Abstract
Understanding the complex and varied mechanisms that regulate gene expression is an important and challenging problem. A fundamental sub-problem is to identify DNA binding sites for unknown regulatory factors, given a collection of genes believed to be co-regulated. We discuss a computational method that identifies good candidates for such binding sites. Unlike local search techniques such as expectation maximization and Gibbs samplers that may not reach a global optimum, the method discussed enumerates all motifs in the search space, and is guaranteed to produce the motifs with greatest z-scores. We discuss the results of validation experiments in which this algorithm was used to identify candidate binding sites in several well studied regulons of Saccharomyces cerevisiae, where the most prominent transcription factor binding sites are largely known. We then discuss the results on gene families in the functional and mutant phenotype catalogs of S.cerevisiae, where the algorithm suggests many promising novel transcription factor binding sites. The program is available at http://bio.cs.washington.edu/software.html.
INTRODUCTION
One of the major challenges facing biologists is to understand the varied and complex mechanisms governing the regulation of gene expression. This paper focuses on one important aspect of this challenge, the identification of binding sites in DNA for the factors involved in regulation. This is a necessary first step in determining which factors regulate the gene and how.
The analysis of non-coding regions in eukaryotic genomes in order to identify regulatory elements is a difficult problem and one that is not yet well solved. Some of the reasons for this difficulty are as follows: (i) binding sites of multiple interacting transcription factors often play a role in the regulation of a single gene; (ii) there can be great variability in the binding sites of a single factor, and the nature of the allowable variations is not well understood; (iii) the regulatory elements may be located quite far from the corresponding coding region, either upstream or downstream or in the introns.
Any algorithm whose goal is to discover novel regulatory elements takes as input a set of regulatory regions of genes, many of which are suspected to contain a common regulatory element. There are many possible sources for such co-regulated genes, including expression microarray experiments, gene knockout experiments and functional classes from the literature. This paper focuses on the regulation of genes in the yeast Saccharomyces cerevisiae, since much is known both about its transcription factors and about the functions of its genes.
A number of algorithms to discover general motifs have been proposed (1–9). Many of these algorithms are designed to find longer or more general motifs than are required for identifying transcription factor binding sites. The price paid for this generality is that many of the cited algorithms are not guaranteed to find globally optimal solutions, since they employ some form of local search, such as Gibbs sampling, expectation maximization or greedy algorithms, that may terminate in a locally optimal solution. There have been some studies that have applied these local search techniques specifically to the problem of identifying transcription factor binding sites in S.cerevisiae, with some success (10–14).
The number of well conserved bases in the collection of binding sites of a single S.cerevisiae transcription factor is typically six to ten (15–16). This number is small enough that, for this particular problem, one need not rely on such general local search heuristics. Instead, one can afford to use enumerative methods that guarantee global optimality. This is the approach taken by the current paper, whose method is most closely allied to those of van Helden et al. (17–19) and Tompa (20). There are also other studies using an enumerative approach to motif finding (21–23).
We review a motif model that is tailored to accurately represent transcription factor binding sites in S.cerevisiae. We then review an enumerative algorithm from Sinha and Tompa (24) called YMF (Yeast Motif Finder) which, given the regulatory regions of several related genes, is guaranteed to produce the motifs with greatest z-scores. The present paper focuses on the application of that method to classes of yeast genes. We first present the results of validation experiments in which YMF was used to identify candidate binding sites in several well studied regulons of S.cerevisiae, where the most prominent transcription factor binding sites are largely known. We then present results on gene families in the functional and mutant phenotype catalogs of S.cerevisiae taken from the MIPS database (25), where YMF suggests many novel transcription factor binding sites. Our goal was to discover motifs in the classes from these catalogs, since genes with common mutant phenotypes or common function may have the same regulatory mechanism and hence may share informative binding sites.
Hughes et al. (11) performed a similar analysis of the MIPS functional catalog using AlignACE, a local search algorithm based on Gibbs sampling. There are a number of differences between their results and ours. Most important is that differences in the motif model and search method (local search heuristic versus enumerative search) lead to different significant motifs. In a separate paper (S.Sinha and M.Tompa, in preparation) we compare the accuracy of YMF and other methods such as AlignACE on both simulated data and on yeast regulons. Those results suggest that YMF may provide more accurate prediction of regulatory elements. A second difference is that Hughes et al. (11) merge motifs found in different functional classes. As a result, the important connection between transcription factor binding site and gene function, necessary for understanding regulatory relationships, is not apparent from their tables, whereas it is explicit in ours. Finally, Hughes et al. (11) report results only on the functional catalog and not on the mutant phenotype catalog.
MATERIALS AND METHODS
Variability among binding site instances
The first question that must be addressed is ‘What constitutes a motif?’ for the application of transcription factor binding sites in S.cerevisiae. An inspection of transcription factor databases such as TRANSFAC (15; http://transfac.gbf-braunschweig.de/TRANSFAC/) and SCPD (16; http://cgsigma.cshl.org/jian/) and of the relevant literature (26–33), particularly Jones et al. (26), which is rich in examples, reveals that there is significant variation among the binding sites of any single transcription factor. Moreover, the nature of the variability itself varies from factor to factor, so that the ‘correct’ motif model is far from clear.
Certain trends that must be incorporated in the motif model do, however, emerge from this literature, particularly from SCPD (see the column labeled ‘Consensus’ in Table 1 for examples). (i) Many of the motifs, such as the Gal4p binding site CGGNNNNNNNNNNNCCG, have spacers varying in length from 1 to 11 bp. The spacers usually occur near the middle of the motif, often because the factors bind as dimers or tetramers. (ii) The number of well conserved bases (not including spacers, of course) is usually in the range 6–10. This number is called the length of the motif. (iii) When there is variation in a conserved motif position, it is often a transition (i.e. the substitution of a purine for a purine or a pyrimidine for a pyrimidine) rather than a transversion. This is because of the similarity in nucleotide size necessary to fit the transcription factor’s fixed DNA-binding domain. Somewhat less often, the variation in a given position may be between a pair of complementary bases. Other positional variations are rarer. (iv) Insertions and deletions among binding sites are uncommon, again because of the fixed structure of the factor’s DNA-binding domain.
Table 1. Performance of YMF on regulons in SCPD with known binding sites.
