Abstract
DNA replication in most organisms is regulated such that all chromosomes are replicated once, and only once, per cell cycle. In rapidly growing Escherichia coli, replication of eight identical chromosomes is initiated essentially simultanously, each from the same origin, oriC. Plasmid-borne oriC sequences (minichromosomes) are also initiated in synchrony with the eight chromosomal origins. We demonstrate that specific inactivation of newly formed, hemimethylated origins (sequestration) was required for the stable co-existence of oriC-dependent replicons. Cells in which initiations were not confined to a short interval in the cell cycle (carrying mutations in sequestration or initiation genes or expressing excess initiator protein) could not support stable co-existence of several oriC-dependent replicons. The results show that such stable co-existence of oriC-dependent replicons is dependent on both a period of sequestration that is longer than the initiation interval and a reduction of the initiation potential during the sequestration period. These regulatory requirements are the same as those required to confine initiation of each replicon to once, and only once, per cell cycle.
Keywords: chromosome replication/E.coli/incompatibility/initiation synchrony/sequestration
Introduction
Most organisms regulate chromosome replication in a strict fashion such that it occurs once, and only once, per cell cycle. So also in the bacterium Escherichia coli, which initiates up to eight chromosomal copies of its origin, oriC, within a short time interval (Skarstad et al., 1986; Boye et al., 1996, 2000). In addition, up to 200 extrachromosomal oriC copies may be initiated in synchrony with the chromosomal copies (Leonard and Helmstetter, 1986; Løbner-Olesen, 1999). Plasmids and other extrachromosomal elements do not regulate replication in this fashion. Replication from plasmid origins is random (del Solar et al., 1998), but the replication control still maintains a certain number of plasmids per cell. It does not, however, keep track of which plasmid origin has been initiated. The regulatory circuit governing initiation of chromosomal origins, on the other hand, keeps track of which origins have been initiated, but does not count how many origins have been initiated. The regulation works, as far as we know, in the following way (Boye et al., 2000): newly replicated origins are inactivated by sequestration (Lu et al., 1994). When all origins in the cell have been initiated and sequestered, the amount of free, active initiator protein, DnaA, gradually decreases because the chromosome is replicated and several hundred initiator-binding sites all around the chromosome are generated (Hansen et al., 1991; Kitagawa et al., 1998). Inactivation of the active ATP form of DnaA by the regulatory inactivation of DnA (RIDA) activity of the replication fork also contributes to this reduction of initiation potential (Katayama et al., 1998). Sequestration lasts for about one-third of a generation and, when origins emerge from sequestration, the ability to initiate replication is low. Normally, one generation must pass before sufficient amounts of active DnaA protein are made to initiate replication again.
Escherichia coli is able to recognize and inactivate new origins because they are hemimethylated (Russell and Zinder, 1987). Dam methyltransferase methylates the adenine of the sequence GATC. Newly replicated GATC sequences stay hemimethylated for a while until the Dam enzyme gets the opportunity to methylate them. The inactivation of origins by sequestration involves binding of the SeqA protein to hemimethylated GATC sites, of which there are 11 in oriC (Campbell and Kleckner, 1990; Lu et al., 1994; von Freiesleben et al., 1994; Slater et al., 1995). Loss of SeqA leads to rapid re-methylation of oriC (Lu et al., 1994) and re-initiation at origins that recently have initiated (Boye et al., 1996).
The exact time of initation in the cell cycle is dependent on the activity of the initiator protein, DnaA. Over expression of DnaA protein leads to an increase in origin concentration and origin number per cell, reflecting initiation at a lower cell mass (Atlung et al., 1987; Løbner-Olesen et al., 1989; Skarstad et al., 1989). DnaA binds to 9mer sequences in oriC as well as at >300 other locations on the chromosome (Kitagawa et al., 1998; Roth and Messer, 1998; Christensen et al., 1999; Morigen et al., 2001). When in the ATP-bound, ‘active’ form (Katayama et al., 1998), DnaA is capable of interacting with 6mer sequences found in the AT-rich left part of oriC, stabilizing this DNA in the single-stranded form, and forming an open complex (Speck and Messer, 2001). In the final stages of initation, DnaA interacts with the DnaB helicase and recruits it to the open complex (Marszalek and Kaguni, 1994; Seitz et al., 2000). Also, architectual elements, IHF and Fis, as well as transcriptional activation are required for proper initiation (Asai et al., 1992; Filutowicz et al., 1992; Boye et al., 1993). In vivo footprint studies indicate that IHF is present at an active initiating origin, while Fis may be present at an inactive origin (Cassler et al., 1995).
Here, we investigate the regulatory requirements for the stable co-existence of eight chromosomal and multiple plasmid-borne origins in wild-type E.coli. It was shown earlier that dam mutant cells are unable to support the stable co-existence of minichromosomes and chromosomes (Messer et al., 1985; Smith et al., 1985; Russell and Zinder, 1987; Løbner-Olesen and von Freiesleben, 1996) It was suggested that because the dam mutants lack sequestration, origins can compete for initiation factors throughout the cell cycle. Therefore, there may be an equal probability of re-initiating a new origin as of initiating an old origin (Løbner-Olesen and von Freiesleben, 1996), as well as an equal probability of initiating a minichromosomal and a chromosomal origin. We show here that not only in sequestration-deficient mutants, such as dam and seqA, but also in initiation-compromised mutants, the regulation breaks down so that origins compete for factors and the stable co-existence of several replicons is no longer possible.
Results
Absence of sequestration in a dam mutant strain leads to origin incompatibility
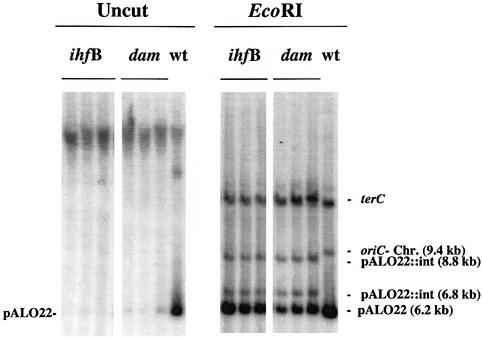
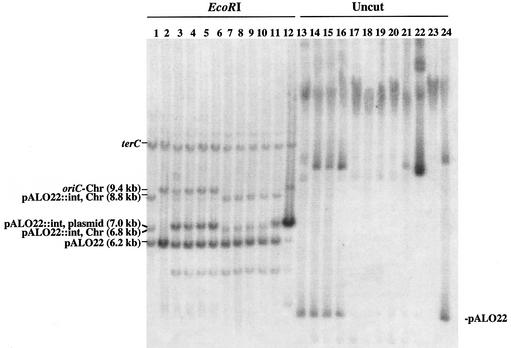
The ability of strains to harbour free pALO22 minichromosomes was investigated by Southern analysis. In the wild-type strain, MG1655, all minichromosomal copies were found to be extrachromosomal as visualized in a blot of uncut DNA hybridized with an oriC-specific probe (Figure 1, lane 7). Digestion of the DNA with EcoRI resulted in a 9.4 kb fragment harbouring the chromosomal origin and a 6.2 kb fragment representing minichromosome pALO22 (Figure 1, lane 14).
