected by electrophoretic mobility shift analysis. Moreover, the affinity of PapI for free Lrp is also low based on protein crosslinking (24) and gel filtration analyses (24). PapI binds specifically with high affinity to Lrp in complex with pap sites 2 and 5, but not to Lrp bound to other pap sites or Lrp binding sites within the ilvIH regulatory sequence (ref. 24 and A.H. and D.L., unpublished data). This sequence selectivity appears to be caused by the presence of a conserved core sequence that includes “ACGATC” in pap sites 2 and 5 (Fig. 6) containing base pairs critical for PapI binding. Our data indicate that Lrp bound at adjacent and partially overlapping sites interacts with PapI, stabilizing PapI–DNA interactions. This is manifested by over 20-fold reduction in the dissociation rate of Lrp from pap DNA (M.K. and D.L., unpublished data) and a 10-fold increase in the affinity of Lrp for sites 2 or 5 (A.H. and D.L., unpublished data). Because binding of Lrp to pap DNA is highly cooperative, PapI increases the affinity of Lrp for these sites by forming PapI–Lrp–DNA complexes at sites 2 or 5.
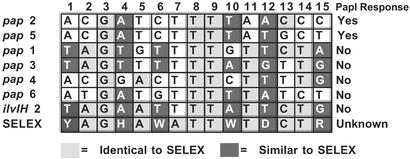
Figure 6.
Sequence comparison of Lrp binding sites. The consensus Lrp binding sequence as determined by selex (43) is shown on the bottom line (Y = C or T, H = not G, W = A or T, D = not C, R = A or G). Lrp binding sites 1 through 6 of pap and ilvIH binding site 2 are shown above the selex sequence. The abilities of PapI to increase the affinity of Lrp for each site is shown at the far right, where “Yes” indicates an increase in Lrp affinity.
Roles of Dam.
The finding that PapI increases the affinity of Lrp for sites 4–6 based on in vitro binding data are consistent with the observation that PapI is required in vivo for methylation protection of GATCdist present in site 5. However, PapI also increases Lrp's affinity for sites 1–3, which raises the problem of how Lrp moves from promoter proximal to promoter distal sites in the phase OFF to ON transition. A possible answer involves DNA methylation. As shown in Fig. 4, wild-type pap is transcriptionally inactive in the absence of Dam. In addition, a normally locked ON GCTCdist mutant also requires Dam for transcription (10) and a GCTCdist GCTCprox double mutant is locked OFF, indicating that methylation of GATCprox is essential for transition to the phase ON state. Apparently, methylation of GATCprox in phase ON cells is not saturating because overproduction of Dam significantly increases transcription in the phase-locked ON GCTCdist mutant (10). Dam, however, is not required for pap transcription when Lrp binding site 3 is disrupted by substitution (Fig. 4), suggesting that methylation at GATCprox might inhibit binding of Lrp and/or PapI-Lrp to sites 1–3. Recent data indicate that PapI-dependent binding of Lrp at GATCprox is blocked by methylation at this site, although Lrp binding is unaffected (A.H. and D.L., in preparation). In contrast, methylation at GATCdist blocks binding of Lrp, but has much less affect on PapI dependent Lrp binding. Based on these data, we conclude that methylation of GATCprox may be required to provide directionality to the switch by reducing the affinity of PapI–Lrp for sites 1–3.
Roles of CAP.
CAP plays important roles in activation at the papBA and papI promoters (14–16, 25). Binding of CAP to a single site located at −215.5 bp from the papBA start and −115.5 bp from the papI start is essential for activation of both papBA and papI transcription. CAP directly activates transcription at the papBA promoter (16). However, recent data suggest that CAP activates papI transcription indirectly by means of expression of PapB regulatory protein because activation of papI transcription occurs in the absence of CAP when papB is expressed from an independent promoter (manuscript in preparation). CAP appears to affect Pap phase switching indirectly by means of its control of PapI transcription because movement of Lrp from sites 1–3 to 4–6 occurs in vivo in the absence of CAP under conditions in which transcription is PapI-independent (16). In addition, although the CAP binding site is centered 36 bp from Lrp binding site 4, Lrp and CAP bind independently to their respective binding sites (16). The mechanism by which PapB activates papI transcription appears to involve binding of PapB at a high affinity binding site adjacent to the CAP binding site (17, 18) (Fig. 1).