| Name | Size | Consensus | l = 6 | Ns | z | pmax | l = 7 | Ns | z | pmax | l = 8 | Ns | z | pmax |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABF1 | 19 | TCRN[6]ACG | TCAN[6]ACG | 29 | 10.56 | 0.00 | TTTYYTT | 180 | 7.96 | 0.01 | AGSCYCGC | 7 | 19.93 | 0.11 |
| CAR1 | 12 | AGCCGCCRa | TTTNNNTTY | 146 | 9.09 | 0.00 | TTTTYTT | 112 | 8.86 | 0.01 | AGCCGCCG | 4 | 14.18 | 0.00 |
| CPF1 | 3 | TCACGTG | CACGWG | 9 | 6.94 | 0.40 | YCACGWG | 8 | 9.66 | 0.10 | CACGTGGC | 3 | 19.49 | 0.00 |
| CSRE | 4 | YCGGAYRRAWGG | CGGN[6]GGA | 9 | 12.43 | 0.00 | CTCCGGG | 3 | 10.22 | 0.04 | CGGGCCCG | 2 | 14.79 | 0.08 |
| GAL4 | 6 | CGGN[11]CCG | CGGN[11]CCG | 28 | 32.72 | 0.00 | CGCWCGG | 6 | 13.11 | 0.00 | CGCACGGA | 3 | 17.14 | 0.00 |
| GCN | 38 | TGANTN | TGACTC | 44 | 12.54 | 0.00 | RTGASTC | 47 | 13.76 | 0.00 | TGASTCAY | 31 | 15.87 | 0.00 |
| GCR1 | 6 | CWTCC | TTTN[4]TYY | 111 | 8.53 | 0.04 | SCAYGTG | 10 | 8.55 | 0.11 | CGGGATTC | 3 | 13.92 | 0.08 |
| HAP1 | 5 | CGGNNNTANCGG | SGGN[6]SGG | 8 | 9.01 | 0.05 | TGSCCCC | 5 | 8.50 | 0.13 | GGGGSCAW | 5 | 11.18 | 0.61 |
| HSE | 6 | GAANNTCC | AACNNNCRG | 14 | 6.27 | 0.52 | TCYAGAA | 12 | 6.98 | 0.65 | ACTCCGTG | 2 | 9.36 | 0.93 |
| MATa2 | 7 | CRTGTWWWW | ATGN[10]CAY | 25 | 6.78 | 0.21 | CATGTWW | 17 | 7.31 | 0.41 | YCACGAAA | 7 | 9.76 | 0.81 |
| MCB | 6 | WCGCGW | ACGCGT | 16 | 11.98 | 0.00 | RACGCGT | 12 | 13.97 | 0.00 | SCGACGCG | 4 | 20.22 | 0.00 |
| MCM1 | 23 | CCNNNWWRGG | ARAN[4]AAR | 317 | 10.66 | 0.00 | TRTRTAT | 81 | 9.01 | 0.00 | ATAYAYAT | 58 | 13.09 | 0.00 |
| MIG1 | 9 | CCCCRNNWWWWW | CGGN[11]CCG | 16 | 14.95 | 0.00 | CCCCRGR | 9 | 8.56 | 0.03 | ARCCGCCG | 5 | 13.20 | 0.05 |
| PDR3 | 7 | TCCGYGGA | CCGNGGA | 28 | 28.39 | 0.00 | CCGYGGA | 28 | 38.56 | 0.00 | TCCGYGGA | 24 | 55.17 | 0.00 |
| PHO4 | 5 | CACGTK | SCACGT | 18 | 12.12 | 0.00 | CACGTGS | 10 | 14.42 | 0.00 | CACGTGGG | 3 | 16.75 | 0.00 |
| RAP1 | 16 | RMACCCA | CSCNNNCRC | 30 | 9.30 | 0.00 | GCAYGTG | 13 | 10.37 | 0.00 | CCCGWYGC | 7 | 11.37 | 0.06 |
| REB1 | 14 | YYACCCG | RAAN[5]AAR | 219 | 9.86 | 0.00 | TTACCCG | 12 | 14.10 | 0.00 | ATTACCCG | 8 | 17.94 | 0.00 |
| ROX1 | 3 | YYNATTGTTY | AAANNAAA | 59 | 11.71 | 0.00 | AAARRAA | 71 | 11.81 | 0.04 | CCGACGTC | 2 | 15.99 | 0.09 |
| SCB | 3 | CNCGAAA | CACGAA | 10 | 9.44 | 0.01 | CACGAAA | 10 | 15.92 | 0.00 | YCACGAAA | 9 | 20.05 | 0.00 |
| SFF | 3 | GTMAACAA | ATTN[9]TTW | 28 | 6.37 | 0.74 | GATCTAT | 4 | 7.18 | 0.86 | ACACTCCG | 2 | 13.25 | 0.28 |
| STE12 | 4 | ATGAAA | TRCN[7]GGW | 17 | 6.56 | 0.44 | ATGAAAC | 9 | 9.44 | 0.07 | ACAARGCC | 5 | 11.93 | 0.48 |
| TBP | 17 | TATAWAW | AAANNNAAA | 127 | 7.92 | 0.00 | TTTTYTT | 130 | 7.74 | 0.02 | AAARAAAA | 89 | 10.46 | 0.14 |
| UASPHR | 17 | CTTCCT | TAYNTAY | 107 | 7.46 | 0.01 | CRRCAAC | 26 | 8.15 | 0.02 | CARCARCA | 25 | 13.49 | 0.01 |
Name, name of the transcription factor; size, number of genes in the regulon; consensus, the known consensus of the binding site, according to SCPD. The remaining columns tabulate the results of YMF for the three runs (l = 6, 7 and 8, respectively) on that regulon. Each column lists the most significant motif found, its total count Ns, its z-score z, and pmax(z). Bold indicates a match to the known consensus.
aThe consensus for CAR1 listed at SCPD is AGCCGCSA, but an alignment of the 12 listed sites suggests the consensus AGCCGCCR.
Based on these observations, a motif for our application is a string of length 6–10 over the alphabet {A,C,G,T,R,Y,S,W}, with 0 or more consecutive N residues inserted at the center, and a limited number of R (purine), Y (pyrimidine), S (strong) and W (weak) characters, also called degenerate symbols. We choose such a consensus model rather than (say) a weight matrix in order to be able to enumerate motifs. An examination of the 50 binding site consensi included in SCPD (16) revealed that the number of consensi that exactly fit this characterization is 34 (68%). About 10 more fit the characterization if very slight differences from the exact consensus are tolerated.
Measure of statistical significance
Given some set of (presumably co-regulated) S.cerevisiae genes, the input to YMF is the corresponding set of promoter regions, each having length 800 bp and having its 3′ end at the gene translation start site. For each motif s, let Ns be the number of occurrences of s in the input sequences, allowing an arbitrary number of occurrences in both orientations per promoter region. A reasonable measure of s as a motif will reflect how unlikely it would be to have Ns occurrences, if the sequences were instead drawn at random according to the background distribution. We use as this measure the statistical significance of the ‘z-score’ of Ns. First, to specify the background distribution, let X be a set of random DNA sequences of the same number and length as the input promoter sequences, but generated by a Markov chain of order m, whose transition probabilities are determined by the (m + 1) mer frequencies in the full complement of 6000+ promoter regions (each of length 800 bp) of S.cerevisiae. In our experiments, we chose m = 3 in order for the background model to account for the TATA, AAAA and TTTT sequences that are ubiquitous throughout the genome’s promoter regions (17). Let the random variable Xs be the number of occurrences of the motif s in these random sequences X and let E(Xs) and σ(Xs) be its mean and standard deviation, respectively. Then the z-score associated with s is
zs = (Ns – E(Xs))/σ(Xs)1
The measure zs is the number of standard deviations by which the observed value Ns exceeds its expectation. See Leung et al. (34) for a detailed discussion of this statistic.
The z-score zs obeys a normal distribution in the asymptotic limit as the total length of the input promoter regions increases (35). If the assumption of normality is inaccurate, it may not be as meaningful to compare the z-scores of different motifs. In view of this, YMF will be most accurate when the total length of the input promoter region is large. The ultimate test of the method, however, is not whether the z-score passes normality tests, but whether YMF successfully predicts true transcription factor binding sites. Therefore, in order to demonstrate the robustness of the method, in the validation experiments on known regulons we report the results on regulons consisting of as few as three genes.
Since YMF enumerates a large motif space, thereby sampling a large number of points from the distribution, it is expected that some motifs will have a high z-score by chance. To address this, we associate with z-score x a significance pmax(x), which measures the probability that the maximum z-score is at least x, if the input sequences were random. This maximum is taken over all motifs of the given length, number of spacers and number of degenerate symbols. We precompute pmax for a variety of motif parameters and input sequence lengths, by simulation. Random sequences of the same length as the input promoter regions are generated according to the Markov model being used, and YMF is run on these random sequences. The maximum z-score reported is recorded. This experiment is repeated 100 times. The fraction of experiments that yielded maximum z-score at least x is used as an estimate of pmax(x).
Algorithm summary
The algorithm used by YMF is summarized here. The inputs to the algorithm are as follows: (i) a set of promoter regions; (ii) the number l of non-spacer characters in the motifs to be enumerated (called the motif length); (iii) the maximum number w of spacers in the motifs; (iv) the transition matrix for a third order Markov chain modeling the background distribution of promoter regions.
The parameters l and w, along with the implicitly assumed motif model, define a search space of all candidate motifs that will be evaluated. This space consists of all motifs that have l characters from {A,C,G,T,R,Y,S,W}, and between 0 and w spacers (N) in the middle. Typically, the maximum number of degenerate symbols (R, Y, S or W) was restricted to 2 for computational efficiency, although YMF can be configured to handle different values of this parameter. YMF first makes a pass over the input sequences, tabulating the number Ns of occurrences of each motif s in either orientation, including overlapping occurrences. For each motif s for which Ns > 0, it then computes the mean and standard deviation of the motif count using a method described by Sinha and Tompa (24). Finally, it uses equation 1 to compute the z-score zs and pmax(zs) and outputs the motifs sorted by z-score.
Because the number of motifs is exponential in l, we can afford this enumerative method only for modest values of l. In contrast, however, the running time is linear in the size of the input sequences, so that the method scales very well to larger gene families and longer promoter regions. The current implementation typically runs in a few seconds for motifs of length 6 on a Pentium processor with 256 MB memory. For length 9 motifs it requires a few minutes.
Both a web interface and the source code for YMF are freely available at http://bio.cs.washington.edu/software.html.
Experimental methods
The maximum number w of spacers allowed in a motif was varied depending on the motif length parameter l. For l = 6, we used w = 11, which means that length 6 motifs were allowed to have between 0 and 11 spacers in the middle. This is in accord with observed motifs from SCPD. However, this introduces an inherent bias in the method toward finding motifs with spacers, since there are 11 times as many motifs with spacers as without. To include some runs without this bias, when YMF was run with l > 6, we used w = 0, i.e. no spacers allowed.
