Abstract
Background
Transgenic proteins expressed by genetically modified food crops are evaluated for their potential allergenic properties prior to marketing, among others by identification of short identical amino acid sequences that occur both in the transgenic protein and allergenic proteins. A strategy is proposed, in which the positive outcomes of the sequence comparison with a minimal length of six amino acids are further screened for the presence of potential linear IgE-epitopes. This double track approach involves the use of literature data on IgE-epitopes and an antigenicity prediction algorithm.
Results
Thirty-three transgenic proteins have been screened for identities of at least six contiguous amino acids shared with allergenic proteins. Twenty-two transgenic proteins showed positive results of six- or seven-contiguous amino acids length. Only a limited number of identical stretches shared by transgenic proteins (papaya ringspot virus coat protein, acetolactate synthase GH50, and glyphosate oxidoreductase) and allergenic proteins could be identified as (part of) potential linear epitopes.
Conclusion
Many transgenic proteins have identical stretches of six or seven amino acids in common with allergenic proteins. Most identical stretches are likely to be false positives. As shown in this study, identical stretches can be further screened for relevance by comparison with linear IgE-binding epitopes described in literature. In the absence of literature data on epitopes, antigenicity prediction by computer aids to select potential antibody binding sites that will need verification of IgE binding by sera binding tests. Finally, the positive outcomes of this approach warrant further clinical testing for potential allergenicity.
Background
Commercial cultivation of genetically modified (GM) crops has increased substantially since their market introduction in the mid-1990's [1]. Most of these crops have been modified with the agronomically important traits, such as herbicide tolerance and insect resistance. Other crops that are still in development and currently field tested may reach the market soon. The transgenic traits that these future crops carry will likely be much more diverse than at present. The safety of new proteins expressed in these crops will be part of the safety assessment that GM crops undergo prior to their market approval by national governments.
One of the main issues in the safety assessment of a genetically modified organism, such as a GM crop, is its potential allergenicity. Genetic modification can affect the allergenicity of the modified organism in two ways: I) by introducing allergens, or II) by changing the level or nature of intrinsic allergens. Allergens can potentially be introduced by the expression of transgenic proteins, because proteins have been found to be the causative agents of food allergies, contact allergies, and inhalant allergies (pollen, fungal spores). Assessment of the potential allergenicity of a newly expressed protein usually follows the consensus decision-tree approach of the joint International Life Sciences Institute – International Food Biotechnology Council (ILSI / IFBC) [2]. The path that will be followed through this decision tree will depend on data and outcomes, such as the allergenicity of the source of the foreign gene, the comparison of the amino acid sequence of the foreign protein to the sequences of known allergens using computer databases, and the stability of the foreign protein to digestive enzymes (most food allergens are stable to digestion). In some cases, further testing with allergy patients' sera, followed by skin prick tests and food challenges may be recommended.
The assessment approach, including this decision tree, is currently discussed within the Codex alimentarius committee of the joint Food and Agriculture Organisation and World Health Organisation (FAO/WHO) in preparation of Codex guidelines [3]. Recent FAO/WHO Expert Consultations in Rome, January 2001, and Vancouver, September 2001, were convened in the frame of these discussions [4,5]. Adoption of the guidelines is expected in the year 2003, and their implementation by Codex Member States will follow suit. In addition, two recent articles review the assessment methodology of potential allergenicity of transgenic proteins [6,7].
It can be anticipated that many of the source organisms that provide candidate proteins for genetic engineering will lack a history of allergenicity. An example is a soil bacterium providing an enzyme that degrades herbicides and, if expressed in crops, would convey herbicide tolerance to these crops. In this case, the first step in the ILSI / IFBC decision tree would be to compare the primary protein structure (i.e. the sequence of amino acid residues) of the novel protein with the primary structures of known allergens. To this end, computer algorithms are used that enable the computer user to align a given protein sequence with the sequences of allergenic proteins stored in a database. Two common algorithms that can be used for these searches are FASTA and BLAST. FASTA compares two sequences and aligns them with each other from the amino-terminus towards the carboxy-terminus, eventually slided with respect to each other, i.e. it compares overall similarity. BLAST on the other hand, does not focus on the overall alignment and therefore can also identify isolated stretches of similarity between two sequences in random order. With the appropriate settings, including the use of an "identity matrix" instead of an "evolutionary matrix", FASTA can also be employed to search for short identical sequences [8]. Publicly accessible Internet websites currently feature the possibility for website visitors to run FASTA and BLAST searches (Table 1). These Internet facilities may provide for an accessible tool to screen protein sequences for identities with allergenic proteins.
Table 1.
Examples of protein sequence databases
| Name | Internet address | Entries |
| EMBL | http://srs.embl-heidelberg.de:8000/srs5/ | EMBL, GenBank, PIR, SwissProt, and others |
| Entrez | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi | GenBank, PDB, PIR, PRF, RefSeq, SwissProt, and others |
| PIR-NREF | http://pir.georgetown.edu/ | GenPept, PDB, PIR-PSD, RefSeq, SwissProt, and TrEMBL |
| SwissProt | http://www.expasy.ch | EMBL and SwissProt |
Identical stretches are selected from the results of the alignment if their size is immunologically relevant, for example eight or more contiguous amino acids in the ILSI / IFBC decision tree approach [2]. Shorter stretches can also be relevant according to recent insights, because, for example, small sequences of four and six amino acids length can be recognised and bound by IgE antibodies from antisera of allergic patients (IgE is the immunoglobulin class associated with allergy) [9]. These stretches represent "continuous" epitopes, i.e. antibody-binding sites consisting of linear amino acid sequences. In addition, it can be envisaged that single or a few mismatches within a stretch of sufficient length may not affect, or even enhance, immunoglobulin binding. This is not discussed at present within the Codex and would also require additional guidance on the acceptability of substitutions of identical amino acids. In the absence of such guidance, some false negatives may be generated.
