Abstract
Background
Chlamydiae are obligate intracellular bacteria, which are important human pathogens. Genome sequences of C. trachomatis and C. pneumoniae have revealed the presence of a Chlamydia specific gene family encoding polymorphic outer membrane proteins, Pmps. In C. pneumoniae the family comprises twenty-one members, which are all transcribed. In the present study, the expression, processing and localisation of the sixteen full-length Pmps in C. pneumoniae strain CWL029 have been further investigated by two-dimensional gel electrophoresis and immunofluorescence microscopy.
Results
Ten Pmps were identified in elementary bodies (EBs). Eight of these were investigated with respect to time dependent expression and all were found to be up-regulated between 36 and 48 hours post infection. Antibodies against Pmp6, 8, 10, 11 and 21 reacted with chlamydiae when infected cells were formalin fixed. Pmp6, Pmp20 and Pmp21 were found in cleaved forms, and the cleavage sites of Pmp6 and Pmp21 were identified.
Conclusions
The Pmps are heavily up-regulated at the time of conversion of RB to EB, and at least ten Pmps are present in EBs. Due to their reaction in formalin fixation it is likely that Pmp6, 8, 10, 11 and 21 are surface exposed. The identified cleavage sites of Pmp6 and Pmp21 are in agreement with the theory that the Pmps are autotransporters.
Background
Chlamydiae are pathogenic gram-negative bacteria of which C. pneumoniae causes upper and lower respiratory tract infections in humans [1]. Going through a developmental cycle the chlamydiae alternate between infective elementary bodies (EBs) and replicative reticulate bodies (RBs) [2]. The bacteria are obligate and intracellular, residing inside a specialized phagosome, named the chlamydial inclusion. The duration of the developmental cycle for C. pneumoniae cultivated in cell culture is about 72 hours [3].
The C. pneumoniae CWL029 genome sequence revealed the presence of a gene family, the pmp family, consisting of 21 members [4] that were paralogous to the pmps found in C. trachomatis [5] and C. psittaci [6-9]. The C. psittaci Pmps have been analysed by two-dimensional electrophoresis in an earlier study [37]. The Pmps are two-domain proteins with similarity to autotransporter proteins [4,10,11]. They are characterized by a high frequency of the two sequences FxxN and GGAI in the N-terminal part (twelve and seven repeats on average, respectively) [4], and their C-terminal part shows the characteristics of a β-barrel [10]. The GGAI motif and the prediction of a C-terminal β-barrel suggest that the Pmps are autotransporters, transporting an N-terminal passenger domain through a pore formed by their C-terminal part [10,12]. Seventeen C. pneumoniae Pmps contain a signal peptidase I cleavage site directing transport over the inner membrane, and two Pmps contain a signal peptidase II cleavage site suggesting lipid modification [4].
The Mw of most Pmps in C. pneumoniae strain CWL029 is predicted to be just below 100 kDa, but three are larger: Pmp6 is 142 kDa, Pmp20 is 178 kDa and Pmp21 is 167 kDa and Pmp12 is only 56 kDa. Four genes (pmp3, 4, 5 and 17) contain a mutation resulting in premature stop. In C. pneumoniae strain AR39 [13] and J138 [14]pmp6 contains 393 less base pairs, and in strain J138 Pmp2 and Pmp4 contain frame shift mutations [14].
Variation of membrane protein expression is thought to provide protection against the immune system of the host [15] and it has been suggested that the Pmps may provide such a protection of chlamydiae. This was indicated by the findings of Birkelund et al. [16] and Pedersen et al. [17] who observed differential expression of Pmp10.
Transcripts have been detected from all Pmp genes in C. trachomatis [18] as well as C. pneumoniae [19]. As part of a large-scale proteome analysis [20], ten Pmps (Pmp2, 6, 7, 8, 10, 11, 13, 14, 20 and 21) or fragments of these were identified by mass spectrometry (MS) in C. pneumoniae CWL029 EBs. The positions of these in the 2-D protein profile can be viewed at http://www.gram.au.dk. In addition to these, Montigiani et al. [21] identified Pmp16 by MS. Grimwood et al. [19] found Pmp2, 6, 7, 8, 9, 11, 13, 14, 15, 16, and 18 to be present in C. pneumoniae CWL029 EBs by immunoblotting (IMB) using antibodies raised against synthetic 20-mer peptides. Pmp1, 19, 20 and 21 were not detected although full-length genes encode these. Pmp6 migrated around 100 kDa although it theoretically should be 144 kDa.
The detection of Pmp6 forty kDa below the predicted value has been interpreted by Henderson and Lam [10] as an indication that an N-terminal passenger domain is cleaved off, which is frequently seen for autotransporters. In strains AR39 and TW183, sharing the 393 base pair deletion, Pmp6 was detected at the resulting theoretical full-length position of 130 kDa by Grimwood et al. [19].
In the study described in this paper, 2-D PAGE was used to elucidate the expression pattern of the C. pneumoniae Pmps in CWL029 at different times during the developmental cycle. Pmp6, Pmp20 and Pmp21 were found to be cleaved and the cleavage sites of Pmp6 and Pmp21 were identified. IMF was used to determine whether Pmps could be detected at the surface of the bacteria.