There are three different types of post-processing steps that were used to produce the most promising candidate binding sites to report. The first is a tool called FindExplanators (36). A set of promoter sequences having bindings sites for a few different transcription factors typically contains hundreds of statistically overrepresented motifs, most of them being minor variations of the true binding site motifs. YMF will report all these overrepresented motifs. For example, suppose a factor binds to TCACGCT in a set of sequences, causing this motif to be overrepresented. Many of its variations, e.g. CACGCTT or TCACGCW, are also likely to be overrepresented, simply because each has its number of occurrences artifically increased by the presence of TCACGCT. FindExplanators is a tool that extracts the few significantly independent motifs from the vast number that are simply artifacts of these few.
Since YMF evaluates a motif based on its total number of occurrences in a set of sequences, a motif may have a high z-score (low pmax) even if it occurs unusually often in only one of the promoters. Such motifs may not be interesting candidates for transcription factor binding sites. Multiple occurrences of a motif in a promoter suggest some significance, but a very large number of occurrences in the same promoter may suggest a repetitive element rather than a regulatory element. Thus motifs are post-processed so that those that have high z-scores due to a large number of occurrences in one or two promoters are not reported. For this purpose we developed a numerical measure that captures the notion of a motif being well distributed among the promoters. Given a set X of promoters and a motif s, we first count the occurrences of s in each promoter. Let X+ be the set of promoters that have at least one occurrence of s and let D = {d1, d2, …, dp} (where p = |X+|) be the distribution of occurrences of s in X+. Intuitively, a well distributed motif is one for which D has a low variance. However, the variance itself is not comparable for sample distributions obtained from different populations, so we normalize it by dividing by the expectation. Thus, our statistic is w = Σi = 1 … p (di – µ)2/µ, where µ is the mean of the distribution D. We call this the w-score of motif s. Lower values of the w-score indicate better distributed motifs. Note that w is identical to the χ2 statistic and we use the χ2 distribution with p – 1 degrees of freedom to compute a significance threshold on the w-score. Notice that we compute w from X+ and not from X, since in general we may not find the binding site present in all the input promoter sequences.
As another means of evaluating the motifs, we introduce a co-expression score, which measures the similarity of the expression profiles of the genes corresponding to X+. This score is computed from data in the database ExpressDB (37), which catalogs mRNA expression level information from several different studies under a common framework. The expression data is normalized across studies by converting them into estimated relative abundancies or ‘ERAs’. Such values are available for all yeast genes under 217 different conditions. For each pair of genes, we compute the correlation coefficient of their ERA values. Given the set X+, we compute the average pairwise correlation coefficient over all pairs of genes in X+. We then estimate a p-value of this average pairwise correlation coefficient, by choosing |X+| random genes and computing the same score for these, and repeating several times. This p-value is called the co-expression score of X+ and a low value indicates that the genes in X+ have an unusually high pairwise correlation coefficient on average.
RESULTS AND DISCUSSION
Validation on known regulons
The SCPD database (16) has a collection of transcription factors and the genes regulated by each factor. Each such set of genes comprises a regulon. For each gene in a regulon, the database lists the experimentally determined binding sites of the transcription factor, and in many cases the consensus sequence of the binding sites in the regulon is also given. It is not always clear from the binding sites alone what their consensus should be, because there is often more than one way to align them, and to choose a consensus with degenerate symbols. Hence, we rely on the consensus listed at SCPD. YMF was run on each regulon in SCPD that has at least three genes and has a cataloged consensus sequence for its binding sites. There are 23 such regulons. The success of YMF was assessed by comparing the top motifs reported with the known consensus for the regulon. The program was run three times on each regulon, to find motifs of length 6, 7 and 8, respectively. For length 6 motifs, a maximum of 11 spacers in the middle was allowed. For lengths 7 and 8, the motif model did not include spacers (see Materials and Methods for details). In all runs, a maximum of two degenerate symbols (R, Y, S or W) was allowed in the candidate motifs.
The results are summarized in Table 1. Each row corresponds to a regulon. For each of the three runs of YMF on that regulon, the motif with greatest z-score is presented, along with its total count in the input promoter regions, its z-score z, and pmax(z). Lower pmax values are indicative of higher statistical significance (see Materials and Methods). Reported motifs that can be superimposed with the known consensus for the regulon without conflicting characters at any position, and that have at least four positions (possibly degenerate symbols) identical to the consensus are considered matches and are typeset in bold.
For 15 of the 23 regulons, the top motif reported (for one or more values of the motif length parameter) was a match. For 14 of the 15 regulons, there was a match with pmax < 0.1, the exception being MATa2. In another regulon, MCM1, the top ranking motifs (for length 6) were variants of the poly(A) element (any motif that can be instantiated to a string of all A residues, e.g. AAAAWAAA), and the first non-poly(A) motif, at rank 11 with pmax 0.01, was CCSNNNNAGG, similar to the known consensus CCNNNWWRGG. For the regulon RAP1, the top motif reported (for length 7) is GCAYGTG, which matches part of the inositol/choline response element (ICRE) with consensus SCAYRTGAARW (we discuss this motif and its connection to the RAP1 regulon below). The first motif reported by YMF (for the same length parameter) that is not a variant of GCAYGTG is RCACCCA, at rank 11 with pmax 0.02. Note that this closely matches the known consensus RMACCCA for the RAP1 regulon. For the regulon HSE,HSTF, the consensus cataloged at SCPD is GAANNTCC. However, an alignment of the known binding sites of this transcription factor, as reported in the same database, reveals a consensus pattern of TCTAGAA. This closely matches the top motif TCYAGAA reported by YMF for length 7. Thus, counting MCM1, RAP1 and HSE,HSTF also as successes, we are left with only five regulons (GCR1, ROX1, SFF, TBP and UASPHR) on which YMF failed to report any match to the known binding site consensus. Note that the 23 regulons represent the typical input for a motif-finder; they are of varying sizes (3–38 genes) and have a variety of known binding sites (length 5–10, with few to many spacers or degenerate symbols). The results thus demonstrate the applicability of the method on a variety of data sets.
In most cases, a match was found in the top three motifs for multiple values of l, indicating that the performance is not crucially dependent on prior knowledge of the motif length. In some cases, YMF found a match even though the known consensus of the binding site does not conform to the motif model YMF uses. For instance, the regulon SCB has the sequence CNCGAAA as its binding site consensus, with an ‘N’ that is not in the middle. Nevertheless, a very similar motif CACGAAA was reported. Similarly, for HAP1 (consensus CGGNNNTANCGG), the motif SGGNNNNNNSGG was discovered.
The regulon ABF1 is an example of a case where multiple occurrences of the binding site are found in the same promoter region. Of the 19 genes in this regulon, eight have two or more occurrences of the motif TCRNNNNNNACG in their promoter region. There are a total of 36 occurrences of the motif, giving it a very high z-score of 10.07. If each of the 19 genes had only one occurrence of the motif, for a total of 19 occurrences, the z-score would have been about 4.03, which is rather low, meaning that the motif would not have been reported as significant.
As noted above, a pmax value of 0.1 or less served as a good indicator of a significant motif, in the sense that most of the matches occurred with pmax < 0.1. We therefore examined all motifs (from Table 1) that are reported to have a pmax value less than this threshold, to see if there are interesting signals in the regulon that are different from the known binding sites, and also to have an idea of the false positive rate. Table 2 summarizes our observations. It includes each motif from Table 1 that has a pmax value <0.1, is not a poly(A), poly(T) or TATA motif and is not a match. There are 13 such motifs. Three of them (CGCWCGG and CGCACGGA in the GAL4 regulon and ARCCGCCG in the MIG1 regulon) occur overlapping with known Gal4 binding sites in the respective promoters. The motif CGGNNNNNNNNNNNCCG in the MIG1 regulon is identical to the Gal4 binding site consensus, and this family contains the genes Gal3, Gal10, Gal1 and Gal4, which are known to contain this binding site. The motif GCAYGTG in the RAP1 regulon matches a prefix of the ICRE consensus SCAYRTGAARW (38,39). Among the genes of this regulon are Fas1, known to contain the ICRE motif (40), and Opi3 and Itr1, both known to have the similar motif CATGTGAA, which is shared by promoters of phospholipid synthetic enzymes such as these two (25). Thus, for five of the motifs in Table 2, there is strong evidence that they correspond to known binding sites of other transcription factors, leaving eight motifs about which we do not have any clear evidence. Even if we regard all these eight motifs as spurious, the resulting false positive rate would be small, considering that a total of 39 motifs in Table 1 meet the criteria of having pmax < 0.1 and not being a poly(A), poly(T) or TATA motif. Some occur at approximately conserved positions relative to the translation start site, strengthening the possibility that they might be targets of other transcription factors.