Continuous (linear) epitopes can be distinguished from "discontinuous" (conformational) epitopes consisting of amino acid residues that occur separated from each other within the primary, one-dimensional protein sequence, but that are within each other's proximity and accessible for antibodies on the surface of the folded, three-dimensional allergenic protein. It may be worth noting that also structural overall similarity with an allergenic protein, i.e. 35% identity within an 80-amino acid long stretch, is being considered to become part of the assessment of potential allergenicity by Codex alimentarius. Furthermore, Hileman et al. [10] concludes that at least 50% overall structural identity would be a good predictor for potential allergenicity, based on 35+% identities that these authors found between random maize proteins and allergenic proteins. A prediction method to pinpoint the amino acid residues that are present within such structural, discontinuous epitopes was recently described [11]. For these predictions, the three-dimensional structure of the specific protein must be either known or predictable from similarity to a known protein structure. At present, this requirement cannot be fulfilled for most of the allergenic- and transgenic-proteins. In addition to linear- and conformational-peptide epitopes, glycans have also been shown to be major IgE binding sites in allergenic glycoproteins [12].
With regard to the prediction of continuous epitopes within transgenic proteins, discussions within the FAO/WHO currently focus on whether the minimal degree of identity should be eight contiguous amino acids, as devised by the ILSI / IFBC decision tree, or six contiguous amino acids.
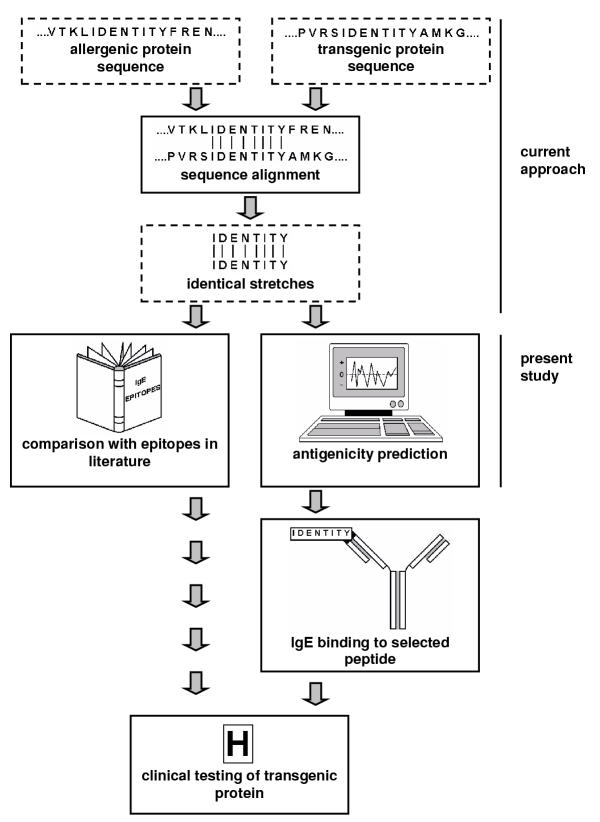
To our knowledge, no foreign protein expressed in commercial genetically modified crops shares identical stretches of eight or more amino acids with allergenic proteins. If six amino acids would, however, be established as the minimum requirement, the chance for identification of identical stretches in transgenic proteins and allergens will likely increase. Many of such positive outcomes will represent "false positives" that do not constitute binding sites (epitopes) for the allergy-associated IgE immunoglobulins. It can be argued, for example, that some sequences, based on their location on the protein surface and on the side chain characteristics of the amino acids, are more likely to be bound than other sequences in the same protein. A high number of false positives will make it impractical to use sequence alignment for assessment of the potential allergenicity of a transgenic protein. Therefore, further steps should enable the risk assessor to select those similarities that constitute more likely an allergenic hazard than others. This need for selection is further underscored by the recent results reported by Hileman et al. [10], who observed that a number of native maize proteins displayed identical stretches of eight or more contiguous amino acids that were also present in allergenic proteins, while transgenic Bacillus thuringiensis proteins displayed stretches of at most seven amino acids. We therefore propose a strategy, in which the sequence alignment is extended with further steps to identify the identical stretches that may contain IgE-epitopes (Figure 1). This strategy is a two-track approach:
Figure 1.
Proposed strategy for identifying potential linear IgE-epitopes in transgenic proteins
• In the first track, sequences of linear epitopes are extracted from literature on a particular allergenic protein and compared to the identical stretches that this protein has in common with a transgenic protein.
• In the other track, the most antigenic site of the protein is predicted by using a computer algorithm for antigenicity prediction. Subsequently, it is verified whether this antigenic site coincides with the sequence that the transgenic protein and allergenic protein have in common. This may provide additional information especially in case no literature data are available on the epitopes within an allergenic protein. Positive outcomes need further verification by IgE-binding assays because antigenic sites are not necessarily allergenic (e.g., IgE) epitopes, as can be inferred, for example, from the fact that IgG- and IgE-immunoglobulins may have different target sites on the same protein.
Transgenic proteins that probably contain epitopes, based on the outcomes of the two tracks, should be further tested clinically to determine the true potential for IgE binding by the transgenic protein and, eventually, skin prick tests and food challenges (Figure 1).