Recently, the mechanism by which CAP stimulates transcription at the papBA promoter has been explored (16). In many respects, activation of papBA transcription by CAP is similar to activation of class I promoters by CAP (lac for example), even though the distance between CAP and the papBA transcription start site (215.5 bp) is much larger than lac (61.5 bp) and other class I operons (see Fig. 1). CAP-dependent activation of transcription of the papBA promoter and class I operons (26) share a requirement for the following: (i) activating region 1 (AR1) of CAP, (ii) α C-terminal domain of RNA polymerase, (iii) helical phase dependence between CAP and RNA polymerase, and (iv) only the promoter–proximal subunit of the CAP homodimer is required. These results clearly show that CAP plays a direct role in activation of papBA transcription by contact with the transcription apparatus. Although a previous study suggested that CAP may activate papBA transcription indirectly by means of antagonism with the histone-like protein H-NS (25), this does not appear to be the case because CAP AR1 is required for transcription even in the absence of H-NS (16).
Role of PapB.
Early studies by Uhlin's laboratory established that PapB plays an essential role in activation of pap transcription (14). There is a least one high affinity PapB binding site, located between the CAP binding site and the papI promoter in each pap operon that has been studied (Fig. 1) (18). Binding of PapB to this high-affinity site is essential for papI transcription and subsequent Pap pili expression. PapB is a 12-kDa regulatory protein that binds to the minor grooves of DNA targets containing the nonamer GACACAAAC (18). PapB appears to bind as a multimer of 8–10 subunits protecting a DNA region of 50–70 nucleotides (18). The role of PapB in activation of Pap transcription is solely due to activation at the papI promoter because PapB is not essential for Pap phase variation under conditions in which PapI is constitutively expressed (5). PapB thus acts as a feedback link between the papBA and papI promoters, a link that is involved in self-perpetuation of the phase ON state (see below).
Role of H-NS.
The H-NS, unlike Lrp, CAP, PapI, Dam, and PapB, is not essential for Pap phase variation (13, 16). However, this 15.5-kDa global regulatory protein does modulate Pap phase switching, and appears to play an essential role in the repression of Pap expression by environmental signals including low temperature (27). At temperatures below 26°C, H-NS appeared to bind to the pap regulatory region as evidenced by methylation protection of the two pap GATC sites (6). This conclusion was supported by the finding that H-NS binds specifically to pap DNA and blocks methylation of GATCdist and GATCprox in vitro (6). Simultaneous analysis of pap transcription and the pap DNA methylation pattern after a shift to low temperature showed that transcription shut off before switching to the OFF state, indicating that H-NS blocks transcription from phase ON cells at low temperature, possibly as a result of the formation of a nucleoprotein repression complex.
At 37°C, H-NS appears to play both positive and negative roles in modulating Pap expression. In the absence of hns, the pap phase OFF to ON switch rate is reduced 2- to 3-fold (7, 13), indicating that H-NS helps promote transition to the phase ON state. In contrast, H-NS was shown to inhibit the basal activity of the papBA promoter in the absence of Lrp (13). The mechanism by which H-NS exerts these affects is not known, but likely involves specific binding within the pap regulatory region. H-NS could positively regulate OFF to ON switching by reducing the affinity of Lrp for sites 1–3, and negatively regulate papBA transcription by altering the interactions of RNA polymerase with the promoter.
The Self-Perpetuating Phase ON State.
Addition of Lrp, cAMP-CAP, and RNA polymerase-σ70 with pap-13 template DNA (pap-13 contains a Lrp binding site 3 mutation making transcription PapI-independent) activates papBA transcription about 12-fold compared with basal transcription measured with RNA polymerase alone (16). This relative activation level is similar to that observed in vivo (13), indicating that Lrp (bound at sites 4–6) and cAMP-CAP are sufficient for the activation of papBA transcription observed in phase ON cells. As described above, a PapB feedback loop ensues because of transcription and expression of PapB, which activates papI transcription. PapI, in turn, will facilitate movement of Lrp to sites 4–6, maintaining the switch in the ON state (Fig. 3E). If this model is correct, then papI and papBA transcription should be coordinated ON or OFF, depending on the phase state. Although this has not yet been tested, analysis of a papI–lacZ fusion shows that the papI promoter is subject to phase variation control with similar OFF and ON rates to the papBA promoter. Moreover, mutations in Lrp binding sites 4 and 5 block both papI and papBA transcription (A. Brinkman, N. Weyand, and D.L., manuscript in preparation). These data support the hypothesis that papI and papBA transcription is coordinated by a PapB feedback loop.