Table 2. Motifs in SCPD regulons that are different from the known principal binding sites.
| Name | Motif | Ns | z | pmax | Comments |
|---|---|---|---|---|---|
| CSRE | CTCCGGG | 3 | 10.22 | 0.04 | Occurs in 2 of 4 genes; roughly conserved in position |
| CGGGCCCG | 2 | 14.79 | 0.08 | Occurs in only one gene | |
| GAL4 | CGCWCGG | 6 | 13.11 | 0.00 | All occurrences overlap Gal4-binding sites (in Gal1, Gal2, Gal7 and Gal10) |
| CGCACGGA | 3 | 17.14 | 0.00 | All occurrences overlap Gal4-binding sites (in Gal1, Gal2 and Gal7) | |
| GCR1 | CGGGATTC | 3 | 13.92 | 0.08 | Occurs in 3 of 6 genes; roughly conserved in position |
| MIG1 | CGGN[11]CCG | 16 | 14.95 | 0.00 | Is identical to the GAL4 motif |
| ARCCGCCG | 5 | 13.20 | 0.05 | Occurs overlapping known Gal4-binding sites in Gal3, Gal10 and Gal1. The other two occurrences are in Fbp and Fps1 | |
| RAP1 | CSCNNNCRC | 30 | 9.30 | 0.00 | Occurs in 13 of 16 genes; roughly conserved in position |
| GCAYGTG | 13 | 10.37 | 0.00 | Is similar to the ICRE motif (39) | |
| CCCGWYGC | 7 | 11.37 | 0.06 | Occurs in 3 of 16 genes; well conserved position | |
| ROX1 | CCGACGTC | 2 | 15.99 | 0.09 | Occurs only in Rox1 gene |
| UASPHR | CRRCAAC | 26 | 8.15 | 0.02 | Occurs in 7 of 17 genes; roughly conserved in position |
| CARCARCA | 25 | 13.49 | 0.01 | Occurs in 3 of 17 genes; not conserved in position |
The table lists the motif sequence, its total count Ns, its z-score z, and pmax(z).
Results on MIPS catalogs
The MIPS database at the Munich Information Center for Protein Sequences (25) catalogs yeast genes classified according to different criteria. One such catalog is based on gene function, while another classifies genes based on phenotypes with which mutant versions of the genes have been implicated. These catalogs will be referred to as the functional and the phenotype catalogs, respectively. Each catalog has a hierarchical organization, the different levels of the hierarchy corresponding to different degrees of specificity of the classification criterion. Our goal was to discover motifs in the classes from these catalogs, since many genes with common mutant phenotypes or common function may have the same regulatory mechanism and hence may share binding sites. We extracted from each catalog the classes that were at or near the bottom of the hierarchy and had five or more genes. YMF was run on the 800 bp long promoter regions of genes in each class, with the same set of parameter values as in the experiments on SCPD regulons. In some cases, a class of genes contains one or more pairs of divergent genes, whose promoter regions overlap. For such pairs, the single promoter region between the two genes replaced two separate promoters. The top 1000 motifs from each of the three runs of YMF were input to the program FindExplanators (see Materials and Methods), which reported the three best independent motifs in its input list of 1000 motifs. For classes with over 100 genes, only the single best motif of the 1000 was reported, for computational efficiency. For each motif obtained from the previous step, the w-score (see Materials and Methods) was computed to measure how well distributed the motif is, and motifs with poor w-scores (at 95% level of significance) were rejected. All motifs with pmax > 0.1, as well as those that are poly(A), poly(T) or TATA repeats, were rejected. Matches of each of the remaining motifs to binding sites of known transcription factors in yeast (as cataloged in the database TRANSFAC) are reported. Also, for each remaining motif, the co-expression score was computed (see Materials and Methods) for the set of genes in the class that contain the motif in their promoters.
Functional catalog. There were 204 classes extracted from the functional catalog and the motif-finding steps reported a total of 465 motifs. Tables 3 and 4 present a selection of the results. This selection was done by manual inspection of the 465 reported motifs, using the following more stringent criteria. Motifs with pmax > 0.05 were eliminated. If a single functional class had motifs of different lengths that were variants of each other, only that with the least pmax value was retained. Motifs that had more than two matches to known binding sites of the same transcription factor are presented in Table 3, while the others are in Table 4, and are good candidates as novel transcription factor binding sites.
Table 3. Significant motifs in classes from the MIPS functional catalog.
| Name | Motif | Ns | z | pmax | Comments |
|---|---|---|---|---|---|
| Regulation of amino acid metabolism | YCACGTGC | 11 | 14.78 | 0.00 | CBF1 (TCACGTG) has role in amino acid metabolism (MIPS) |
| Nitrogen and sulfur metabolism | TCACGTG | 18 | 7.94 | 0.00 | CBF1 (TCACGTG) has role in nitrogen and sulfur metabolism (MIPS) |
| Tricarboxylic acid pathway | TCACGTG | 11 | 8.45 | 0.00 | CBF1 (TCACGTG) |
| Other transcription activities | TCACGTG | 16 | 7.65 | 0.00 | CBF1 (TCACGTG) |
| Mitochondrial transport | TCACGTG | 19 | 7.41 | 0.00 | CBF1 (TCACGTG) |
| Anion transporters (Cl, PO4 etc.) | CACGTG | 20 | 6.89 | 0.00 | PHO4 (CACGTK) has role in phosphate metabolism (MIPS) |
| Phosphate metabolism | CACGTGSG | 12 | 19.09 | 0.00 | PHO4 (CACGTK) has role in phosphate metabolism (MIPS) |
| Homeostasis of phosphate | CACGTGSG | 5 | 17.53 | 0.00 | PHO4 (CACGTK) has role in phosphate metabolism (MIPS) |
| DNA synthesis and replication | ACGCGW | 124 | 16.04 | 0.00 | MCBF (WCGCGW) is involved in DNA synthesis (45) |
| DNA repair | ACGCGWW | 49 | 7.77 | 0.00 | MCBF (WCGCGW) binds to MCB in DNA replication genes (TRANSFAC) |
| Deoxyribonucleotide metabolism | ACGCGY | 27 | 11.85 | 0.00 | MCBF (WCGCGW) is involved in DNA synthesis (45) |
| Cellular import | YCCCCAC | 27 | 7.69 | 0.00 | MIG1 (CCCCRNNWWWWW) is known to regulate some of the HXT genes in this class (43) |
| C-compound, carbohydrate transport | CYCCRC | 77 | 10.43 | 0.00 | MIG1 (CCCCRNNWWWWW) has role in C-compound metabolism (MIPS) |
| C-compound, carbohydrate transporters | CCCCRC | 40 | 9.39 | 0.00 | MIG1 (CCCCRNNWWWWW) has role in C-compound metabolism (MIPS) |
| Meiosis | TAGCCGCC | 23 | 23.54 | 0.00 | Repressor-of-CAR1 (AGCCGCCR) has role in meiosis (MIPS) |
| Amino acid transporters | GCCGCCGA | 5 | 12.81 | 0.00 | Repressor-of-CAR1 (AGCCGCCR) has role in amino acid metabolism (MIPS) |
| Homeostasis of metal ions | GSACCC | 46 | 7.60 | 0.00 | Rap1 (RMACCCA) |
| Cation transporters | GSACCC | 42 | 6.71 | 0.00 | Rap1 (RMACCCA) |
| Regulation of amino acid metabolism | RTGN[5]GTR | 93 | 8.91 | 0.00 | Matches 9 RAP1 binding sites |
| Ribosome biogenesis | AYCCRTAC | 104 | 28.87 | 0.00 | Matches 4 RAP1 binding sites. Rap1 controls transcription of most ribosome protein genes (MIPS) |
| Assembly of protein complexes | TTANCCG | 52 | 7.21 | 0.00 | REB1 (YYACCCG) |
| Cytoplasmic and nuclear degradation | TTACCCG | 28 | 10.93 | 0.00 | REB1 (YYACCCG) |
| Vesicular transport (Golgi network) | TACCCGG | 22 | 9.20 | 0.00 | REB1 (YYACCCG) |
| Cellular communication mechanism | TTACCCG | 17 | 8.62 | 0.00 | REB1 (YYACCCG) |