Algorithms are available to predict the antigenicity, i.e. the antibody binding, of peptide sequences (reviewed by [13]). Such algorithms are used in, for example, the design of peptide vaccines. One commonly employed algorithm is that of Hopp and Woods [14], in which the antigenicity of a point in the protein sequence is determined by averaging the antigenicity values of this point and the amino acids flanking this point. Hydrophilic and acidic amino acids, for example, have high antigenicity values. The window size used for the calculation, i.e. the total number of residues that are averaged, can be varied. Hopp and Woods [14] concluded that a window size of six amino acids would be most reliable. In many cases, however, a window size of seven amino acids is used, probably because the outcome can be assigned to the middle (fourth) amino acid within this window. The point with the highest score can be predicted with high probability to be part of an antigenic determinant of the protein. The Hopp and Woods method is accessible through Internet (Table 2).
Table 2.
Internet sites that offer free access to antigenicity prediction algorithms
| Host | Internet address | Methods |
| Colorado State University | http://arbl.cvmbs.colostate.edu/molkit/hydropathy/index.html | Hopp & Woods, Kyte & Doolittle |
| Expasy | http://us.expasy.org/cgi-bin/protscale.pl | Hopp & Woods, Kyte & Doolittle, and others |
| Weizman Institute | http://bioinformatics.weizmann.ac.il/hydroph/ | Hopp & Woods, Kyte & Doolittle |
Other antigenicity prediction methods have also been developed. Some of these calculate the hydrophilicity / hydrophobicity of peptide stretches, like the Hopp and Woods algorithm, whereas others take the predicted secondary structure (helix, sheet, turns) and protein segment mobility into account. Combinations of such algorithms are also used, as described, for example, by Jameson and Wolf [15]. As an example of prediction with the aid of combined algorithms, the antigenicity of peptides derived from potato virus Y coat protein has been found to correlate well with beta turns, hydrophilicity, and protein segment mobility [16].
Van Regenmortel and Pellequer [17] tested 22 algorithms and found that they all scored within the 50–60 % range of correct epitope predictions. It should be noted that these and other authors have used the algorithms to assign multiple epitopes within a protein, whereas Hopp and Woods [14] recommended to predict one epitope, i.e. the one containing the highest scoring point of the antigenicity plot. This point can be part of either a linear or a conformational epitope [18].
Antigenicity prediction algorithms have been successfully employed to predict IgE epitopes in allergenic proteins. IgE epitopes were correctly predicted with the Hopp and Woods algorithm, for example, in the housedust mite allergen Der p 2 (window size 7) [19] and in the cow's milk allergens β-lactoglobulin and α-lactalbumin [20].
A single IgE-epitope, however, does not make a protein an allergen. Binding of an allergenic protein containing multiple IgE epitopes to IgE on the surface of mast cells will lead to cross-linking of these IgE molecules. This clustering of IgE molecules on the cell surface will trigger the mast cell to release mediators, such as histamine and cytokines, which cause the symptoms of allergic reactions ("anaphylaxis"). Peptides and proteins containing only one IgE-epitope, however, will neither crosslink IgE nor provoke an allergic reaction, and are used as antagonists in therapy of allergic disease [21].
So far, antigenicity prediction has not been used for the safety assessment of transgenic proteins prior to marketing. Such a prediction may prove helpful if a transgenic protein shares with allergenic proteins identical stretches for which it is unknown if they are part of an epitope.
In the present work, it has been investigated if foreign proteins expressed in market-approved transgenic crops share identical peptides of at least six contiguous amino acids with known allergens. It has been verified whether these identical stretches constitute linear IgE binding epitopes by searching literature on allergenic epitopes. In addition, the antigenicity of the identical stretches has been predicted by the Hopp and Woods method.
Results
Sequence similarities with allergens
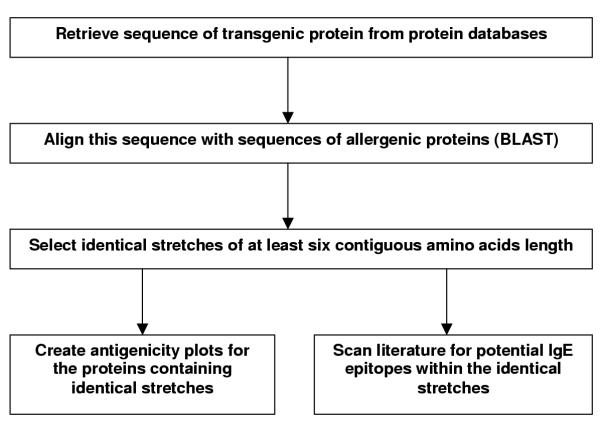
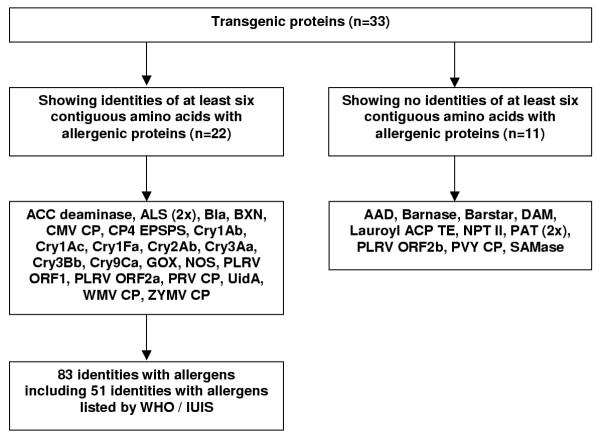
The procedure and results of this investigation are summarised in Figures 2 and 3, respectively. For detailed results, see additional file 1. Two-thirds of the thirty-three aligned transgenic proteins displayed identical stretches of at least six contiguous amino acids with allergenic proteins. The size of the identical peptides shared by transgenic proteins and allergenic proteins was in 75 out of 83 cases six amino acids, and seven amino acids in the remaining eight cases (Figure 3). Not all of the allergenic proteins appear on the official list of allergens composed by the Allergen Nomenclature Subcommittee of the joint World Health Organisation and International Union of Immunological Societies (WHO / IUIS; Table 3). This is in some cases due to the recent discovery of a particular allergenic protein that has not been listed yet.