Results
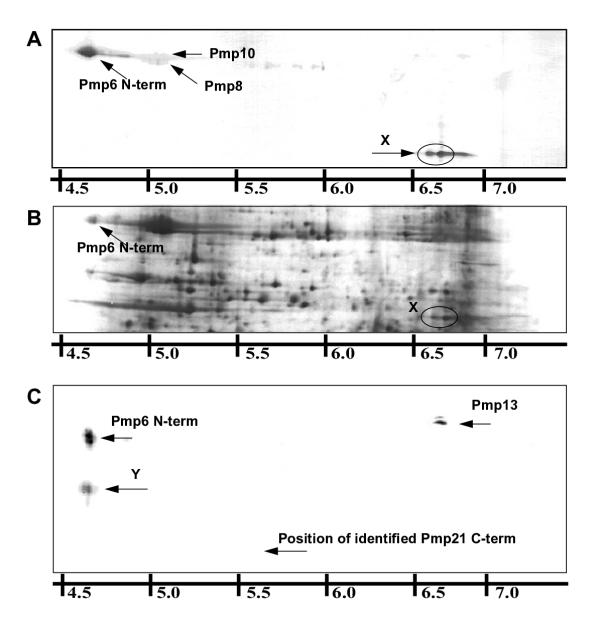
The proteome study by Vandahl et al. [20] was not conclusive on the absence of any Pmps. Hence, we performed 2-D immnoblotting (IMB) to identify the 2-D PAGE position of expressed Pmps not earlier mapped. Pmp3, 4, 5, 12 and 17 were omitted from the study because these are truncated. Polyclonal antibodies (pAbs) were obtained by immunization of rabbits with recombinant proteins produced in E. coli (table 1). Antigens were raised against the N-terminal part of Pmp6, 7, 8, 10, 13, 14, 15, 16, 18, 19, 20 and 21 because antibodies raised against full length Pmps showed cross-reactivity. The N-terminal part is the most variable and has been detected at the surface for C. psittaci Pmps [9]. All the obtained pAbs reacted in 1-D IMB with the recombinant Pmps to which they were raised (data not shown). The antigen for IMB was EB lysate. Reacting spots (figure 1A) were identified by comparison to a reference gel upon autoradiography of the blot (figure 1B) [22] or by MS analysis of spots excised from corresponding gels.
Table 1.
Overview of Pmps, antibodies and immunoblot reactions
| pAb | Pmp | Cl. | Fam. | L | aa | cloned | MS | G | M | IMB | MeOH | Form. |
| 204 | 1 | I | G | I | 923 | 26–923 | - | - | I | 1?2 6 78 10 13 14 20 21 | X | - |
| 207 | 2 | I | G | I | 842 | 20–842 | X | S | M | 2 6 8 10 | X | - |
| - | 3* | I | G | I | ||||||||
| - | 4* | I | G | I | ||||||||
| - | 5* | I | G | I | ||||||||
| 221 | 6 | II | G | I | 1408 | 18–905 | N+C | S | M | 6 8 10X | X | X |
| 220 | 7 | II | G | I | 937 | 25–545 | X | S | 2 6 7 8 10 11 13 | X | - | |
| 201 | 8 | II | G | I | 931 | 21–931 | X | S | M | 8 10 | X | X |
| 208 | 9 | II | G | I | 929 | 27–929 | - | S | I | 2 6 7 8 9? 10 11 13 14 20 | X | u |
| 203 | 10 | II | G | II | 929 | 20–929 | X | - | M | 10 | X | X |
| 195 | 11 | II | G | II | 929 | 1–929 | X | W | M | 6 11 | X | X |
| - | 12# | II | - | I | ||||||||
| 222 | 13 | II | G | I | 974 | 20–542 | X | S | M | not performed | X | - |
| 228 | 14 | II | H | I | 979 | 25–492 | X | S | M | not performed | X | - |
| 229 | 15 | III | EF | I | 939 | 18–478 | - | W | I | - | - | - |
| 230 | 16 | III | EF | N | 935 | 1–472 | - | S | M | 8 10 | - | - |
| - | 17* | III | EF | I | ||||||||
| 231 | 18 | III | EF | N | 893 | 1–566 | - | P | 8 10 | - | - | |
| 234 | 19 | IV | A | I | 948 | 22–501 | - | - | - | - | - | |
| 236 | 20 | IV | BC | I | 1724 | 22–1272 | X | - | M | 20 13 | - | - |
| 237 | 21 | D | I | 1610 | 52–1129 | N+C | - | M | 6 13Y | X | X |
pAb: Antibody number; Pmp: Pmp number: *, genes containing a premature stop; #, naturally short gene; shading indicates either * or #. Cl.: Cluster in the C. pneumoniae genome.L: Predicted leader sequences: I, signal peptidase I cleavage site; II, signal peptidase II cleavage site; -, no predicted cleavage site. Fam.: Closest related C. trachomatis Pmp. aa: Amino acids in full-length Pmp. cloned: First and last amino acid in the recombinant protein. MS: Mass spectrometry identification by: X, peptides dispersed along the entire sequence; N, peptides localized to the N-terminal part; C, peptides localized to the C-terminal part; -, not identified by MS. G: Detection in 1D IMB by Grimwood et al.: S, strong reaction; W, weak reaction; P, possible reaction; -, no reaction. M: Detection by Montigiani et al. [24]: M, MS and immunoblot identification; I, immunoblot detection alone. IMB: Immunoblotting results: bold numbers indicate strong reaction; X is a C-terminal fragment of Pmp6; Y is an N-terminal fragment of Pmp21; results marked ? were inconclusive. MeOH: MeOH fixation IMF at 72 hpi: X, reaction; -, no reaction. Form.: Formalin fixation IMF at 72 hpi: X specific reaction; u, unspecific reaction; -, no reaction.
Figure 1.
2-D immunoblotting of EB proteins Radiolabeled proteins from EBs purified 72 hpi were separated by 2-D PAGE and electroblotted onto PVDF membranes. The membranes were reacted with polyclonal rabbit antibodies raised against recombinant Pmp. Bound antibodies were visualized by color reagents and membranes were afterwards exposed to X-ray films so that reacting spots could be identified by comparison to an annotated gel. A: PVDF blot of EB proteins reacted with Pmp6-pAb221, X shows a C-terminal fragment of Pmp6 recognized by Pmp6-pAb221; B: Autoradiography of A; C: PVDF blot of EB proteins reacted with pAb237-rPmp21, Y shows an N-terminal fragment of Pmp21 recognized by pAb237-rPmp21.