| Cell growth/morphogenesis | TYACCCG | 30 | 7.86 | 0.00 | REB1 (YYACCCG) |
| Intracellular transport vesicles | TACCCGG | 11 | 8.65 | 0.00 | REB1 (YYACCCG) |
| Vacuole or lysosome | TTACCCG | 15 | 7.51 | 0.00 | REB1 (YYACCCG) |
| Cytoskeleton | TTACCCG | 26 | 9.45 | 0.00 | REB1 (YYACCCG) |
| General transcription activities | TTACCCG | 19 | 8.95 | 0.00 | REB1 (YYACCCG) |
| Purine ribonucleotide metabolism | TGACTC | 31 | 6.90 | 0.00 | GCN4 (TGANTN) regulates general control in response to purine starvation (MIPS) |
| Amino acid transporters | TSASTC | 54 | 6.40 | 0.03 | GCN4 (TGANTN) is a transcriptional activator of amino acid biosynthetic genes (MIPS) |
| Drug transporters | CSGN[9]CGS | 40 | 9.24 | 0.00 | Matches 4 GAL4 binding sites |
| Cell wall | TCCGAA | 33 | 7.51 | 0.00 | Matches 5 GAL4 binding sites |
| Directional cell growth (morphogenesis) | CRYN[6]CGA | 44 | 6.06 | 0.05 | Matches 5 TAF binding sites |
| Intracellular transport vesicles | CGTN[7]GAY | 40 | 6.25 | 0.00 | Matches 11 BAF1 binding sites. Baf1 is a multifunctional protein involved in transcriptional regulation of various genes (MIPS) |
| Extracellular/secretion proteins | CCTAATT | 12 | 7.32 | 0.05 | Matches 3 MCM1 binding sites. Three (Mfα 1 and 2, HSP150) of the five genes that have this motif are known to be regulated by MCM1 (SCPD) |
| Nitrogen and sulfur utilization | GATAAG | 52 | 9.58 | 0.00 | GATA box (GATAAG). The four GATA-binding factors (Gtz3, Dal80, Gln3, Gat1) regulate the expression of nitrogen catabolic genes (MIPS) |
| Nitrogen and sulfur metabolism | AAGATAAG | 23 | 10.76 | 0.00 | GATA box (GATAAG). The four GATA-binding factors (Gtz3, Dal80, Gln3, Gat1) regulate the expression of nitrogen catabolic genes (MIPS) |
| Other cation transporters | AGAYAAG | 32 | 7.66 | 0.00 | GATA box (GATAAG) |
| Lipid and fatty-acid transport | TCCGCGGR | 12 | 18.19 | 0.00 | PDR1/PDR3 (TCCGYGGA) |
| Homeostasis of metal ions | TCCGYGGA | 13 | 9.18 | 0.02 | PDR1/PDR3 (TCCGYGGA) are implicated in transcription of two of the four genes in this class that have the motif (46) |
| Transport mechanism | CCGYGGA | 24 | 8.48 | 0.00 | PDR1/PDR3 (TCCGYGGA) are implicated in transcription of five of the nine genes in this class that have the motif (46) |
| C-compound and carbohydrate transporters | TCCGYGS | 26 | 8.39 | 0.00 | PDR3 (TCCGYGGA) binds Hxt11 |
| Cation transporters | YGSACCC | 32 | 9.16 | 0.00 | AFT1 (TRCACCC) is involved in homeostasis of iron (MIPS) |
| Metabolism of energy reserves (glycogen, trehalose) | CCCCTGA | 13 | 10.20 | 0.00 | STRE (CCCCT) is a stress response regulatory element. Three (TPS1, GSY1, PGM2) of the 11 genes in this class that have this motif are induced by stress |
| Proteolytic degradation | GGTGGCAA | 38 | 17.71 | 0.00 | RPN4 (GGTGGCAAA) |
| Peroxisome | TYGGRGT | 30 | 7.36 | 0.00 | ADR1 (TYGGRG) regulates peroxisomal genes (MIPS) (47) |
The last column is the name of the known transcription factor binding site whose consensus sequence is similar to the found motif. The consensus sequence was obtained from SCPD (16) for all except the following: the consensus for MET was obtained from van Helden et al. (17); the consensus for RPN4 from Hughes et al. (11).
Table 4. Significant novel motifs in classes from the MIPS functional catalog.
| Name | Size | Motif | Ns | z | pmax |
|---|---|---|---|---|---|
| Amino acid transport | 23 | GCCGTRCS | 13 | 17.70 | 0.00 |
| GYCGCCGA | 7 | 13.47 | 0.00 | ||
| GAWAGCG | 19 | 9.08 | 0.00 | ||
| Amino acid degradation (catabolism) | 35 | CGGN[10]YCG | 29 | 8.94 | 0.00 |
| GACTSCGS | 14 | 14.99 | 0.00 | ||
| Regulation of nitrogen and sulfur utilization | 29 | CGGN[10]SGS | 27 | 6.63 | 0.01 |
| Purine ribonucleotide metabolism | 45 | GGCTAGGA | 7 | 10.69 | 0.01 |
| Deoxyribonucleotide metabolism | 11 | CGCN[8]GYG | 17 | 8.87 | 0.00 |
| Polynucleotide degradation | 27 | CTYATCGC | 9 | 10.23 | 0.02 |
| Nucleotide transport | 14 | CGCGSGC | 10 | 13.11 | 0.00 |
| Phosphate transport | 10 | CGGN[4]GSS | 19 | 9.22 | 0.00 |
| Regulation of lipid, fatty acid and isoprenoid metabolism | 20 | CGSN[6]CCS | 23 | 6.83 | 0.00 |
| Biosynthesis of vitamins, cofactors and prosthetic groups | 63 | CTGN[5]GAC | 33 | 6.66 | 0.00 |
| Glycolysis and gluconeogenesis | 35 | TASGTAW | 46 | 8.52 | 0.00 |
| CTCWSCCC | 14 | 11.59 | 0.01 | ||
| CGTSSGG | 16 | 8.31 | 0.00 | ||
| Tricarboxylic acid pathway citrate cycle, Krebs cycle, TCA cycle | 25 | CGGCGCCG | 8 | 17.92 | 0.00 |
| GCWN[5]RGC | 54 | 6.92 | 0.00 | ||
| Glyoxylate cycle | 6 | CCGN[5]SSG | 18 | 11.66 | 0.00 |
| Other energy generation activities | 16 | CGCACCGC | 4 | 13.47 | 0.00 |
| DNA restriction or modification | 32 | CACN[11]WCC | 38 | 7.33 | 0.00 |
| rRNA transcription | 109 | CGATGAG | 50 | 17.00 | 0.00 |
| tRNA transcription | 83 | GATGAGS | 45 | 9.11 | 0.00 |
| Translation | 64 | AAWTTTTY | 170 | 9.95 | 0.00 |
| Cytoplasmic and nuclear degradation | 99 | TTGCCAC | 51 | 13.08 | 0.00 |
| Mitochondrial transport | 80 | SGCCSGG | 23 | 7.59 | 0.00 |
| G-protein-mediated signal transduction | 12 | CCCN[7]CGS | 13 | 8.52 | 0.00 |
| Homeostasis of anions | 13 | TCGN[7]SCR | 28 | 7.62 | 0.01 |
| Perception of nutrients and nutritional adaptation | 25 | CCSN[4]CCS | 30 | 8.25 | 0.00 |
| Cell death | 10 | GSCN[4]CCS | 19 | 8.05 | 0.01 |
| Nucleus | 31 | ATCACST | 21 | 7.05 | 0.01 |
| GCGGATCC | 5 | 11.49 | 0.00 | ||
| Cell wall | 38 | AGATCTCG | 11 | 15.37 | 0.00 |
| GGYCCST | 19 | 7.30 | 0.00 | ||
| Centrosome | 31 | TTWSGCG | 27 | 6.86 | 0.03 |
| Chromosome | 44 | ACTCGCCG | 5 | 10.42 | 0.03 |
| Regulator of G-protein signaling | 13 | CTACTCG | 6 | 7.98 | 0.04 |
| Target of regulation | 13 | CTACTCG | 6 | 7.98 | 0.04 |
| Cation transporters | 62 | CGCN[6]CGS | 31 | 7.40 | 0.00 |
| Ion transporters | 79 | CGSSCGC | 27 | 8.20 | 0.00 |
| C-compound and carbohydrate transporters | 46 | CGGAGWWA | 28 | 14.17 | 0.00 |
| Amino acid transporters | 25 | GATAGCGA | 6 | 10.29 | 0.02 |
| Allantoin and allantoate transporters | 9 | CGCNCGC | 9 | 10.13 | 0.00 |
| Drug transporters | 35 | CGACAGG | 10 | 8.37 | 0.00 |
| CGGCGCTA | 6 | 11.62 | 0.01 |
Most of the motifs in Table 3 match the known binding site consensus of some transcription factor, in which case the name of the factor is reported along with the consensus. Some of the motifs in this table do not match a known consensus, but do match two or more binding sites of a single transcription factor. For such motifs, we report the name of the factor, along with the number of matching binding sites. In either case, it would be interesting to pursue, for each of the motifs in the table, whether the transcription factor whose binding sites it matches has some regulatory role for the genes in that functional class. For instance, the motif CACGTGSG, which matches the Pho4 consensus, is found to be significant in the functional class ‘phosphate metabolism’ (Table 3), and it may be verified from the literature that the Pho4 transcription factor indeed regulates many of the genes in this class that have the motif in their promoters. Many other similar connections can be found in the comments column of Table 3.
