Abstract
Organ bending through differential growth represents a major mechanism by which plants are able to adaptively alter their morphology in response to local changes in the environment. Two plant hormones, auxin and ethylene, have been implicated as regulators of differential growth responses; however, the mechanisms by which they elicit their effects remain largely unknown. Here, we describe isolation of the NPH4 gene of Arabidopsis, which is conditionally required for differential growth responses of aerial tissues, and we report that NPH4 encodes the auxin-regulated transcriptional activator ARF7. The phenotypes of nph4 mutants, which include multiple differential growth defects associated with reduced auxin responsiveness, including impaired auxin-induced gene expression, are consistent with the predicted loss of function of a transcriptional activator, and these phenotypes indicate that auxin-dependent changes in gene transcription are prerequisite for proper organ bending responses. Although NPH4/ARF7 appears to be a major regulator of differential growth, it is not the sole regulator because phenotypes of nph4 null mutants were suppressed by application of ethylene. This latter finding illustrates the intimate connection between auxin and ethylene in the control of growth in higher plants.
INTRODUCTION
Plants have evolved movement strategies that involve organ bending to respond adaptively to environmental signals. Dramatic and rapid changes in plant morphology can result from differential growth, that is, unequal cellular growth in one position of an organ relative to an opposing position. Examples of differential growth responses include stem and root tropisms, modification of apical hook structures, and nastic movements of leaves (reviewed in Darwin and Darwin, 1896; Palmer, 1985). Each of these examples of stimulus-driven organ bending represents a process by which plants maximize the positive attributes of their environment while minimizing the negatives.
Two plant hormones, auxin and ethylene, have been implicated as regulators of differential growth responses (Went and Thimann, 1937; Davies, 1987; Kaufman et al., 1995). Although each of these hormones is capable of modulating growth when applied externally, the relative contribution of each in response to changes in their endogenous concentrations, and the sensitivities to either, has been difficult to reconcile (Davies, 1987). Much of this ambiguity stems from functional overlap between the auxin and ethylene signal and response, as well as their biosynthetic, pathways. For example, auxin stimulates ethylene production (Yang and Hoffman, 1984), which in turn stimulates the expression of genes, such as HOOKLESS 1 (HLS1; Lehman et al., 1996), that are involved in auxin homeostatic processes. Results from recent genetic and molecular studies suggest that auxin may be the major regulator of differential growth responses, with ethylene modifying the auxin responses (Romano et al., 1993; Lehman et al., 1996; Chen et al., 1998; Luschnig et al., 1998; Madlung et al., 1999).
The mechanism by which auxin modulates plant growth is still not understood; however, the gene activation hypothesis proposes that auxin regulates the transcription of specific mRNAs that encode proteins necessary for growth control (Key, 1969). Numerous target genes for auxin action, as well as regulatory loci controlling auxin-dependent gene expression, have now been identified (reviewed in Abel and Theologis, 1996; Sitbon and Perrot-Rechenmann, 1997; Guilfoyle et al., 1998a, 1998b). Although auxin-regulated transcription of proteins directly involved in growth control, such as α-expansins (Cosgrove, 1999), only recently has begun to be explored (Hutchison et al., 1999), considerable effort has been dedicated to the study of regulatory proteins. Two families of proteins that regulate auxin-dependent gene expression, the Aux/IAA proteins (Abel and Theologis, 1996; Guilfoyle, 1998) and the auxin response factors (ARFs; Guilfoyle et al., 1998a), have received the most attention.
The Aux/IAA genes are expressed in response to auxin and encode short-lived, small hydrophilic nuclear proteins that function as transcriptional repressors in transient assay systems (reviewed in Abel and Theologis, 1996; Guilfoyle, 1998). Like the Aux/IAA proteins, ARF proteins also function as transcriptional regulators (reviewed in Guilfoyle et al., 1998a); however, at least some appear to be transcriptional activators (Ulmasov et al., 1999a). Unlike the Aux/IAA genes, ARF gene expression does not appear to be sensitive to auxin (Ulmasov et al., 1999b). The Aux/IAA and ARF proteins are related to each other through two domains of conserved sequences that appear to be necessary for dimerization within and between these two classes of proteins (Kim et al., 1997; Ulmasov et al., 1999b). Because only the ARF proteins have been shown to exhibit clear DNA binding activity, auxin has been proposed to modulate gene expression through modification of ARF activity by way of cell- and tissue-specific auxin-dependent ARF–ARF and ARF–Aux/IAA dimerization (Ulmasov et al., 1999a, 1999b).
The functional roles of Aux/IAA and ARF proteins are beginning to be elucidated through the molecular analysis of Arabidopsis mutants found to contain lesions in Aux/IAA or ARF genes. For example, mutations in the SHORT HYPOCOTYL 2 (SHY2)/IAA3 and AUXIN RESISTANT 3 (AXR3)/IAA17 loci have been shown to have effects on similar auxin-dependent responses, including root elongation and proliferation, root gravitropism, hypocotyl elongation in light-grown seedlings, and inflorescence development (Leyser et al., 1996; Rouse et al., 1998; Tian and Reed, 1999). Mutations in two ARF genes, ETTIN (ETT)/ARF3 and MONOPTEROS (MP)/ARF5, suggest that auxin-regulated transcription is essential for proper pattern formation. Specifically, ett mutations alter regional identity within floral meristems to disrupt proper patterning of reproductive organs (Sessions et al., 1997), whereas mp mutations disrupt body axis formation and vascular patterning within the embryo (Hardtke and Berleth, 1998). Together, analyses of these mutants provide genetic evidence that auxin-dependent changes in gene expression are important factors regulating plant development. The shy2 and axr3 mutants also provide compelling support for the gene activation hypothesis for regulation of differential growth, because both disrupt at least one bending response: root gravitropism (Leyser et al., 1996; Tian and Reed, 1999). However, because the phenotypes of these mutants are highly pleiotropic, the altered gravitropic responses may be the result of earlier effects on development rather than direct effects of changes in gene expression on growth. Conclusive evidence in support of the gene activation hypothesis would require the identification of an aux/iaa or arf mutant that specifically alters differential growth while leaving the overall developmental program intact.
Analyses of the nph4 mutants of Arabidopsis suggest that the NPH4 protein may represent a specific regulator of differential growth (Liscum and Briggs, 1996; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998; Watahiki et al., 1999). Originally identified by their dramatically reduced phototropic responses (Figures 1A and 1B; Liscum and Briggs, 1995), the nph4 mutants have been found to exhibit several additional differential growth defects, including altered stem gravitropism, phytochrome-dependent stem curvature, apical hook maintenance, and abaxial/adaxial leaf blade expansion (Liscum and Briggs, 1996; Stowe-Evans et al., 1998; Watahiki et al., 1999). The severely impaired auxin-induced hypocotyl bending, hypocotyl growth inhibition, and gene expression responses in the nph4 mutant background have led to the hypothesis that the NPH4 protein functions as a modulator of auxin-dependent differential growth (Stowe-Evans et al., 1998). In this study, we show that NPH4 encodes the auxin response factor ARF7 and that this transcriptional activator is conditionally required for the modulation of differential growth of aerial tissues in Arabidopsis. These findings provide support for the gene activation hypothesis. Furthermore, the findings that ethylene can suppress the phenotypic abnormalities of nph4 mutants illustrate the functional overlap occurring between auxin and ethylene response pathways, findings that should provide an ideal system for future studies of this interaction.
Figure 1.
Phototropic Response of 3-Day-Old Dark-Grown Wild-Type (Col) and nph4 Seedlings.