Figure 2.
Procedure followed in this investigation
Figure 3.
Outcome of the sequence alignment of transgenic proteins to allergenic proteins
Table 3.
Allergen Databases on the Internet
| Name | Webadress | Allergen type | Information |
| Agmobiol | http://ambl.lsc.pku.edu.cn | Food, pollen | Protein databank accessions, literature references |
| CSL | http://www.csl.gov.uk/allergen (free registration required) | All | Protein databank accessions, epitopes literature references |
| Farrp | http://www.allergenonline.com (free registration required) | All | Protein databank accessions of 658 allergens |
| NCFST | http://www.iit.edu/~sgendel/fa.htm | All, plus coeliac | Protein databank accessions |
| Protall | http://www.ifr.bbsrc.ac.uk/protall | Plant | Protein databank accessions, biochemical and clinical data |
| SDAP | http://129.109.73.75/SDAP/ | All | Protein databank accessions, allergenic protein sequences, search facility for identical and similar peptide sequences1 |
| SwissProt | http://us.expasy.org/cgi-bin/lists?allergen.txt | All | Protein databank accessions |
| WHO/IUIS | http://www.allergen.org | All | Nomenclature, protein databank accessions |
1 The SDAP database and the peptide similarity search algorithm are described by Ivanciuc et al. [24]
Antigenicity prediction by computer
Table 4 features the identical stretches between a transgenic- and an allergenic-protein that were predicted by the Hopp and Woods method to be antigenic in either one. It should be noted that, particularly, positive predictions of antigenicity for sequences in allergenic proteins warrant further investigation. The window size of six amino acids has been recommended for this method. The additional positive outcomes using a window size of seven amino acids are also shown, which indicate the effect of changing the window size.
Table 4.
Identical sequences with positive antigenicity predictions
| Sequence | Transgenic protein (1) | Allergen (2) |
Antigenic? (highest peak) |
Part of IgE epitope? (literature) |
|||||
| name | source | name | source |
Transgenic protein |
Allergens | ||||
| w = 6 | w = 7 | w = 6 | w = 7 | ||||||
| PRKGSD | Acetolactate synthase II (mutant S4-Hra) |
Tobacco Nicotiana tabacum | Amb a 1.4 | Ragweed Ambrosia artemisiifolia | - | Yes | - | - | - |
| TSRRRR | Coat protein | Cucumber mosaic virus | ABA-1 | Roundworms Ascaris lumbricoides and A. suum |
Yes | Yes | - (3) | - (3) | No (4) |
| EKQKEK | Coat protein | Papaya ringspot virus P | ABA-1 | Roundworms Ascaris lumbricoides and A. suum |
- (5) | - (5) | Yes (6) | Yes | - |
| VKSEDG | Enoylpyruvate shikimate phosphate synthase |
Agrobacterium CP4 | Der p 7 | Housedust mite Dermatophagoides pteronyssinus | Yes | Yes | - | - | - |
| LAEEAD | Glyphosate oxidoreductase | Achromobacter LBAA | Pan s 1 | Lobster Panulirus stimpsoni | - | - | - | Yes (7) | Yes (8) |
(1) Accessions: ALS: gi124369, CMV CP: gi593495, PRV CP: gi593497, CP4 EPSPS: gi8469107, GOX: gi1252836 (2) Accessions: Amb a 1.4: gi113478, gi539050, gi166445; ABA-1 (TSRRRR): gi159653, gi477301, gi2498099, gi2735096, gi2735098, gi2735100, gi2735102, gi2735104, gi2735106, gi2735108, gi2735110, gi2735112, gi2970629, gi7494507; ABA-1 (EKQKEK): gi2735108, gi2735110, gi2735112, gi2735114, gi2735116, gi2735118, gi2970629, gi7494507; Der p 7: gi1352240, gi1045602; Pan s 1: gi14285797, gi3080761 (3) Calculation not possible because TSRRRR is C-terminal sequence of the ABA-1 proteins. (4) The sequence RRRR of these allergens probably does not occur in vivo in the allergen as it would be split off from the allergen by proteases [25] (5) Sequence EKQKEK corresponds to a plateau slightly below the highest peak(s) in the antigenicity plots for the papaya ringspot virus coat protein (6) Highest score was shared by two or three peaks in the antigenicity plot for each ABA-1 protein (w = 6) (7) Highest score was shared by five peaks in the antigenicity plot for the Pan s 1 protein (window 7 amino acids) (8) Sequence LAEEAD is part of a 9-mer peptide from the shrimp tropomyosin allergen Pen i 1 that is bound by sera from shrimp allergic patients [26,27]. This sequence has also been part of two 15-mer peptides from the shrimp allergen Pen a 1 tested for sera binding. One peptide is bound by sera from allergic individuals, whereas the other peptide is not [28].