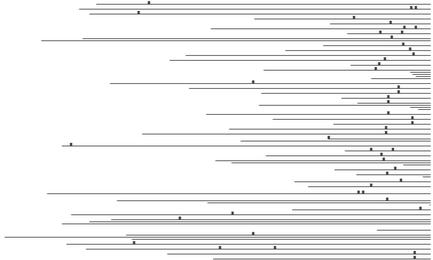
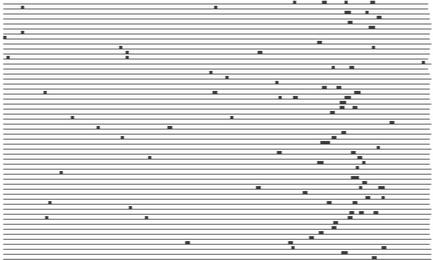
We will now discuss some of the most interesting observations from Table 4, showing that some of these motifs are excellent candidates as novel transcription factor binding sites. The 7mer CGATGAG is highly overrepresented in the promoters of the functional class ‘rRNA transcription’. This motif was also discovered by Hughes et al. (11), who recognized it as the PAC box (41), an element for which neither function nor binding factor has been identified. It occurs a total of 50 times in 45 of the 109 promoters in the class, with a z-score of 17.00 and pmax < 0.01. It is a very well distributed motif, its 50 occurrences being spread over 45 promoters. Moreover, these 45 promoters belong to genes that are highly co-expressed. Their co-expression score is 0.04, which means that the average pairwise correlation coefficient of their expression data has a p-value of 0.04. Another property that makes this motif a very compelling candidate for a binding site is its extremely high conservation in position in the promoter sequences. Figure 1 illustrates this point. It shows a plot of the occurrences of the motif in the 45 promoters, the 3′ end being on the right. We also plotted this motif in promoter regions of orthologs of the 45 yeast genes in other yeast species (Fig. 2). The orthologous genes considered here belong to other yeast strains and the orthology information was obtained from Paul Cliften (personal communication). We see that the motif occurs frequently and is conserved in position in these orthologous promoters also, even though the orthologous genes were identified based on their protein sequences. Moreover, a very similar motif GATGAGS is found to be significant in the related functional class ‘tRNA transcription’. This motif occurs 45 times in 34 promoters of the class, with a z-score of 9.11 (pmax < 0.01).
Figure 1.
Occurrences of motif CGATGAG in 45 promoters of the MIPS functional class ‘rRNA transcription’. Each horizontal line represents a promoter, the right end being at the translation start site. Vertical bars represent motif occurrences.
Figure 2.
Occurrences of motif CGATGAG in orthologous promoters of genes in the MIPS functional class ‘rRNA transcription’. The orthology information was obtained from Paul Cliften (personal communication). The sequences are plotted with their 3′ ends on the left, contrary to the convention followed in other similar plots in this paper.
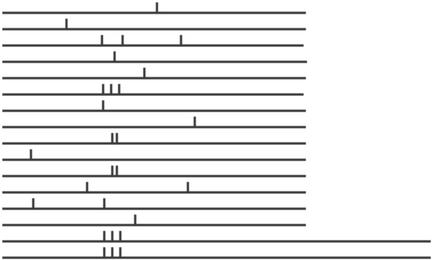
Another motif worth special mention is the 8mer CGGAGWWA, which occurs in the functional class ‘C-compound and carbohydrate transporters’ that has 46 genes. It occurs a total of 28 times in 16 different promoters of this class, whose corresponding genes have a co-expression score of 0.05. This motif is significantly well conserved in its location (Fig. 3), although not as strongly as the previous motif. Included in the 16 promoters of the class that contain the motif are nine of the glucose transporting HXT genes (Hxt2, Hxt3, Hxt5, Hxt8, Hxt11, Hxt13, Hxt15, Hxt16 and Hxt17). The regulation of these genes has been the subject of detailed biological studies. One study by Theodoris and Bisson (42) shows that ‘DNA sequence dependent suppressing elements’ (DDSEs) located in the promoters of HXT genes affect glucose sensing, and the authors further hypothesize that the DDSE region contains binding sites for the Rgt1p transcriptional repressor/activator. Rgt1p is believed to bind to promoters of Hxt2, Hxt3 and Hxt4. However, the Rgt1p binding site they propose for the HXT genes is TTTCAC GGAAAATTATATTTTG, which does not match our motif CGGAGWWA. A review by Ozcan and Johnston (43) describes another mechanism that represses transcription of some HXT genes in high glucose conditions through Mig1, which is a transcription factor (repressor) known to bind to the promoter of Hxt2 and Hxt4 genes. Again, we verified that the known binding sites of Mig1 (consensus CCCCRNN WWWWW) do not match the motif CGGAGWWA. Only limited information is available on the expression of Hxt5 and Hxt8 to Hxt17. In fact, it is not even certain if these are involved in glucose transport; they could act as transporters for other sugars. Hxt11 is bound by the transcription factor PDR3, although the PDRE (the binding site for PDR3, with consensus TCCGYGGA) does not match our motif CGGAGWWA. The promoter of Hxt13 was obtained in a screen for targets of the transcription factor Hap2, whose binding sites are quite different from CGGAGWWA. Gal2, a galactose permease that is >60% similar to the HXT proteins, is also one of the 16 genes whose promoters contain the motif under investigation. However, it is well established that Gal2 is regulated by the Gal4p transcription factor, which binds to the element CGGNNNNNNNNNNNCCG. In summary, while much is known about the transcriptional regulation of the glucose transporting genes, none of the known mechanisms seem to explain the presence of such a strong shared motif, which is therefore worth investigating further.
Figure 3.
Occurrences of motif CGGAGWWA in 16 promoters of the MIPS functional class ‘C-compound and carbohydrate transporters’.
The motif AAWTTTTY occurs 170 times in 52 of the 64 promoters of the class ‘translation’, with a z-score of 9.95 (pmax < 0.01). These 52 genes are highly co-expressed, as indicated by a co-expression score of 0.01. Another compelling feature of the motif is its high conservation in location in the promoters, as revealed by Figure 4. The class ‘amino acid transport’ has a significant motif GCCGTRCS, which occurs 13 times in nine promoters of the class, with a very high z-score of 17.70 (pmax < 0.01). Among the nine promoters that have this motif are TAT1, DIP5, GAP1 and GNP1. SPS-initiated signals are known to modulate the expression of these four genes (44), and it would be interesting to find out if the discovered motif is related to this known regulation.
Figure 4.
Occurrences of motif AAWTTTTY in 52 promoters of the MIPS functional class ‘translation’.
Phenotype catalog. The phenotype catalog from MIPS yielded 138 classes. All motifs reported by the motif-finding steps described above were examined. There were a total of 265 such motifs. Tables 5 and 6 present a selection from these motifs. Once again, we find among these motifs both known binding sites (Table 5) and novel motifs (Table 6) that may be good candidates for experimental verification. The motifs in Table 5 match the binding sites of the transcription factors Reb1, Mcb, repressor of Car1, Cbf1, Rap1, Mata1 and Pho4. It would be interesting to find out if these transcription factors are known to regulate some of the genes in the respective phenotype classes.