(A) Seedlings exposed to 8 hr of unilateral blue light (BL; 0.1 μmol m−2 sec−1) from the left. Col, Columbia ecotype.
(B) Quantitative analysis of hypocotyl phototropism in 3-day-old dark-grown seedlings exposed to 8 hr of unilateral blue light. Data represent the mean response of a minimum of 20 seedlings for each genotype. Error bars indicate sd.
RESULTS
Physical Mapping and Cloning of NPH4
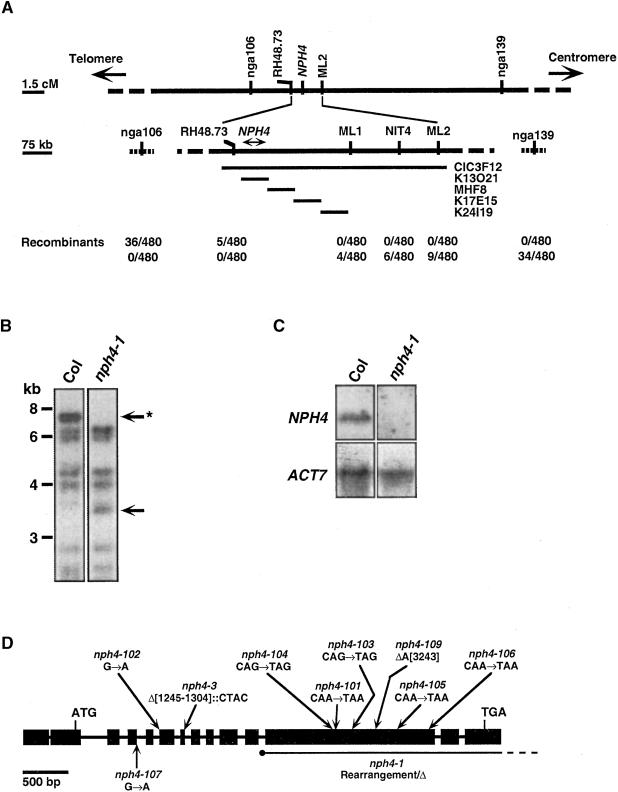
Previous work has shown that the NPH4 locus mapped to the proximal arm of chromosome 5, between simple sequence length polymorphism (SSLP) markers nga106 and nga139 (Ruegger et al., 1997; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998; Figure 2A). We used a combination of polymerase chain reaction (PCR)–based marker systems to define the rough physical location of the NPH4 locus within this genetic interval to a region of ∼250 kb represented by the proximal half of yeast artificial chromosome clone CIC3F12 (Figure 2A). DNA hybridization analysis using the transformation-competent artificial chromosome clone K13O21 (Figure 2A) as a probe identified a 6.9-kb fragment in EcoRI-XbaI double-digested DNA from a wild-type plant that was absent in DNA from the fast neutron–generated nph4-1 mutant (Figure 2B). When cloned and used as a probe of RNA gel blots, this 6.9-kb fragment hybridized with a transcript of ∼5 kb from 2.5-day-old etiolated wild-type seedlings that was undetectable in nph4-1 seedlings (Figure 2C), suggesting that this DNA fragment might contain the NPH4 locus.
Figure 2.
Map-Based Cloning and Structure of the NPH4 Gene.
(A) Mapping of the NPH4 locus on the proximal arm of chromosome 5. Genetic linkage map is shown at top; physical linkage map is shown in the middle; and numbers of recombinant chromosomes out of the total examined are shown at bottom. nga106, nga139, and ML2 are SSLP markers (Bell and Ecker, 1994); NIT4 is a cleaved amplified polymorphic sequence marker (Bartel and Fink, 1994; modified as described in Methods); ML1 is a simple nucleotide polymorphism marker (Cho et al., 1999); and RH48.73 is an amplified fragment length polymorphism marker (Liscum, 1999). CIC3F12 is a yeast artificial chromosome clone (Creusot et al., 1995; see Methods); MHF8 is a P1 phagemid clone (Liu et al., 1995); and K13O21, K17E15, and K24I19 are transformation-competent artificial chromosome clones (Liu et al., 1999). Arrows indicate that the chromosome continues beyond the region shown. cM, centimorgans.
(B) DNA gel blot made from Columbia (Col) and nph4-1 genomic DNA double digested with EcoRI and XbaI and hybridized with 32P-labeled K13O21 (see [A]). Arrows indicate polymorphic bands. The asterisk indicates the band cloned from wild-type DNA that was used as a probe in (C). Numbers at left denote size markers in kilobases.
(C) RNA gel blot made with total RNA from 7-day-old Col and nph4-1 seedlings and hybridized with the 6.9-kb DNA fragment described in (B) or with ACT7 (McDowell et al., 1996) as a control.
(D) Structure of the NPH4 gene and position of nph4 mutations. The locations of start (ATG) and stop (TGA) codons are indicated. Exon (boxes) and intron (lines) positions were determined by comparing the genomic DNA sequence with the sequences of ARF7 and BIPOSTO cDNAs. The position and identity of various nph4 mutations are shown. For purposes of nucleotide numbering, the adenine of the start codon is considered position 1. The dashed line indicates that the end of the rearrangement is unknown.
Searches of the GenBank library with sequence from one end of the 6.9-kb DNA fragment identified several related cDNAs that represent various members of the ARF family of transcriptional regulators (Ulmasov et al., 1999a, 1999b). The closest sequence identity was observed with a pair of cDNAs—ARF7 and BIPOSTO (GenBank accession numbers AF022368 and AF042195, respectively). The sequences of the ARF7 and BIOPOSTO cDNAs differ by just 12 bp and are predicted to encode proteins differing in sequence by only eight amino acids. Because these cDNAs were isolated from the same genetic background, they appear to represent independent isolates of the same gene, with the differences arising from errors in sequencing. The sequence of genomic DNA representing ∼2.3 kb of upstream untranslated sequence, the ARF7/BIPOSTO open reading frame, and ∼200 bp of downstream untranslated sequence has been determined for the wild-type gene (GenBank accession number AF186466) and several nph4 mutants, and we have found mutations within the open reading frame in each mutant allele (Figure 2D). Comparisons of NPH4 genomic sequence with ARF7 and BIPOSTO cDNA sequences also have allowed us to identify sequencing errors in both cDNAs. Of nine total single-base discrepancies identified, only two result in amino acid sequence differences relative to the published ARF7 sequence (Ulmasov et al., 1999a), namely, A-627 to S and H-638 to Q. A third amino acid difference, the addition of a W at position 271, results from the addition of one codon (GTG) at the exon 9/intron 8 junction.
nph4 Mutations and Their Predicted Effects on the NPH4/ARF7 Protein
Although we have not determined the precise nature of the nph4-1 lesion, PCR analysis indicates that a rearrangement, most likely an inversion with an internal deletion, breaks the coding sequence between exons 11 and 12 (Figures 2B and 2D; R.M. Harper and E. Liscum, unpublished results). We believe that nph4-1 represents a null allele because no NPH4 mRNA has been observed by either RNA gel blot (Figure 2C) or reverse transcription (RT)–PCR (Figure 3A) analysis. A second fast neutron–generated allele, nph4-3, also carries a complex mutation: a 59-bp deletion/4-bp insertion occurring across the junction between exon 7 and intron 7 (Figure 2D). This mutant produces a nph4 mRNA at a reduced level relative to NPH4 (Figure 3A) that is predicted to encode a protein corresponding to the first third of the DNA binding domain of NPH4/ARF7. The nph4-3 protein, if present, would be 192 amino acids long, the first 182 amino acids being identical to the wild-type protein and the last 10 amino acids being novel (Figure 3B). A third fast neutron–generated allele, nph4-109, contains a single-base deletion (Figure 2D) that results in a frameshift and premature stop codon at position 709. Each of the remaining seven alleles that have been sequenced contain a single-base substitution: in nph4-101, nph4-103, nph4-104, nph4-105, and nph4-106, this results in premature stop codons within the Q-rich middle region of the NPH4/ARF7 protein, and those in nph4-102 and nph4-107 disrupt splice site junctions within the DNA binding domain coding region (Figures 2D and 3B). The aphototropic phenotype of the latter two mutant alleles is weaker than that of the homozygous nph4-1 null alleles (Figure 1B; Stowe-Evans et al., 1998), suggesting that the former plants are likely to express nph4 mRNA that is alternatively spliced to give rise to partially functional protein. Although the nph4-107 allele has not been examined, RT-PCR results indicate that the nph4-102 splice site mutant does in fact contain transcript. However, the expression or stability of the nph4-102 mRNA is dramatically reduced relative to the wild type (Figure 3A). We have not determined how the nph4-102 mRNA is spliced.