Discussion
A comparison was made between the molecular structures of 33 transgenic proteins and those of allergenic proteins by alignment of their amino acid sequences obtained from public protein databanks. This comparison yielded 83 identical stretches of at least six contiguous amino acids length in 22 transgenic proteins. These results confirm previous reports by Gendel [8] and Hileman et al. [10] in which identical stretches of at most seven amino acids between a limited number of transgenic proteins and allergenic proteins were found. For many of these stretches, it remains unknown if they are true epitopes that bind IgE antibodies from sera of patients allergic to the specific allergen.
Table 5 lists four identical stretches that are assumed relevant based on at least one of the following criteria:
Table 5.
Identical sequences between transgenic- and allergenic proteins of special interest
| Transgenic protein (1) | Identical peptide | Allergens (2) | Remark |
| Insecticidal protein Cry1Ac (Bacillus thuringiensis kurstaki HD-73) | GNAAPQ GSTGITI | Cedar pollen allergens Cup a 1, Jun a 1, Jun o 1, Juniperus virginiana 1–1, and Juniperus virginiana 1–2 | Two sequences shared with same allergens, potential crosslinking of IgE if bound by both sequences |
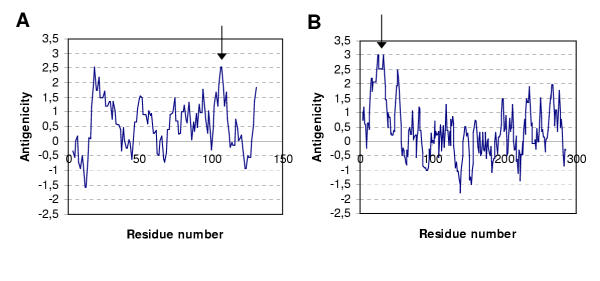
| Papaya Ringspot Virus coat protein | EKQKEK | Nematode allergen ABA-1 (Ascaris suum, A. lumbricoides) | Sequence predicted to be antigenic determinant of allergenic protein (Figure 4) |
| Acetolactate synthase (GH50 mutant, Arabidopsis thaliana) | KVLENR (3) | Shrimp allergen Met e 1 Lobster allergens Hom a 1 and Pan s 1 Crab allergen Cha f 1 | KVLENR is part of 15-mer peptides that are recognised by sera from allergic patients [28,29] |
| Glyphosate oxidoreductase (Achromobacter LBAA) | LAEEAD | Shrimp allergen Met e 1 Lobster allergens Hom a 1 and Pan s 1 Crab allergen Cha f 1 Fish parasite allergen Ani s 3 | LAEEAD is part of 9-mer peptide from the shrimp tropomyosin allergen Pen i 1 that is bound by sera from shrimp allergic patients [26,27] (4) |
(1) Accessions: Cry1Ac: gi117547; PRV CP: gi593497; ALS: gi124372; GOX: gi1252836 (2) Accessions: Cup a 1: gi19069497, gi9087167, gi6562326; Jun a 1: gi9087152, gi4138877, gi4138879; Jun o 1: gi15139849; Juniperus virginiana major pollen allergens 1–1 and 1–2: gi8843917, gi8843921; ABA-1: gi2735108, gi2735110, gi2735112, gi2735114, gi2735116, gi2735118, gi2970629, gi7494507; Met e 1: gi607633, gi6094504; Hom a 1: gi14285796; Pan s 1: gi3080761, gi14285797; Cha f 1: gi7024506, gi14285800; Ani s 3: gi14423976 (3) Sequence KVLENR immediately flanks the highest peak in the antigenicity plot of acetolactate synthase. This peak is located between the arginine residu (KVLEN R) and the adjacent C-terminal residue (4) Sequence LAEEAD is also part of two 15-mer peptides from the shrimp allergen Pen a 1 tested for sera binding. One peptide is bound by sera from allergic individuals, whereas the other peptide is not [28].
• Predicted antigenicity within the allergenic protein indicating potential binding of the stretch by IgE from allergic patients.
• Binding of IgE to peptides containing the identical stretch as reported by literature.
• Sharing of two or more stretches of identity by a transgenic protein with an allergenic protein. In the "worst case" scenario, these stretches are true IgE-epitopes and can therefore bind at least two IgE molecules on the surface of mast cells in allergic individuals. Such "cross-linking" of IgE is known to trigger the release of histamine and cytokines from the mast cells, leading to anaphylaxis.
Cry1Ac, for example, shares two identical peptides, GNAAPQ and GSTGITI with cedar pollen allergens. Hopp and Wood's prediction method does not indicate, however, pronounced antigenicity for the GNAAPQ sequence in the cedar pollen allergens and yields a negative score for the GSTGITI sequence. It therefore appears that no further testing would be needed. In contrast, the peptide EKQKEK shared by Papaya Ringspot Virus coat protein with nematode allergens can be classified as probably antigenic based on the same prediction method (Figure 4). For the EKQKEK sequence, no further data have been found on the potential IgE-binding. Confirmation of IgE binding to peptides containing the identical stretch would therefore be the next phase in the proposed strategy (Figure 1). Finally, literature reports describe the binding of sera from shrimp-allergic patients to peptides containing the KVLENR sequence of transgenic acetolactate synthase and the LAEEAD sequence of glyphosate oxidoreductase, which are shared with tropomyosin allergens from various organisms. From these literature reports, it also became apparent that not all tropomyosins (e.g., Pen a 1, Pen i 1) containing these identical sequences had been retrieved from the protein database during the alignment. The fact that the KVLENR and LAEEAD sequences are part of sequences that have been shown to react with patients' sera warrants further clinical investigation into the potential allergenicity of the transgenic proteins (Figure 1). This would include the screening of binding of sera from allergic patients to the transgenic proteins.
