Abstract
Inoculation of turnip crinkle virus (TCV) on the resistant Arabidopsis ecotype Dijon (Di-17) results in the development of a hypersensitive response (HR) on the inoculated leaves. To assess the role of the recently cloned HRT gene in conferring resistance, we monitored both HR and resistance (lack of viral spread to systemic tissues) in the progeny of a cross between resistant Di-17 and susceptible Columbia plants. As expected, HR development segregated as a dominant trait that corresponded with the presence of HRT. However, all of the F1 plants and three-fourths of HR+ F2 plants were susceptible to the virus. These results suggest the presence of a second gene, termed RRT, that regulates resistance to TCV. The allele present in Di-17 appears to be recessive to the allele or alleles present in TCV-susceptible ecotypes. We also demonstrate that HR formation and TCV resistance are dependent on salicylic acid but not on ethylene or jasmonic acid. Furthermore, these phenomena are unaffected by mutations in NPR1. Thus, TCV resistance requires a yet undefined salicylic acid–dependent, NPR1-independent signaling pathway.
INTRODUCTION
Plants resisting pathogen attack frequently activate a variety of defense responses that are initiated by the direct or indirect interaction between the products of a plant-encoded resistance (R) gene and its corresponding pathogen-encoded avirulence (Avr) gene. A majority of such interactions also result in the induction of host cell death at the site of pathogen infection, a phenomenon known as the hypersensitive response (HR). Subsequent to the HR, the uninoculated tissues usually develop a long-lasting, enhanced resistance to further attacks by the same or even unrelated microbial pathogens; this is referred to as systemic acquired resistance (SAR) (Ryals et al., 1996; Durner et al., 1997; Dempsey et al., 1999).
Although the HR is one of the earliest manifestations of disease resistance, it remains unclear whether this phenomenon is a prerequisite for gene-for-gene-mediated disease resistance or whether it simply results from the activation of multiple defense signaling pathways. For example, the HR does not develop in potato during Rx-mediated resistance against potato virus X (Köhm et al., 1993) or in the Arabidopsis dnd1 mutant after pathogen infection (Yu et al., 1998). Similarly, in barley, the HR is dispensable in Mlg-specified resistance to the powdery mildew fungus (Koga et al., 1990; Schiffer et al., 1997). Several studies on tobacco infected with tobacco mosaic virus (TMV) have also indicated that localization of the virus is dependent on accumulation of salicylic acid (SA) and not cell death (Bi et al., 1995; Mittler et al., 1996; Ryals et al., 1996; Mur et al., 1997). In contrast, an HR is required for Mla-specified resistance to powdery mildew in barley (Koga et al., 1990; Schiffer et al., 1997). Therefore, the relationship between HR and disease resistance (cause or consequence) most probably depends on the specifics of the interaction between the resistance and avirulence gene products and the downstream components activated as a result of their interaction.
Many studies have demonstrated that SA is an important component of the signal transduction pathway leading to disease resistance (Durner et al., 1997; Dempsey et al., 1999). For example, plants unable to accumulate SA because of the expression of a bacterial nahG gene encoding salicylate hydroxylase fail to develop SAR and exhibit heightened susceptibility to pathogen infection (Gaffney et al., 1993; Delaney et al., 1994). In addition to SA, ethylene and jasmonic acid (JA) serve as important signals for the induction of various defense responses. JA and ethylene have been implicated in (1) resistance to several fungi (Knoester et al., 1998; Thomma et al., 1998; Vijayan et al., 1998), (2) induced systemic resistance triggered by Pseudomonas fluorescens (Pieterse et al., 1998; Pieterse and Van Loon, 1999), (3) activation of genes encoding thionin and defensin (Epple et al., 1995; Penninckx et al., 1996, 1998), and (4) activation of enzymes involved in phytoalexin synthesis (Boller et al., 1983; Ecker and Davis, 1987; Mauch and Staehelin, 1989). Interestingly, ethylene and JA work in conjunction with SA to signal some but not all defense responses (Dong, 1998).
Genetic analysis of the SA-regulated pathway leading to disease resistance has revealed that a key signaling component is encoded by the NPR1/NIM1 gene (Cao et al., 1994, 1997; Delaney et al., 1995; Glazebrook et al., 1996; Ryals et al., 1997; Shah et al., 1997). Arabidopsis mutants lacking a functional NPR1/NIM1 gene are unable to express the pathogenesis-related (PR) genes in response to SA or its functional analogs 2,6-dichloroisonicotinic acid and benzothiadiazole. In addition, npr1/nim1 plants show enhanced susceptibility to both bacterial and fungal pathogens (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). Conversely, overexpression of NPR1 confers resistance against both bacterial and fungal pathogens in a dosage-dependent manner (Cao et al., 1998). Whereas NPR1 plays a critical role in resistance to various bacterial and fungal pathogens, its involvement in viral resistance is not known.
In Arabidopsis, resistance to most viral pathogens does not involve an HR (Ishikawa et al., 1991; Leisner et al., 1993; Lee et al., 1994; Callaway et al., 1996). However, inoculation of turnip crinkle virus (TCV) (Morris and Carrington, 1988) on plants from the resistant ecotype Dijon (Di-0 or Di-17) results in both an HR and the induction of PR gene expression (Simon et al., 1992; Dempsey et al., 1993, 1997; Uknes et al., 1993). In contrast, TCV-susceptible ecotypes, including Columbia (Col-0), fail to mount an HR, exhibit delayed and weak PR gene expression, and develop systemic disease symptoms (Li and Simon, 1990; Dempsey et al., 1993). Genetic analyses revealed that HR development is conferred by a single dominant gene termed HRT (for HR to TCV) (Dempsey et al., 1997). HRT also appears to be required for resistance to TCV infection; all of the HR− progeny from crosses between resistant and susceptible ecotypes developed systemic disease symptoms. However, HRT alone may not be sufficient for complete resistance because many of the HR+ progeny also succumbed to infection. In this study, we show that TCV resistance is influenced by a second locus, named RRT (for regulates resistance to TCV). Furthermore, we demonstrate that the HR and resistance are dependent on SA but independent of NPR1-, ethylene-, and JA-mediated defense signaling.
RESULTS
Defense Gene Expression during the Resistance Response
Several families of PR genes have been shown to be activated in Di-17 plants that resist TCV infection (Dempsey et al., 1993; Uknes et al., 1993). To determine whether other known defense genes are induced during this resistance response, we extracted total RNA from the TCV-inoculated and mock-inoculated leaves of Di-17 and Col-0 plants at 1 to 3 days postinoculation (DPI). The accumulation of gene transcripts for PR-1, phenylalanine ammonia lyase (PAL) (Wanner et al., 1995), glutathione S-transferase (GST1 and GST6) (Chen et al., 1996; Yang et al., 1999), defensin (PDF1.2 ) (Penninckx et al., 1996), thionin (THI2.1) (Epple et al., 1995), ascorbate peroxidase (APX1 and APX2) (Kubo et al., 1993; Karpinski et al., 1997), lipoxygenase (LOX2) (Bell and Mullet, 1993), NPR1 (Cao et al., 1997), 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (Van der Straeten et al., 1992), and alternative oxidase (AOX) (Kumar and Soll, 1992) were then analyzed by reverse transcription–polymerase chain reaction (RT-PCR) and RNA gel blot analysis (Figure 1 and data not shown). Whereas the transcripts for ACC synthase and AOX accumulated to very low amounts (data not shown), transcripts for PR-1 and GST1 accumulated to high amounts in the TCV-inoculated leaves of Di-17 plants (Figure 1). Expression of the other genes was not induced in these leaves (Figure 1 and data not shown). In contrast, the TCV-inoculated leaves of Col-0 plants showed only basal-level expression for all of the defense genes assayed (Figure 1).