Figure 3.
Expression of NPH4/ARF7 in nph4 Mutant Backgrounds.
(A) RT-PCR analysis of steady state amounts of NPH4/ARF7 transcript in 2.5-day-old dark-grown wild-type (Col) and nph4 mutants. RT-PCR products were detected by DNA gel blot analysis using 32P-labeled NPH4/ARF7 (top) or PHYTOCHROME E (PHYE; bottom). Genomic DNA from Col (Col-DNA) was used as a control template for both genes. Because the amplimers from genomic DNAs contain intron sequences, they are larger than the amplimers from RNA templates.
(B) Structure of wild-type (WT) NPH4/ARF7 (Ulmasov et al., 1999b) and nph4-3/arf7 proteins predicted from nph4-3 sequence. Amino acid changes in nph4-3/arf7 (top line) are shown relative to the wild-type sequence (bottom line). C, C terminus; DBD, DNA binding domain; N, N terminus; Q-Rich, glutamine rich; III/IV, motifs homologous with domains III and IV of the C-terminal protein–protein interaction domain of Aux/IAA proteins (Guilfoyle, 1998).
Aphototropism of the nph4-1 Null Mutant Is Suppressed by Ethylene Action
As described earlier, the nph4 mutants of Arabidopsis represent a class of mutants that is disrupted with respect to several auxin-dependent differential growth responses. However, many of these phenotypes are conditional, depending on particular growth conditions. For example, air-grown nph4 seedlings are partially hookless in appearance, whereas siblings exposed to ethylene exhibit an exaggerated apical hook typical of ethylene-treated wild-type seedlings (Stowe-Evans et al., 1998). This ethylene-dependent suppression of the phenotype of nph4 mutants prompted us to examine whether ethylene action could influence other mutant phenotypes of nph4 plants.
As shown in Figure 4A, a considerable phototropic response was recovered when seedlings homozygous for the nph4-1 null allele were grown in ethylene (50 μL/L) instead of unsupplemented air. In contrast, the aphototropic phenotype of the phototropism photoreceptor mutant nph1-5 (Christie et al., 1998) was not affected by ethylene treatment (Figure 4A). Interestingly, ethylene caused a depressed phototropic response in wild-type seedlings (Figure 4A). Because high concentrations of ethylene induce a typical “triple response” in etiolated Arabidopsis (Guzman and Ecker, 1990), the reduced phototropic curvature observed in ethylene-grown wild-type seedlings probably resulted from decreased overall growth capacity. This conclusion is supported by the finding that an ethylene receptor mutant, etr1-1 (Bleecker et al., 1988; Schaller and Bleecker, 1995), and signal transduction mutant ein2-1 (Alonso et al., 1999) exhibited no change in phototropic response when grown in ethylene (Figure 4A).
Figure 4.
Effects of Ethylene on Phototropism of Wild-Type (Col) and nph4 Seedlings.
(A) Phototropic curvature of 3-day-old seedlings grown in darkness under ambient air conditions (filled bars) or exposed to 50 μL/L ethylene (open bars) and then exposed to 8 hr of unilateral blue light (0.1 μmol m−2 sec−1). Data represent the mean response of at least 40 seedlings from at least two replicate experiments. Error bars indicate se.
(B) Dose–response curve for phototropism of wild-type (filled circles), nph4-1 (open circles), nph1-5 (open triangles), and etr1-1 (open diamonds) seedlings grown on ACC. Seedlings were grown in darkness for 3 days on medium containing the indicated concentration of ACC and then exposed to 8 hr of unilateral blue light (0.1 μmol m−2 sec−1). Data represent the mean response of at least 40 seedlings from at least two replicate experiments. Error bars indicate se. Because errors are small, many error bars are not visible.
(C) Ethylene production in wild-type and nph4 mutant seedlings. Seedlings were grown in sealed glass vials on a nutrient medium–agar plug for 72 hr in darkness. For auxin treatments, the nutrient medium was supplemented with the indicated concentration of IAA. Ethylene accumulation was measured as described in Methods. Values represent the mean response from three replicate experiments. Error bars indicate sd.
Etiolated nph4 seedlings exhibit wild-type ethylene sensitivity with respect to hypocotyl elongation (Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998); thus, growth inhibition probably explains why the recovered phototropic response of nph4-1 seedlings did not exceed the response of ethylene-treated wild-type seedlings but was merely the same as the wild-type response. Ethylene-dependent growth inhibition does not, however, explain why the aphototropic phenotype of the nph4 mutants was suppressed. Because nph4-1 seedlings have wild-type concentrations of ethylene (Woeste et al., 1999), both in the presence and absence of auxin (Figure 4C), the trivial explanation that nph4 seedlings are aphototropic in the absence of exogenous ethylene because they fail to properly synthesize ethylene is untenable.
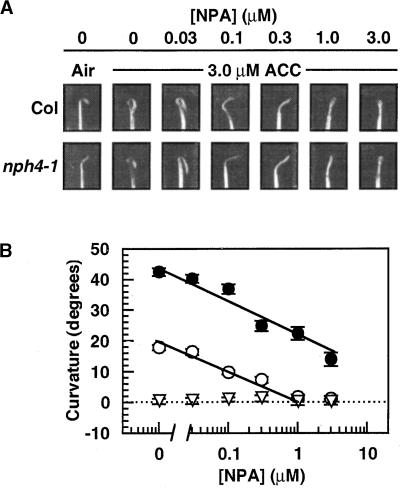
We also have examined the phototropic response of wild-type and mutant seedlings grown on various concentrations of 1-aminocyclopropane-1-carboxylic acid (ACC), the immediate precursor of ethylene (Yang and Hoffman, 1984), to determine whether the positive and negative effects of ethylene on phototropism could be separated on the basis of ethylene dose responses. Consistent with their behavior in high ethylene conditions (Figure 4A), etr1-1 and nph1-5 mutants exhibited no change in phototropic responsiveness when grown on ACC (Figure 4B). Wild-type seedlings grown on concentrations of ACC ⩾1 μM exhibited an inhibition of phototropic curvature (Figure 4B) equivalent to that observed in 50 μL/L ethylene (Figure 4A). This inhibition response correlates well with an effective dose (1 μM) required for half-maximal ACC-dependent inhibition of hypocotyl growth in both wild-type and nph4 seedlings (E.L. Stowe-Evans, unpublished results). Maximal recovery of phototropism in nph4 seedlings was observed over a range of ACC concentrations similar to that required for inhibition of phototropism in the wild type. However, the threshold for the recovery response was 10 to 30 times lower than the threshold for the inhibition response in the wild type, indicating that the recovery response in nph4 seedlings is not associated with growth inhibition.