The PapB feedback loop is subject to autoregulation, which may prevent spiraling up of papI and papB transcription (17). Titration of PapB showed that, at low levels, papBA transcription was enhanced, whereas at higher levels transcription was inhibited. Autoregulation appears to be caused by the presence of a low-affinity PapB binding site located overlapping the −10 hexamer RNA polymerase binding site in the papBA promoter (Fig. 1), though this has not been directly shown. Under certain conditions, PapI might also act as an autoregulator. Overexpression of PapI in normally phase-locked ON cells containing a pap GCTCdist mutation results in a switch phenotype in which both phase ON and OFF colonies are present after plating on solid media, indicating that PapI induces some cells to turn off (unpublished data). This could occur by means of PapI-assisted binding of Lrp to sites 1–3, which normally is inhibited by methylation of GATCprox. After DNA replication, the fully methylated GATCprox in phase ON cells becomes hemimethylated for a short period before remethylation by Dam. It is possible that a single methyl group on the top or bottom pap GATCprox site may not inhibit PapI-dependent binding of Lrp at sites 1–3 when high PapI levels are present. This hypothesis could be tested by titration of PapI with analysis of phase switch rates by fluorescent activated cell sorting using a fluorescent reporter for papBA expression. High PapI levels should increase the switch rate to the OFF phase.
Switch Inputs
Pap pili-phase variation is controlled by a variety of environmental stimuli (7, 14). Growth of E. coli in glucose reduces the OFF to ON switch rate by 35-fold as a result of lowered cAMP level, which prevents CAP-dependent activation of papBA and subsequent papI transcription (23). Pap pilin transcription is also significantly repressed by growth at low temperature (<26°C) (27, 28) and rich medium (LB broth) (7). It is not clear whether these growth conditions directly alter the phase switch itself or alternatively inhibit pap transcription in phase ON cells. The mechanism by which these conditions repress pap transcription is not known, but appears to involve H-NS because introduction of an hns mutation partially relieves repression (7, 27).
In addition to negative regulatory inputs, recently a positive input to pap transcription was described by the Silhavy and Hultgren laboratories (29, 30). The CpxAR two-component system consists of the CpxA membrane sensor and the CpxR response regulator. This sensor system appears to be activated by misfolded proteins in the periplasm such as unchaperoned Pap pilin subunits as well as binding of Pap pili to solid surfaces (29), which initiate a phosphotransfer relay from CpxA to CpxR. Once phosphorylated, CpxR-P controls transcription of a number of genes, including degP and the pap operon. It was shown that when the Cpx pathway is activated, papI and papBA transcription was enhanced 2-fold. Moreover, Pap pili were expressed under conditions of catabolite repression (+ glucose) when the Cpx pathway was activated (30). Recent data from our laboratory have indicated that under conditions of constitutive phosphorylation of CpxR, the phase OFF to ON rate increases 3-fold. Our data show that CpxR-P binds specifically to the pap site 1–3 region, which could increase phase ON switching by reduction of the affinity of Lrp for binding sites 1–3, similar to the phenotypes of mutations in pap sites 2 and 3 (see Fig. 1) (P. Engelbert and D.L., in preparation).
A number of non-pap pili operons including sfa (S pili), daa (F1845 pili), and fae (K88 pili) in E. coli and pef (Pef pili) in S. typhimurium share common regulatory features with pap (4). These include conserved GATCdist and GATCprox sites as well as papI and papB homologues. Although some of these regulatory sequences (sfa for example) contain a conserved CAP binding site at the same position as that in pap, others such as fae and pef do not (4). This finding raises the possibility that additional regulatory inputs could control these pili operons. This is the case with pef expression, which, though normally heavily biased to the OFF expression state, is induced by growth under acidic conditions to switch ON (pH 4.5) (4, 31). The mechanism by which Pef pili are induced by low pH is unknown, but induction cannot be caused by activation of PefI expression (a PapI homologue) because PefI acts as a negative regulator of pef transcription (31). Compared with pap, the organization of pef may be reversed so that the binding of Lrp to pef sites 4–6 occurs with the highest affinity, with PefI facilitating transition to the OFF state by movement of Lrp to sites 1–3 (31).
Outputs
As a result of the PapB feedback loop, phase ON cells expressing Pap pili also express relatively high levels of PapB and PapI, which control other genes in E. coli besides pap. Many uropathogenic E. coli contain multiple pili operons, many of which share the core control mechanisms of the pap operon including cross-complementing PapI and PapB homologues. Analysis of the pap-related fimbiae (prf) operon of the uropathogenic E. coli strain 536 showed that deletion of the prfI and prfB genes, homologues of papI and papB, reduced the expression of S pili encoded by the unlinked sfa operon (32). Introduction of constitutively expressed PrfB or PrfI restored S pili expression, indicating that cross-activation of sfa was occurring. In contrast, little if any cross-talk between the pap-17 and pap-21 pili operons in E. coli C1212 appears to occur based on comparison of Pap-17 and Pap-21 pili expression on individual cells when only one versus both operons are present (33). Further work needs to be done to understand the extent of cross-talk between pap-like operons.