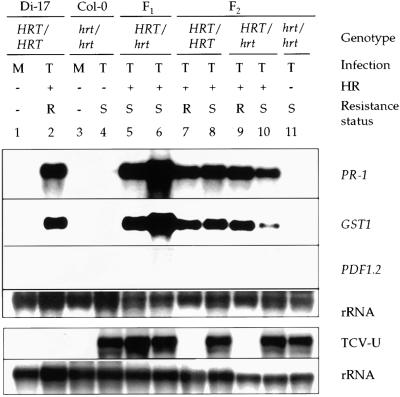
Figure 1.
PR-1, GST1, and PDF1.2 Expression and Systemic Spread of TCV in Di-17 × Col-0 F1 and F2 Plants.
Expression of PR-1, GST1, and PDF1.2 in the inoculated leaves of TCV (T)- and mock (M)-infected Di-17 and Col-0 plants and the F1 and F2 progeny obtained from a cross between them. The inoculated leaves were monitored for the presence (+) or absence (−) of the HR, and the RNA was extracted from the inoculated leaves at 4 DPI. The replication and spread of the virus were analyzed by extracting RNA from the uninoculated (TCV-U) bolt tissue at 10 DPI. Two F1 and five F2 plants were analyzed. The genotype of the F2 plants at the HRT locus was determined by CAPS analysis; both homozygous and heterozygous plants were analyzed for the expression of defense genes in inoculated leaves, the presence of TCV RNA in the uninoculated bolt tissue, and the appearance (S) or absence (R) of disease symptoms. The blot (1 to 11) was sequentially probed for the indicated genes (PR-1, GST1, and PDF1.2). RNA extracted from the systemic tissues was run on a separate gel; rRNA was used as an internal control for gel loading and transfer for both blots.
Genetic Characterization of Resistance against TCV
We previously showed that the HR in TCV-infected plants is dependent on the dominant HRT gene (Dempsey et al., 1997). However, because many progeny from a Di-17 × Col-0 cross are HR+ but TCV susceptible, this gene may not be sufficient to confer complete resistance. To further characterize the role of HRT in TCV-infected plants, we crossed Di-17 and Col-0 plants. All of the F1 progeny developed an HR on the TCV-inoculated leaves (Table 1) and accumulated increased amounts of PR-1 and GST1 transcripts (Figure 1). Nonetheless, these F1 plants allowed systemic spread of the virus (Figure 1) and developed disease symptoms (crinkled leaves and drooping bolts; Table 1). One explanation for the TCV-susceptible phenotype of these F1 plants might be that resistance requires homozygosity for the HRT allele. Alternatively, a second recessive gene, in addition to HRT, might regulate resistance.
Table 1.
Segregation of the HR and Resistance in a Cross between Di-17 and Col-0 Plants
| Phenotype of Plants
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Crossa | Generation | Total No. of Plants | HR+/Sc | HR+/Rd | HR−/S | HR−/R | χ2b | P |
| Di-17 × Col-0 | F1 | 6 | 6 | 0 | 0 | 0 | ||
| F2 | 458 | 280 | 85 | 93 | 0 | 0.01 | >0.8 | |
The pollen-accepting plant is indicated first and the pollen donor second.
Calculation based on a 13:3 segregation.
Susceptible, disease symptoms include crinkling of leaves and drooping of the bolt.
Resistant, no disease symptoms.
To determine whether resistance depends on HRT dosage or the presence of a second gene, we analyzed F2 progeny from the Di-17 × Col-0 cross. Of the 458 F2 plants, 373 were susceptible and 85 were resistant. The ratio between susceptible and resistant plants fits very closely the 13:3 segregation expected if resistance is due to at least one dose of HRT and homozygosity for a recessive gene at an unlinked locus (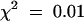 ; 0.95 > P > 0.8), a gene we have named RRT. In contrast, if resistance were dependent on two doses of HRT, ∼33% of the HR+ F2 plants (122) should have been resistant. Because the data obtained do not fit the Mendelian ratio of 2:1 (
; 0.95 > P > 0.8), a gene we have named RRT. In contrast, if resistance were dependent on two doses of HRT, ∼33% of the HR+ F2 plants (122) should have been resistant. Because the data obtained do not fit the Mendelian ratio of 2:1 (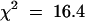 ; P < 0.01), we can reject this possibility. As expected, the HR segregated as a dominant trait (Table 1; 365 HR+/93 HR− plants;
; P < 0.01), we can reject this possibility. As expected, the HR segregated as a dominant trait (Table 1; 365 HR+/93 HR− plants; 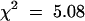 ; 0.05 > P > 0.01), although the number of HR+ plants (365) was somewhat higher than expected (344). This greater number of HR+ plants could be the result of differential transmission at the HRT locus, and indeed, genotype analysis showed that this was the case (see below).
; 0.05 > P > 0.01), although the number of HR+ plants (365) was somewhat higher than expected (344). This greater number of HR+ plants could be the result of differential transmission at the HRT locus, and indeed, genotype analysis showed that this was the case (see below).
To provide additional support for our hypothesis that both HRT and RRT regulate resistance to TCV, we monitored the HRT genotype, the ability to form an HR, and viral resistance in a population of 355 F2 plants derived from a cross between Di-17 and Col-0 plants (Table 2). Using a cleaved amplified polymorphic sequence (CAPS) marker that can differentiate between the HRT allele of Di-17 and the hrt allele of Col-0 (Cooley et al., 2000), we identified 119 plants as HRT/HRT, 164 as HRT/hrt, and 72 as hrt/hrt. All of the HRT/− plants developed HR at 3 DPI, whereas none of the hrt/hrt plants showed any visible lesions, confirming the requirement of HRT for the HR. As was previously observed, the number of HR+ plants (283) was somewhat more than what was expected (267). Moreover, substantially more of these plants were homozygous for the HRT allele (119) than would be predicted (89) by a Mendelian ratio of 1:2:1 (Table 2). Thus, recovery of the dominant HRT allele (57%) in the F2 is significantly higher than that of the recessive allele (43%) (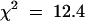 ; P < 0.01). Similar results were obtained when selfing other F1 plants, suggesting preferential transmission of HRT by one or both gametophytes in this specific F1 hybrid.
; P < 0.01). Similar results were obtained when selfing other F1 plants, suggesting preferential transmission of HRT by one or both gametophytes in this specific F1 hybrid.
Table 2.
Segregation of the HRT Locus and Resistance in a Cross between Di-17 and Col-0 Plants
| Genotypeb at the HRT Locus
|
Resistantc Plants
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Crossa | Generation | Total No. of Plants | HRT/HRT | HRT/hrt | hrt/hrt | HRT/HRT | HRT/hrt | hrt/hrt |
| Di-17 × Col-0 | F2 | 355 | 119 | 164 | 72 | 42 | 19 | 0 |
The pollen-accepting plant is indicated first and the pollen donor second.
The genotype at HRT was determined by CAPS analysis.
Resistant, no disease symptoms.
Further analysis of these F2 plants confirmed that HRT is also required for resistance. All 72 of the hrt/hrt plants not only failed to develop an HR but also were susceptible to TCV infection. If resistance were not dependent on HRT but required only rrt, then ∼18 hrt/hrt plants should have been resistant. In addition, resistance does not correlate with the presence of two doses of the HRT allele; 42 of the 119 HRT/HRT plants and 19 of 164 HRT/hrt plants were TCV resistant (Table 2). Rather, the data are more consistent with the numbers predicted if a second locus, in addition to HRT, is required for resistance. In this scenario, 25% of the HRT/HRT plants (30) and 25% of the HRT/hrt plants (41) should be resistant. The number of HRT/HRT–resistant plants (42) was somewhat more than expected, and the number of HRT/hrt–resistant plants (19) was only half of that expected. A likely explanation for the discrepancy between the observed and expected numbers is that the increased dosage of HRT enhances the durability of resistance or reduces the extent to which environmental factors affect the expressivity of HRT. Further supporting this possibility is the observation that increased expression of HRT in transgenic Col-0 plants leads to enhanced resistance to TCV (Cooley et al., 2000).