Figure 4.
Examples of antigenicity plots created with the Hopp and Woods method, window size six amino acid A Antigenicity plot for one of the ABA-1 allergen proteins from the nematode Ascaris lumbicoides that share the peptide sequence EKQKEK (arrow) with the transgenic protein Papaya Ringspot Virus coat protein. B Antigenicity plot for the transgenic protein Papaya Ringspot Virus coat protein. The sequence EKQKEK is part of a plateau slightly below the two highest peaks (arrow)
In short, twenty-two transgenic proteins were found to have identical stretches in common with allergenic proteins. Merely two proteins (glyphosate oxidoreductase, acetolactate synthase) of these twenty-two proteins contain identical stretches that may be IgE binding epitopes according to literature (Table 5). For the other twenty transgenic proteins, either no or negative indications for IgE binding of the identical stretches could be found in literature, while for one of these proteins (Papaya Ringspot Virus coat protein), the calculated point of highest antigenicity of the allergenic protein coincided with the identical stretch. The minimum length of six amino acids was chosen for this study following the recommendation made by a recent FAO/WHO Expert Consultation. This consultation recommended that transgenic proteins with positive outcomes in the alignment procedure should be considered likely allergenic [4]. This item is currently discussed within FAO/WHO Codex alimentarius in preparation of guidelines for the risk assessment of foods derived through biotechnology. The results of this study indicate that, if the recommended six-amino-acids threshold is applied, the outcomes of sequence alignments of transgenic proteins to allergenic proteins may not be conclusive about potential allergenicity. The six-amino-acids threshold therefore reflects a precautionary approach.
Our results extend previous observations made by Hileman et al. [10], who investigated the sequence similarities that transgenic proteins originating from Bacillus thuringiensis, non-allergenic proteins, and endogenous maize proteins shared with allergenic proteins. Hileman et al. [10] concluded among others that a threshold size of six amino acids will not distinguish allergenic from non-allergenic proteins and recommended to set a minimum threshold of eight amino acids in order to reduce the number of false positives. Interestingly, the eight-amino-acid threshold proposed by Hileman et al. [10] is consistent with the recommendation made by ILSI/IFBC in 1996 in their decision tree approach, which has since then been internationally recognised by GM food safety assessors. In this study, we propose an alternative approach to reduce false positives by identification of potential IgE binding epitopes among the identical stretches identified during the sequence alignment of transgenic proteins with allergenic proteins (Figure 1). This alternative approach allows to search for identical stretches with a minimum length of six amino acids, which is sufficient for some IgEs to bind. In this respect it is noteworthy that the two identical stretches LAEEAD and KVLENR, which have been identified in this study as potential IgE epitopes based on literature data (Table 5), would have been missed if the eight-amino-acids threshold were applied. Care should therefore be taken not only to reduce false positives, but also to reduce the likelihood of false negatives in further refinement of methods to screen for potential IgE epitopes in transgenic proteins.
For further refinement, additional criteria may be employed. One example of an additional criterion is the "foreignness", i.e. the non-similarity, of a protein of interest compared to human proteins. The underlying theory is that the less similar the studied protein is to human proteins, the more likely it represents an allergen [22]. This approach appears to be applicable to overall structures of transgenic- and allergenic-proteins. However, application of this approach to potential linear IgE epitopes in transgenic proteins may create false negatives, because in theory, human proteins may contain single IgE epitopes without eliciting clinical symptoms.
Another criterion would be the "similarity" of peptide sequences with certain permissible amino acid substitutions. This criterion is more flexible than the current requirement for identicalness of peptide sequences. It has been observed that IgE binding to peptides carrying linear IgE epitopes of the shrimp allergen Pen a 1 was not impaired, and in some cases even enhanced by various specific substitutions of amino acids within these peptides [22,23].
Conclusions
Internet-hosted facilities allow the genetic engineer to screen transgenic proteins for the presence of linear epitopes of allergenic proteins. These facilities include the alignment of protein sequences by using the Protein BLAST and prediction of the antigenicity of peptide sequences by the Hopp and Woods method. It should be noted that, for transgenic proteins from host organisms without a history of allergenicity, the search for sequence identity with allergenic proteins will be one of the first steps in the assessment of the potential allergenicity. Based on the outcome of this search, further steps may be required to assess the potential allergenicity. As shown by the results of this investigation, many transgenic proteins have six- and seven-amino acid stretches in common with allergenic proteins. If the threshold of six contiguous amino acids would be lowered to five or four amino acids, the number of outcomes can be expected to increase substantially over the present output. Many of these outcomes, however, can be expected to be "false positives". Antigenicity prediction methods, such as the Hopp and Woods method, may reduce the number of false positives.
Alternatively, the transgenic protein sequence can be aligned directly with the sequences of known linear epitopes of allergens such that false positives will be precluded from the outcome. For this purpose, a database with linear epitopes would be helpful, but still needs to be constructed. In addition, supplementary methods are needed for the prediction of conformational epitopes and glycan-containing epitopes. In cases where multiple potential epitopes have been identified within a transgenic protein, methods to estimate the protein's ability to cross-link IgE molecules on mast cell surfaces would enable prediction of allergic reactions due to mast cell stimulation by the particular protein.