Fig. 1. Integration of minichromosomes into the chromosomal oriC region of ihfB and dam mutants. The wild-type strain (MG1655) harbouring the minichromosome pALO22 was transduced with mutant allelles ihfB::Cm and dam16::Km. Transductants were cultured, DNA extracted and Southern hybridization performed with an oriC probe and a ter probe (Materials and methods). Left panel: uncut DNA from three different ihfB transductants (lanes 1–3), three different dam transductants (lanes 4–6) and wild type (lane 7). Right panel: DNA from the same ihfB transductants (lanes 8–10), dam transductants (lanes 11–13) and wild type (lane 14) as in the left panel, cut with EcoRI, which linearizes the 6.2 kb pALO22 and yields a 9.4 kb chromosomal oriC fragment and two chromosomal pALO22::int fragments of 6.8 and 8.8 kb. The nature of the unmarked band in the uncut wild type is not known, but is assumed to be dimer of pALO22.
Strain MG1655, originally harbouring free pALO22, transduced with the dam– gene and grown for ∼60 generations, no longer maintained significant amounts of the minichromosomes in the free state (Figure 1; Table I). The integration of minichromosomes at the chromosomal origin was demonstrated by the lack of free pALO22 in the Southern blot of uncut DNA (Figure 1, lanes 4–6) and by the replacement of the 9.4 kb oriC-containing fragment with two fragments, 6.8 and 8.8 kb, in the blot of EcoRI-digested DNA (Figure 1, lanes 11–13). This oriC fragment pattern represents integration of pALO22 at the chromosomal oriC site. The presence of the 6.2 kb fragment indicated that more than one copy of pALO22 was integrated (see also figure 1 in Løbner-Olesen and von Freiesleben, 1996).
Table I. Minichromosomes in dam mutant cells.
| Host strain | Minichromosome | dam allelea | No. of clones tested | Fraction of cells with integrated minichromosomes (%)b |
|---|---|---|---|---|
| MG1655 | pALO22 | Wild type | >20 | 0 |
| MG1655 | pALO22 | dam16::Km | 11 | 100 |
| MG1655 | pOC24 | Wild type | 4 | 0 |
| MG1655 | pOC24 | dam16::Km | 12 | 100 |
| CM1671 (ΔoriC, Hfr) | pOC24 | Wild type | 4 | 0 |
| CM1671 (ΔoriC, Hfr) | pOC24 | dam16::Km | 17 | 0 |
aThe dam16::Km allele was introduced into minichromosome-containing cells by P1 transduction.
bData from Southern blots of DNA digested with EcoRI or AseI.
The results suggested that total lack of sequestration led to a situation where all origins in a cell, chromosomal or minichromosomal, competed for initiation factors (presumably ATP–DnaA, IHF and other positively acting factors) throughout the cell cycle (i.e. origin incompatibility). Only in cells where the minichromosome had integrated into the chromosome would each initiation event lead to replication of the chromosome as well as the minichromosome.
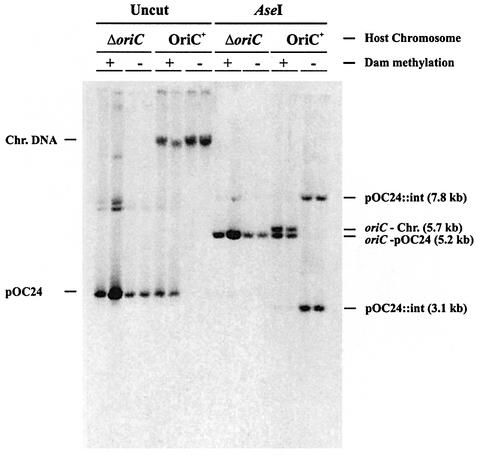
We wished to test whether origin incompatibility is the correct explanation for the observed inability of dam strains to harbour free minichromosomes. This was done by replacing the chromosomal origin with a plasmid origin. In this situation, different factors are limiting for initiation of the chromosome and the minichromosome. They should therefore be compatible even when methylation and sequestration are lacking. The chromosome of strain CM1671 has a deletion of oriC and is therefore dependent on the origin of an integrated F plasmid to replicate its chromosome. This strain was transformed with the minichromosome pOC24 conferring ampicillin resistance (strain CM1671 carries a tetracycline resistance gene; therefore, minichromosome pALO22 which confers tetracycline resistance could not be used). Mini chromosome pOC24 replicated autonomously in strain CM1671 as well as in the wild-type MG1655, as seen by Southern blot analysis of uncut DNA (Figure 2). Digestion with restriction enzyme AseI resulted in a 5.7 kb fragment harbouring the chromosomal origin of strain MG1655 and a 5.2 kb fragment repesenting minichromosome pOC24. The copy number of the pOC24 minichromosome was about one per chromosomal origin in strain MG1655 (oriC+) and similar in CM1671 (ΔoriC). The reason for the lower copy number compared with that of pALO22 (which is ∼10 per chromosomal origin) may be lack of the mioC region in pOC24. This region previously has been shown to affect the copy number of minichromosomes (Stuitje and Meijer, 1983; Løbner-Olesen et al., 1987).
Fig. 2. Replication of minichromosomes in dam mutant strains is autonomous when chromosome replication is not dependent on oriC. The wild-type strain (MG1655) or ΔoriC strain (CM1671) harbouring minichromosome pOC24 was transduced with the mutant allele dam16::Km. Transductants were cultured, DNA extracted and Southern hybridization with an oriC probe performed with uncut DNA and with DNA treated with restriction enzyme AseI, which linearizes the 5.2 kb pOC24, and yields a 5.7 kb chromosomal oriC fragment and two chromosomal pOC24::int fragments of 7.8 and 3.1 kb. Two clones of each are shown. The nature of the unmarked double band in the uncut dam+ΔoriC lane is not known, but is assumed to be dimer of pOC24.
The dam– allele was transduced into strains CM1671 and MG1655, both containing pOC24, and transductants investigated. All ΔoriCdam– transductants contained free minichromosomes with a copy number comparable with that of the Dam+ situation (Figure 2; Table I). In all oriC+dam– transductants, however, the minichromosome pOC24 was integrated into the oriC region of the chromosome (Figure 2; Table I). The integration of a single copy of pOC24 resulted in oriC containing AseI restriction fragments of 7.8 and 3.1 kb instead of the 5.7 kb chromosomal fragment and the 5.2 kb pOC24 fragment (Figure 2; Table I).
It may be argued that the minichromosome harboured in the ΔoriCdam– strain might have difficulty integrating, since the region of homology is deleted, and that there might be a competition situation but the assay will not detect it. To provide additional evidence for the lack of competition, the growth rate of the ΔoriCdam– strain with and without minichromosome was measured. If there were a competition situation, but the minichromosome were absolutely prevented from integration, a reduced culture growth rate should be seen, since cells in which replication of only the chromosome or only the minichromosome was favoured should die. Such a reduction in growth rate was not seen. The ΔoriCdam– and ΔoriCdam– /pOC24 strains had growth rates of 52 and 54 min, while the ΔoriC and ΔoriC/pOC24 strains had growth rates of 41 and 46 min, respectively. Thus, there was a slight reduction in growth rate due to segregational instability of the minichromosome and a slight reduction in growth rate due to the dam– allele, but no reduction in growth rate due to competition for factors.
These data show that when the chromosome and the minichromosome depend on different factors for initiation of replication, free minichromosomes may also be maintained in the dam mutant strain. The inability of dam mutant strains to harbour free minichromosomes was therefore not due to plasmid mis-segregation or other conceivable deficiencies, but to competition for initation factors, or, in other words, origin incompatibility.