More recently, it has been shown that PapB greatly reduces the expression of type 1 pili by a form of regulatory cross-talk (34, 35). Type 1 pili are regulated by a DNA inversion-mediated phase variation switch catalyzed by two DNA recominbinases, FimB and FimE. The former enzyme enhances both OFF to ON and ON to OFF switching, whereas the latter enzyme mediates ON to OFF switching only (36, 37). PapB was found to increase FimE-dependent ON to OFF switching by 2-fold, and inhibited FimB-dependent switching by over 50-fold, thus blocking switching to the ON phase while speeding up switching to the OFF phase (34). Although it has not been directly shown, these data suggest that expression of Pap pili and type 1 pili is mutually exclusive.
Biological Relevance.
As described above, the Pap pili phase variation mechanism is highly complex and tightly regulated by many components that contribute to reversible OFF–ON switching of Pap pili. One reason for this complexity may be that it allows for global (Lrp, CAP, Dam, H-NS) as well as local (PapI, PapB) regulatory inputs that provide a means for environmental factors to regulate Pap phase switch rates (see above). If only a small fraction of E. coli express Pap pili in a specific environment and if that confers a selective advantage on E. coli, then the cell population will rapidly convert to phase ON cells because of the heritable nature of Pap pili expression states. Thus, environmental stimulation of Pap phase OFF to ON switching in the appropriate milieu should accelerate E. coli's colonization of that environmental niche. In addition, environmental control would help to conserve cell resources when pili are not needed because pili expression requires a significant fraction of the cells energy.
Pap pili phase variation can be thought of as a “simple” developmental switch mechanism that controls cell differentiation. Not only do pap phase ON cells express Pap pili at their surface, but type 1 pili expression is turned off as a result (34). Although the physiological relevance of this effect is not known, Pap and type 1 pili bind to different receptors and have distinct roles in pathogenesis: type 1 pili are required for colonization of the lower urinary tract by uropathogenic E. coli (38), whereas Pap pili play an essential role in colonization of the upper urinary tract (39). Because inappropriate expression of pili may be deleterious to E. coli by enhancing detection by the immune system, PapB-mediated shut-off of type 1 pili may be important in the biology of E. coli. Recent data suggest that pap gene expression may also negatively control cell motility by means of inhibition of flagellar expression (40). This regulatory mechanism might facilitate colonization of mucosal surfaces of the intestines or urinary tract by cycling between motile and sessile states that could counteract bacterial removal by peristalsis and urine output, respectively. Comparative microarray analysis of gene expression in Pap phase OFF and phase ON cells should reveal whether additional regulatory outputs are present.
Acknowledgments
We are grateful to former laboratory members Natalia Kozak, Patrick Engelberts, Arjen Brinkman, and Nathan Weyand for unpublished work. We thank the National Institutes of Health for continuing support of this project (Grant AI23345 to D.L.).
Abbreviations
- Lrp
leucine-responsive regulatory protein
- Pap
pyelonephritis-associated pili
- Dam
DNA adenine methylase
- CAP
catabolite gene activator protein
- H-NS
histone-like nucleoid structuring protein
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Fulks K A, Marrs C F, Stevens S P, Green M R. J Bacteriol. 1990;172:310–316. doi: 10.1128/jb.172.1.310-316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomfield I C, Kulasekara D H, Eisenstein B I. Mol Microbiol. 1997;23:705–717. doi: 10.1046/j.1365-2958.1997.2241615.x. [DOI] [PubMed] [Google Scholar]
- 3.Mehr I J, Seifert H S. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 4.Krabbe M, Weyand N, Low D. In: Bacterial Stress Responses. Storz G, Hengge-Aronis R, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 305–321. [Google Scholar]
- 5.van der Woude M, Braaten B, Low D. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 6.White-Ziegler C A, Angus Hill M L, Braaten B A, van der Woude M W, Low D A. Mol Microbiol. 1998;28:1121–1137. doi: 10.1046/j.1365-2958.1998.00872.x. [DOI] [PubMed] [Google Scholar]