The proteins detected by each antibody are listed in table 1 using bold for strong reaction and plain for weaker reaction. PAb203-rPmp10 was mono-specific for Pmp10. The antibodies against rPmp2, rPmp7, rPmp8 and rPmp11 recognized these Pmps (respectively), and additional Pmps as listed in table 1. PAb221-rPmp6 recognized the 91 kDa spot that was earlier identified as originating from Pmp6 [20] and in addition another Pmp6 spot at 60 kDa marked X in figure 1A. PAb236-rPmp20 recognized a spot located just above the 90 kDa Pmps, which was earlier identified as originating from Pmp20 [20]. PAb237-rPmp21 recognized the 91 kDa Pmp6 spot, Pmp13 and a 48 kDa Pmp21 fragment marked Y in figure 1C. The results with antibodies against rPmp1 and rPmp9 were inconclusive and MS identification of these Pmps could not be obtained. Antibodies against rPmp16 and rPmp18 both reacted with Pmp8 and Pmp10 but no other proteins. Antibodies against rPmp15 and rPmp19 did not react with any EB proteins. We were thus not able to detect Pmp 15, 16, 18 and 19 by IMB using EB lysates as antigen.
Identification of cleaved Pmps
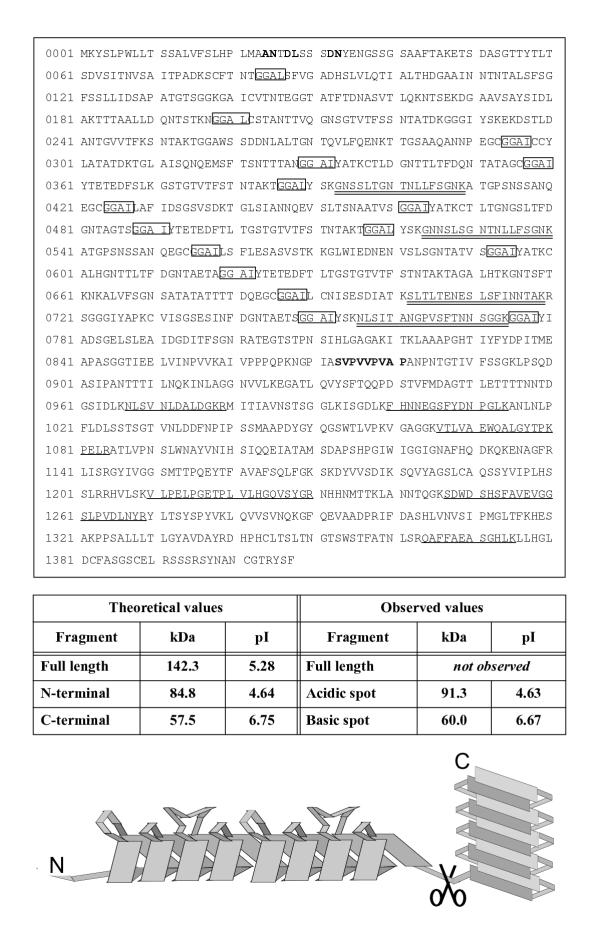
The antibodies against the high molecular weight Pmps (Pmp6, Pmp20 and Pmp21) recognized protein spots at lower molecular weight than expected. PAb221-rPmp6 recognized a set of spots, X, at 60 kDa (figure 1A) in addition to the earlier identified 91 kDa fragment of Pmp6. MS analysis of a tryptic digest of X revealed peptides from the C-terminal part of Pmp6 (singly underlined peptides, figure 2). In contrast, the mass spectra from the Pmp6 spot at 91 kDa was found to contain peptides solely from the N-terminal part of Pmp6 (doubly underlined peptides, figure 2). To verify the identifications, gels were electroblotted onto PVDF membranes and N-terminal amino acid sequences of the proteins were assessed by Edman degradation. The sequence obtained from X (xVPVVPVAP) confirmed that it was a C terminal fragment of Pmp6 as it matched an internal sequence of Pmp6 (872SVPVVPVAP) (figure 2). The sequence obtained from the N-terminal fragment (xNTDLxSSD) matched a sequence of Pmp6 (24ANTDLSSSD) that confirmed cleavage at the signal peptidase I cleavage site predicted by Signal-P [23,24] (figure 2). As seen in figure 2, these cleavage sites explain the observed pI and Mw coordinates. Also in figure 2, a model of Pmp6 is shown where the internal cleavage site is mapped relative to the predicted domains.
Figure 2.
Pmp6 fragments Peptides identified from the acidic protein spot by MS are doubly underlined. Peptides identified from the basic spot are singly underlined. Amino acids from N-terminal sequences obtained by Edman degradation of the fragments are shown in bold. GGAI/L repeats are boxed. Notice that all repeats are located N-terminally of the cleavage site. Theoretical pI and Mw values of fragments were calculated by the Compute pI/Mw tool http://www.expasy.ch/tools/pi_tool.html. A schematic drawing of the predicted domains is shown at the bottom, the pair of scissors representing the internal cleavage site.
PAb237-rPmp21 reacted with a novel spot pattern, Y, at 66 kDa (figure 1C). A tryptic digest of Y was found to contain solely N-terminal peptides of Pmp21 by MS analysis (amino acids 122–130, 181–202, 203–213, 251–268, 493–510, and 530–548). The earlier identified Pmp21 spot at 48 kDa [20] was found to contain solely C-terminal peptides (amino acids 1205–1223, 1251–1273, 1279–1289, 1295–1311, 1312–1326, 1315–1326, 1400–1417, 1447–1457, 1464–1483, 1465–1483, 1484–1494, 1502–1521, 1504–1521, 1525–1540, 1594–1606). No reaction was observed with the latter spot (figure 1C) explained by the fact that pAb237-rPmp21 was raised against a recombinant protein covering only the N-terminal part of Pmp21, up to amino acid 1129 (table 1). Edman degradation of the 48 kDa spot revealed a sequence matching an internal sequence of Pmp21 (1185SSPTPNKDKA) thereby identifying the cleavage site. Cleavage at this site results in a fragment of a theoretical Mw of 51 kDa, which is in good agreement with the observed Mw of 48 kDa. For the protein spot at 66 kDa containing the N-terminal peptides, cleavage at the signal peptidase cleavage site predicted by Signal-P was confirmed at the amino acid 30 (30AHSLHSSELD). The theoretical molecular weight of a fragment starting at this position and ending at the cleavage site at amino acid 1185 is 116 kDa. As no peptides were identified at the C-terminal side of amino acid 548 in the N-terminal fragment, further cleavage or degradation from the C-terminal end will be a likely explanation of the discrepancy between expected and observed values.