To determine whether HRT dosage has any effect on the extent of viral spread and defense gene expression, we monitored these phenomena in a subset of the Di-17 × Col-0 F2 progeny. All of the plants exhibiting disease symptoms were found to contain viral RNA in the uninoculated tissue. These plants accumulated similar amounts of viral RNA, regardless of whether they contained zero, one, or two doses of the HRT allele (Figure 1). In contrast, PR-1 and GST1 expression in the TCV-inoculated leaves occurred only in F2 progeny containing at least one dose of the HRT allele and independent of their resistance phenotype. Comparable expressions of these genes were generally detected in homozygous and heterozygous plants, suggesting that these events are not sufficient to confer resistance (Figure 1).
Role of SA in HR Development and Resistance to TCV
We have previously shown that in Arabidopsis, increases in SA and its glucoside (SAG) correlate with resistance to TCV (Dempsey et al., 1997). To further explore the role of SA in TCV resistance, we first tested whether exogenously applied SA confers resistance to plants from the susceptible ecotypes Col-0 and Nössen (Nö). Two days after treatment with SA, plants were infected with TCV and then monitored for development of an HR, the appearance of disease symptoms, and the presence of TCV in uninoculated tissue. At 3 DPI, no macroscopic HR was evident on the TCV-inoculated leaves of SA-treated Col-0 plants. However, by 10 DPI, disease symptoms were evident (data not shown), and viral RNA had accumulated to very high amounts in the systemic leaves (Figure 2). No differences in the amounts of viral RNA were observed between the SA-treated and water-treated control Col-0 plants. To ensure that the SA treatment was effective in inducing defense signaling, we monitored PR-1 expression in the inoculated and systemic tissues at 10 DPI. SA treatment was observed to induce PR-1 expression in these plants (Figure 2); however, that was insufficient to confer resistance to TCV. Similar results were obtained with Nö plants (data not shown).
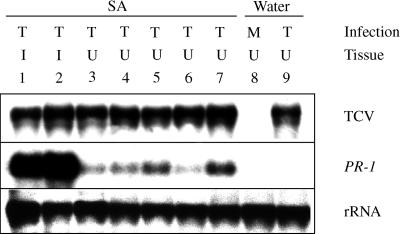
Figure 2.
PR-1 Expression and Systemic Spread of TCV in SA-Treated Plants.
Three-week-old Col-0 plants were treated with SA (500 μM) or water and then TCV (T)- or mock (M)-inoculated 2 days after treatment. The expression of PR-1 and the accumulation of viral genome were analyzed in both inoculated (I) and uninoculated (U) tissues at 10 DPI. The blot (1 to 9) was sequentially probed for TCV, PR-1, and rRNA (the rRNA served as an internal control for gel loading and transfer).
We next determined whether constitutively increased amounts of endogenous SA would confer TCV resistance. Several Arabidopsis mutants have been isolated that exhibit enhanced resistance to bacterial and fungal pathogens, constitutively express PR genes, and have increased SA/SAG (Durner et al., 1997; Yang et al., 1997). Of these, the cpr5 (Bowling et al., 1997), cep (Silva et al., 1999), ssi1 (Shah et al., 1999), ssi2 (J. Shah, P. Kachroo, and D.F. Klessig, unpublished results), and cpr22 (K. Yoshioka, F. Tsui, P. Kachroo, S.B. Sharma, and D.F. Klessig, unpublished results) mutants were analyzed for resistance to TCV. All of these mutants are derived from the TCV-susceptible Col-0, Nö, or Wassilewskija ecotypes. When infected, all exhibited severe disease symptoms and accumulated TCV RNA in the uninoculated tissues in amounts similar to those observed in the respective TCV-infected wild-type parents (data not shown). Thus, increased endogenous SA/SAG and the constitutive expression of PR genes are not sufficient to confer resistance to TCV.
Although the preceding studies demonstrate that increased amounts of SA do not confer resistance to susceptible plants, they do not address whether SA is involved in activating resistance in Di-17 plants. Therefore, we tested the effect of the nahG-encoded salicylate-degrading enzyme salicylate hydroxylase on HR development and TCV resistance. Reciprocal crosses were made between Di-17 and NahG transgenic (ecotype Nö) plants. Because both HRT and the nahG transgene are dominant, the phenotypes were monitored in two F1 progeny from each cross (Table 3). As a control, HR development and resistance to TCV also were assayed in F1 progeny from a cross between Di-17 and wild-type Nö plants. All four F1 plants derived from the Di-17 × NahG Nö cross accumulated transcripts for the nahG transgene and failed to express the prototypical SA-induced PR-1 gene after TCV infection (Figure 3 and Table 3). These plants, like the parental NahG Nö transgenic and wild-type Nö plants, failed to develop an HR after TCV infection. In contrast, the parental Di-17 plants and three F1 plants derived from Di-17 × Nö cross developed an HR after TCV inoculation (Figure 3 and data not shown). Based on these results, increased amounts of SA are required for the HR to TCV. All four NahG-containing F1 plants also exhibited disease symptoms and allowed viral replication and spread from the inoculated leaves into the uninoculated tissues (Figure 3). However, because the F1 progeny from the Di-17 × Nö cross were TCV susceptible, the Nö ecotype did not appear to contain the recessive rrt allele required for resistance. Thus, analyses of the F2 progeny were performed so that the importance of SA in resistance could be assessed.
Table 3.
Segregation of the HR and Resistance in a Cross between Di-17 and a NahG Transgenic Plant
| Crossa | Generation | Total No. of Plants | NahG+/HR+ | NahG+/HR− | NahG−/HR+ | NahG−/HR− |
|---|---|---|---|---|---|---|
| NahG Nö × Di-17 | F1 | 2 | 0 | 2 Sb | 0 | 0 |
| Di-17 × NahG Nö | F1 | 2 | 0 | 2 S | 0 | 0 |
| F2 | 139 | 0 | 92 S | 21 Rc; 23 S | 3 S |
The pollen-accepting plant is indicated first and the pollen donor second.
S, susceptible; disease symptoms include crinkling of leaves and drooping of the bolt.
R, resistant; no disease symptoms.
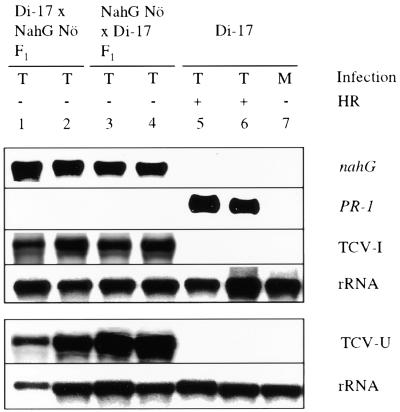
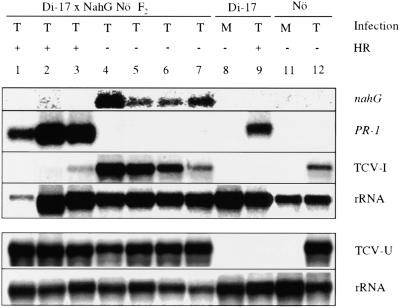
Figure 3.
PR-1 Expression and Systemic Spread of TCV in Di-17 × nahG F1 Plants.
Two F1 plants each from a cross of Di-17 and NahG Nö and its reciprocal cross were inoculated with TCV and monitored for the presence (+) or absence (−) of the HR. TCV (T)- or mock (M)-inoculated Di-17 parental plants were used as controls. Expression of the PR-1 gene in the inoculated leaves was analyzed at 4 DPI. The systemic spread and replication of the virus (TCV-U) were determined by analyzing RNA extracted from the uninoculated bolt tissues at 10 DPI. The blot (1 to 7) was sequentially probed for PR-1, the nahG transgene, and TCV genome. RNA extracted from the systemic tissues was run on a separate gel. rRNA was used as an internal control for gel loading and transfer for both blots.