Methods
Transgenic protein sequences
The procedure applied for this study is summarised in Figure 2. Sequences of transgenic proteins expressed in market-approved genetically modified crops could be retrieved from protein databases hosted on the Internet (Table 1). The sequence of the Potato Virus Y coat protein has been obtained from the literature [30]. Sequences from the Cry2Ab, Cry3Aa, and Cry3Bb proteins, which are present in pre-commercial crops, have also been included. Transgenic proteins that are mutants of host proteins, such as maize EPSPS expressed in GM maize, as well as hypothetical proteins that could arise from engineered antisense genes have been excluded from this investigation. For Genbank accession numbers of transgenic proteins and data on truncations and amino acid substitutions of certain proteins, see additional file 1.
Sequence alignment
Alignments of the transgenic sequences with sequences of allergenic proteins were carried out with the BLAST tool to search "short nearly exact matches" on the NCBI website http://www.ncbi.nlm.nih.gov/BLAST/, while limiting the aligned sequences using the limit query "allergen". It should be noted, however, that many, but not all, allergens will be retrieved by the query limit "allergen". In addition, some proteins retrieved by this query limit may not be true allergens, such as allergen binding antibodies or sequences that resemble those of allergens. These non-allergenic proteins should not be further considered.
The search has not been limited to food allergens, as other types of allergens may also be relevant. Some aeroallergens, for example, are cross-reactive with food allergens (e.g. birch pollen and apple, respectively). Moreover, next to food consumption, inhalation is another route of exposure to a genetically modified crop, such as through pollen and dust from crop processing.
Antigenicity prediction
Antigenicity prediction plots have been created with the graphic interface on the Colorado State University's website (Table 2) for the sequences of the transgenic protein and the allergenic protein that share the identical peptides according to the Hopp and Woods method, using a window size of six amino acids [14]. Additional Internet facilities where this calculation can be run are listed in Table 2.
Literature search
Literature has been checked for data on IgE epitopes in allergenic proteins that might coincide with the identical peptides that were identified in the alignment. For that purpose, PubMed, an on-line version of the medical bibliography Medline, has been used to explore literature references http://www.ncbi.nlm.nih.gov/entrez/query.fcgi. In addition, information on allergenic proteins, including literature references, is provided by a number of on-line databases (Table 3).
Authors' contributions
Author GK carried out the sequence alignment, antigenicity prediction, literature search, and participated in manuscript drafting. Author AP reviewed the methodology, analysed the results, and participated in manuscript drafting
Supplementary Material
This Annex gives a detailed account of the method and the results in table format. The alignments of six or more identical amino acids between transgenic- and allergenic-proteins are listed, together with the outcomes of the Hopp and Woods antigenicity prediction method and literature search on linear IgE epitopes.
Acknowledgments
Acknowledgements
The authors gratefully acknowledge financial support from the Ministry of Agriculture, Nature Management and Fisheries, scientific programs 378 and 390.
Contributor Information
Gijs A Kleter, Email: g.a.kleter@rikilt.wag-ur.nl.
Ad ACM Peijnenburg, Email: a.a.c.m.peijnenburg@rikilt.wag-ur.nl.
References
- James C. Global Review of Commercialized Transgenic Crops: 2001. Ithaca, International Service for the Acquisition of Agri-biotech Applications. 2001. http://www.isaaa.org/publications/briefs/Brief_24.htm
- Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit Rev Food Sci Nutr. 1996;36:S165–S186. doi: 10.1080/10408399609527763. [DOI] [PubMed] [Google Scholar]
- FAO/WHO Draft Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants (ALINORM 01/34A). Rome, Codex Alimentarius Committee, Food and Agriculture Organisation of the United Nations. 2002. http://www.who.int/fsf/GMfood/codex_index.htm
- FAO/WHO Joint FAO/WHO Expert Consultation on Foods Derived from Biotechnology – Allergenicity of Genetically Modified Foods – Rome, 22 – 25 January 2001. Rome, Food and Agriculture Organisation of the United Nations. 2001. http://www.fao.org/es/esn/food/pdf/allergygm.pdf
- FAO/WHO Report of the Working Group 12 September 2001, Ad Hoc Open-Ended Working Group on Allergenicity, Vancouver, 10–12 September 2001, Hosted by the Government of Canada. Rome, Food and Agriculture Organisation of the United Nations. 2001. ftp://ftp.fao.org/codex/ccfbt3/wgreport_e.pdf
- Oehlschlager S, Reece P, Brown A, Hughson E, Hird H, Chisholm J, Atkinson H, Meredith C, Pumphrey R, Wilson P, Sunderland J. Food allergy – towards predictive testing for novel foods. Food Addit Contam. 2001;18:1099–1107. doi: 10.1080/02652030110050131. [DOI] [PubMed] [Google Scholar]
- Taylor SL. Protein allergenicity assessment of foods produced through agricultural biotechnology. Annu Rev Pharmacol Toxicol. 2002;42:99–112. doi: 10.1146/annurev.pharmtox.42.082401.130208. [DOI] [PubMed] [Google Scholar]