Pmp20 was identified just above the 90 kDa Pmps, but as no molecular weight markers were present above 90 kDa, the exact molecular weight of the protein cannot be deduced. However, it is found much lower than the expected 178 kDa and the earlier obtained MS identification of this protein spot was based on peptides located in the C-terminal part of Pmp20 (residues 1057–1069, 1387–1400, 1421–1432, 1506–1527, 1557–1571, 1587–1597, and 1644–1654) indicating that also Pmp20 is cleaved.
2-D PAGE analysis of time dependent expression
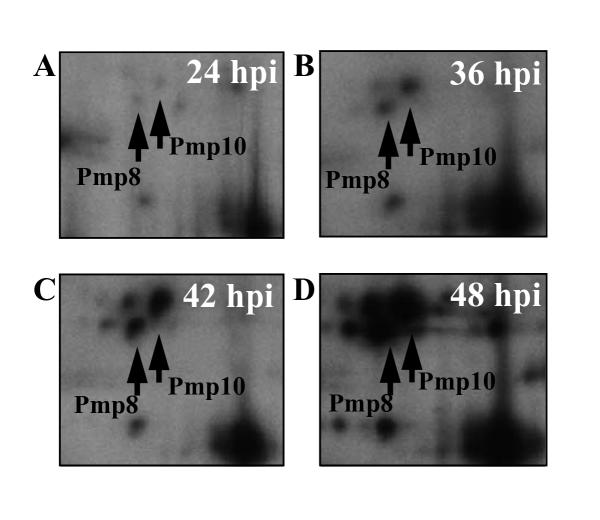
The time of expression of the Pmps identified in the 2-D protein profile was investigated by pulse labeling with [35S]methionine for two-hour-periods followed by 2-D PAGE and autoradiography. Samples were labeled at 12, 24, 36, 42 and 48 hours post infection (hpi). The earliest time for detection of Pmps was 24 hpi, at which time the highest abundant Pmps in the gels (Pmp8 and Pmp10) were only barely visible (figure 3A). The amount of Pmps expressed in a two-hour labeling period increased during the developmental cycle as illustrated for Pmp8 and Pmp10 in figure 3A,3B,3C,3D. In figure 4 the volumes of Pmp spots (Pmp2, 7, 8, 10, 11, 13, 14 and the C-terminal part of Pmp6) are plotted as percentages of the total spot volumes of the gels at 24, 36 and 48 hpi. The remaining identified Pmps were omitted from this analysis since they could not be confidently quantified due to overlapping spots. Spot volumes were calculated as density integrated over Gaussian area of spots visualized by autoradiography and detected by use of the Melanie II software. The volume of each spot was divided by the number of methionines of the protein. The corrected volume percentages are arbitrary, as the total spot volumes of the gels could not be corrected for methionine content of every protein present in the gel. However, the numbers do reflect the mutual proportion of the measured proteins and the time dependent change in their contribution to the total protein synthesis. Over time, the expression of all the Pmps in figure 4 increased from below 0.05 units at 24 hpi to between 0.15 and 1.35 units at 48 hpi. The most pronounced increase was from 36 to 48 hpi. Pmp10 was found to be expressed at the highest rate (1.5 units at 48 hpi) followed by Pmp8 (0.6 units at 48 hpi). It must be noted that the apparent level of expression is influenced by the properties of the proteins with respect to separation by 2-D PAGE. However, the Pmps must be expected to exhibit comparable behaviour.
Figure 3.
Visualization of Pmp8 and Pmp10 by two-hour labeling and 2-D PAGE Sections from autoradiographs of 2-D PAGE separated proteins from C. pneumoniae infected cells labeled with [35S]methionine by two-hour cultivation in a [35S]methionine medium added cycloheximide.
Figure 4.
Time dependent expression of Pmps Histogram showing the contribution of selected Pmps to the total amount of protein synthesized in a two-hour labeling period. Chlamydia proteins were selectively labeled with [35S]methionine under the addition of cycloheximide to stop host cell protein synthesis, separated by 2-D PAGE and visualized by autoradiography. Infected cells were labeled at 24, 36 and 48 hours post infection. Spots were detected by using the MelanieII software. Spot volumes were calculated as density integrated over Gaussian area, and the percentage of total spot volume in each gel was calculated. The percentages were corrected for the methionine content of each protein. The corrected spot volume percentages are given in arbitrary units at the Y-axis. Only Pmps located in the gel at positions where no overlapping protein spots hindered quantification are depicted. MOMP and DnaK are included as controls.
A parallel analysis of the expression of MOMP and DnaK was performed for comparison to the Pmps (figure 4). MOMP is expressed during the growth fase of Chlamydiae [25]. DnaK is an early protein [26] and has been suggested to be RB specific and hence to show decreased expression late in the developmental cycle [27]. The expression of MOMP was found to increase at the same rate from 24 to 36 hpi as from 36 to 48 hpi (figure 4). The expression of DnaK was similar to that of MOMP at 24 hpi, but showed no increase from 24 to 36 hpi and decreased from 36 to 48 hpi (figure 4).
It can be concluded that all Pmps investigated with respect to time dependent regulation (Pmp2, 6, 7, 8, 10, 11, 13, 14) were found to be strongly up-regulated between 36 and 48 hpi, coinciding with the conversion of RB to EB. This is in agreement with what has been reported for C. trachomatis and C. psittaci POMPS [28,29]. At 48 hpi Pmp8 and Pmp10 were several fold higher expressed than any other Pmp.
Immunofluorescence microscopy
In order to investigate whether the Pmps absent from EBs could be detected during growth in cell culture, the antibodies against these Pmps were used in IMF on C. pneumoniae infected HEp-2 cells. Cells cultivated on coverslips were infected and fixed in methanol at 72 hpi. The investigated antibodies were visualized with a FITCH conjugated secondary antibody. The antibodies against rPmp15, 16, 18 and 19, which did not react in IMB, did not react in IMF either (table 1). Furthermore, Pab236-rPmp20 was found not to react in IMF (table 1) although Pmp20 was identified in EB gels by IMB and MS. Examples are shown in figure 5.
Figure 5.