Initiation-compromised mutants initiate asynchronously and show origin incompatibility
The simultaneousness (or synchrony) of initiation may be measured by flow cytometry (Skarstad et al., 1986). Mutants compromised in either initiation or sequestration [dnaA(Ts), ihf, dam or seqA mutants] show a phenotype of initiation asynchrony (see below; Skarstad et al., 1988; Boye and Løbner-Olesen, 1990; Boye et al., 1993; Lu et al., 1994; von Freiesleben et al., 2000b). The reason for the asynchrony presumably differs in the different mutants. In the initiation-compromised mutants, dnaA(Ts) and ihf, initiation is limiting, leading to underinitiation (too few initiations per mass), and asynchrony is most likely to be due to initiation of replication at individual origins at different times throughout the cell cycle. Thus, the window during which initiation occurs in these mutants (the initiation period) will extend over a substantial part of the cell cycle (Skarstad et al., 1986). In the sequestration-deficient mutants, dam and seqA, the asynchrony is probably due to re-initiation of origins already initiated once in that generation and therefore due to overinitiation (too many initiations per mass).
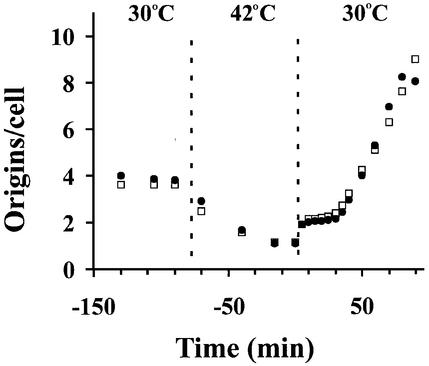
We wished to determine whether dnaA(Ts) and ihf mutants could support the stable co-existence of minichromosomes and chromosomes. The products of these genes have not been implicated in origin inactivation and are therefore expected to be sequestration proficient. To verify this, the duration of the sequestration period was investigated indirectly in one of the mutants, ihfB, by measuring the length of the eclipse period (von Freiesleben et al., 2000a). This method takes advantage of accumulation and release of initiation potential in a dnaA46 mutant. Initiation potential was accumulated in dnaA46 and dnaA46 ihfB mutants by incubation at the non-permissive temperature (42°C) for 90 min. Most cells then contained one fully replicated chromosome (Figure 3). After shift to the permissive temperature (30°C), all origins were initiated and cells contained two origins per cell. In both strains, the new origins were inert to initiation of replication for ∼30 min (the eclipse), and then re-initiated. Because sequestration of newly formed origins seems to be the basis for the eclipse (von Freiesleben et al., 2000a), we conclude that, at least for the ihf mutant, the sequestration period is similar to that of wild type (Ihf+).
Fig. 3. Measurement of the eclipse period in the ihfB mutant. Strains CM735dnaA46 (filled circles) and CM735dnaA46 ihfB::Cm (open squares) growing exponentially at 30°C were shifted to 42°C for 90 min to accumulate cells with a single origin. The temperature was then shifted to 30°C, allowing all origins to be initiated simultanously. The number of origins per cell was measured by flow cytometry after treatment with rifampicin and cephalexin, and replication run-out.
Minichromosomes were, however, found integrated in the oriC region of the chromosome of ihfB, dnaA46 and dnaA204 (Figure 1; Table II). These results indicate that mutants with a disability in initiation of replication that leads to asynchrony of initiation also are unable to maintain free minichromosomes, irrespective of whether sequestration is intact or not. We suggest that sequestration occurs at each individual origin independently, so that the first origin initiated may be released from sequestration before the last origin has been initiated. A mutant with an initiation period lasting longer than the sequestration period will not be able to distinguish between the new pair of origins released from sequestration, and an old origin not yet initiated. Consequently, some origins are initiated twice within a generation and some are not initiated at all. Again, only in cells in which the minichromosome had integrated into the chromosome would an initiation event ensure the replication of both the chromosome and the minichromosome.
Table II. Minichromosomes in initiation-compromised cells.
| Host strain | Minichromosome | Mutation | No. of clones tested | Fraction of cells with integrated minichromo somes aftergrowth for<50 generations (%)c | Fraction of cells with integrated minichromo-somes after growth for >60 generations (%)c |
|---|---|---|---|---|---|
| MG1655 | pALO22 | Wild type | >20 | 0 | 0 |
| MG1655 | pALO22 | ihfB::Cma | 8 | 50 | 100 |
| CM735 | pALO22 | Wild type | 1 | 0 | 0 |
| CM735 | pALO22b | dnaA204 | 4 | 100 | 100 |
| CM735 | pALO22b | dnaA46 | 3 | 100 | 100 |
aThe ihfB::Cm allele was introduced into minichromosome-containing cells by P1 transduction.
bMinichromosome pALO22 was introduced into dnaA(Ts) cells by transformation.
cData from Southern blots of DNA digested with EcoRI.
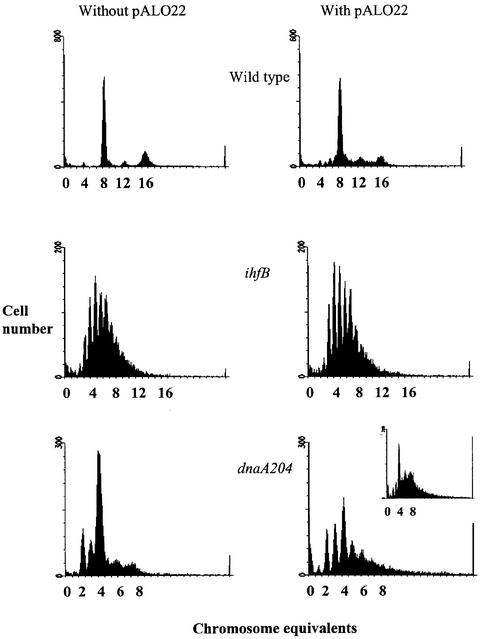
The two dnaA(Ts) mutants we investigated exhibit either a high (dnaA46) or a low (dnaA204) degree of asynchrony (Skarstad et al., 1988), and we found it rather surprising that the dnaA204 mutant was unable to maintain free minichromosomes when the asynchrony is slight. The initiation window (J) in this mutant is 0.2–0.3τ (Skarstad et al., 1986, 1988), while the sequestration period should be at least 0.3τ (Lu et al., 1994). We therefore investigated whether the degree of chromosomal initiation synchrony changed in the dnaA204 mutant when minichromosome pALO22 was introduced into the cells. The dnaA204 strain with no minichromosome exhibited a moderate degree of asynchrony (Figure 4). The same strain harbouring pALO22 showed more severe asynchrony (J >0.5τ) and a 20–30% reduced growth rate initially (Figure 4). After ∼100 generations of growth, which led to a selection of cells with pALO22 integrated in the chromosome (Table II), the asynchrony was again more moderate (Figure 4, small panel) and the normal growth rate was restored. Thus, the dnaA204 strain seems to be capable of fairly synchronous initiation only when few copies of oriC are present. The ihfB and dnaA46 mutants exhibited an origin distribution representative of severely asynchronous initiation both before and after transformation with the minichromosome (Figure 4; data not shown).