Altered Hypocotyl Gravitropism of nph4 Is Suppressed by Ethylene Action
In addition to exhibiting a reduced gravitropic curvature in response to a reorientation stimulus (Liscum and Briggs, 1996; Watahiki and Yamamoto, 1997), hypocotyls of nph4 seedlings exhibit randomized growth orientation that tends toward horizontal when grown unsupported in the absence of a reorientation stimulus (Figure 5A; Watahiki et al., 1999). Although the growth orientation of wild-type seedlings was unaffected, normal growth orientation of nph4 seedlings was recovered in the presence of ACC (Figure 5B). The randomized growth orientation phenotype in both nph4 mutants examined was suppressed with an ACC threshold of <0.1 μM, again indicating that simple growth inhibition does not represent the causal mechanism for suppression of the mutant phenotypes by ethylene.
Figure 5.
Effects of Ethylene on Gravity-Dependent Growth Orientation of Wild-Type (Col) and nph4 Seedlings.
(A) Seedlings grown on horizontally oriented plates in the presence or absence of ACC.
(B) Dose–response curve for hypocotyl gravitropic growth orientation of wild-type (filled circles), nph4-1 (open circles), and nph4-3 (open inverted triangles) seedlings grown on ACC. Seedlings were grown in darkness for 3 days on vertically oriented plates containing the indicated concentration of ACC. Data represent the mean response of at least 45 seedlings from at least two replicate experiments. Error bars indicate se. Because errors are small, many error bars are not visible.
Auxin Transport and Response Systems Are Required for the Ethylene-Dependent Suppression of the Mutant Phenotypes of nph4 Mutants
Results from our studies on phototropism and gravity-dependent growth orientation presented here (Figures 4 and 5) suggest that ethylene, at concentrations suboptimal for hypocotyl growth inhibition, enhances the activity or abundance (or both) of some component required for differential growth, a component that is limiting in the absence of NPH4/ ARF7. Because ethylene can have several effects on auxin physiology (Burg and Burg, 1966, 1967; Lieberman and Knegt, 1977; Schwark and Schierle, 1992; Visser et al., 1996), perhaps the action of ethylene somehow enhances auxin responsiveness in the nph4 mutants, thereby suppressing the mutant phenotypes. If this hypothesis is correct, any conditions that disrupt auxin responses independently of NPH4/AFR7 action would be expected to negate the effects of ethylene. Alternatively, if ethylene action is completely separable from auxin action, additional defects in auxin responsiveness would be without effect on suppression of the phenotypes of the nph4 mutants.
To test these hypotheses, we examined the effects of N-1-naphthylphthalamic acid (NPA), an inhibitor of polar auxin transport (Lomax et al., 1995), on the ability of ethylene to suppress the hookless and aphototropic phenotypes of nph4-1 seedlings. As shown in Figure 6A, although air-grown nph4-1 exhibited a partially hookless phenotype, siblings grown on ACC exhibited an exaggerated apical hook similar to that of the ACC-grown wild-type seedlings. However, this ACC-dependent exaggerated apical hook was negated by NPA for both wild-type and nph4-1 seedlings, with similar dose dependencies in each case. As shown in Figure 6B, NPA also negated the phototropic response of nph4-1 conditioned by growth on ACC (Figure 4B). Despite differences in their basal phototropic responses in the absence of NPA, wild-type and nph4 seedlings appeared to exhibit similar sensitivities with respect to the effects of NPA on phototropism, as indicated by their parallel dose–response curves (Figure 6B). These similarities in sensitivities of wild-type and nph4-1 seedlings to NPA are consistent with the hypothesis that ethylene exerts its effects on nph4 seedlings by influencing auxin responsiveness.
Figure 6.
Effects of the Polar Auxin Transport Inhibitor NPA on Ethylene-Dependent Changes in Apical Hook Structure and Phototropism in Wild-Type (Col) and nph4-2 Seedlings.
(A) Hook regions of 3-day-old seedlings grown in darkness on 3.0 μM ACC or in the absence of ACC (Air), with various concentrations of NPA.
(B) Dose–response curve for phototropism of wild-type (filled circles), nph4-1 (open circles), and nph1-5 (open inverted triangles) seedlings grown on NPA. Seedlings were grown as described in Figure 1B, except that they were grown in the presence of 0.3 μM ACC and the indicated concentration of NPA. Data represent the mean response of at least 33 seedlings from at least two replicate experiments. Error bars indicate se. Because errors are small, many error bars are not visible.
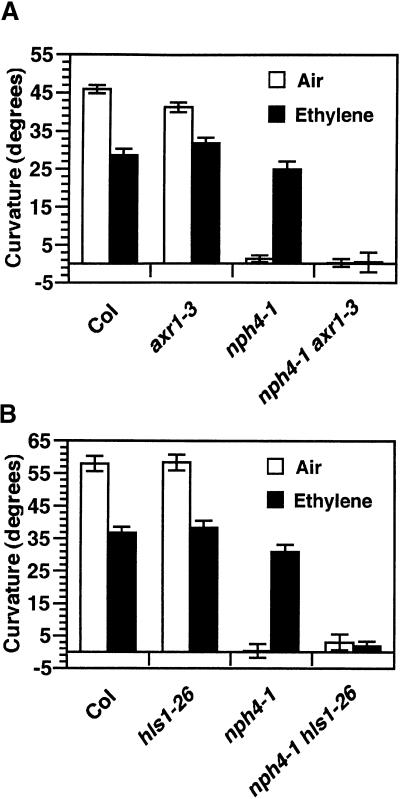
Although NPA generally is accepted as being a specific inhibitor of polar auxin transport, the molecular target or targets and modes of action of NPA remain largely unknown (reviewed in Lomax et al., 1995). As such, with regard to the role of ethylene in auxin-mediated processes, conclusions based solely on pharmacological studies should be viewed skeptically. Hence, we also used a genetic approach to address the role of auxin in ethylene-dependent recovery of phototropism in nph4-1 seedlings; that is, we disrupted additional genes involved in auxin responsiveness. As shown in Figures 7A and 7B, seedlings homozygous for either the weak axr1-3 allele (Lincoln et al., 1990) or the strong hls1-26 allele (Hou et al., 1993) were phototropically indistinguishable from wild-type seedlings when grown in air or ethylene. However, when either mutation was combined with the nph4-1 null allele as a double homozygote, the effects of ethylene on the aphototropic response of nph4 seedlings were negated (Figures 7A and 7B). Thus, although the auxin responsiveness appears not to be limiting with respect to phototropism in arx1-3 and hls1-26 single-mutant backgrounds, it becomes limiting when NPH4 is also mutated, providing additional support for the hypothesis that the effects of ethylene on the phenotypes of the nph4 mutants occur because of changes in auxin responsiveness.
Figure 7.
Effects of Auxin Response Mutations on Phototropism.
(A) Phototropism in 3-day-old wild-type (Col), axr1-3, nph4-1, and nph4-1 axr1-3 double mutant seedlings grown in ambient air (open bars) or 50 μL/L ethylene (filled bars), then exposed to 8 hr of unilateral blue light (0.1 μmol m−2 sec−1). Data represent the mean response of at least 41 seedlings from at least two replicate experiments. Error bars indicate se.
(B) Phototropism in 3-day-old Col, hls1-26, nph4-1, and nph4-1 hls1-26 double mutant seedlings treated the same as the seedlings in (A), except that 10 hr of unilateral blue light was given. Data represent the mean response of at least 26 seedlings from two replicate experiments. Error bars indicate se.