Immunofluorescence microscopy of C. pneumoniae infected HEp-2 cells Immunofluorescence microscopy pictures of infected HEp-2 cells fixed in methanol or 3.7% formaldehyde at 72 hours post infection (hpi). Bound antibody was detected with a FITCH conjugated secondary antibody. Fixed cells reacted with 1: Monoclonal antibody recognizing DnaK (MAb18.1), 2: pAb116-rMOMP, 3: pAb110-rOmp2, 4: pAb237-rPmp21, 5: pAb234-rPmp19. 6: Competition IMF. Reaction with pAb201-rPmp8 in the presence of: rPmp8: rPmp8, to which the antibody was raised, rPmp10: rPmp10, to which the antibody cross reacted in IMB, control: no recombinant proteins. As rPmp8 prevents reaction of pAb201-rPmp8 whereas rPmp10 does not affect the reaction, we conclude that the observed reaction is specific for Pmp8.
All Pmp antibodies were used in IMF of infected cells that were formalin fixed at 72 hpi. Formalin cross-linking of outer membrane proteins prevents antibodies from penetrating the bacteria so that only proteins in the outer membrane are accessible. MAb18.1-DnaK was included as a control of the impermeability as described earlier [30]. As the DnaK epitope recognized by MAb18.1 is formalin insensitive [30], the lack of reaction of MAb18.1 (figure 5, row 1) must be due to the prevention of reaction of interior proteins by the fixation.
The pAbs raised against recombinant Pmp1, 2, 7, 13, 14, 15, 16, 19 and 20 did not react after formalin fixation. Due to the described cross-reaction of the Pmp antibodies, the reacting antibodies were absorbed with recombinant proteins. The Pmp antibodies were absorbed with rPmps corresponding to the Pmps to which they cross-reacted in IMB and with the rPmps to which they were raised, respectively. The reaction of pAb208-rPmp9 could be prevented by addition of any of the Pmps 6, 8, 10, 11 or 21 (table 2), and hence it must be considered non-specific. The pAbs raised against rPmp6, 8, 11 and 21 were found to react specifically in formalin fixation (table 2), as their reaction was prevented by the recombinant Pmp that they were raised agains, but not by the ones to which they cross reacted in IMB. Pmp8 is shown as an example in figure 5 (6). Pab203-rPmp10 was not investigated by competition IMF as it was shown to be mono-specific for Pmp10 by IMB.
Table 2.
Competition immunofluorescence
| none | rPmp6 | rPmp11 | rPmp8 | rPmp9 | rPmp21 | rPmp7 | rPmp10 | |
| pAb221-rPmp6 | +++ | 0 | +++ | |||||
| pAb195-rPmp11 | +++ | + | 0 | |||||
| pAb201-rPmp8 | +++ | 0 | +++ | |||||
| pAb208-rPmp9 | ++ | 0 | 0 | 0 | 0 | 0 | ||
| pAb237-rPmp21 | +++ | +++ | 0 |
The results of competition IMF in which the recombinant Pmp listed in the top row was added to an IMF reaction using the polyclonal antibodies listed in the first column. The reactions were carried out only for the shaded table cells: 0, no reaction; +, weak reaction; ++, moderate reaction; +++, strong reaction.
Pmp10 [30,17] and Pmp11[30] have earlier been shown to be surface exposed and the results indicate that this is also the case for Pmp6, 8 and 21. Other Pmps may also be at the surface, but in that case the epitopes recognized by our antibodies have been destroyed by the fixation or in some way masked.
PAb116-rMOMP did not react after formalin fixation (figure 5, row 2), which is in agreement with earlier reports on the lack of surface exposed linear epitopes of C. pneumoniae MOMP [31]. Pab110-rOmp2 did react (figure 5, row 3) indicating the presence of surface exposed epitopes of Omp2 in C. pneumoniae in opposition to C. trachomatis where Omp2 is probably located at the inner surface of the outer membrane complex [32,33].
Discussion
In the present study, cleavage sites of Pmp6 and Pmp21 have been identified, and it is suggested that also Pmp20 is cleaved. Cleavage of the these Pmps explain why they are found at different molecular weight in different studies. Grimwood et al. [19] detected the N-terminal fragment of Pmp6 at 90 kDa that was also identified by Vandahl et al. [20], whereas Montigiani et al. [21] found the lower molecular weight C-terminal fragment of Pmp6. Only a C-terminal fragment of Pmp21 has been described previously [20,21].
There is no similarity in amino acid sequence between the cleavage sites in Pmp6 and Pmp21, but both sites are located between the C-terminal predicted β-barrel [10] and the N-terminal predicted parallel β-helix fold [34]. This position is in agreement with the theory that the Pmps may be autotransporters; as such often cleave off their N-terminal part [10]. However, if the N-terminal part is liberated it would not be detected in IMB using proteins from purified EBs as antigens, and as the N-terminal fragment is detected in EB gels for Pmp6 and Pmp21 it must be concluded that it remains bound to the EB.
Pab236-rPmp20 was raised against a fragment of Pmp20 covering the N-terminal part and some of the C-terminal fragment, but only the C-terminal part could be detected in IMB (data not shown), suggesting that the N-terminal part may be liberated. Pab236-rPmp20 did not react in IMF. It was raised against unfolded recombinant protein and may not recognize the correctly folded protein, especially not if the N-terminal part is liberated or degraded.
The fragments of Pmp6, Pmp20 and Pmp21 were detected after a labeling period of two hours, and no increase in the amount of cleavage products was observed after a chase period of six hours (data not shown). This suggests that the cleavage occurs rapidly after synthesis. We consider it unlikely that the cleavage should be an artificial phenomenon as the infected cells were harvested in a lysis solution containing 7 M urea, 2 M thiourea and reducing agents, and as the results were highly reproducible. The lack of detection of full-length products of the high molecular weight Pmps may be caused by low resolution in 2-D PAGE of such proteins, but full-length products of these were also absent in 1-D IMB by Grimwood et al. [19]. Interestingly, Grimwood et al. [24] detected Pmp6 from strains TW183 and AR39 at a Mw of 130 kDa, which is the theoretical Mw of Pmp6 in these strains, due to a deletion of the bases encoding amino acids 429 to 559, which does not include the cleavage site.