After self-pollination of an F1 plant, 139 F2 progeny were analyzed for HR formation and resistance to TCV as well as the presence of the nahG transgene (Table 3). Consistent with our hypothesis that SA is required for the activation of defense responses (including the HR) and resistance to TCV, all of the NahG+ plants were HR− and all of the HR+ progeny were NahG− (Table 3, and Figures 4 and 5). Moreover, all of the NahG+ F2 progeny were susceptible to TCV infection.
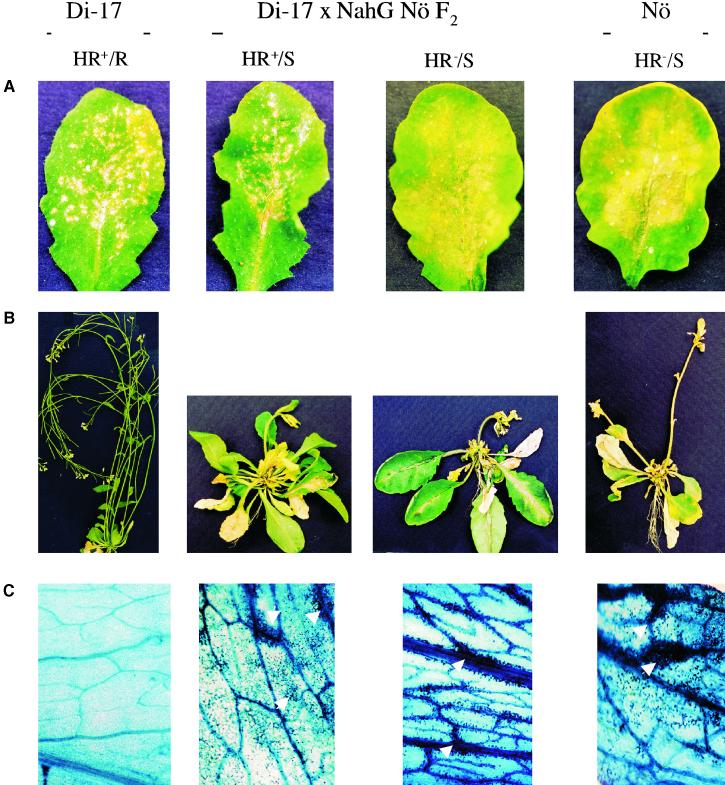
Figure 4.
Morphological Phenotypes of TCV-Infected Di-17, Nö, and F2 Plants from a Di-17 × NahG Nö Cross.
(A) TCV-inoculated leaves at 4 DPI. Both of the F2 plants shown contain at least one allele of HRT. The HR+ F2 plant lacks the nahG transgene, whereas the HR− plant contains the nahG transgene, which leads to suppression of HR.
(B) The morphological phenotypes of the corresponding TCV-inoculated plants at 14 DPI. Except for the Di-17–resistant parent (R, resistant), the rest of the plants show severe crinkling of their leaves and stunted, drooping bolts (S, susceptible).
(C) This series shows systemic cell death induced by TCV in the corresponding TCV-infected susceptible plants. Uninoculated leaves were removed from the bolt at 14 DPI and stained with trypan blue. The leaves of the plants exhibiting disease symptoms show intensely stained areas of dead cells (marked by arrowheads), which are concentrated around the veins and gradually spread to the rest of leaf. Cell death was not evident in the uninoculated leaves of resistant Di-17 infected with TCV (left).
Figure 5.
PR-1 Expression and Systemic Spread of TCV in Di-17 × nahG F2 Plants.
F2 plants derived from a Di-17 × NahG Nö cross and the Di-17 and Nö parents were TCV (T)- or mock (M)-inoculated and scored for the presence (+) or absence (−) of an HR at 4 DPI. RNAs extracted at 4 DPI from the inoculated leaves of three HR+ and four HR− F2 plants (chosen at random) and their parents were analyzed for the expression of PR-1, the nahG transgene, and the replication of TCV in the inoculated leaves (TCV-I) (1 to 12). The systemic spread and replication of TCV in the uninoculated tissue (TCV-U) were determined by analyzing RNA extracted from bolt tissues at 10 DPI. RNA extracted from the systemic tissues was run on a separate gel, and rRNA was used as an internal control for gel loading and transfer for both blots.
To ensure that the HR− susceptible phenotype in at least a portion of the F2 plants was the result of SA deficiency rather than absence of the HRT allele, the genotypes at the nahG and HRT loci were tested by PCR and CAPS analysis in 101 of the 139 F2 plants. From this analysis, 70 NahG+ plants were identified; 52 of these were either homozygous or heterozygous for the HRT allele, and all were susceptible to TCV. Given that rrt segregates as an independent locus, ∼13 of these 70 plants should have been rrt/rrt and resistant to TCV if SA were not required. Of the 31 NahG− plants, all contained at least one copy of the HRT allele and developed an HR after TCV infection. In addition, 16 of these 31 plants were resistant to TCV. This number of resistant plants is more than expected (eight). However, that result would be explained if the NahG transgene is loosely linked to the RRT allele in the Nö parents; selecting for NahG− progeny would thereby increase the number of individuals carrying the rrt allele associated with resistance. Taken together, these results indicate that both SA and HRT are required but not sufficient for resistance to TCV.
Role of NPR1 in the HR and Resistance to TCV
NPR1 plays an essential role in the SA-mediated signal transduction pathway that leads to the activation of PR genes (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). Because SA appears to be required for HR development and resistance, we asked whether NPR1 plays a role in signaling these phenomena by crossing Di-17 with npr1-1 (Col-0 ecotype; Cao et al., 1994) or npr1-5 (Nö ecotype; Shah et al., 1997) plants. Both of these npr1 alleles are recessive compared with the wild-type NPR1 allele. As expected, the two F1 plants derived from the Di-17 × npr1-1 cross exhibited an HR after TCV infection but were still susceptible to TCV (Table 4). Using CAPS analysis, we identified F2 progeny homozygous for npr1-1 or npr1-5. These plants were subsequently monitored for their ability to develop an HR and resist TCV infection. Of 13 npr1-1/npr1-1, HRT/− segregants, all exhibited an HR and two were TCV resistant (Table 4). Likewise, of 31 npr1-5/npr1-5, HRT/− F2 segregants, all were HR+ (Figure 6) and nine were TCV resistant (Table 4). Resistance in these nine plants was confirmed by RNA gel blot analysis; no TCV genomic RNA was detected in the uninoculated tissues (data not shown). Because the HR was not blocked by the presence of the npr1-1 or npr1-5 alleles and because the number of TCV-resistant progeny expected (based on digenic segregation of HRT and rrt) was unaffected in npr1 homozygous plants, NPR1 is not required for the HR or resistance to TCV, even though SA is. This surprising result argues that resistance to TCV requires a yet to be characterized SA-dependent, NPR1-independent signaling pathway.
Table 4.
Segregation of the HR and Resistance to TCV in a Cross between Di-17 and npr1-1, npr1-5, etr1-1, or coi1-1 Mutantsd
| Crossa | Generation | Total No. of Plants | Genotype at NPR1, ETR1 or COI1 Locib |
Phenotype of Plantsc
|
χ2d | Pe | |||
|---|---|---|---|---|---|---|---|---|---|
| HR+/Sf | HR+/Rg | HR−/S | HR−/R | ||||||
| Di-17 × npr1-1 (Col-0) | F1 | 2 | NPR1/npr1-1 (2) | 2 | 0 | 0 | 0 | ||
| F2 | 121 | npr1-1/npr1-1 (15) | 11 | 2 | 2 | 0 | |||
| Di-17 × npr1-5 (Nö) | F2 | 237 | npr1-5/npr1-5 (41) | 22 | 9 | 10 | 0 | 0.274 | >0.8 |
| etr1-1 (Col-0) × Di-17 | F1 | 4 | ETR1/etr1-1 (4) | 4 | 0 | 0 | 0 | ||
| F2 | 102 | ETR1/etr1-1 plus | |||||||
| etr1-1/etr1-1 (74) | 44 | 14 | 16 | 0 | 0.462 | >0.8 | |||
| coi1-1 (Col-0) × Di-17 | F1 | 4 | COI1/coi1-1 (4) | 4 | 0 | 0 | 0 | ||
| F2 | 137 | coi1-1/coi1-1 (38) | 21 | 6 | 11 | 0 | 0.413 | >0.8 | |
The pollen-accepting plant is indicated first and the pollen donor second.