Fig. 4. Flow cytometry DNA histograms of rifampicin/cephalexin- treated cultures of strains MG1655 (wild type), MG1655/pALO22, MG1655ihfB, MG1655ihfB/pALO22, CM735dnaA204 and CM735dna A204/pALO22. Cultures were sampled 60 generations after transformation. The latter strain was also sampled after ∼100 generations (small panel).
These results suggest that asynchrony of initiation will lead to incompatibility of origins if the total initiation period lasts longer than the sequestration period.
Shortening of the sequestration period resulted in integration of minichromosomes
If the above explanation of origin incompatibility in asynchronous mutants is correct, cells with a too short sequestration period should also display origin incompatibility. Shortening the sequestration period, by increasing the level of Dam methylase or by deleting the seqA gene, was shown previously to result in asynchrony of initiation (Boye and Løbner-Olesen, 1990; Lu et al., 1994). This asynchrony is in both cases accompanied by an increase in the number of origins per cell and is likely to result from re-initiation of some origins within the same generation, or, in other words, overinitiation (initiation of too many origins per mass) (Boye and Løbner-Olesen, 1990; Boye et al., 1996).
Cells harbouring minichromosomes were transformed with plasmid pdam118, which resulted in ∼2-fold overproduction of Dam methylase (Arraj et al., 1990). In these cells, the duration of the eclipse was reduced by at least 50% (von Freiesleben et al., 2000a) and the sequestration period should therefore be ∼0.1–0.15τ, while the initiation window is normal, i.e. 0.05–0.1τ (Skarstad et al., 1986). Minichromosomes were found integrated in the chromosomal oriC region in ∼25% of the cells after 60 generations (Table III). In a seqA deletion mutant, the length of the sequestration period is shortened by two-thirds to ∼0.1τ (Lu et al., 1994), while the initiation window presumably is normal (0.05–0.1τ). SeqA-less cells were transformed with pALO22. After ∼60 generations of growth, 40% of the cells contained integrated minichromosomes (Table III), and after further growth all cells contained integrated minichromosomes (Table V, mock P1 transduction; data not shown).
Table III. Minichromosomes in cells with a shortened sequestration period.
| Recipient strain | Transformed with plasmid | No. of clones tested | Fraction of cells with integrated minichromosomes after growth for 60 generations (%)a |
|---|---|---|---|
| MG1655/pALO22 | None | >20 | 0 |
| MG1655/pALO22 | pdam118 | 8 | 25 |
| MG1655seqA10 | pALO22 | 6 | 40 |
aData from Southern blots of DNA digested with EcoRI.
Table V. Absence of SeqA alleviates origin incompatibility in initiation-compromised mutants.
| Recipient strain | Minichromosome | Mutationa | No. of cultures tested | Fraction of cells with minichromosomes integrated (%)c |
|---|---|---|---|---|
| MG1655 | pALO22 | dam16::Km | 11 | 100 |
| MG1655seqA10 | pALO22b | dam16::Km | 10 | 70 |
| MG1655 | pALO22 | ihfB::Cm | 8 | 100 |
| MG1655seqA10 | pALO22b | ihfB::Cm | 6 | 50 |
| MG1655seqA10 | pALO22b | Mock P1 transduction | 2 | 100 |
aThe mutant alleles were introduced into minichromosome-containing cells by P1 transduction.
bClone of MG1655seqA10 containing free pALO22 minichromosomes.
cData from Southern blots of DNA digested with EcoRI.
These results suggest that, at least in some cells in the cultures, the duration of the sequestration period was reduced sufficiently to cause incompatibility of origins in otherwise wild-type cells.
The kinetics of the integration process
The rate of integration of minichromosomes was found to vary somewhat for the different mutants, indicating that the competition for initiation factors was more severe in some cases than in others. Only dam and dnaA mutant cultures contained no chromosomal oriC regions without integrated minichromosomes after <50 generations (Table II). The ihf mutant cultures had, at this stage, ∼50% cells with integrated minichromosomes, indicating that the loss of IHF led to a milder condition (Table II). Even less severe was the situation in the seqA mutant and in cells where Dam methylase was overproduced (Table III). It is reasonable to assume that the rate of integration in the latter two cases was low because the window of sequestration was large enough to cover the window of initiation some of the time. In the dam mutants which lack sequestration, and dnaA mutants that have a very large J, the rate of integration was high, probably because the inadequacy of sequestration was severe.
Overinitiation of replication leads to origin incompatibility
We investigated whether incompatibility of chromosomal and minichromosomal origins is found in cells producing excess amounts of DnaA protein, which results in overinitiation of replication (Atlung et al., 1987; Løbner-Olesen et al., 1989; Skarstad et al., 1989; Atlung and Hansen, 1993; Hansen and Atlung, 1995). Cells harbouring minichromosome pALO22 were transformed with plasmids pFH539 or pFH871, giving 6- and 10-fold overproduction of DnaA, respectively (data not shown). Transformants were grown for ∼60 generations and the DNA analysed by Southern blot (Figure 5). DNA rearrangements were found to consist of either: (i) minichromosomes integrated into the chromosomal oriC region, yielding 8.8 and 6.8 kb fragments when cut with EcoRI; or (ii) minichromosomes integrated into the DnaA-producing plasmid at regions of homology with vector regions of the minichromosome, yielding a 7.0 kb fragment (Figure 5). In the latter case, the chromosomal origin region was intact, yielding the 9.4 kb fragment (Figure 5). Incompatibility was found to be surprisingly severe in both cases of DnaA overproduction (Figure 5; Table IV). No cultures with both an intact chromosomal oriC region and an original DnaA-producing plasmid were found. In a complementary experiment, cells already harbouring pFH539 were transformed with pALO22. Here, all transformants were found to harbour the minichromosomes integrated in pFH539 (Table IV). Control experiments were performed with plasmid pTAC1300, which, instead of the entire dnaA gene, carries a dnaA gene with an internal 120 bp deletion. In the strain harbouring pTAC1300, subsequently transformed with pALO22, no integration of minichromosomes was seen (Table IV). However, in the control experiment using pTAC1300 to transform a minichromosome-containing strain, incompatibility was found in two out of five cultures analysed (Table IV). The reason for this is not understood, but could be due to negative complementation.
Fig. 5. Integration of minichromosomes into the chromosomal oriC region or the DnaA-producing plasmid. The wild-type strain (MG1655) harbouring the minichromosome, pALO22, was transformed with DnaA-producing plasmid pFH539. Several transformants were cultured, DNA extracted and Southern hybridization performed with an oriC probe and a ter probe. DNA from 10 transformants (lanes 3–12), a dam transductant from Figure 1 (lane 1) and a wild type (lane 2), cut with EcoRI, which linearizes the 6.2 kb pALO22, yields a 9.4 kb chromosomal oriC fragment, two chromosomal pALO22::int fragments of 6.8 and 8.8 kb and a 7.0 kb plasmid pALO22::int fragment. Uncut DNA from the same 10 transformants (lanes 13–22), a dam transductant from Figure 1 (lane 23) and a wild type (lane 24). The nature of the unmarked band in the uncut wild type is not known, but is assumed to be dimer of pALO22. The nature of other unmarked bands is not known.