In the present study we did not detect any of the Pmps15-18, originating from cluster III. Pmp16 and Pmp18 are the only Pmps lacking a leader sequence. Pmp17 is truncated and was thus not investigated. Montigiani et al. [21] have reported identification of Pmp16 in C. pneumoniae from a clinical isolate, but Grimwood et al. [19] found that both Pmp16 and Pmp18 were unstable in CWL029, which may explain that these Pmps were not observed in our study. However, it could also be speculated whether differences have been introduced through laboratory passages. We observe expression of Pmp10 in strain CWL029 whereas it was not detected in strain CWL029 by Grimwood et al. [19] due to a frame shift mutation. Grimwood et al. [19] have described differences in the expression of Pmp1 and Pmp3 between strains CWL029 and TW183 as determined by immunoblotting, meaning that at least some Pmps are differentially expressed between strains.
Pmp6, Pmp8, Pmp10, Pmp11 and Pmp21 were detected at the surface of the bacteria by formalin fixation IMF. Apart from Pmp21, all the Pmps detected at the surface show greatest similarity to PmpG [4], a constituent of the outer membrane of C. trachomatis L2 [35,28]. An explanation of the expansion of the number of C. pneumoniae Pmps similar to PmpG could be that C. pneumoniae varies its surface to escape the immune defence of the host by changing the expression of Pmps. However, if Pmp expression is changed in order to vary the surface, this is most likely a consequence of the surface localization of Pmps rather than the very reason for the existence of Pmps.
The most highly expressed among all the investigated Pmps at all points in time is Pmp10, which is known to contain surface exposed epitopes [30]. Pmp10 is differentially expressed in infected cell culture [17] and mice [16]. Pmp10 still being the most highly expressed Pmp suggests that it carries out an important function. This function would presumably have to be supplemented in bacteria not expressing Pmp10. Pmp10 contains a predicted signal peptidase II cleavage site directing lipid modification and pmp11, which is located next to pmp10 (but in the opposite direction), encodes the only other predicted lipid modified Pmp. If the function of Pmp10 depends on lipid modification, and this function is needed in bacteria lacking Pmp10, Pmp11 would be the obvious alternative.
The finding that all Pmps are heavily upregulated at the time of conversion of RBs into EBs indicates that the function of these is structural but they may also be needed with respect to attachment or entry of EBs. An N-terminal triangular beta-layer motif could provide the bacteria with a shielding lattice and ensure proper spacing to a host cell or an epitope exposed to the complement system. If the lipid modifications of Pmp10 and Pmp11 are used as anchors inserted into the host cell membrane, subsequent action of other entry molecules would probably depend on proper spacing. However, all theories on functions of the Pmps remain very speculative.
Conclusions
The Pmps investigated with respect to time dependent regulation (Pmp2, Pmp6, Pmp7, Pmp8, Pmp10, Pmp11, Pmp13, and Pmp14) were found to be up-regulated late in the developmental cycle. Due to their reaction in formalin fixation we propose that Pmp6, Pmp8, Pmp10, Pmp11, and Pmp21 are surface exposed. Pmp6, Pmp20 and Pmp21 were found in cleaved forms and the identified cleavage sites of Pmp6 and Pmp21 are in agreement with the theory that the Pmps are autotransporters.
Methods
Organisms and cultivation
C. pneumoniae CWL029 (also known as VR1310) (ATTC) was cultivated in semi confluent monolayers of Hep-2 cells (ATCC) as described [20]. Elementary bodies were purified 72 hpi essentially as described [30] with the exception that Visipaque replaced Urografin for gradient making.
PCR and cloning
PCR enzymes were Expand™ High Fidelity (Roche) and reaction conditions were as recommended by the manufacturer. As the Ligation-Independent Cloning (LIC) kit (Novagen) was used for cloning of PCR products, all primers (DNA Technology) were designed with LIC specific 5'-sequences: Forward primer: 5' GAC GAC GAC AAG AT pmp-sequence 3'. Reverse primer: 5' GAG GAG AAG CCC GGT pmp-sequence 3'. Primers for amplification of pmp genes were placed as listed in table 1 and the sequences were obtained from GeneBank (AE001363). In those cases where only the N-terminal of the Pmp gene was cloned a stop codon was introduced. The pET 30Ek LIC vector (Novagen) was used for cloning and expression of recombinant protein according to the protocol provided by the manufacturer. Cloning was performed in E. coli NovaBlue.
Protein expression and purification
Plasmid DNA was prepared from NovaBlue cells by the alkaline lysis method and used for transformation of E. coli BL21(DE3) by eletroporation. Clones were tested for correct insert sequence by PCR with vector specific primers. The PCR product was used for sequence reactions using the Terminator Ready Reaction Mix (Perkin Elmer), and sequencing performed on an ABI PRISMTM 377 DNA Sequencer (Perkin Elmer). IPTG (1 mM) induction of expression of His-tagged fusion proteins was performed for two hours at 37°C when A600 reached 0.4. Recombinant protein was purified from pelleted cells using HiTrap Ni2+ columns (Amersham Pharmacia). The purity of the recombinant proteins was tested by SDS PAGE and the protein concentration measured by use of the Bradford Protein Assay (BioRad).
Production of antibodies
New Zealand White rabbits were immunized: i) intramuscularly on days 1, 8 and 15 using 10 μg of protein in PBS and 50% Freunds incomplete adjuvant; ii) intravenously on days 29, 36 and 43 using 10 μg of protein in PBS. The rabbits were bled on day 60.
Labeling
Pulse labeling of infected cell cultures was performed by using 100 μCi/mL radioactive methionine (Amersham Pharmacia) in a methionine-free RPMI 1640 medium as described previously [20]. 40 μg/mL cycloheximide was added to inhibit host cell protein synthesis during labeling. Cell cultures were labeled for two-hour-periods at 6, 12, 24, 26, 42, 48 and 54 hpi and harvested immediately after labeling by scraping off in lysis solution (7 M urea, 2 M thiourea, 4% w/v CHAPS, 40 mM Tris base, 65 mM DTE and 2% v/v Pharmalyte 3–10 (Amersham Pharmacia)). Before purification of EB the same labeling periods were used to ensure incorporation of radioactivity into proteins synthesized at all times during the developmental cycle.