The genotype at the NPR1, COI1, and ETR1 loci were determined by CAPS analysis. The total number of plants of the given genotype that were analyzed for the HR and resistance to TCV is given in parentheses.
CAPS analysis indicated that all TCV-resistant plants contained at least one copy of the dominant HRT allele, whereas all HR−-susceptible plants were homozygous for the recessive hrt allele. Except for the Di-17 × npr1-1 cross, which showed skewed segregation of the HRT locus similar to the Di-17 × Col-0 cross (Table 2), the rest of the other crosses showed 1:2:1 Mendelian segregation of the HRT locus (data not shown).
Calculation based on a 9:3:4 ratio.
Two degrees of freedom.
S, susceptible; disease symptoms include crinkling of leaves and drooping of the bolt.
R, resistant; no disease symptoms.
Figure 6.
The HR in TCV-Inoculated Leaves of F2 Progeny from Various Crosses.
A comparison (at 4 DPI) of TCV-inoculated leaves from F2 progeny derived from the crosses between Di-17 and wild-type Col-0, npr1-5, etr1-1, or coi1-1 plants. The F2 progeny from the crosses between Di-17 and npr1-5, etr1-1, or coi1-1 were homozygous for the mutant alleles and had at least one copy of the HRT gene. The small specks seen on the inoculated leaves of the Di-17 × Col-0 hrt/hrt F2 plant are damaged tissue or dried inoculation buffer. Those are also seen on mock-inoculated plants (data not shown; Dempsey et al., 1993).
Because activation of PR-1 expression in Arabidopsis plants that resist TCV infection correlates with increases in SA contents (Figure 1; Uknes et al., 1993; Dempsey et al., 1997), we tested whether this phenomenon depended on NPR1. RNA gel blot analysis of npr1-5/npr1-5 and npr1-1/npr1-1 F2 plants that were homozygous or heterozygous for HRT indicated that PR-1 expression was activated in the inoculated leaves (Figure 7). Although the amount of expression in these leaves was substantial, it was not as strong as that detected in the inoculated leaves of NPR1/NPR1 F2 plants or in the Di-17 controls. Thus, PR-1 expression appears to be regulated by both NPR1-dependent and -independent pathways in TCV-inoculated resistant plants. A similar conclusion has been drawn from studies with the npr1 mutants and bacterial pathogens (Glazebrook et al., 1996; Shah et al., 1997).
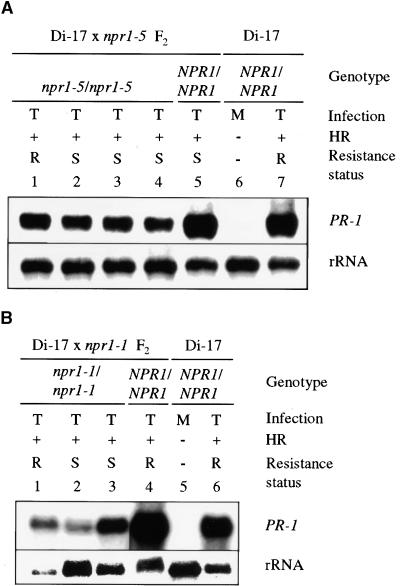
Figure 7.
PR-1 Expression in F2 Progeny from the Crosses between Di-17 and npr1 Plants.
(A) F2 plants from a Di-17 × npr1-5 cross.
(B) F2 plants from a Di-17 × npr1-1 cross.
The resistant Di-17 parents were inoculated with TCV (T) or inoculation buffer (M) and monitored for the presence (+) or absence (−) of an HR at 4 DPI. The plants were subsequently scored as resistant (R) or susceptible (S) at 14 DPI, based on the absence or presence of disease symptoms, respectively. The genotype at the NPR1 locus was determined by CAPS analysis. RNA gel blot analysis was performed with RNA extracted from the inoculated leaves at 4 DPI. The blots were hybridized with probes for PR-1 and rRNA (1 to 7 in [A] and 1 to 6 in [B]), with the latter serving as an internal control for gel loading and transfer.
Role of Ethylene and JA in Signaling HR Development and Resistance to TCV
The role of ethylene and JA signaling in HR development and resistance to TCV was determined by using Arabidopsis mutants insensitive to either of these defense signaling molecules. To analyze the role of ethylene, Di-17 plants were crossed with the dominant ethylene-insensitive etr1-1 (ecotype Col-0) mutant (Chang et al., 1993). All of the F1 progeny derived from this cross exhibited an HR but were susceptible to TCV (Table 4). Because the etr1-1 mutation is dominant, this result suggests that ethylene signaling is not required for the HR to TCV. To determine the effect of etr1-1 on resistance, 58 HRT/− F2 progeny containing at least one dose of the dominant etr1-1 allele were identified by CAPS analysis and then infected with TCV. All of these plants exhibited an HR (Figure 6; 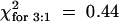 ;
; 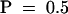 ), and 14 of these were resistant to TCV (Table 4;
), and 14 of these were resistant to TCV (Table 4; 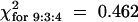 ; 0.95 > P > 0.8). Resistance in these plants was confirmed by RNA gel blot analysis, which failed to detect TCV genomic RNA in the uninoculated tissues (Figure 8). Because the number of HR+ TCV-resistant plants conformed to previously observed digenic segregation ratios, despite the presence of etr1-1, we conclude that this mutation and therefore ethylene signaling had no effect on either HR formation or TCV resistance.
; 0.95 > P > 0.8). Resistance in these plants was confirmed by RNA gel blot analysis, which failed to detect TCV genomic RNA in the uninoculated tissues (Figure 8). Because the number of HR+ TCV-resistant plants conformed to previously observed digenic segregation ratios, despite the presence of etr1-1, we conclude that this mutation and therefore ethylene signaling had no effect on either HR formation or TCV resistance.
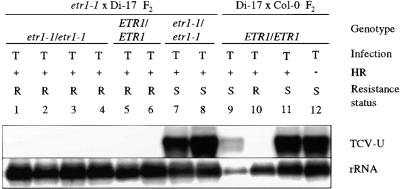
Figure 8.
Systemic Spread of TCV in etr1-1 × Di-17 F2 Plants.
F2 plants from an etr1-1 × Di-17 or a Di-17 × Col-0 cross were inoculated with TCV (T) and scored for the presence (+) or absence (−) of an HR. The genotype at the ETR1 locus was determined by CAPS analysis. The plants were scored as resistant (R) or susceptible (S) at 14 DPI, based on the absence or presence of disease symptoms, respectively. RNA was extracted from the uninoculated bolt tissues (TCV-U) at 14 DPI and analyzed for the presence of viral genome (1 to 12). This blot also was probed for rRNA as an internal control for gel loading and transfer.
The involvement of JA in the activation of defense responses against TCV was assessed by crossing the jasmonate-insensitive coi1-1 mutant (ecotype Col-0) (Xie et al., 1998) with a Di-17 plant. The F1 progeny all developed an HR but were susceptible to TCV. To determine the effect of the recessive coi1-1 mutation on HR and resistance, 27 F2 plants homozygous for the coi1-1 allele and containing at least one copy of the HRT allele were identified by CAPS analysis. All of these plants developed an HR (Figure 6; 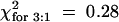 ; 0.8 > P > 0.5), and six were resistant to TCV (Table 4;
; 0.8 > P > 0.5), and six were resistant to TCV (Table 4; 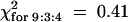 ; 0.95 > P > 0.8). The above ratios conform to what would be expected if the coi1-1 mutation had no effect on either the HR or resistance. Hence, JA, like ethylene, is not required for development of the HR or resistance to TCV.