- Gendel SM. The use of amino acid sequence alignments to assess potential allergenicity of proteins used in genetically modified foods. Adv Food Nutr Res. 1998;42:45–62. doi: 10.1016/s1043-4526(08)60092-3. [DOI] [PubMed] [Google Scholar]
- Becker WM. Sequence homology and allergen structure (Topic 4). Joint FAO/WHO Expert Consultation on Foods Derived from Biotechnology – Allergenicity of Genetically Modified Foods – Rome, 22 – 25 January 2001. Rome, Food and Agriculture Organisation of the United Nations. 2001. http://www.fao.org/es/esn/food/pdf/bi06al.pdf
- Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, Astwood JD, Hefle SL. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int Arch Allergy Immunol. 2002;128:280–291. doi: 10.1159/000063861. [DOI] [PubMed] [Google Scholar]
- Kolaskar AS, Kulkarni Kale U. Prediction of three-dimensional structure and mapping of conformational epitopes of envelope glycoprotein of Japanese Encephalitis Virus. Virology. 1999;261:31–42. doi: 10.1006/viro.1999.9859. [DOI] [PubMed] [Google Scholar]
- Garcia Casado G, Sanchez Monge R, Chrispeels MJ, Armentia A, Salcedo G, Gomez L. Role of complex asparagine-linked glycans in the allergenicity of plant glycoproteins. Glycobiology. pp. 471–477. [DOI] [PubMed]
- Pellequer JL, Westhof E, Van Regenmortel MHV. Predicting location of continuous epitopes in proteins from their primary structures. Methods Enzymol. 1991;203:176–201. doi: 10.1016/0076-6879(91)03010-e. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. CABIOS. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- Vuento M, Paananen K, Vihinen Ranta M, Kurppa A. Characterization of antigenic epitopes of potato virus Y. Biochim Biophys Acta. 1993;1162:155–160. doi: 10.1016/0167-4838(93)90142-E. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel MH, Pellequer JL. Predicting antigenic determinants in proteins: looking for unidimensional solutions to a three-dimensional problem? Pept Res. 1994;7:224–228. [PubMed] [Google Scholar]
- Hopp TP. Protein surface analysis. Methods for identifying antigenic determinants and other interaction sites. J Immunol Meth. 1986;88:1–18. doi: 10.1016/0022-1759(86)90045-1. [DOI] [PubMed] [Google Scholar]
- Smith AM, Chapman MD. Localization of antigenic sites on Der p 2 using oligonucleotide-directed mutagenesis targeted to predicted surface residues. Clin Exp Allergy. 1997;27:593–599. doi: 10.1046/j.1365-2222.1997.460852.x. [DOI] [PubMed] [Google Scholar]
- Adams SL, Barnett D, Walsh BJ, Pearce RJ, Hill DJ, Howden MEH. Human IgE-binding synthetic peptides of bovine β-lactoglobulin and α-lactalbumin. In vitro cross-reactivity of the allergens. Immunol Cell Biol. 1991;69:191–197. doi: 10.1038/icb.1991.28. [DOI] [PubMed] [Google Scholar]
- Ganglberger E, Sponer B, Scholl I, Wiedermann U, Baumann S, Hafner C, Breiteneder H, Suter M, Boltz Nitulescu G, Scheiner O, Jensen Jarolim E. Monovalent fusion proteins of IgE mimotopes are safe for therapy of type I allergy. FASEB J. 2001;15:2524–2526. doi: 10.1096/fj.00-0888fje. [DOI] [PubMed] [Google Scholar]
- Lehrer SB, Ayuso R, Reese G. Current understanding of food allergens. Ann N Y Acad Sci. 2002;964:69–85. doi: 10.1111/j.1749-6632.2002.tb04133.x. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Reese G, Leong Kee S, Plante M, Lehrer SB. Molecular basis of arthropod cross-reactivity: IgE-binding cross-reactive epitopes of shrimp, house dust mite and cockroach tropomyosins. Int Arch Allergy Immunol. 2002;129:38–48. doi: 10.1159/000065172. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18:1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- McReynolds LA, Kennedy MW, Selkirk ME. The polyprotein allergens of nematodes. Parasitol Today. 1993;9:403–406. doi: 10.1016/0169-4758(93)90046-I. [DOI] [PubMed] [Google Scholar]
- Shanti KN, Martin BM, Nagpal S, Metcalfe DD, Rao PV. Identification of tropomyosin as the major shrimp allergen and characterization of its IgE-binding epitopes. J Immunol. 1993;151:5354–5463. [PubMed] [Google Scholar]
- Subba-Rao PV, Rajagopal D, Ganesh KA. B- and T-cell epitopes of tropomyosin, the major shrimp allergen. Allergy. 1998;53:44–47. doi: 10.1111/j.1398-9995.1998.tb04959.x. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Lehrer SB, Reese G. Identification of continuous, allergenic regions of the major shrimp allergen Pen a 1 (tropomyosin). Int Arch Allergy Immunol. 2002;127:27–37. doi: 10.1159/000048166. [DOI] [PubMed] [Google Scholar]
- Reese G, Ayuso R, Carle T, Lehrer SB. IgE-binding epitopes of shrimp tropomyosin, the major allergen Pen a 1. Int Arch Allergy Immunol. 1999;118:300–301. doi: 10.1159/000024108. [DOI] [PubMed] [Google Scholar]
- Lawson C, Kaniewski W, Haley L, Rozman R, Newell C, Sanders P, Tumer NE. Engineering resistance to mixed virus infection in a commercial potato cultivar: resistance to potato virus X and potato virus Y in transgenic Russet Burbank. Biotechnology. 1990;8:127–134. doi: 10.1038/nbt0290-127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This Annex gives a detailed account of the method and the results in table format. The alignments of six or more identical amino acids between transgenic- and allergenic-proteins are listed, together with the outcomes of the Hopp and Woods antigenicity prediction method and literature search on linear IgE epitopes.