Electrophoresis
One-dimensional SDS gels were run as described [30]. The protocol for two-dimensional gel electrophoresis was as described [20]. First dimension was carried out using non-linear Immobiline Drystrips pH 3–10 (Amersham Pharmacia). Proteins were focused in the strips using an IPGphor (Amersham Pharmacia) applying a voltage of 20 V for rehydration of strips and 120 kVh for focusing. For second dimension 9–16% T gradient polyacrylamide SDS gels were used. Comparative gels were loaded with 300.000 cpm as determined by TCA precipitation and scintillation counting. Gels for N-terminal sequencing were loaded with 600 μg protein from purified unlabeled EB. Gels for MS identification were loaded with 3.000.000 cpm and a total of 600 μg protein – all from purified EB. Gels for IMB were loaded with 75 μg protein from purified unlabeled EB and 300.000 cpm labeled EB protein. Protein spots in gels were visualized by autoradiography as described [20]. Comparative gels were treated with Amplify (Amersham Pharmacia) before exposure to BioMax MR X-ray films (Kodak).
Immunoblotting
Two-dimensional gels were washed for five min. in double-distilled water and soaked for 30 min. in transfer buffer. PVDF membranes (Immobilon-P, pore size 0.45 μm, Millipore) were soaked for one min. in methanol and then for 30 min. in transfer buffer. The transfer buffer contained: 50 mM Tris, 50 mM boric acid, 0.02% SDS and 10% methanol. The transfer was performed at 90 V and 10°C for four hours. Membranes were blocked in a buffer containing 20 mM Tris, 150 mM NaCl, and 3% gelatine, pH 7.5, washed in washing buffer (20 mM Tris, 500 mM NaCl, 0.05% tween-20) and incubated with polyclonal antibodies (pAbs) diluted 1/300 in antibody buffer (washing buffer added 0.2% gelatine) for one hour at 37°. After three washing steps membranes were incubated with secondary antibody (goat-anti-rabbit-IgG-AP-conjugate, BioRad) diluted 1/2000 for another hour at 37°C. After washing the blots, bound antibody was visualized with 270 μL NBT (50 mg/mL in 70% DMF) and 270 μL BCIP (25 mg/mL in 100% DMF) in 45 mL buffer containing 100 mM NaCl, 5 mM MgCl and 100 mM Tris-HCl at pH 9.5. As a fraction of the EB proteins was radiolabeled, reacting spots could be identified by comparison to a reference gel [20] upon autoradiography of the PVDF membrane [22].
Mass spectrometry
Protein spots excised from preparative gels were identified by MALDI TOF mass spectrometry as described [20].
N-terminal sequencing
Protein spots excised and pooled from eight Coomassie Brilliant Blue stained PVDF membranes each loaded with 600 μg of protein from purified EB were analyzed using an Applied Biosystems 404 protein sequencer (Perkin Elmer).
Immunofluorescence microscopy
IMF was performed as described [36] analyzing infected cells fixed in methanol or formalin at 72 hpi. Formalin fixed cells were permeabilized with 0.2% Triton X-100 for 10 minutes at room temperature. Bound primary antibody was visualized with a secondary FITCH conjugated antibody (DAKO), which was absorbed to prevent cross-reaction.
Authors' contributions
B.B.V. carried out the 2-D PAGE, IMB and IMF experiments, analyzed the Edman degradation results and drafted the manuscript. A.S.P. made the recombinant Pmps. K.G. and J.V. carried out the MS analysis. A.H. carried out the Edman degradation. G.C and S.B. participated in design and coordination of the study. All authors read and approved the final manuscript
Acknowledgments
Acknowledgements
We are grateful to Karin Skovgaard Sørensen, Charlotte Holm, Inger Andersen and Lisbet Wellejus Pedersen for excellent technical assistance. The work was supported financially by the European Commision (grant QLRT-1999-31536), the Danish Health Research Council (grants 9700659, 9900750), Helga and Peter Kornings Foundation, Aarhus University Research Foundation, the Concerted Research Actions (GOA) from the Flemish Community and the Fund for Scientific Research – Flanders (Belgium) (F.W.O.-Vlaanderen) of which Kris Gevaert is a Postdoctoral Fellow.
Contributor Information
Brian Berg Vandahl, Email: vandahl@medmicro.au.dk.
Anna Sofie Pedersen, Email: annas@biobase.dk.
Kris Gevaert, Email: kris.gevaert@rug.ac.be.
Arne Holm, Email: arho@kvl.dk.
Joël Vandekerckhove, Email: joel.vandekerckhove@rug.ac.be.
Gunna Christiansen, Email: gunna@medmicro.au.dk.
Svend Birkelund, Email: chlam@medmicro.au.dk.
References
- Kuo CC, Jackson LA, Campbell LA, Grayston JT. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Fischer E, Hackstadt T. Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect Immun. 2000;68:2379–2385. doi: 10.1128/IAI.68.4.2379-2385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood J, Stephens RS. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb Comp Genomics. 1999;4:187–201. doi: 10.1089/omi.1.1999.4.187. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Cevenini R, Donati M, Brocchi E, De Simone F, La Placa M. Partial characterization of an 89-kDa highly immunoreactive protein from Chlamydia psittaci A/22 causing ovine abortion. FEMS Microbiol Lett. 1991;65:111–115. doi: 10.1016/0378-1097(91)90481-O. [DOI] [PubMed] [Google Scholar]
- Souriau A, Salinas J, De Sa C, Layachi K, Rodolakis A. Identification of subspecies- and serotype 1-specific epitopes on the 80- to 90-kilodalton protein region of Chlamydia psittaci that may be useful for diagnosis of chlamydial induced abortion. Am J Vet Res. 1994;55:510–514. [PubMed] [Google Scholar]
- Longbottom D, Russell M, Dunbar SM, Jones GE, Herring AJ. Molecular cloning and characterization of the genes coding for the highly immunogenic cluster of 90-kilodalton envelope proteins from the Chlamydia psittaci subtype that causes abortion in sheep. Infect Immun. 1998;66:1317–1324. doi: 10.1128/iai.66.4.1317-1324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longbottom D, Findlay J, Vretou E, Dunbar SM. Immunoelectron microscopic localisation of the OMP90 family on the outer membrane surface of Chlamydia psittaci. FEMS Microbiol Lett. 1998;164:111–117. doi: 10.1016/S0378-1097(98)00187-6. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp.--autotransporters beyond the Proteobacteria. TRENDS in Microbiology. 2001;9:573–578. doi: 10.1016/S0966-842X(01)02234-X. [DOI] [PubMed] [Google Scholar]
- Birkelund S, Christiansen G, Vandahl B, Pedersen ASH. Proceedings of the Tenth International Symposium on Human Chlamydial Infection. 2002. pp. 551–554.