Ethylene Does Not Appear to Mediate Changes in Gross Auxin Sensitivity in the nph4 Mutant Background
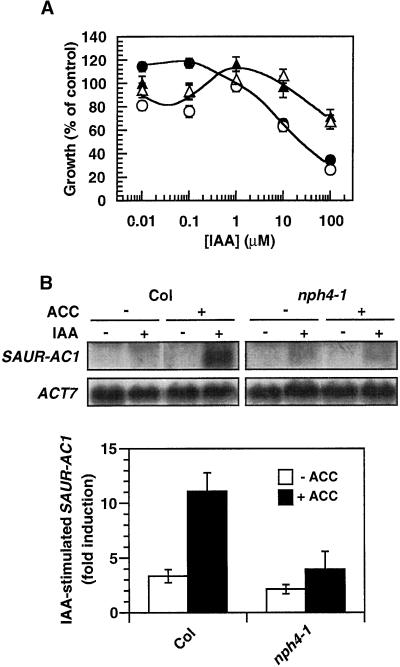
The studies discussed earlier in which NPA (Figure 6) and auxin response mutations (Figure 7) were used to disrupt auxin responsiveness suggest that ethylene-dependent changes in auxin responsiveness account for the ethylene-mediated suppression of the phenotypes of the nph4 mutants. However, these studies did not address how ethylene might alter auxin responsiveness. In an initial attempt to answer this question, we have examined whether ethylene treatment resulted in dramatic changes in auxin responsiveness of wild-type and nph4 seedlings by measuring auxin-dependent changes in hypocotyl growth and gene expression (Figures 8A and 8B). Although some increase in auxin responsiveness was observed in wild-type seedlings exposed to ACC, for both hypocotyl growth at low concentrations of auxin (Figure 8A) and the expression of auxin-induced genes (Figure 8B; data not shown), no dramatic changes in responsiveness were observed with similarly treated nph4-1 seedlings (Figure 8). These results suggest that the effects of ethylene on the phenotypes of the nph4 mutants result from subtle rather than dramatic changes in auxin responsiveness.
Figure 8.
Effects of Ethylene on Auxin-Sensitive Growth and Gene Expression.
(A) Dose–response curve for auxin-dependent hypocotyl growth inhibition of seedlings grown in ambient air (filled circles, wild type; filled triangles, nph4-1) or in the presence of 0.3 μM ACC (open circles, wild type; open triangles, nph4-1). Data represent the mean response of at least 29 seedlings from at least three replicate experiments. Error bars indicate se.
(B) Effects of ethylene on auxin-induced SAUR-AC1 expression in wild-type (Col) and nph4-1 seedlings. Seedlings were grown and treated with IAA, and RNA was analyzed as described previously (Stowe-Evans et al., 1998), except that one set of seedlings was grown in the presence of 0.3 μM ACC. Representative RNA gel blots probed with SAUR-AC1 (Gil et al., 1994) and ACT7are shown at top. At bottom are quantitative data representing the mean response (n-fold induction by IAA relative to no IAA control) of three replicate RNA gel blots analyzed by densitometry. All data are normalized relative to an ACT7 control. Error bars indicate sd. Similar results were obtained with IAA6 and IAA13 (E.L. Stowe-Evans and E. Liscum, unpublished results). (–), no added hormone; (+), hormone added.
DISCUSSION
Most of the phenotypic alterations in the nph4 mutants appear to be associated with auxin-dependent differential growth responses in aerial tissues (Liscum and Briggs, 1996; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998). With the cloning of the NPH4 gene presented here, a hypothesis for the molecular basis for the phenotypes of nph4 mutants now can be developed. This hypothesis must take into account the findings that multiple phenotypes of the nph4 null mutant can be suppressed by exposure to ethylene.
NPH4/ARF7–Induced Gene Expression Is an Important Component of Auxin-Dependent Differential Growth in Aerial Tissues
It has long been known that auxins can modulate plant growth (reviewed in Went and Thimann, 1937); however, the molecular mechanisms by which auxins elicit these effects have remained more elusive. As discussed earlier, a gene activation hypotheses has been proposed to explain the effects of auxin on plant growth (Key, 1969). Despite evidence in support of this hypotheses (reviewed in Abel and Theologis, 1996), a direct causal relationship between auxin-mediated changes in gene expression and changes in specific growth processes had not been demonstrated. Cloning the NPH4 locus represents a considerable step toward demonstrating a causal link between auxin-induced gene expression and control of growth.
NPH4 encodes the auxin response factor ARF7, which is a member of a large family of auxin-responsive transcriptional regulators in Arabidopsis (reviewed in Guilfoyle et al., 1998a). In general, ARF proteins are composed of three domains: an N-terminal DNA binding domain, a C-terminal protein–protein interaction domain, and a middle region of variable length and amino acid composition that separates the DNA binding domain and the C-terminal domain (see Figure 3B; Ulmasov et al., 1999a, 1999b). The middle region of an ARF determines whether the protein functions as a transcriptional activator or a repressor (Ulmasov et al., 1999a). Not surprisingly, given the observed reductions in auxin-induced gene expression in the nph4 background (Stowe-Evans et al., 1998), the middle region of NPH4/ARF7 has been shown to function as an activator domain in transient expression assays (Ulmasov et al., 1999a). This latter finding, together with the differential growth-specific loss-of-function phenotypes of nph4 null mutants, indicates that activation of gene expression by auxin-induced NPH4/ARF7 function is an important component of auxin-dependent localized growth responses.
nph4 Mutants Reveal Functional Properties of the N-Terminal DNA Binding and C-Terminal Protein–Protein Interaction Domains of NPH4/ARF7
Analyses of the nph4 mutants also provide useful information for deciphering the biological functions of the domains of NPH4/ARF7 identified by biochemical studies (Ulmasov et al., 1999a, 1999b). When assayed in a carrot transient expression system, ARF proteins modulate auxin-dependent transcription either through direct DNA binding by way of the DNA binding domain or through assumed associations with ARFs already bound to DNA by way of the C-terminal protein–protein interaction domain (Ulmasov et al., 1999a). NPH4/ARF7, in particular, was shown to function as a transcriptional activator, whether as a full-length protein or as a truncated protein in which the DNA binding domain had been deleted (Ulmasov et al., 1999a), suggesting that the C-terminal protein–protein interaction domain plays a critical role in NPH4/ARF7 function in the plant.
The loss-of-function phenotypes of nph4 mutants carrying alleles containing premature stop codons that truncate the NPH4/ARF7 protein upstream of the C-terminal domain (i.e., nph4-101, nph4-103, nph4-104, nph4-105, and nph4-106) provide support for the aforementioned hypothesis. However, findings that the phenotypes of such mutants are less severe than those of the apparent null mutant nph4-1 (Figure 1B; Stowe-Evans et al., 1998) indicate that NPH4/ARF7 retains some activity in the absence of the C-terminal protein–protein interaction domain. A similar conclusion can be drawn from analyses of mutant alleles of the MP locus. Although true null alleles of MP are presumed lethal (Hardtke and Berleth, 1998), which precludes any direct comparison between partial and complete loss-of-function alleles, a variation in phenotypic severity has been observed among existing mp alleles that contain premature stop codons preceding the ARF5 C-terminal domain (Berleth and Jürgens, 1993; Hardtke and Berleth, 1998).