Table IV. Minichromosomes in strains overinitiating DNA replication.
| Strain | Transformed with plasmida | No. of clones tested | No. of cultures containing cells with pALO22 integrated into the chromosomal oriC regionb | No. of cultures containing cells with pALO22 integrated into pFH539, pTAC1300 or pFH871b | Fraction of cells with pALO22 integrated (%)b |
|---|---|---|---|---|---|
| MG1655/pALO22 | None | >20 | 0 | NA | 0 |
| MG1655/pALO22 | pFH539 | 10 | 6 | 4 | 100 |
| MG1655/pALO22 | pTAC1300 | 5 | 2 | 0 | 40 |
| MG1655/pFH539 | pALO22 | 7 | 0 | 7 | 100 |
| MG1655/pTAC1300 | pALO22 | 5 | 0 | 0 | 0 |
| MG16655/pALO22 | pFH871 | 7 | 5c | 5c | 100 |
aCells containing pFH539 or pFH871 had a 6- or 10-fold concentration, respectively, of DnaA protein. Plasmid pTAC1300 produced a truncated version of the DnaA protein.
bData from Southern blots of DNA digested with EcoRI.
cMixed populations in three out of seven cultures.
These results indicate that a high initiation potential throughout the cell cycle, obtained by overproduction of DnaA, enables an origin to be initiated again as soon as it is released from sequestration, and therefore leads to a situation of incompatibility between chromosomal and minichromosomal origins.
Absence of SeqA protein alleviates origin incompatibility in initiation-compromised mutants
It was observed earlier that combining the seqA2 mutation with the dam deletion resulted in cells capable of maintaining free minichromosomes (Løbner-Olesen and von Freiesleben, 1996). This was a surprising result since the sequestration-compromised seqA2 strain as well as the double mutant would have been expected to have the same deficiency as the dam mutant in maintaining free minichromosomes.
Mutant alleles dam and ihfB were transduced individually into the minichromosome-containing seqA10 strain. Care was taken to use a clone of seqA10 shown to contain cells with free minichromosomes when preparing competent cells. The resulting double mutant cultures contained 30 and 50%, respectively, of cells harbouring free minichromosomes after ∼60 generations of growth (Table V). Thus, the ability of both mutants to maintain free minichromosomes improved when combined with the seqA10 mutation. This result could be due to the fact that SeqA-less cells overinitiate replication, thus compensating for limiting initiation in the initiation-compromised mutants (Lu et al., 1994; von Freiesleben et al., 1994; Boye et al., 1996). The dam mutant thus seems not only to lack a sequestration period, but also to have problems with initiation. This could be due to a lower than wild-type expression of DnaA protein in dam– cells (Braun et al., 1985).
Likewise, the ability of the seqA10 mutant to maintain free minichromosomes improved when combined with either dam or ihfB mutations (Table V, compare double mutants with mock P1 transduction), indicating that reduced overinitiation alleviates origin incompatibility. This suggests that both lack of sequestration and overinitiation contribute to the severe origin incompatibility in the seqA single mutant.
Discussion
Origin incompatibility occurs when the sequestration interval becomes shorter than the initiation interval
In this work, we have investigated the compatibility between chromosomal and minichromosomal origins in the bacterium E.coli. The ability to maintain several copies of autonomously replicating minichromosomes was found to rely on three partly overlapping factors: (i) origin sequestration; (ii) synchronous initiation of replication; and (iii) sufficient reduction in initiation potential during sequestration.
It was observed previously that dam mutant cells are unable to maintain free minichromosomes (Messer et al., 1985; Løbner-Olesen and von Freiesleben, 1996). With the discovery of sequestration (Ogden et al., 1988; Campbell and Kleckner, 1990; Lu et al., 1994; von Freiesleben et al., 1994), a possible explanation was suggested: in dam mutant cells, lacking sequestration, origins experience a competition for initiation factors, such that any origin that accumulates sufficient DnaA and other initiation factors to form a functional initiation complex will initiate independently of the other origins (Løbner-Olesen et al., 1994). As soon as two new origins are formed, they immediately will compete for the available initiation factors and have the same probability of being initiated as the old origins (Figure 6B). The results presented here show that autonomous minichromosomes may be maintained in dam mutant cells by substituting the chromosomal origin with a plasmid origin. Thus, competition for initiation factors is the most plausible explanation for the inability of dam mutant cells to harbour automously replicating minichromosomes.
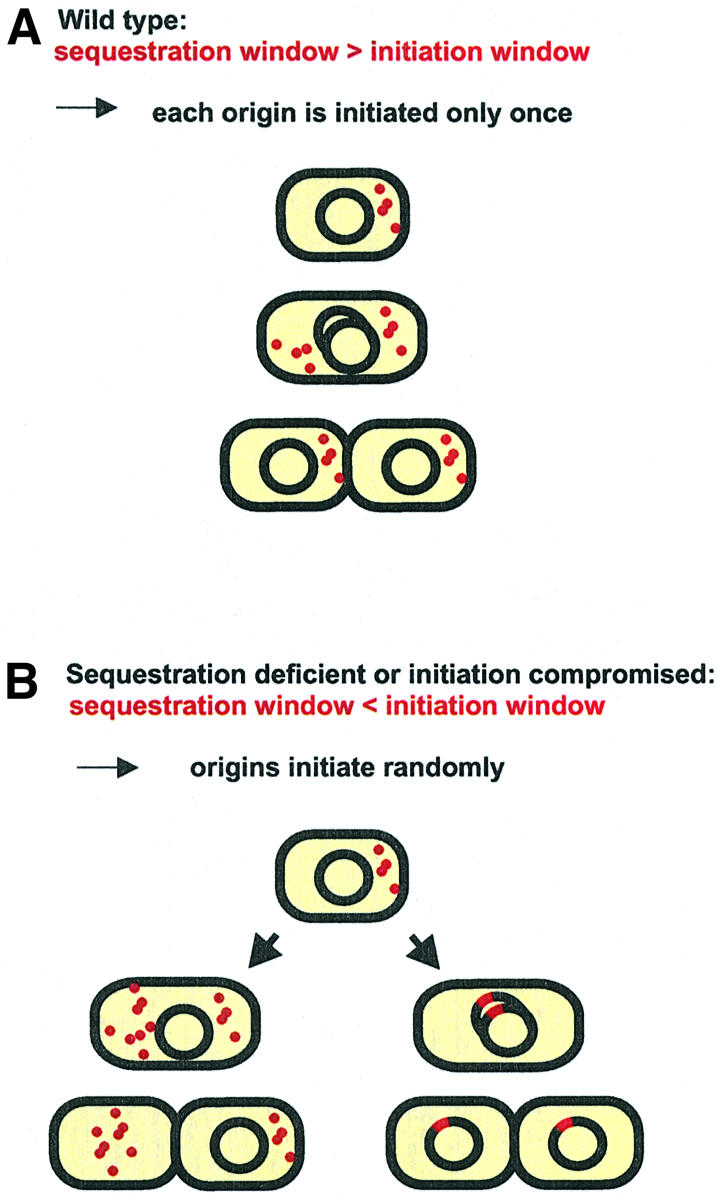
Fig. 6. Origin compatibility is obtained only if the initiation potential drops during the period of sequestration, so that the initiation window is kept shorter than the sequestration window. This situation results in only one initiation event per origin (A). If sequestration is inadequate, competition occurs between uninitiated origins and already initiated origins, and a stable situation can only be obtained by integration of all essential origins into the same contiguous replicon (B). The representation is simplified, showing replication of only one chromosomal and four minichromosomal origins.