; 0.95 > P > 0.8). The above ratios conform to what would be expected if the coi1-1 mutation had no effect on either the HR or resistance. Hence, JA, like ethylene, is not required for development of the HR or resistance to TCV.
In the previous section, we showed that although TCV-induced PR-1 expression is SA dependent, it can be activated by way of NPR1-dependent and -independent pathways (Figure 7). Because ethylene and JA are the two other major signals for activation of defense responses after pathogen attack, we asked whether PR-1 expression after TCV inoculation was affected by the etr1-1 or coi1-1 mutations. Analysis of at least 10 HRT F2 plants insensitive to either ethylene or JA (Figure 9) failed to detect any important difference in the amounts of TCV-induced PR-1 expression in comparison with that observed in similar F2 progeny from a cross between Di-17 and Col-0. Thus, the pathway leading to PR-1 expression after TCV infection appears to require neither ethylene nor JA but only SA.
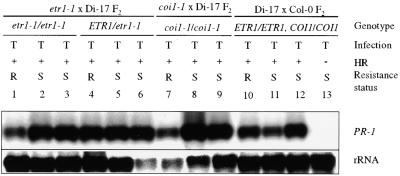
Figure 9.
PR-1 Expression in etr1-1 × Di-17, coi1-1 × Di-17, and Di-17 × Col-0 F2 Plants.
F2 plants from crosses between Di-17 and etr1-1, coi1-1, or Col-0 plants were inoculated with TCV (T) and scored for the presence (+) or absence (−) of an HR at 4 DPI. The genotype at the ETR1 or COI1 locus was determined by CAPS analysis. The plants were scored as resistant (R) or susceptible (S) at 14 DPI, based on absence or presence of disease symptoms, respectively. RNA was extracted from the inoculated leaves at 4 DPI and analyzed for PR-1 expression (1 to 13). The blot was also probed for rRNA as an internal control for gel loading and transfer.
DISCUSSION
In this article, we demonstrate that the dominant HRT allele is necessary and sufficient for development of an HR after TCV infection. HRT also is required for resistance to TCV; however, the results of our genetic analyses argue that a homozygous recessive allele of a second unlinked gene, termed rrt, is also required. HRT was recently cloned and shown to encode a classic leucine zipper/nucleotide binding site/leucine rich repeat–containing protein (Cooley et al., 2000). In contrast, the structure and function of rrt are unknown. The recessive rrt allele found in Di-17 plants could encode a protein that is unable to suppress an active resistance pathway. This situation appears to exist in wheat, where an allele of the LrT2 locus confers resistance to stem rust by encoding a nonfunctional form of protein that otherwise suppresses resistance conferred by other R genes (Dyck, 1987). Alternatively, the recessive rrt allele might encode a protein that directly or indirectly interacts with HRT or a downstream HRT-activated factor to transduce the signal leading to TCV resistance. In this scenario, the recessive nature of the rrt allele could be explained if it encodes a protein that functions as a multimer, and the allele carried by the susceptible ecotypes encodes a dominant-negative variant that destroys the activity of a multimeric complex. We currently are unable to distinguish between these possibilities.
Many studies have demonstrated that SA is a critical signal for the activation of defense responses and disease resistance (Durner et al., 1997; Dempsey et al., 1999). Indeed, although susceptibility to TCV was unaffected by treatment with exogenous SA or in mutants exhibiting high constitutive amounts of SA and PR gene expression, both HR development and TCV resistance were blocked by the presence of nahG-encoded salicylate hydroxylase, which prevents accumulation of SA (Table 3 and Figures 2 to 5). The possibility that the inability to accumulate SA reduces the size of TCV-induced lesions (resulting in micro-HR formation rather than loss of an HR) was ruled out by trypan blue staining of TCV-inoculated leaves from NahG+/− HRT/− F2 progeny (data not shown). These results argue that SA alone is insufficient to confer resistance to TCV in the absence of HRT; it is required, however, for the HRT-mediated activation of an HR and resistance.
Because NPR1 encodes a critical component of the SA-mediated signal transduction pathway leading to PR gene expression and disease resistance (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997), we tested whether mutations in this gene would affect PR expression, HR development, or disease resistance after TCV infection. Plants carrying the npr1-1 or npr1-5 mutation exhibited delayed and decreased expression of PR-1 after TCV infection. In contrast, PR-1 induction was completely inhibited in TCV-infected plants expressing the nahG transgene. Thus, PR-1 expression appears to be regulated by both an SA-dependent, NPR1-independent pathway and an SA- and NPR1-dependent pathway (Figure 10). Earlier studies using npr1 mutants and bacterial pathogens also suggested the existence of an NPR1-independent pathway leading to PR expression (Glazebrook et al., 1996; Shah et al., 1997; Reuber et al., 1998).
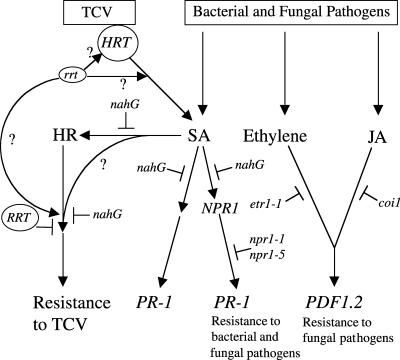
Figure 10.
Model for Induction of the HR and Resistance to TCV.
Inoculation of TCV on resistant Arabidopsis results in an HR at the point of inoculation and the accumulation of SA (Dempsey et al., 1997). Development of the HR is mediated by the dominant HRT gene. Resistance is regulated by HRT and a second gene, RRT. The product of the dominant RRT allele or alleles present in susceptible ecotypes may act as a suppressor (T-bars) of the HRT-mediated resistance pathway downstream of SA and HR. Alternately, the protein encoded by the recessive rrt allele in Di-17 may directly or indirectly interact with HRT or a component downstream in the HRT-mediated defense pathway to facilitate transduction of the signal leading to TCV resistance. Although HRT is required for resistance, it is unclear if the HR is necessary also (question marks). Initiation of the HR and the signaling steps leading to resistance are blocked (T-bars) in plants expressing the nahG gene encoding salicylate hydroxylase. SA-mediated signaling leading to PR-1 expression is transduced by both NPR1-dependent and NPR1-independent pathways, which are blocked by the expression of nahG. The NPR1-dependent pathway, utilized for the activation of resistance to some bacterial and fungal pathogens, is not required for the HR or resistance to TCV. Ethylene- or JA-mediated signaling that leads to PDF1.2 expression and resistance to some fungal and bacterial pathogens is not necessary for the HR or resistance to TCV because mutations resulting in insensitivity to ethylene (etr1-1) or JA (coi1-1) do not affect either of these phenotypes. Thus, the pathways required for resistance to TCV differ from those for bacterial or fungal pathogens.
In contrast, neither HR development nor resistance to TCV was altered in npr1-1 or npr1-5 mutant plants. Consistent with these results, the HR was shown to develop normally in npr1-1 mutant plants infected with an avirulent bacterial pathogen (Cao et al., 1994). However, these same npr1 mutations caused increased susceptibility to bacterial and fungal pathogens (Cao et al., 1994; Shah et al., 1997). Given that both of the npr1 mutant alleles used in this study contain a point mutation in the ankyrin repeats, perhaps they fail to disrupt a function necessary for HR formation and TCV resistance. Alternatively, these phenomena may be mediated by an NPR1-independent pathway that is distinct from the NPR1-dependent pathways leading to fungal and bacterial resistance (Figure 10).