Although removal of the C-terminal domain from several ARFs disrupts stable binding of those ARFs to artificial palindromic auxin response elements in vitro, presumably by abolishing dimerization of the ARF proteins (Ulmasov et al., 1999b), nph4 alleles lacking the C-terminal domain demonstrate that the DNA binding and the middle region activation domains are sufficient for partial biological response—suggesting either that NPH4/ARF7 dimerization is unnecessary for partial auxin-dependent transcriptional activation or that dimerization can occur at some site outside the C-terminal protein–protein interaction domain in vivo. Many native auxin response elements are actually composite elements, composed of the TGTCTC auxin response element and a constitutive or coupling element rather than simple palindromes (Guilfoyle et al., 1998b). Hence, truncated nph4/arf7 proteins present in the nph4 alleles that lack the C-terminal domain may be able to form partially functional heterodimers with factors bound to coupling elements.
The nph4-3 allele, which contains a premature stop codon within the DNA binding domain, is a particularly interesting allele. As might be expected, the phenotypes of this mutant were similar to those of the nph4-1 null mutant, except for showing a stronger randomized gravitropic growth orientation under all conditions (Figure 5). If we assume that the mRNA shown to be present in the nph4-3 allele produces a truncated nph4 protein, this observation suggests that the truncated nph4-3/arf7 DNA binding domain can function as a dominant negative inhibitor of auxin-dependent growth. That a nph4-3/arf7 protein would be able to act as a competitive repressor of auxin-induced transcription by direct binding to auxin response elements seems unlikely, however, because the nph4-3 truncation occurs in the middle of the subdomain shown to be essential for DNA binding of ARF proteins (Ulmasov et al., 1999a). The most plausible explanation for the apparent dominant negative effects of the nph4-3 mutation is that the truncated nph4-3/arf7 protein interacts with (and thus removes from action) an additional transcriptional activator, which might be either another ARF or a coupling factor, thereby preventing auxin-induced transcription. Although no studies have specifically addressed whether the DNA binding domain also can mediate protein–protein interactions, analysis of the DNA binding properties of ARF1 suggested that such a property might exist. Specifically, the ARF1 protein that lacks the C-terminal protein–protein interaction has been shown to bind more stably to palindromic auxin response elements than to auxin response element half-sites, suggesting that this truncated ARF is binding as a dimer, with the dimerization occurring through the DNA binding domain (Ulmasov et al., 1999b). However, exactly how a partial NPH4/ARF7 DNA binding domain can function in an apparent dominant negative fashion remains to be determined.
Ethylene Acts as a Modulator of Auxin-Dependent Differential Growth
In this study, we have shown that ethylene is able to suppress multiple differential growth defects of nph4 mutants. These findings suggest that ethylene enhances the activity or sensitivity of a redundant system that is capable of mediating differential growth, presumably a system that involves changes in gene expression. In fact, this redundant system appears to be an auxin response system because further disruptions of auxin responsiveness, whether through mutation or pharmacological manipulation, negated the suppressive influence of ethylene on phenotypes of the nph4 mutants. For instance, the effects of ethylene on the aphototropic response of nph4 seedlings were counteracted by mutations in the AXR1 locus (Figure 7; E.L. Stowe-Evans and E. Liscum, unpublished results). AXR1 encodes a component of a ubiquitin-activating enzyme (E1) complex that activates ubiquitin-like proteins believed to be involved in the degradation of repressors of auxin responses (reviewed in del Pozo and Estelle, 1999). Previous studies suggested that NPH4/ARF7 and AXR1 function in independent pathways to regulate tropic responses (Watahiki et al., 1999); the results presented here are consistent with those findings and further suggest that AXR1-dependent regulation of differential growth is partially redundant to the NPH4/ARF7–dependent process.
We also failed to observe ethylene-dependent normalization of the phenotypes of the nph4 mutants when either polar auxin transport or HLS1 activity was disrupted. HLS1 encodes a putative N-acetyltransferase proposed to function as a regulator of local auxin concentrations (Lehman et al., 1996); thus, ethylene-dependent increases in active auxin pools within target cells/tissues might stimulate the activity of a redundant differential growth modulating system. However, activation of such a redundant system did not appear to grossly alter auxin responsiveness in the nph4 background, because neither auxin-dependent growth inhibition nor gene expression was much enhanced in response to ethylene treatment.
Previous studies have shown that ethylene can influence both polar (Burg and Burg, 1967; Suttle, 1988; Schwark and Schierle, 1992) and lateral (Burg and Burg, 1966; Lee et al., 1990; Schwark and Schierle, 1992) auxin transport. Moreover, a direct interaction between ethylene and auxin transport recently has been established with the cloning of the AGRAVITROPIC 1 (AGR1)/ETHYLENE-INSENSITIVE ROOT 1 (EIR1)/PIN FORMED 2 (PIN2) gene of Arabidopsis (hereafter referred to as AGR1), which encodes a transmembrane protein that probably represents a component of a polar auxin efflux system in primary roots (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998). Mutations in the AGR1 locus, aside from disrupting polar auxin transport (Chen et al., 1998; Luschnig et al., 1998), result in decreased sensitivity of the root to both endogenous and exogenous ethylene (Bell and Maher, 1990; Roman et al., 1995; Chen et al., 1998). Hence, one target for ethylene action in root growth inhibition appears to be AGR1 function (Palme and Gälweiler, 1999; Rosen et al., 1999).
Presumably, an interaction between ethylene and auxin transport also might be occurring in aerial tissues. If so, ethylene-dependent changes in auxin transport could result in the subtle changes in auxin responsiveness that appear to be necessary for the suppression of differential growth defects in the nph4 mutants. For example, an increase in local auxin concentration as a result of ethylene-dependent changes in auxin transport might allow for the partial activation of an ARF having a lower sensitivity to auxin. However, because no data are currently available to definitively address this hypothesis of auxin transport, it is equally plausible that the ethylene effects on nph4 might reflect an enhanced auxin sensitivity of a redundant ARF system through changes in expression or activity of that ARF protein. Both hypotheses ultimately predict that in the absence of ethylene, NPH4/ARF7 is the predominant ARF regulating gene expression in response to auxin in cells of aerial tissues exhibiting differential growth, whereas in the presence of ethylene, one or more additional ARFs also may be functioning.
In summary, we have cloned the NPH4 gene of Arabidopsis, which when mutated disrupts multiple differential growth responses within aerial tissues, and we found that the gene encodes the auxin-responsive transcriptional activator ARF7. The cloning of this locus and various analyses of mutant alleles provide evidence directly linking auxin-induced gene transcription to auxin-dependent changes in growth. Despite the clear requirement for NPH4/ARF7 in the regulation of differential growth, the conditional nature of the nph4-1 null allele indicates that one or more partially redundant systems must exist. We anticipate that the nph4 mutants may be very helpful in elucidating the mechanism by which ethylene and other environmental conditions, such as phytochrome activation (Liscum and Briggs, 1996; E.L. Stowe-Evans and E. Liscum, unpublished results), stimulate the activity of a redundant ARF system(s). In particular, we should be able to identify factors involved in the suppression of the phenotypes of the nph4 mutants through isolation and characterization of second-site mutations that convert conditional phenotypes into constitutive phenotypes; in turn, we can directly ask whether any such factors are involved in signaling, auxin-transport processes, transcriptional regulation, or other aspects of auxin-mediated differential growth.
METHODS
Plant Materials, Growth Conditions, and Analyses of Seedling Responses
For all experiments, seeds of Arabidopsis thaliana were surface-sterilized and plated on nutrient medium solidified with 1.0% agar (w/v), as described previously (Liscum and Briggs, 1995). Murashige and Skoog nutrient medium (Murashige and Skoog, 1962) at one-half strength and without sucrose was used for all experiments except the auxin-dependent hypocotyl growth inhibition and gene expression studies, for which full-strength Murashige and Skoog medium supplemented with 2.0% (w/v) sucrose was used. Cold treatment and exposure to red light to induce uniform germination were as described previously by Liscum and Briggs (1995). Unless otherwise noted, ethylene and auxin treatments were performed as described previously (Stowe-Evans et al., 1998).