- Veiga E, Sugawara E, Nikaido H, de Lorenzo V, Fernandez LA. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 2002;21:2122–2131. doi: 10.1093/emboj/21.9.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai M, Hirakawa H, Ouchi K, Tabuchi M, Kishi F, Kimoto M, Takeuchi H, Nishida J, Shibata K, Fujinaga R, Yoneda H, Matsushima H, Tanaka C, Furukawa S, Miura K, Nakazawa A, Ishii K, Shiba T, Hattori M, Kuhara S, Nakazawa T. Comparison of outer membrane protein genes omp and pmp in the whole genome sequences of Chlamydia pneumoniae isolates from Japan and the United States. J Infect Dis. 2000;181:S524–S527. doi: 10.1086/315616. [DOI] [PubMed] [Google Scholar]
- Brunham RC, Plummer FA, Stephens RS. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–2276. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkelund S, Knudsen K, Madsen AS, Falk E, Mygind P, Christiansen G. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. 1998. pp. 275–278.
- Pedersen AS, Christiansen G, Birkelund S. Differential expression of Pmp10 in cell culture infected with Chlamydia pneumoniae CWL029. FEMS Microbiol Lett. 2001;203:153–159. doi: 10.1016/S0378-1097(01)00341-X. [DOI] [PubMed] [Google Scholar]
- Lindquist EA, Stephens RS. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. 1998:259–262. [Google Scholar]
- Grimwood J, Olinger L, Stephens RS. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect Immun. 2001;69:2383–2389. doi: 10.1128/IAI.69.4.2383-2389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandahl BB, Birkelund S, Demol H, Hoorelbeke B, Christiansen G., Vandekerckhove J, Gevaert K. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis. 2001;22:1204–1223. doi: 10.1002/1522-2683()22:6<1204::AID-ELPS1204>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Montigiani S, Falugi F, Scarselli M, Finco O, Petracca R, Galli G, Mariani M, Manetti R, Agnusdei M, Cevinini R, Donati M, Nogarotto R, Norais N, Garaguso I, Saletti G, Rosa D, Ratti G, Grandi G. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Pedersen AS, Hjernø K, Vandahl B, Birkelund S. Potential relevance of Chlamydia pneumoniae surface proteins to an effective vaccine. J Infect Dis. 2000;181:S528–S537. doi: 10.1086/315633. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- http://www.cbs.dtu.dk/services/SignalP-2.0/
- Hatch TP, Miceli M, Sublett JE. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol. 1986;165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundemose AG, Birkelund S, Larsen PM, Fey SJ, Christiansen G. Characterization and identification of early proteins in Chlamydia trachomatis serovar L2 by two-dimensional gel electrophoresis. Infect Immun. 1990;58:2478–2486. doi: 10.1128/iai.58.8.2478-2486.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkelund S. The molecular biology and diagnostics of Chlamydia trachomatis. Dan Med Bull. 1992;39:304–320. [PubMed] [Google Scholar]
- Tanzer RJ, Hatch TP. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J Bacteriol. 2001;183:2686–2690. doi: 10.1128/JB.183.8.2686-2690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer RJ, Longbottom D, Hatch TP. Identification of polymorphic outer membrane proteins of Chlamydia psittaci 6BC. Infect Immun. 2001;69:2428–2434. doi: 10.1128/IAI.69.4.2428-2434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen K, Madsen AS, Mygind P, Christiansen G, Birkelund S. Identification of two novel genes encoding 97- to 99-kilodalton outer membrane proteins of Chlamydia pneumoniae. Infect Immun. 1999;67:375–383. doi: 10.1128/iai.67.1.375-383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LA, Kuo CC, Grayston JT. Structural and antigenic analysis of Chlamydia pneumoniae. Infect Immun. 1990;58:93–97. doi: 10.1128/iai.58.1.93-97.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MW, Lambden PR, Everson JS, Clarke IN. Immunoreactivity of the 60 kDa cysteine-rich proteins of Chlamydia trachomatis, Chlamydia psittaci and Chlamydia pneumoniae expressed in Escherichia coli. Microbiol. 1994;140:2003–2011. doi: 10.1099/13500872-140-8-2003. [DOI] [PubMed] [Google Scholar]
- Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Cowen L, Menke M, King J, Berger B. BETAWRAP: successful prediction of parallel beta -helices from primary sequence reveals an association with many microbial pathogens. Proc Natl Acad Sci USA. 2001;98:14819–14824. doi: 10.1073/pnas.251267298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mygind PH, Christiansen G, Roepstorff P, Birkelund S. Membrane proteins PmpG and PmpH are major constituents of Chlamydia trachomatis L2 outer membrane complex. FEMS Microbiol Lett. 2000;186:163–169. doi: 10.1016/S0378-1097(00)00135-X. [DOI] [PubMed] [Google Scholar]
- Clausen JD, Christiansen G, Holst HU, Birkelund S. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol. 1997;25:441–449. doi: 10.1046/j.1365-2958.1997.4591832.x. [DOI] [PubMed] [Google Scholar]
- Giannikopoulou P, Bini L, Simitsek PD, Pallini V, Vretou E. Two-dimensional electrophoretic analysis of the protein family at 90 kDa of abortifacient Chlamydia psittaci. Electrophoresis. 1997;18:2104–2108. doi: 10.1002/elps.1150181137. [DOI] [PubMed] [Google Scholar]