Table 5. Significant motifs in classes from the MIPS phenotype catalog.
| Name | Motif | Ns | z | pmax | Comments |
|---|---|---|---|---|---|
| G1 arrest | ATTACCC | 13 | 7.33 | 0.02 | REB1 (YYACCCG) |
| Mating efficiency | TTACCCG | 12 | 8.81 | 0.00 | REB1 (YYACCCG) |
| Bud localization | TTACCCG | 12 | 8.15 | 0.00 | REB1 (YYACCCG) |
| Osmotic sensitivity | TTACCCG | 13 | 7.79 | 0.00 | REB1 (YYACCCG) |
| Cytoskeleton mutants | TTACCCG | 27 | 9.53 | 0.00 | REB1 (YYACCCG) |
| Secretory mutants | TTACCCG | 20 | 10.21 | 0.00 | REB1 (YYACCCG) |
| Carbohydrate and lipid biosynthesis defects | CGCGWCG | 11 | 7.19 | 0.00 | MCBF (WCGCGW) |
| DNA repair mutants | ACGCGW | 83 | 10.98 | 0.00 | MCBF (WCGCGW) is involved in DNA synthesis (45) |
| DNA replication mutants | ACGCGW | 40 | 9.13 | 0.00 | MCBF (WCGCGW) binds to MCB in DNA replication genes (TRANSFAC) |
| Respiratory deficiency | GSCGCCGA | 18 | 11.46 | 0.00 | Repressor of CAR1 (AGCCGCCR) |
| Vanadate resistance | GYCGSCG | 7 | 9.53 | 0.04 | Repressor of CAR1 (AGCCGCCR) |
| Recombination mutants | TAGCCGCC | 8 | 11.02 | 0.00 | Repressor of CAR1 (AGCCGCCR) |
| Divalent cations and heavy metals | GYSGCCG | 27 | 6.51 | 0.05 | Repressor of CAR1 (AGCCGCCR) |
| Methionine auxotrophy | TCACGTGC | 5 | 17.73 | 0.00 | CBF1 (TCACGTG) null mutant is methionine auxotroph (MIPS) |
| Elongated cell and bud morphologies | TCCGTAC | 9 | 6.69 | 0.05 | Matches 3 RAP1 binding sites |
| CAANNNCAR | 73 | 6.01 | 0.04 | Matches 3 Mata-1 binding sites | |
| Divalent cations and heavy metals | CACGTGS | 27 | 8.00 | 0.00 | PHO4 (CACGTK) |
The last column is the name of the known transcription factor binding site whose consensus sequence is similar to the found motif. The consensus sequence was obtained from SCPD (16).
Table 6. Significant novel motifs in classes from the MIPS mutant phenotypes catalog.
| Name | Size | Motif | Ns | z | pmax |
|---|---|---|---|---|---|
| Rapamycin | 7 | CCTGCTTC | 6 | 14.68 | 0.04 |
| Sensitivity to immunosuppressants | 14 | GTYGCCG | 9 | 8.30 | 0.01 |
| G2/M arrest | 38 | ACCCGCCC | 5 | 10.68 | 0.02 |
| Mating efficiency | 31 | CCGN[4]GGS | 20 | 6.95 | 0.00 |
| CGTCGGTA | 5 | 10.04 | 0.00 | ||
| CTSCCCSG | 8 | 9.56 | 0.01 | ||
| Methionine auxotrophy | 9 | CCGN[8]CCG | 8 | 9.95 | 0.00 |
| GGSCGGG | 6 | 9.95 | 0.00 | ||
| CCTN[4]CYC | 20 | 9.55 | 0.00 | ||
| ACGGGCGC | 3 | 14.24 | 0.02 | ||
| Galactose fermentation | 14 | GGGGCCCS | 5 | 13.99 | 0.00 |
| CCGN[6]CGG | 10 | 7.34 | 0.02 | ||
| Other carbon utilization defects | 27 | CCGTAGAC | 6 | 13.96 | 0.00 |
| CCRNNNCGS | 33 | 6.44 | 0.02 | ||
| Nitrogen utilization | 8 | GGCNNGCC | 12 | 8.83 | 0.02 |
| TGCGGCG | 5 | 9.47 | 0.01 | ||
| Other auxotrophies | 20 | GTGACYC | 12 | 9.23 | 0.00 |
| Bud localization | 35 | GTCGGGTA | 5 | 10.16 | 0.02 |
| TAYGTRT | 45 | 7.90 | 0.00 | ||
| ACSCGA | 29 | 6.49 | 0.01 | ||
| Elongated cell and bud morphologies | 32 | RGGN[7]GGW | 53 | 6.80 | 0.00 |
| TCCGTAC | 9 | 6.69 | 0.05 | ||
| Papulacandin B sensitivity | 22 | GCTGCTGY | 12 | 11.10 | 0.00 |
| Killer toxin resistance | 21 | CCGCACRC | 5 | 10.30 | 0.04 |
| Killer toxin | 29 | CSGN[5]CGC | 17 | 6.69 | 0.01 |
| GCGN[9]ASR | 46 | 6.59 | 0.01 | ||
| Zymolyase sensitivity | 31 | CCGN[5]SGY | 31 | 6.23 | 0.02 |
| Hygromycin B sensitivity | 26 | GRAGATG | 26 | 7.07 | 0.04 |
| Spindle mutants | 26 | TTTGYTT | 50 | 7.24 | 0.02 |
| Benomyl sensitivity | 30 | GTRN[11]TGW | 86 | 6.43 | 0.01 |
| Other tubulin cytoskeleton mutants | 15 | CGGNNSCG | 14 | 7.04 | 0.03 |
| CCCGCGAG | 3 | 12.29 | 0.03 | ||
| CGGGTCYG | 4 | 12.03 | 0.03 | ||
| Secretory mutants | 61 | CGAN[9]CGA | 30 | 7.36 | 0.00 |
| Mitochondrial mutants | 37 | CGGN[5]CSG | 23 | 7.15 | 0.00 |
| CCCGGRC | 9 | 7.19 | 0.00 | ||
| WAASCTG | 42 | 7.11 | 0.00 | ||
| pH sensitivity | 9 | GCGGCSGC | 8 | 20.96 | 0.00 |
| CCYN[8]GGC | 14 | 7.75 | 0.03 | ||
| Vacuolar mutants | 65 | GRRTTTG | 60 | 6.86 | 0.03 |
| Starvation sensitivity | 26 | ACAN[9]ATA | 47 | 6.25 | 0.04 |
| Other oxidizing agents | 15 | ARGN[6]AGG | 36 | 8.64 | 0.00 |
| Oxidizing agents | 32 | ACCNNACY | 35 | 6.30 | 0.01 |
| Divalent cations and heavy metals resistance | 13 | GGCN[7]GSC | 20 | 8.55 | 0.00 |
| Divalent cations and heavy metals sensitivity | 69 | GCSGCCGY | 14 | 8.78 | 0.05 |
| ACWNNNACA | 128 | 8.23 | 0.00 | ||
| Carbohydrate and lipid biosynthesis defects | 47 | CGYN[9]CGY | 58 | 8.05 | 0.00 |
| Other DNA repair mutants | 32 | TATGTAY | 29 | 7.27 | 0.00 |
| Silencing mutants | 26 | CTGGGGTC | 4 | 9.98 | 0.03 |
| Recombination mutants | 55 | ARGNNAAG | 128 | 6.93 | 0.00 |
| DNA replication mutants | 30 | CGTGCGCC | 4 | 10.74 | 0.00 |
| Silencing mutants | 26 | CCGGTAGA | 6 | 11.53 | 0.00 |
| Recombination mutants | 55 | CRGTCGG | 12 | 6.99 | 0.02 |
| Staurosporine sensitivity | 7 | CGGN[7]ACC | 8 | 8.40 | 0.03 |
| Caffeine sensitivity | 34 | CARN[10]AGC | 51 | 6.48 | 0.01 |
We now discuss some of the most interesting motifs reported in Table 6. The motif GTYGCCG occurs a total of nine times (z-score 8.30, pmax 0.01) in seven of the 14 promoters of the class ‘sensitivity to immunosuppressants’. The seven promoters belong to highly co-expressed genes, as indicated by the low co-expression score of 0.015. The motif does not match any known transcription factor binding site.
The motif CTSCCCSG, found in the class ‘mating efficiency’, deserves special mention. It occurs eight times in seven different promoters of the class, with a z-score of 9.56 (pmax 0.01). An interesting feature of this motif is that its instances occur, with up to one mismatch, overlapping Gal4p binding sites in five of the six genes regulated by this transcription factor. Though it does not match the Gal4p consensus CGGNNNNNNNNNNNCCG, this coincidence seems worth investigating.
Another interesting motif is CCGCACRC, found in four of the 21 promoters of the class ‘killer toxin resistance’. It occurs a total of five times in these promoters, with a z-score of 10.30 (pmax 0.04). The occurrences of the motif are conserved in position in the four promoters, as Figure 5 reveals. Moreover, the four corresponding genes are highly co-expressed, having a co-expression score of 0.025.
Figure 5.
Occurrences of motif CCGCACRC in four promoters of the MIPS phenotype class ‘killer toxin resistance’.
Unclassified proteins. The MIPS database also has a table of 230 ORFs with strong sequence similarity to known proteins. YMF was run on sets of genes that have similarity to the same protein or family of proteins, and in some cases significant motifs were reported. For instance, there are seven ORFs with a strong similarity to members of the SRP1/TIP1 family. YMF reported two strong motifs AGGCAY (pmax < 0.01) and TCGTWYA (pmax < 0.01) in this set. Both these motifs were found to be significant when YMF was run on the eight members of the SRP1/TIP1 family, thereby furthering evidence for a relation between the seven unclassified ORFs and the SRP1/TIP1 family.