Phototropic responses were measured as described previously for hypocotyls of dark-grown Arabidopsis seedlings (Liscum and Briggs, 1995). Hypocotyl growth orientation was measured in dark-grown seedlings as described by Liscum and Hangarter (1993). Auxin-dependent hypocotyl growth inhibition was determined as described previously (Stowe-Evans et al., 1998), except that where appropriate, 1-aminocyclopropane-1-carboxylic acid (ACC) was included during the first 3 days of growth, after which time all seedlings were transferred to ACC-free plates containing indole-acetic acid (IAA).
Ethylene Measurement
For each treatment, seedlings were grown on a 10-mL Murashige and Skoog nutrient medium–agar plug inside a 44-mL glass vial sealed with a screw cap containing a Teflon septum. After cold treatment and red light exposure to induce uniform germination of seeds (Liscum and Briggs, 1995), the vials were wrapped in foil and transferred to a light-tight box. Seedlings were allowed to grow for 3 days, after which 30 mL of headspace air was removed from each vial and simultaneously replaced with ethylene-free air. The sample removed was concentrated in a cold trap, released by heating with boiling H2O, and passed onto a gas chromatograph for ethylene determination (Spollen et al., 2000). The number of seedlings present in each vial was determined and used to calculate the data presented in Figure 4C.
Fine-Structure Mapping and Cloning of the NPH4 Locus
A recombinant population was generated by crossing the nph4-1 mutant in the Columbia (Col) ecotype to wild-type Landsberg erecta (Ler), allowing the F1 plants to self-pollinate, and selecting aphototropic plants (nph4-1/−) from the resulting F2 progeny. Because nph4 segregates as a semidominant trait (Stowe-Evans et al., 1998), the phototropic response was rechecked in the F3 generation to eliminate heterozygotes from the mapping population. Genomic DNA was prepared (Edwards et al., 1991) from 240 recombinant nph4-1/nph4-1 homozygotes and then used for polymerase chain reaction (PCR)–based mapping as described.
Amplified fragment length polymorphism (AFLP)–based mapping was performed as described elsewhere (Liscum, 1999). The tightly linked AFLP fragment RH48.73 (see Figure 2A) was isolated from acrylamide gels (Sambrook et al., 1989) and reamplified with oligonucleotides homologous with the AFLP adapters but having 5′ extensions containing a restriction enzyme recognition site that had been engineered into them. EcoRI-adapter sequences were primed with an oligonucleotide containing an EcoRI site (5′-GCGGAATTCCTC-GTAGACTGCGTACCAATTC-3′), and MseI-adapter sequences were primed with an oligonucleotide containing an XbaI site (5′-CGTCTAGAGACGATGAGTCCTGAGTAA-3′). The resulting PCR products were double-digested with EcoRI and XbaI, subcloned into compatible sites within pBluescript SK+ (Stratagene, La Jolla, CA), and sequenced. RH48.73-specific oligonucleotides (5′-GTAAAGCTGTGT-TGATGATA-3′ and 5′-GAATACAAATATCTATCTGAGC-3′) then were generated and used to screen a pooled yeast artificial chromosome DNA library (Creusot et al., 1995) by PCR.
All additional mapping utilized the newly identified ML1–simple nucleotide polymorphism and ML2–simple sequence length polymorphism (SSLP) markers and a modified NIT4-cleaved amplified polymorphic sequence marker. ML1 was identified by comparing the sequence of Ler genomic DNA with the corresponding Col sequence represented by the proximal end of the P1 clone, MTM4 (see http://www.kazusa.or.jp/arabi/chr5/pmap/P1_map_7.html and http://www.kazusa.or.jp/arabi/endseq/). Ecotype-specific ML1 products were amplified by PCR (26 cycles with 30 sec of denaturation, annealing, and extension and an annealing temperature of 56°C) by using ecotype-specific forward primers differing by just the simple nucleotide polymorphism at the 3′ terminal position (Col-specific oligonucleotide: 5′-CACATAATCGAGCTGCCTCC-3′; Ler-specific oligonucleotide: 5′-CACATAATCGAGCTGCCTCG-3′) and a common reverse primer (CCATAGGCCATCGAGAGTTTC). ML2 represents a CA dinucleotide repeat identified on the distal end of P1 clone, MQJ16 (see http://www.kazusa.or.jp/arabi/chr5/pmap/P1_map_7.html and http://www.kazusa.or.jp/arabi/endseq/), which is polymorphic between the Col and Ler ecotypes. PCR amplification with ML2-specific primers (5′-GAGGTTTATGGATTCGTAGACA-3′ and 5′-TTAGGAACAAAAGCA-GGATTAG-3′) and subsequent product resolution on 4.0% (w/v) agarose gels were performed as described previously for other SSLP markers (Bell and Ecker, 1994). New NIT4-specific primers (5′-GATTTCAACTGCTCCACAAGAC-3′ and 5′-TTGATGATGAACGGA-AACTATAAA-3′) were designed to allow for the amplification of a 405-bp PCR product (containing a single MboII site) in Ler products that is missing in Col products.
RNA preparation and gel blot analyses were performed as described by Ausubel et al. (1995). DNA gel blot analysis was performed as described by Evola et al. (1986). Sequence comparisons of NPH4 genomic sequence (GenBank accession number AF186466) with database entries were performed by using GappedBLAST (Altschul et al., 1997). Sequence analyses, cDNA and genomic sequence alignments, and amino acid sequence predictions were performed using the DNASTAR software package (DNASTAR, Madison, WI).
Reverse Transcription–PCR Analysis
For expression analysis, total RNA was extracted from 2.5-day-old dark-grown seedlings by using an RNeasy plant minikit (Qiagen, Chatsworth, CA). NPH4 and PHYTOCHROME E (PHYE; Clack et al., 1994) transcripts were assayed simultaneously in separate tubes with their respective gene-specific primers by using identical RNA aliquots. Reverse transcription–PCR (RT-PCR) reactions by using 100 ng of total RNA per sample (or 10 ng of genomic DNA as a control) were performed with the Access RT-PCR system (Promega), according to the manufacturer's directions. Amplifications were done for 30 cycles with an annealing temperature of 54°C and an extension time of 4 min. Products then were separated by electrophoresis on an 0.8% (w/v) agarose gel, blotted, and hybridized with 32P-labeled gene-specific DNA probes.
Acknowledgments
We thank members of the Liscum laboratory, Drs. Jim Birchler, Jane Murfett, Tom Guilfoyle, Gretchen Hagen, and Tobias Baskin for critical reading of the manuscript. Thanks to Drs. Bill Spollen and Bob Sharp for measurement of ethylene production. We also thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for seed stocks (axr1-3, ein2-1, etr1-1, and hls1-26), the pooled CIC yeast artificial chromosome library, and Arabidopsis cDNA libraries; Mitsui Plant Biotechnology Research Institute (MPBRI) and Kazusa DNA Research Institute for TAC clones; and MPBRI and the Research Institute of Innovative Research for the Earth for P1 clones. This work was supported by National Science Foundation Grant No. MCB-9723124 and University of Missouri Research Board Grant No. RB96-055 to E.L., and grants-in-aid from the Japanese Ministry of Education, Science, and Culture to K.Y. R.M.H. was partially supported by the University of Missouri Food for the 21st Century Program. E.L.S.-E. was supported by a predoctoral fellowship from the University of Missouri Maize Biology Training Program, a unit of the Department of Energy/National Science Foundation/U.S. Department of Agriculture Collaborative Research in Plant Biology Program. D.R.L. was a University of Missouri, College of Arts and Sciences, Undergraduate Research Fellow.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Millar, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D, Seidman, J.G., Smith, J.A, and Struhl, K., eds (1995). Short Protocols in Molecular Biology. (New York: John Wiley).