Some precedence for an NPR1-independent viral resistance pathway comes from the studies of Carr and colleagues (Chivasa et al., 1997; Chivasa and Carr, 1998; Murphy et al., 1999), who demonstrated that in tobacco, resistance to TMV is mediated by an SA-dependent pathway that appears to require AOX. Strikingly, although resistance to TMV is blocked by salicylhydroxamic acid (SHAM), an inhibitor of AOX activity, the TMV-induced activation of PR expression is not. In addition, SHAM treatment does not affect resistance to a bacterial or a fungal pathogen (Chivasa et al., 1997). Thus, tobacco appears to contain at least two distinct SA-regulated defense pathways. One pathway presumably is NPR1 dependent and mediates PR expression and resistance to bacterial and fungal pathogens, whereas the other is probably NPR1 independent and activates resistance to a virus.
In addition to SA, the secondary signals ethylene and JA have been implicated in the induction of various defense genes and the activation of resistance to certain pathogens (Xu et al., 1994; Epple et al., 1995; Penninckx et al., 1996, 1998; Dong, 1998; Pieterse and Van Loon, 1999). Thus, their role in establishing resistance to TCV was assessed. The observation that the ethylene- or JA-inducible defense genes PDF1.2 and THI2.1 are not expressed in TCV-infected Di-17 plants raised the possibility that neither signal is involved in activating TCV resistance. This conclusion was confirmed by genetic studies with ethylene (etr1-1)- and JA (coi1-1)-insensitive mutants. Neither mutation affected the HRT-mediated development of an HR (Figure 6) or resistance to TCV (Table 4 and Figure 8). In addition, these mutations had no detectable effect on PR-1 gene induction by TCV (Figure 9). However, because SA is the primary signal for HR development and PR-1 expression, small contributions of ethylene or JA to these phenomena may have gone undetected in these studies.
In conclusion, we have identified a new gene, designated RRT, whose recessive allele works in conjunction with HRT to signal TCV resistance by way of an SA-dependent pathway that is independent of NPR1, ethylene, and JA (Figure 10). In view of these features, the TCV resistance pathway differs from the SA-dependent, NPR1-dependent and the SA-independent, ethylene- or JA-dependent pathways involved in activating resistance to bacterial and fungal pathogens. Strikingly, however, the HRT/rrt–mediated resistance pathway shares many similarities with the SA-dependent, ethylene-independent N gene–regulated pathway for TMV resistance in tobacco (Knoester et al., 1998; Murphy et al., 1999). These discoveries provide further evidence that viral resistance in plants is activated via a mechanism distinct from those used for other microbial pathogens. Future studies should help clarify the relationship between the two viral resistance pathways as well as provide new insights into how plants protect themselves from pathogens, particularly viruses.
METHODS
Conditions for Plant Growth and Viral Infections
Plants (Arabidopsis thaliana) were grown in growth chambers as described by Shah et al. (1999). Transcripts synthesized in vitro from a cloned cDNA of turnip crinkle virus (TCV) genome using T7 RNA polymerase were used for viral infections (Dempsey et al., 1993; Oh et al., 1995). For inoculations, the viral transcript was suspended at a concentration of 0.05 μg/μL in inoculation buffer, and the infection was performed as described earlier (Dempsey et al., 1993). The hypersensitive response (HR) was determined visually 3 to 5 days postinoculation (DPI). Tissues (leaves or bolt) were removed at various times for RNA extraction and analysis of defense gene activation and viral replication by RNA gel blot hybridization.
Histochemistry and Microscopy
Microscopic visualization of HR was conducted by staining TCV-inoculated leaves at 3 to 4 DPI with trypan blue. Samples were processed and analyzed as described by Bowling et al. (1997). Systemic cell death was studied by staining the uninoculated leaf at 14 DPI.
Chemical Treatment of Plants
Three-week-old plants were sprayed and subirrigated with a solution of 500 μM salicylic acid (SA). Control plants were treated with water, and at 2 DPI three leaves per plant were inoculated with TCV RNA.
RNA Extraction and Gel Analysis
Small-scale RNA extractions was performed with TRIzol reagent (Gibco BRL, Rockville, MD), according to the manufacturer's instructions. RNA gel blot analysis and synthesis of random primed probes were performed as described earlier (Shah et al., 1999). RNA gel blot hybridization was performed as described previously (Kachroo et al., 1995).
Cleaved Amplified Polymorphic Sequence Analyses for HRT, NPR1, ETR1, and COI1
The genotypes of the F2 plants at the NPR1, ETR1, COI1, and HRT loci were determined by conducting cleaved amplified polymorphic sequence (CAPS) analysis. The polymerase chain reaction (PCR) primers used for amplifying the NPR1 locus were as described by Shah et al. (1999), and the plants containing npr1-1 and npr1-5 alleles were identified by digesting the amplified product with NlaIII or NlaIV, respectively. Because the etr1-1 (Chang et al., 1993) mutation does not lead to an alteration of any restriction site, we used the dCAPS technique (Neff et al., 1998) to generate polymorphism between the wild type and etr1-1 alleles. PCR primers were designed to amplify a 100-bp region encompassing the mutant base, where the reverse primer contained two altered bases at the 3′ end such that the presence of the mutant base in etr1-1 allele results in loss of an ApaLI restriction site. Therefore, digestion of the PCR product from plants homozygous for the wild-type allele yielded two bands at 75 and 25 bp, whereas the digested PCR product amplified from etr1-1 DNA generated only a single band at 100 bp. The PCR reaction (ETR1 Fwd, 5′-CTTTGTGAAGAAATCAGCCGTGT-3′; ETR1 Rev, 5′-CCATAAGTTAATAAGATGAGTTGGTGCA-3′) was performed at an annealing temperature of 56°C for 50 cycles. The PCR-based analysis to determine the genotype at the COI1 locus was done as described by Xie et al. (1998). The genotype at the HRT locus was determined by producing a 950-bp PCR product, followed by digestion with AluI (Cooley et al., 2000). After digestion, the PCR product of Dijon (Di-17) HRT allele generated two bands of ∼100 and 850 bp. The 850-bp fragment was further cleaved into two fragments of 300 and 550 bp when the PCR products were derived from hrt/hrt Columbia (Col-0) or Nössen (Nö) ecotypes.
Genetic Analysis
Reciprocal crosses of Di-17 plants with Nö plants homozygous for the nahG transgene were performed. The success of the crosses was determined by CAPS analysis and by analyzing expression of the nahG gene. To facilitate easy identification of F2 plants containing the nahG transgene, two PCR primers (5′-GGCTTGCGCATCGGTATCGTCGGC-3′ and 5′-GCCATGGGCCCGATAGGCTTCTCG-3′) were designed to yield a 500-bp amplification product. PCR amplification of nahG was performed at an annealing temperature of 60°C for 35 cycles. Crosses of Di-17 with npr1-1 or npr1-5 plants were performed by using pollen from the mutant plants to pollinate the Di-17 flowers. The crosses with etr1-1 and coi1-1 plants were made by pollinating the flowers from these plants with pollen from Di-17. The F1 plants were analyzed with NPR1-, ETR1-, or COI1- specific CAPS and with random CAPS or simple sequence length polymorphism (SSLP) markers from the genome to ensure that progeny indeed resulted from the crosses. The F2 plants were first analyzed by gene-specific CAPS markers and subsequently were inoculated with TCV. DNA for PCR was isolated either from leaf or bolt tissue by the method of Konieczny and Ausubel (1993). CAPS or SSLP amplifications were performed as described earlier (Konieczny and Ausubel, 1993; Bell and Ecker, 1994).
NOTE ADDED IN PROOF
Interestingly, RPP8-dependent resistance to Peronospora parasitica has recently been shown to be largely independent of SA as well as NPR1 (McDowell, J.M., Dangl, J.L., and Holub, E.B. [2000]. Plant J., in press).