In addition to sequestration-deficient mutants, some sequestration-proficient mutants exhibited incompatibility of origins. The sequestration-proficient mutants were compromised in initiation of replication and exhibited asynchronous initiation of replication. We suggest that the reason for the incompatibility of origins in these mutants is that the deficiencies in formation of an initiation complex lead to a low probability of initiation even though all components required are present and initiation normally should have occurred. This leads to a large time window in the division cycle during which initiation is occurring. A normal sequestration interval would then be short in comparison with the initiation interval, resulting in a situation where newly formed origins, already released from sequestration, could compete with the origins not yet initiated, for initiation components.
In summary, it seems that if the interval of initiation is longer than the sequestration interval, incompatibility of origins will be the consequence. Thus, for compatibility of several oriCs to be maintained, it is crucial that the initiation period is over before the end of sequestration at any of the origins present (Figure 6).
Cells provided with excess Dam methylase or deleted for seqA also showed an incompatibility phenotype. This indicates that re-initiaton of origins will occur if the sequestration period is short enough. The mechanism may be the same as suggested for the initiation-compromised mutants, namely that the latest initiations have not yet occurred when the first origins are released from sequestration. An alternative explanation in the wild-type situation is that initiations occur essentially synchronously but that the initiation capacity or potential is considerably higher than required for initiation of the chromosomal origins present, as indeed indicated by the capacity also to initiate many minichromosomes. Re-initiation after shortening of the sequestration period will, according to this explanation, occur because the sequestration period ends before the initiation potential has dropped sufficiently.
Overinitiation leads to incompatibility of origins
In cells where the initiation potential was kept high by overproduction of the DnaA protein, severe incompatibility occurred. These cells probably maintained a normal sequestration interval. Because of the constant overproduction of DnaA, origins would be able to re-initiate as soon as they emerged from sequestration. These results show that it is crucial that the initiation potential drops sufficiently during the period of sequestration to make initiation impossible.
In such a scenario, it is not obvious why and how extra initiations would cause a problem in strains harbouring minichromosomes, since strains without minichromosomes tackle overinitiation surprisingly well, initiating earlier in the cell cycle, but in reasonable synchrony (Løbner-Olesen et al., 1989; Skarstad et al., 1989). Many of the chromosomal replication forks resulting from excess initiation are, however, aborted or stalled (Atlung et al., 1987; Skarstad et al., 1989; Atlung and Hansen, 1993). A possible explanation is, therefore, that a larger proportion of initiations of the minichromosome leads to duplication, compared with initiations of the chromosome. If large numbers of minichromosomes accumulate, sequestration may become inadequate. Indications have been found that there may be a limit to the number of origins that can be handled by the sequestration apparatus. A study in which the cellular number of minichromosomes was increased by progressively increasing the concentration of selective drug showed that at >200 oriC copies per cell, free minichromosomes could no longer be maintained (Løbner-Olesen, 1999). Thus, when a large enough number of origins have been duplicated and newly formed origins no longer can be sequestered, the situation will be the same as in the dam mutants, namely that origins compete for initiation factors and initiation occurs randomly at any origin, new or old.
In overinitiating cells, minichromosomes integrated either in the chromosome or in the DnaA-producing plasmid. It is not clear why integration into DnaA-producing plasmids would alleviate incompatibility. We confirmed that the integration event did not turn off the overproduction of DnaA protein (data not shown). It is possible that the likelihood of replication forks stalling or aborting is higher in the larger hybrid plasmid. This would hinder an escalation in the number of oriCs present. Alternatively, there could exist plasmid sequestration mechanisms not yet characterized.
DnaA protein and the timing of initiation
During wild-type steady-state growth, the production of DnaA through the cell cycle will be unchanged from generation to generation and will therefore double when the bulk of cell constituents have doubled. From this follows that the accumulation of sufficient DnaA at each origin will occur at the same point in every cell cycle, generation after generation. It was shown earlier that a properly timed replication cycle could be obtained even when production of DnaA was constant, maintained solely by dnaA transcription from the lac promoter on a plasmid (Løbner-Olesen et al., 1989). This showed that a fluctuating production of DnaA protein was not required for proper timing of initiation. Here, we demonstrate that the amount in the cell, of free, active DnaA protein must, on the contrary, fluctuate. This fluctuation is constituted by the reduction in the amount of free, active DnaA protein during sequestration. This critical decrease in the amount of free DnaA protein is caused by the generation of new DnaA-binding sites by replication (Hansen et al., 1991; Christensen et al., 1999), and is sufficient to prevent new origins from being re-initiated.
Asynchrony of replication in the ihf mutant
The asynchrony of replication in ihf mutants has been questioned recently (von Freiesleben et al., 2000b). It was observed that after rifampicin treatment, residual rounds of replication occurred. Thus, there is a possibility that the asynchrony in this mutant is only apparent and due to asynchrony of rifampicin-resistant initiations rather than asynchrony of initiations prior to the addition of rifampicin. The results found here, namely that minichromosomal and chromosomal origins show incompatibility, suggest that the initiation interval in the ihfB mutant is extended and that initiation in ihf mutants probably is also asynchronous before the addition of rifampicin.
Materials and methods
Strains
Escherichia coli K.12 strains used were MG1655 (λ–F–; Guyer et al., 2001), CM735 (metE46, trp-3, his-4, thi-1, galK2, lacY1 or lacZ4, mtl-1, ara-9, tsx-3, ton-1, rps-8 or rps-9, supE44 λ– (Hansen and von Meyenburg, 1979), CM735dnaA204, CM735dnaA46 (Hansen et al., 1984, 1992) and CM1671 (ΔoriC1671, Hfr::ilv, asnA::Tn10, asnB32, relA1, spoT1, thi-1, fuc, lysA, l- (von Meyenburg and Hansen, 1980). Mutants carrying dam16 or ihfB were constructed by P1 transduction of the mutant allelles from strains LJR24 dam16 (dam16::Km; Løbner-Olesen et al., 1992) or R949 (ihfB::Cm; Flamm and Weisberg, 1985), respectively. MG1655ΔseqA was constructed by P1 co-transduction of the pBIP Kan marker, with the sacB and seqAΔ10 genes (Slater and Maurer, 1993; Slater et al., 1995), into MG1655, generating a co-integrant carrying both alleles of seqA. Selection for the crossing out of sacB, the Kan marker and the wild-type or the seqAΔ10 allele was done by plating on 5% sucrose. The resulting wild-type or seqAΔ10 strain was identified by PCR.
Plasmids
Plasmid pALO22 consists of the chromosomal origin region including oriC, mioC, asnC and asnA together with the Tet gene from Gerdes et al. (1986). Plasmid pOC24 consists of the oriC region on a 2.3 kb HindIII–XhoI fragment carrying oriC ligated to a 2.9 kb HindIII–XhoI fragment carrying the blaZ gene of Staphylococcus aureus plasmid pI258 (Messer et al, 1978). Plasmid pdam118 consists of the pBR322 origin, the bla gene and the dam gene (Brooks et al., 1983). Plasmids pFH539 (von Meyenburg et al., 1985) and pFH871 (Atlung et al., 1985) consist of the dnaA gene with promoter, cloned as a ClaI–XhoI fragment into the same sites of plasmids pBR322 and pACYC184, respectively. Plasmid pTAC1300 consists of the dnaA gene with a 120 bp deletion cloned into pBR322 (Atlung et al., 1985).
Plasmids were purified using JET star (Genomed). Unmethylated plasmid pALO22 was obtained by growth in a seqAdam strain.