Further research
In Tables 3 and 5, it would be interesting to pursue connections (if not already described in these tables) between the known transcription factor listed in the last column and the gene family in which it was found, to determine whether the transcription factor plays a role in the regulation of genes in this family.
Even more interesting would be to pursue the novel motifs listed in Tables 2, 4 and 6 to see whether they lead to transcription factors of heretofore unknown function.
Finally, the motif model described in Materials and Methods was developed from a study of S.cerevisiae transcription factor binding sites. It would be very interesting to understand whether this model is also suitable for the discovery of transcription factor binding sites in other organisms and, if not, how it should be modified. It is relatively easy to extend our algorithm to handle motif models where a motif corresponds to a fixed set of strings over the alphabet {A,C,G,T,N}. The mismatch model, where the motif is a string over this alphabet, and its occurrences may have some fixed number of mismatches from the consensus, belongs to this category of models.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Linda Breeden and Chris Roberts for sharing their insights on transcription factor binding sites, Mathieu Blanchette and Rimli Sengupta for numerous thorough and helpful discussions, and Jacques van Helden for helpful comments. This material is based upon work supported in part by the National Science Foundation and DARPA under grant DBI-9601046, in part by the National Science Foundation under grant DBI-9974498, and in part by a Microsoft Fellowship.
REFERENCES
- 1.Bailey T.L. and Elkan,C. (1995) Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learn., 21, 51–80. [Google Scholar]
- 2.Fraenkel Y.M., Mandel,Y., Friedberg,D. and Margalit,H. (1995) Identification of common motifs in unaligned DNA sequences: application to Escherichia coli Lrp regulon. Comput. Appl. Biosci., 11, 379–387. [DOI] [PubMed] [Google Scholar]
- 3.Galas D.J., Eggert,M. and Waterman,M.S. (1985) Rigorous pattern-recognition methods for DNA sequences: analysis of promoter sequences from Escherichia coli. J. Mol. Biol., 186, 117–128. [DOI] [PubMed] [Google Scholar]
- 4.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence C.E. and Reilly,A.A. (1990) An expectation maximization (EM) algorithm for the identification and characterization of common sites in unaligned biopolymer sequences. Proteins Struct. Funct. Genet., 7, 41–51. [DOI] [PubMed] [Google Scholar]
- 7.Rigoutsos I. and Floratos,A. (1998) Motif discovery without alignment or enumeration. In RECOMB98: Proceedings of the Second Annual International Conference on Computational Molecular Biology. ACM Press, New York, NY, pp. 221–227.
- 8.Rocke E. and Tompa,M. (1998) An algorithm for finding novel gapped motifs in DNA sequences. In RECOMB98: Proceedings of the Second Annual International Conference on Computational Molecular Biology. ACM Press, New York, NY, pp. 228–233.
- 9.Staden R. (1989) Methods for discovering novel motifs in nucleic acid sequences. Comput. Appl. Biosci., 5, 293–298. [DOI] [PubMed] [Google Scholar]
- 10.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 11.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 12.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 13.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Systematic determination of genetic network architecture. Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 15.Wingender E., Dietze,P., Karas,H. and Knüppel,R. (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res., 24, 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J. and Zhang,M.Q. (1999) SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 17.van Helden J., André,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 18.van Helden J., Olmo,M. and Pérez-Ortin,J. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Helden J., Rios,A. and Collado-Vides,J. (2000) Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res., 28, 1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tompa M. (1999) An exact method for finding short motifs in sequences, with application to the ribosome binding site problem. In Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 262–271. [PubMed]
- 21.Brāzma A., Jonassen,I., Vilo,J. and Ukkonen,E. (1998) Predicting gene regulatory elements in silico on a genomic scale. Genome Res., 15, 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagot M. (1998) Spelling approximate repeated or common motifs using a suffix tree. In Latin ’98: Theoretical Informatics, Springer-Verlag Lecture Notes in Computer Science no. 1380. Springer-Verlag, Heidelberg, Germany, pp. 111–127.
- 23.Pavesi G., Mauri,G. and Pesole,G. (2001) An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics, 17, S207–S214. [DOI] [PubMed] [Google Scholar]
- 24.Sinha S. and Tompa,M. (2000) A statistical method for finding transcription factor binding sites. In Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 344–354. [PubMed]
- 25.Mewes H.W., Albermann,K., Heumann,K., Liebl,S. and Pfeiffer,F. (1997) MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res., 25, 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones E.W., Pringle,J.R. and Broach,J.R. (eds) (1992) The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Blaiseau P.-L., Isnard,A.-D., Surdin-Kerjan,Y. and Thomas,D. (1997) Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell. Biol., 17, 3640–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mai B. and Breeden,L. (1997) Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol., 17, 6491–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McInerny C.J., Partridge,J.F., Mikesell,G.E., Creemer,D.P. and Breeden,L.L. (1997) A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev., 11, 1277–1288. [DOI] [PubMed] [Google Scholar]
- 30.Nurrish S.J. and Treisman,R. (1995) DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol., 15, 4076–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima Y., Nobuo,O. and Harashima,S. (1996) Regulation of phosphatase synthesis in Saccharomyces cerevisiae – a review. Gene, 179, 171–177. [DOI] [PubMed] [Google Scholar]
- 32.Roulet E., Bucher,P., Schneider,R., Wingender,E., Dusserre,Y., Werner,T. and Mermod,N. (2000) Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J. Mol. Biol., 297, 833–848. [DOI] [PubMed] [Google Scholar]
- 33.Wemmie J.A., Szczypka,M.S., Thiele,D.J. and Moye-Rowley,W.S. (1994) Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCS1. J. Biol. Chem., 269, 32592–32597. [PubMed] [Google Scholar]
- 34.Leung M.-Y., Marsh,G.M. and Speed,T.P. (1996) Over- and underrepresentation of short DNA words in herpesvirus genomes. J. Comput. Biol., 3, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicodème P., Salvy,B. and Flajolet,P. (1999) Motif statistics. In 7th Annual European Symposium on Algorithms—ESA99. Springer-Verlag Lecture Notes in Computer Science no. 1643. Springer-Verlag, Heidelberg, Germany, pp. 194–211.
- 36.Blanchette M. and Sinha,S. (2001) Separating real motifs from their artifacts. Bioinformatics, 17, S30–S38. [DOI] [PubMed] [Google Scholar]
- 37.Aach J., Rindone,W. and Church,G.M. (2000) Systematic management and analysis of yeast gene expression data. Genome Res., 10, 431–445. [DOI] [PubMed] [Google Scholar]
- 38.Bachhawat N., Ouyang,Q. and Henry,S.A. (1995) Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem., 270, 25087–25095. [DOI] [PubMed] [Google Scholar]
- 39.Schüller H.-J., Richter,K., Hoffman,B., Ebbert,R. and Schweizer,E. (1995) DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor; structural requirements as defined by saturation mutagenesis. FEBS Lett., 370, 149–152. [DOI] [PubMed] [Google Scholar]
- 40.Wagner C., Blank,M., Strohmann,B. and Schüller,H.-J. (1999) Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast, 15, 843–854. [DOI] [PubMed] [Google Scholar]
- 41.Dequard-Chablat M., Riva,M., Carles,C. and Sentenac,A. (1991) RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J. Biol. Chem., 266, 15300–15307. [PubMed] [Google Scholar]
- 42.Theodoris G. and Bisson,L.F. (2001) DDSE: downstream targets of the SNF3 signal transduction pathway. FEBS Microbiol. Lett., 197, 73–77. [DOI] [PubMed] [Google Scholar]
- 43.Ozcan S. and Johnston,M. (1999) Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev., 63, 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsberg H., Gilstring,C.F., Zargari,A., Martinez,P. and Ljungdahl,P.O. (2001) The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol., 42, 215–228. [DOI] [PubMed] [Google Scholar]
- 45.Lowndes N.F., Johnson,A.L., Breeden,L. and Johnston,L.H. (1992) SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature, 357, 505–508. [DOI] [PubMed] [Google Scholar]
- 46.Kean L.S., Grant,A.M., Angeletti,C., Mahe,Y., Kuchler,K., Fuller,R.S. and Nichols,J.W. (1997) Plasma membrane translocation of fluorescent-labeled phosphatidylethanolamine is controlled by transcription regulators, PDR1 and PDR3. J. Cell Biol., 138, 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng C., Kacherovsky,N., Dombek,K.M., Camier,S., Thukral,S.K., Rhim,E. and Young,E.T. (1994) Identification of potential target genes for Adr1p through characterization of essential nucleotides in UAS1. Mol. Cell. Biol., 14, 3842–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]