Acknowledgments
We thank Mike Cooley and Hui-Ju Wu for sharing their unpublished results and for providing the HRT-specific CAPS data. We thank Xinnian Dong for providing the cpr5 and npr1-1 seeds and acknowledge Elliot Meyerowitz and John Turner for providing the etr1-1 and coi1-1 seeds, respectively. We also gratefully acknowledge D'Maris Dempsey and Rob Noad for many useful discussions and D'Maris Dempsey for critical reading of the manuscript. This work was funded by grants from the National Science Foundation (MCB-9723952) and the U.S. Department of Agriculture (99-35303-8087).
References
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bell, E., and Mullet, J.E. (1993). Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 103, 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, Y.-M., Kenton, P., Mur, L., Darby, R., and Draper, J. (1995). Hydrogen peroxide functions upstream of salicylic acid in the induction of PR protein expression. Plant J. 8, 235–245. [DOI] [PubMed] [Google Scholar]
- Boller, T., Gehri, A., Mauch, F., and Vögeli, U. (1983). Chitinase in bean leaves: Induction by ethylene, purification, properties, and possible function. Planta 157, 22–31. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, A., Liu, W., Andrianov, V., Stenzler, L., Zhao, J., Wettlaufer, S., Jayakumar, P., and Howell, S.H. (1996). Characterization of cauliflower mosaic virus (CaMV) resistance in virus-resistant ecotypes of Arabidopsis. Mol. Plant-Microbe Interact. 9, 810–818. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product of two-component regulators. Science 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Chen, W., Chao, G., and Singh, K.B. (1996). The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 10, 955–966. [DOI] [PubMed] [Google Scholar]
- Chivasa, S., and Carr, J. (1998). Cyanide restores N gene–mediated resistance to tobacco mosaic virus in trangenic tobacco expressing salicylic acid hydroxylase. Plant Cell 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa, S., Murphy, A.M., Naylor, M., and Carr, J. (1997). Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid–sensitive mechanism. Plant Cell 9, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M., Pathirana, S., Wu, H.-J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A.. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D.A., Wobbe, K.K., and Klessig, D.F. (1993). Resistance and susceptible responses of Arabidopsis thaliana to turnip crinkle virus. Phytopathology 83, 1021–1029. [Google Scholar]
- Dempsey, D.A., Pathirana, M.S., Wobbe, K.K., and Klessig, D.F. (1997). Identification of an Arabidopsis locus required for resistance to turnip crinkle virus. Plant J. 11, 301–311. [DOI] [PubMed] [Google Scholar]
- Dempsey, D., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Dyck, P. (1987). The association of a gene for leaf rust resistance with the chromosome 7D suppressor of stem rust resistance in common wheat. Genome 29, 467–469. [Google Scholar]
- Ecker, J.R., and Davis, R.W. (1987). Plant defense genes are regulated by ethylene. Proc. Natl. Acad. Sci. USA 84, 5202–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M., Obata, F., Kumagai, T., and Ohno, T. (1991). Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol. Gen. Genet. 230, 33–38. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Leong, S.A., and Chattoo, B.B. (1995). Mg-SINE: A short interspersed nuclear element from the rice blast fungus, Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 92, 11125–11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 4, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoester, M., Van Loon, L.C., Heuvel, J.V.D., Hennig, J., Bol, J.F., and Linthorst, H.J.M. (1998). Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc. Natl. Acad. Sci. USA 95, 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, H., Bushnell, W.R., and Zeyen, R.J. (1990). Specificity of cell type and timing of events associated with papilla formation and the hypersensitive reaction in leaves of Hordeum vulgare attacked by Erysiphe graminis f. sp. hordei. Can. J. Bot. 68, 2344–2352. [Google Scholar]
- Köhm, B.A., Goulden, M.G., Gilbert, J.E., Kavanagh, T.A., and Baulcombe, D.C. (1993). A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kubo, A., Saji, H., Tanaka, K., and Kondo, N. (1993). Genomic DNA structure of a gene encoding cytosolic ascorbate peroxidase from Arabidopsis thaliana. FEBS Lett. 3, 313–317. [DOI] [PubMed] [Google Scholar]
- Kumar, A.M., and Soll, D. (1992). Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc. Natl. Acad. Sci. USA 89, 10842–10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Stenger, D.C., Bisaro, D.M., and Davis, K.R. (1994). Identification of loci in Arabidopsis that confer resistance to geminivirus infection. Plant J. 6, 525–535. [DOI] [PubMed] [Google Scholar]
- Leisner, S., Turgeon, R., and Howell, S.H. (1993). Effects of host plant development and genetic determinants on the long-distance movement of cauliflower mosaic virus in Arabidopsis. Plant Cell 5, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.-H., and Simon, A.E. (1990). Symptom intensification on cruciferous hosts by the virulent satellite RNA of turnip crinkle virus. Phytopathology 80, 238–242. [Google Scholar]
- Mauch, F., and Staehelin, L.A. (1989). Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3-glucanase in bean leaves. Plant Cell 1, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., Seskar, M., and Lam, E. (1996). Inhibition of programmed cell death in tobacco plants during a pathogen-induced hypersensitive response at low oxygen pressure. Plant Cell 8, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, T.J., and Carrington, J.C. (1988). Carnation mottle virus and viruses with similar properties. In The Plant Viruses: Polyhedral Virions with Monopartite RNA genomes (New York: Plenum Press), pp. 73–113.
- Mur, L.A.J., Bi, Y.-M., Darby, R.M., Firek, S., and Draper, J. (1997). Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 12, 1113–1126. [DOI] [PubMed] [Google Scholar]
- Murphy, A.M., Chivasa, S., Singh, D.P., and Carr, J.P. (1999). Salicylic acid–induced resistance to viruses and other pathogen: A parting of the ways? Trends Plant Sci. 4, 155–160. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Oh, J.-W., Kong, Q., Song, C., Carpenter, C.D., and Simon, A.E. (1995). Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant-Microbe Interact. 8, 979–987. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanz, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., and Van Loon, L.C. (1999). Salicylic acid–independent plant defense pathways. Trends Plant Sci. 4, 52–57. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., Van Wees, S.C.M., Van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and Van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.A. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.-Y., Johnson, J., Delaney, T.P., Jesse, T., Voss, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer, R., Görg, R., Jarosch, B., Beckhove, U., Bahrenberg, G., Kogel, K.-H., and Schulze-Lefert, P. (1997). Tissue dependence and differential cordycepin sensitivity of race-specific resistance responses in the barley–powdery mildew interaction. Mol. Plant-Microbe Interact. 10, 830–839. [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid–insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, H., Yoshioka, K., Dooner, H.K., and Klessig, D.F. (1999). Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol. Plant-Microbe Interact. 12, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Simon, A.E., Li, X.H., Lew, J.E., Stange, R., Zhang, C., Polacco, M., and Carpenter, C.D. (1992). Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol. Plant-Microbe Interact. 5, 496–503. [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S., Winter, A.M., Delaney, T., Vernooij, B., Morse, A., Friedrich, L., Nye, G., Potter, S., Ward, E., and Ryals, J. (1993). Biological induction of systemic acquired resistance in Arabidopsis. Mol. Plant-Microbe Interact. 6, 692–698. [Google Scholar]
- Van der Straeten, D., Rodrigues-Pousada, R.A., Villarroel, R., Hanley, S., Goodman, H.M., and Van Montagu, M. (1992). Cloning, genetic mapping, and expression analysis of an Arabidopsis thaliana gene that encodes 1-aminocyclopropane-1-carboxylate synthase. Proc. Natl. Acad. Sci. USA 89, 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, L.A., Li, G., Ware, D., Somssich, I.E., and Davis, K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27, 327–338. [DOI] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1093. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Chang, P.F.L., Liu, D., Narasimhan, M.L., Raghothanma, K.G., Gasegawa, P.M., and Bressan, R.A. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K.Y., Kim, E.Y., Kim, K.S., Guh, J.O., Kim, K.C., and Cho, B.H. (1999). Characterization of a glutathione S-transferase gene ATGST1 in Arabidopsis thaliana. Plant Cell Rep. 17, 700–704. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]