Transduction and transformation
Transduction was by P1 (Miller, 1992) and transformation was by electroporation. Transductants and transformants routinely were restreaked once.
Growth conditions
Growth was in LB or AB minimal medium (Clark and Maaløe, 1967) supplemented with glucose (0.2%) and casamino acids (0.5%), at 37°C for strains MG1655 and CM1671 and derivatives, and at 30°C for strain CM735 and derivatives. Selection for the presence of minichromosomes was maintained throughout experiments. Antibiotics were used at the following concentrations: tetracycline (10 µg/ml), kanamycin (50 µg/ml), chloramphenicol (25 µg/ml) and ampicillin (50 µg/ml). Mass growth was monitored by measuring optical density at 450 and 600 nm in AB and LB medium, respectively.
Southern blot analysis
Total cellular DNA was prepared from 15 ml of exponentially growing cells in LB at an OD of 0.5. Cells were harvested and lysed by treatment with 300 µg/ml lysozyme in 50 mM Tris–HCl, 50 mM EDTA pH 8.0 for 15 min at room temperature prior to incubation with 1% SDS and 100 µg/ml RNase for 30 min at 37°C, followed by two phenol and two chloroform extractions, isopropanol and ethanol precipitation. After digestion with appropriate restriction enzymes (New England Biolabs), the fragments were separated on 0.8% agarose gels, transferred by capillary transfer to Hybond-N+ membranes (Amersham Pharmacia Biotech.) and probed with the oriC-containing 463 bp AvaI fragment from pGO46 (Ogden et al, 1988) and a terC-containing probe generated by PCR (Morigen et al, 2001). Probes were labelled with 32P or 35S (Amersham Pharmacia) using the Random Primer system (‘Prime-a-gene’, Promega).
Treatment with rifampicin and cephalexin
Rifampicin (300 µg/ml; Fluka) and cephalexin (10 µg/ml) (‘Keflex’, Lilly) were added to exponentially growing cells (OD450 = 0.15) to inhibit initiation of DNA replication and cell division, respectively (Skarstad et al., 1986; Boye and Løbner-Olesen, 1991). Incubation continued for three to four doublings to complete ongoing rounds of replication. By inhibiting initiation of replication, allowing ongoing rounds of replication to finish, and then measuring the number of chromosomes per cell with flow cytometry, a measure of the number of origins in each cell at the time of drug addition was obtained. Rapidly growing wild-type cells initiate replication at eight origins and essentially simultanously. This is seen by the presence of either eight or 16 origins in >95% of the cells (Figure 4, upper left panel). A few cells had nine or 12 origins, indicating either that the synchrony is not absolute or that the rifampicin effect is not exactly the same at all origins.
Fixation
Exponentially growing cells (OD450 = 0.15) or cells treated with rifampicin and cephalexin were washed and resuspended in TE buffer and then diluted 10-fold in 77% ethanol for fixation.
Staining for measurement in the FACS Star+ flow cytometer
Cells were washed in 0.1 M phosphate buffer pH 9.0, and stained overnight at 4°C in 1.5 µg/ml fluorescein isothiocyanate (FITC) in the same buffer (made from a 3 mg/ml stock solution) (Wold et al., 1994). This dye binds covalently to cellular proteins. Cells were washed twice in 0.02 M phosphate-buffered saline pH 7.5, to remove unbound dye, and resuspended in the same buffer. The DNA was stained by adding an equal volume of 3 µg/ml Hoechst 33258 in the same buffer (final concentration 1.5 µg/ml).
Staining for measurement in the Bryte flow cytometer
Cells were washed in 10 mM Tris pH 7.4 containing 10 mM MgCl2 and stained with 100 µg/ml mitramycin (Pfizer) and 20 µg/ml ethidium bromide in the same buffer.
Flow cytometry
Cells were analysed for DNA (Hoechst fluorescence) and protein content (FITC fluorescence) in the FACS Star+ flow cytometer (Beckton Dickinson). Cells with known DNA content and not stained with FITC were added to each sample to calibrate the DNA axis (Torheim et al., 2000).
Cells used in the eclipse experiment, Figure 3, were analysed for DNA (ethidium bromide fluorescence, from energy transfer by excitation of mithramycin) and light scattering in the Bryte HS flow cytometer (Bio Rad).
Acknowledgments
Acknowledgements
We thank Anne Wahl and Anne Vad for excellent technical assistance, Kirsti Solberg at the Department of Biophysics’ flow cytometry facility for expert assistance with the flow cytometry, Erik Boye for critical reading of the manuscript, and Walter Messer for providing minichromosome pOC24. This work was supported by the Norwegian Research Council (K.S.), the Norwegian Cancer Society (A.L.-O. and K.S), the Danish Natural Sciences Research Council (A.L.-O.), the Carlsberg foundation (A.L.-O.) and the Novo Nordisk foundation (A.L.-O.).
References
- Arraj J.A., Wu,T.-H. and Marinus,M.G. (1990) Expression of a DNA methylation (dam) gene in Escherichia coli. Curr. Microbiol., 20, 133–136. [Google Scholar]
- Asai T., Chen,C.P., Nagata,T., Takanami,M. and Imai,M. (1992) Transcription in vivo within the replication origin of the Escherichia coli chromosome: a mechanism for activating initiation of replication. Mol. Gen. Genet., 231, 169–178. [DOI] [PubMed] [Google Scholar]
- Atlung T. and Hansen,F.G. (1993) Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J. Bacteriol., 175, 6537–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Clausen,E.S. and Hansen,F.G. (1985) Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet., 200, 442–450. [DOI] [PubMed] [Google Scholar]
- Atlung T., Løbner-Olesen,A. and Hansen,F.G. (1987) Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol. Gen. Genet., 206, 51–59. [DOI] [PubMed] [Google Scholar]
- Boye E. and Løbner-Olesen,A. (1990) The role of dam methyltransferase in the control of DNA replication in E.coli. Cell, 62, 981–989. [DOI] [PubMed] [Google Scholar]
- Boye E. and Løbner-Olesen,A. (1991) Bacterial growth control studied by flow cytometry. Res. Microbiol., 142, 131–135. [DOI] [PubMed] [Google Scholar]
- Boye E., Lyngstadaas,A., Løbner-Olesen,A., Skarstad,K. and Wold,S. (1993) Regulation of DNA replication in Escherichia coli. In Fanning,E., Knippers,R. and Winnacker,E.-L. (eds), DNA Replication and the Cell Cycle. Springer, Berlin, Germany pp. 15–26.