- 7.White-Ziegler C A, Villapakkam A, Ronaszeki K, Young S. J Bacteriol. 2000;182:6391–6400. doi: 10.1128/jb.182.22.6391-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nou X, Braaten B, Kaltenbach L, Low D A. EMBO J. 1995;14:5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nou X, Skinner B, Braaten B, Blyn L, Hirsch D, Low D. Mol Microbiol. 1993;7:545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 10.Braaten B A, Nou X, Kaltenbach L S, Low D A. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 11.Braaten B A, Blyn L B, Skinner B S, Low D A. J Bacteriol. 1991;173:1789–1800. doi: 10.1128/jb.173.5.1789-1800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Woude M, Hale W B, Low D A. J Bacteriol. 1998;180:5913–5920. doi: 10.1128/jb.180.22.5913-5920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Woude M W, Kaltenbach L S, Low D A. Mol Microbiol. 1995;17:303–312. doi: 10.1111/j.1365-2958.1995.mmi_17020303.x. [DOI] [PubMed] [Google Scholar]
- 14.Baga M, Goransson M, Normark S, Uhlin B E. EMBO J. 1985;4:3887–3893. doi: 10.1002/j.1460-2075.1985.tb04162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goransson M, Forsman P, Nilsson P, Uhlin B E. Mol Microbiol. 1989;3:1557–1565. doi: 10.1111/j.1365-2958.1989.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 16.Weyand N J, Braaten B A, van der Woude M, Tucker J, Low D A. Mol Microbiol. 2001;39:1504–1522. doi: 10.1046/j.1365-2958.2001.02338.x. [DOI] [PubMed] [Google Scholar]
- 17.Forsman K, Goransson M, Uhlin B E. EMBO J. 1989;8:1271–1277. doi: 10.1002/j.1460-2075.1989.tb03501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Y, Forsman K, Jass J, Uhlin B E. Mol Microbiol. 1998;30:513–523. doi: 10.1046/j.1365-2958.1998.01080.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Hao Z, Bieniek E, Calvo J M. J Mol Biol. 2001;314:1067–1075. doi: 10.1006/jmbi.2000.5209. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Rosner M H, Calvo J M. J Mol Biol. 2001;312:625–635. doi: 10.1006/jmbi.2001.4955. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Calvo J M. EMBO J. 1993;12:2495–2501. doi: 10.1002/j.1460-2075.1993.tb05904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyand N J, Low D A. J Biol Chem. 2000;275:3192–3200. doi: 10.1074/jbc.275.5.3192. [DOI] [PubMed] [Google Scholar]
- 23.Blyn L B, Braaten B A, White-Ziegler C A, Rolfson D H, Low D A. EMBO J. 1989;8:613–620. doi: 10.1002/j.1460-2075.1989.tb03416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltenbach L S, Braaten B A, Low D A. J Bacteriol. 1995;177:6449–6455. doi: 10.1128/jb.177.22.6449-6455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsman K, Sonden B, Goransson M, Uhlin B E. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busby S, Ebright R H. J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 27.Goransson M, Sonden B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin B E. Nature (London) 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 28.Goransson M, Uhlin B E. EMBO J. 1984;3:2885–2898. doi: 10.1002/j.1460-2075.1984.tb02225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto K, Silhavy T J. Proc Natl Acad Sci USA. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung D L, Raivio T L, Jones C H, Silhavy T J, Hultgren S J. EMBO J. 2001;20:1508–1518. doi: 10.1093/emboj/20.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson B, Low D. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 32.Morschhauser J, Vetter V, Emody L, Hacker J. Mol Microbiol. 1994;11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 33.Low D, Robinson E N, Jr, McGee Z A, Falkow S. Mol Microbiol. 1987;1:335–346. doi: 10.1111/j.1365-2958.1987.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 34.Xia Y, Gally D, Forsman-Semb K, Uhlin B E. EMBO J. 2000;19:1450–1457. doi: 10.1093/emboj/19.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holden N J, Uhlin B E, Gally D L. Mol Microbiol. 2001;42:319–330. doi: 10.1046/j.1365-2958.2001.02656.x. [DOI] [PubMed] [Google Scholar]
- 36.Gally D L, Leathart J, Blomfield I C. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 37.McClain M S, Blomfield I C, Eisenstein B I. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez J J, Mulvey M A, Schilling J D, Pinkner J S, Hultgren S J. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts J A, Marklund B I, Ilver D, Haslam D, Kaack M B, Baskin G, Louis M, Mollby R, Winberg J, Normark S. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Rasko D A, Lockatell C V, Johnson D E, Mobley H L. EMBO J. 2001;20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mead D A, Szczesna-Skorupa E, Kemper B. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 42.Becker M M, Grossmann G. In: Footprinting of Nucleic Acid–Protein Complexes. Revzin A, editor. San Diego: Academic; 1993. pp. 129–157. [Google Scholar]
- 43.Cui Y, Wang Q, Stormo G D, Calvo J M. J Bacteriol. 1995;177:4872–4880. doi: 10.1128/jb.177.17.4872-4880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]