- Bartel, B., and Fink, G.R. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91, 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Maher, E.P. (1990). Mutants of Arabidopsis thaliana with abnormal gravitropic response. Mol. Gen. Genet. 220, 289–293. [Google Scholar]
- Berleth, T., and Jürgens, G. (1993). The role of the MONOPTEROS gene in organizing the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C.R., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Burg, S.P., and Burg, E.A. (1966). The interaction between auxin and ethylene and its role in plant growth. Proc. Natl. Acad. Sci. USA 55, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg, S.P., and Burg, E.A. (1967). Inhibition of polar auxin transport by ethylene. Plant Physiol. 42, 1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95, 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R.J., et al. (1999). Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat. Genet. 23, 203–207. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). NPH1 has the properties of a blue light photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequence and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Creusot, F., et al. (1995). The CIC library: A large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 8, 763–770. [DOI] [PubMed] [Google Scholar]
- Darwin, C., and Darwin, F. (1896). The Power of Movement in Plants. (New York: D. Appleton and Co.).
- Davies, P.J. (1987). Plant Hormones and Their Role in Plant Growth and Development. (Boston: Martinus Nijhoff).
- del Pozo, J.C., and Estelle, M. (1999). Function of the ubiquitin–proteosome pathway in auxin response. Trends Plant Sci. 4, 107–112. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evola, S.V., Burr, F.A., and Burr, B. (1986). The suitability of restriction fragment length polymorphisms as genetic markers in maize. Theor. Appl. Genet. 71, 765–771. [DOI] [PubMed] [Google Scholar]
- Gil, P., Yang, L., Orbivic, V., Verkamp, E., Poff, K.L., and Green, P.J. (1994). Characterization of the auxin inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1998). Aux/IAA proteins and auxin signal transduction. Trends Plant Sci. 3, 205–207. [Google Scholar]
- Guilfoyle, T.J., Ulmasov, T., and Hagen, G. (1998. a). The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J., Hagen, G., Ulmasov, T., and Murfett, J. (1998. b). How does auxin turn on genes? Plant Physiol. 118, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., von Arnim, A.G., and Deng, X.W. (1993). A new class of Arabidopsis constitutive photomorphogenic genes involved in regulation of cotyledon development. Plant Cell 5, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, K.W., Singer, P.B., McInnis, S., Diaz-Sala, C., and Greenwood, M.S. (1999). Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol. 120, 827–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, P.B., Wu, L.-L., Brock, T.G., and Kim, D. (1995). Hormones and orientation of growth. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 547–571.
- Key, J.L. (1969). Hormones and nucleic acid metabolism. Annu. Rev. Plant Physiol. 20, 449–474. [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Chang, W.-K., and Evans, M.L. (1990). Effects of ethylene on the kinetics of curvature and auxin redistribution in gravistimulated roots of Zea mays. Plant Physiol. 94, 1770–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85, 183–194. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Lieberman, M., and Knegt, E. (1977). Influence of ethylene on indole-3-acetic acid concentration in etiolated pea epicotyl tissue. Plant Physiol. 60, 475–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E. (1999). Amplified fragment length polymorphism: Studies on plant development. In PCR Applications: Protocols and Functional Genomics, M.A. Innis, D.H. Gelfand, and J.J. Sninsky, eds (San Diego, CA: Academic Press), pp. 505–519.
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1996). Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Hangarter, R.P. (1993). Genetic evidence that the red-absorbing form of phytochrome B modulates gravitropism in Arabidopsis thaliana. Plant Physiol. 103, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Vazquez-Tello, A., and Whittier, R.F. (1995). Generation of a high-quality P1 library of Arabidopsis suitable for chromosome walking. Plant J. 7, 351–358. [Google Scholar]
- Liu, Y.-G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax, T.L., Muday, G.K., and Rubery, P.H. (1995). Auxin transport. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 509–530.
- Luschnig, C., Gaxiola, R.A, Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung, A., Behringer, F.J., and Lomax, T.L. (1999). Ethylene plays multiple nonprimary roles in modulating the gravitropic response in tomato. Plant Physiol. 120, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Huang, S., McKinney, E.C., An, Y.-Q., and Meagher, R.B. (1996). Arabidopsis thaliana contains ten actin genes encoding six ancient protein subclasses. Genetics 142, 587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Guan, C., Gälweiler, L., Tänzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Plame, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Palme, K., and Gälweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2, 375–381. [DOI] [PubMed] [Google Scholar]
- Palmer, J.H. (1985). Epinasty, hyponasty, and related topics. In Encyclopedia of Plant Physiology, R.P. Pharis and D.M. Reid, eds (Berlin: Springer-Verlag), pp. 139–168.
- Roman, G., Lubarsky, B., Keiber, J.J., Rothenberg, M., and Ecker, J. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, C.P., Cooper, M.L., and Klee, H.J. (1993). Uncoupling auxin and ethylene effects in transgenic tobacco and Arabidopsis plants. Plant Cell 5, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, E., Chen, R., and Masson, P.H. (1999). Root gravitropism: A complex response to a simple stimulus? Trends Plant Sci. 4, 407–412. [DOI] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an Aux/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schaller, G.E., and Bleecker, A.B. (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811. [DOI] [PubMed] [Google Scholar]
- Schwark, A., and Schierle, J. (1992). Interaction of ethylene and auxin in the regulation of hook growth. I. The role of auxin in different growing regions of the hypocotyl hook of Phaseolus vulgaris. J. Plant Physiol. 140, 562–570. [Google Scholar]
- Sessions, A., Nemhauser, J.L., McColl, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sitbon, F., and Perrot-Rechenmann, C. (1997). Expression of auxin-regulated genes. Physiol. Plant. 100, 443–455. [Google Scholar]
- Spollen, W.G., LeNoble, M.E., Samuels, T.D., Bernstein, N., and Sharp, R.E. (2000). ABA accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Harper, R.M., Motchoulski, A.V., and Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle, J.C. (1988). Effect of ethylene treatment on polar IAA transport, net IAA uptake and specific binding of N-1-naphthylphthalamic acid in tissues and microsomes isolated from etiolated pea epicotyls. Plant Physiol. 88, 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 1–11. [DOI] [PubMed] [Google Scholar]
- Utsuno, K., Shikanai, T., Yamada, Y., and Hashimoto, T. (1998). AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 39, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Visser, E.J.W., Cohen, J.D., Barendse, G.W.M., Blom, C.W.P.M., and Voesenek, L.A.C.J. (1996). An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palusris Sm. Plant Physiol. 112, 1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki, M.K., and Yamamoto, K.T. (1997). The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol. 115, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki, M.K., Tatematsu, K., Fujihira, K., Yamamoto, M., and Yamamoto, K.T. (1999). The MSG1 and AXR1 genes of Arabidopsis are likely to act independently in growth-curvature responses of hypocotyl. Planta 207, 362–369. [DOI] [PubMed] [Google Scholar]
- Went, F.W., and Thimann, K.V. (1937). Phytohormones. (New York: Macmillan).
- Woeste, K.E., Ye, C., and Kieber, J.J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Phyisol. 119, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]