- Boye E., Stokke,T., Kleckner,N. and Skarstad,K. (1996) Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA, 93, 12206–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E., Løbner-Olesen,A. and Skarstad,K. (2000) Limiting DNA replication to once and only once. EMBO Rep., 1, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R.E., O’Day,K. and Wright,A. (1985) Autoregulation of the DNA replication gene dnaA in E.coli K-12. Cell, 40, 159–169. [DOI] [PubMed] [Google Scholar]
- Brooks J.E., Blumenthal,R.M. and Gingeras,T.R. (1983) The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res., 11, 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.L. and Kleckner,N. (1990) E.coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell, 62, 967–979. [DOI] [PubMed] [Google Scholar]
- Cassler M.R., Grimwade,J.E. and Leonard,A.C. (1995) Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J., 14, 5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B.B., Atlung,T. and Hansen,F.G. (1999) DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J. Bacteriol., 181, 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.J. and Maaløe,O. (1967) DNA replication and the division cycle in Escherichia coli. J. Mol. Biol., 23, 99–112. [Google Scholar]
- del Solar G., Giraldo,R., Ruiz-Echevarria,M.J., Espinosa,M. and Diaz-Orejas,R. (1998) Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev., 62, 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Ross,W., Wild,J. and Gourse,R.L. (1992) Involvement of Fis protein in replication of the Escherichia coli chromosome. J. Bacteriol., 174, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E.L. and Weisberg,R.A. (1985) Primary structure of the hip gene of Escherichia coli and of its product, the β subunit of integration host factor. J. Mol. Biol., 183, 117–128. [DOI] [PubMed] [Google Scholar]
- Gerdes K. et al. (1986) Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E.coli relB operon. EMBO J., 5, 2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M.S., Reed,R.R., Steitz,J.A. and Low,K.B. (2001) Identification of a sex-factor affinity site in E.coli as γδ. Cold Spring Harbor Symp. Quant. Biol., 45, 135–140. [DOI] [PubMed] [Google Scholar]
- Hansen F.G. and Atlung,T. (1995) Initiation of chromosome replication after induction of DnaA protein synthesis in a dnaA(nuII) rnh mutant of Escherichia coli. Mol. Microbiol., 15, 149–154. [DOI] [PubMed] [Google Scholar]
- Hansen F.G. and von Meyenburg,K. (1979) Characterization of the dnaA, gyrB and other genes in the dnaA region of the Escherichia coli chromosome on specialized transducing phages λ tna. Mol. Gen. Genet., 175, 135–144. [DOI] [PubMed] [Google Scholar]
- Hansen E.B., Atlung,T., Hansen,F.G., Skovgaard,O. and von Meyenburg,K. (1984) Fine structure genetic map and complementation analysis of mutations in the dnaA gene of Escherichia coli. Mol. Gen. Genet., 196, 387–396. [DOI] [PubMed] [Google Scholar]
- Hansen F.G., Christensen,B.B. and Atlung,T. (1991) The initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol., 142, 161–167. [DOI] [PubMed] [Google Scholar]
- Hansen F.G., Koefoed,S. and Atlung,T. (1992) Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol. Gen. Genet., 234, 14–21. [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E.coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Ozaki,T., Moriya,S. and Ogawa,T. (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev., 12, 3032–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A.C. and Helmstetter,C.E. (1986) Cell cycle-specific replication of Escherichia coli minichromosomes. Proc. Natl Acad. Sci. USA, 83, 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A. (1999) Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J., 18, 1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A. and von Freiesleben,U. (1996) Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J., 15, 5999–6008. [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Atlung,T. and Rasmussen,K.V. (1987) Stability and replication control of Escherichia coli minichromosomes. J. Bacteriol., 169, 2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad,K., Hansen,F.G., von Meyenburg,K. and Boye,E. (1989) The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell, 57, 881–889. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Boye,E. and Marinus,M.G. (1992) Expression of the Escherichia coli dam gene. Mol. Microbiol., 6, 1841–1851. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Hansen,F.G., Rasmussen,K.V., Martin,B. and Kuempel,P.L. (1994) The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J., 13, 1856–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Campbell,J.L., Boye,E. and Kleckner,N. (1994) SeqA: a negative modulator of replication initiation in E.coli. Cell, 77, 413–426. [DOI] [PubMed] [Google Scholar]
- Marszalek J. and Kaguni,J.M. (1994) DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem., 269, 4883–4890. [PubMed] [Google Scholar]
- Messer W., Bergmans,H.E., Meijer,M., Womack,J.E., Hansen,F.G. and von Meyenburg,K. (1978) Mini-chromosomes: plasmids which carry the E.coli replication origin. Mol. Gen. Genet., 162, 269–275. [DOI] [PubMed] [Google Scholar]
- Messer W., Bellekes,U. and Lother,H. (1985) Effect of dam methylation on the activity of the E.coli replication origin, oriC. EMBO J., 4, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Morige n, Boye,E., Skarstad,K. and Løbner-Olesen,A. (2001) Regulation of chromosomal replication by DnaA protein availability in Escherichia coli: effects of the datA region. Biochim. Biophys. Acta, 1521, 73–80. [DOI] [PubMed] [Google Scholar]
- Ogden G.B., Pratt,M.J. and Schaechter,M. (1988) The replicative origin of the E.coli chromosome binds to cell membranes only when hemimethylated. Cell, 54, 127–135. [DOI] [PubMed] [Google Scholar]
- Roth A. and Messer,W. (1998) High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol., 28, 395–401. [DOI] [PubMed] [Google Scholar]
- Russell D.W. and Zinder,N.D. (1987) Hemimethylation prevents DNA replication in E.coli. Cell, 50, 1071–1079. [DOI] [PubMed] [Google Scholar]
- Seitz H., Weigel,C. and Messer,W. (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol., 37, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Skarstad K., Boye,E. and Steen,H.B. (1986) Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J., 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., von Meyenburg,K., Hansen,F.G. and Boye,E. (1988) Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol., 170, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Løbner-Olesen,A., Atlung,T., von Meyenburg,K. and Boye,E. (1989) Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol. Gen. Genet., 218, 50–56. [DOI] [PubMed] [Google Scholar]
- Slater S. and Maurer,R. (1993) Simple phagemid-based system for generating allele replacements in Escherichia coli. J. Bacteriol., 175, 4260–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S., Wold,S., Lu,M., Boye,E., Skarstad,K. and Kleckner,N. (1995) E.coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell, 82, 927–936. [DOI] [PubMed] [Google Scholar]
- Smith D.W., Garland,A.M., Herman,G., Enns,R.E., Baker,T.A. and Zyskind,J.W. (1985) Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J., 4, 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C. and Messer,W. (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J., 20, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuitje A.R. and Meijer,M. (1983) Maintenance and incompatibility of plasmids carrying the replication origin of the Escherichia coli chromosome: evidence for a control region of replication between oriC and asnA. Nucleic Acids Res., 11, 5775–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torheim N.K., Boye,E., Løbner-Olesen,A., Stokke,T. and Skarstad,K. (2000) The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol. Microbiol., 37, 629–638. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen,K.V. and Schaechter,M. (1994) SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol., 14, 763–772. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Krekling,M.A., Hansen,F.G. and Løbner-Olesen,A. (2000a) The eclipse period of Escherichia coli. EMBO J., 19, 6240–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen,K.V., Atlung,T. and Hansen,F.G. (2000b) Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol., 37, 1087–1093. [DOI] [PubMed] [Google Scholar]
- von Meyenburg K. and Hansen,F.G. (1980) The origin of replication, oriC, of the E.coli chromosome: genes near oriC and construction of oriC deletion mutations. ICN–UCLA Symp. Mol. Cell. Biol., 19, 137–159. [Google Scholar]
- von Meyenburg K. et al. (1985) Facets of the chromosomal origin of replication, oriC, of Escherichia coli. In Schaechter,M., Neidhardt,F.C., Ingraham,J. and Kieldgaard,N.O. (eds), Molecular Biology of Bacterial Growth. Jones and Bartlett, Boston, MA, pp. 260–281. [Google Scholar]
- Wold S., Skarstad,K., Steen,H.B., Stokke,T. and Boye,E. (1994) The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J., 13, 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]